Members of the domain Archaea contribute about 0.3 to 3.3% of the microbial small subunit (16S and 18S) rRNA in the rumen (22, 39, 60). Archaea have a range of different metabolisms and are found in many habitats (6), but those known to exist in the rumen are strictly anaerobic methanogens. Yanagita et al. (59) observed that 2.8 to 4.0% of ruminal microorganisms displayed autofluorescence characteristic of F420, a methanogen cofactor, able to be seen under UV illumination during microscopy. Taken together with the small subunit rRNA abundance data, this suggests that a large part of the archaeal population is made up of methanogens. Most species of methanogens can grow using H2 and often formate as their energy sources and use the electrons derived from H2 (or formate) to reduce CO2 to CH4. Some species can grow with methyl groups, oxidizing some to CO2 to produce electrons that are used to reduce further methyl groups to methane. A few species can grow with acetate, effectively dissimilating acetate to CH4 and CO2. However, acetate is not metabolized to CH4 to any significant extent in the rumen (13). This is probably because the rate of passage of rumen contents through the rumen is greater than the growth rate of acetate-utilizing methanogens (53).
In a normally functioning rumen, proteins and polymeric carbohydrates, which usually make up the largest part of the incoming feed, are fermented by a mixed microbial community to volatile fatty acids (VFAs), NH4+, CO2, and H2. The hydrogen is metabolized by the methanogens. The VFAs are taken up by the animal across the rumen wall and serve as major carbon and energy sources for the ruminant. A part of the VFAs, undigested feed components, and microbial cells leave the rumen and enter the rest of the animal's digestive tract. The central role of H2 in the rumen fermentation (12) means that, although methanogenic archaea make up only a small part of the rumen microbial biomass, they play an important role in rumen function and animal nutrition. Efficient H2 removal leads to a nutritionally more favorable pattern of VFA formation and to an increased rate of fermentation by eliminating the inhibitory effect of H2 on the microbial fermentation (26, 53).
The rumen can be simplistically described as an open system with discontinuous solid (feed) and liquid (saliva and drinking water) inputs and multiple fractions that have different turnover rates (53). The methanogens in the rumen are found free in the rumen fluid, attached to particulate material and rumen protozoa, associated as endosymbionts within rumen protozoa, and attached to the rumen epithelium. The methanogens associated with these different fractions can be expected to have different growth rates since they will be removed from the rumen at different rates. In addition, the animal itself and the feed also influence the rate of passage of digesta through the rumen system (25). These different habitats may allow niche division among the methanogens and may explain some of the observed phylogenetic diversity of rumen archaea.
Cultured methanogens from the rumen.
Methanogens have been classified into 28 genera and 113 species (11), but many more species can be expected to occur in nature (6). Surprisingly few methanogens have been isolated from the rumen. Those that have been cultured are assigned to only seven species. These are Methanobacterium formicicum (33), Methanobacterium bryantii (16), Methanobrevibacter ruminantium (43), Methanobrevibacter millerae (36), Methanobrevibacter olleyae (36), Methanomicrobium mobile (35), and Methanoculleus olentangyi (16). Methanosarcina spp. have also been cultured from the rumen (1, 34) but are not normally a major part of the archaeal community. In addition, Methanobrevibacter smithii (16) has been reported as being isolated from the rumen, but this is likely to be a strain more closely related to M. millerae (M. Kirs and P. H. Janssen, unpublished data).
Cultivation-based studies usually fail to uncover the full extent of microbial diversity. This is because some species are more readily culturable than others and because the size of any single cultivation-based survey is usually too small to give good insight into the community structure (15). The random isolation of methanogens in small-scale cultivation-based studies is useful for obtaining isolates for detailed investigation and also indicates the presence of a species. However, the abundance of different methanogens in different systems cannot be assessed comprehensively using data from the cultivation-based studies that have been made over the past 50 years.
Cultivation-independent surveys of ruminal methanogens.
Surveys of methanogens and total archaea in the rumen have been made by using PCR to amplify the 16S rRNA genes of archaea, followed by a cloning step in Escherichia coli to separate the different gene variants within the mixed amplicon. An assessment of the dominant groups of archaea in the rumen can be made by comparative sequence analysis and phylogenetic placement of the 16S rRNA genes recovered in such surveys. In some of the studies, not all of the 16S rRNA genes were sequenced, but, instead, a method of screening, such as restriction fragment analysis, was used to group similar clones before determining the sequences of representative clones. In other studies, not all 16S rRNA gene sequences were deposited in publicly accessible databases. However, a number of publications provide enough detail to allow an accurate assessment of the number of occurrences of each sequence type and thus make estimating the abundance of different archaeal groups possible. Comparison of all sequences with a reference data set of almost full-length 16S rRNA gene sequences allows phylogenetic placement of all sequences with some degree of confidence.
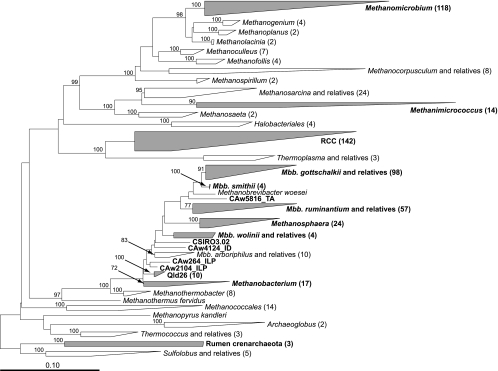
The studies analyzed in this synthesis have been separated into two major types: studies of total rumen archaea and studies of protozoan-associated archaea (Table 1). Nine studies surveyed total rumen archaea, including free-living cells, particle-associated cells, and cells associated with other rumen microbes. Two of these studies (54, 56) reported multiple, distinctly different libraries, so that the nine studies were divided into 12 data sets containing a combined 294 useful sequences that represented 1,026 cloned 16S rRNA genes. Five other studies investigated protozoan-associated archaea. These formed five data sets containing a total of 187 sequences that represented 2,717 cloned 16S rRNA genes. 16S rRNA gene sequences were downloaded from GenBank databases (2) into a global data set curated using ARB (23). In addition, 138 16S rRNA gene sequences from a wide range of cultured archaea, largely methanogens, were included as reference sequences. These reference sequences were all >1,300 nucleotides (nt) long. Evolutionary analyses of these sequences resulted in construction of a dendrogram (Fig. 1), which allowed final assignment of all sequences and associated clones to clades of rumen archaea (Table 2).
TABLE 1.
Numbers of clones and sequences in the studies summarized in this synthesis
| Study subject and authors (reference no.) | No. of data sets | No. of sequences
|
No. of clones | |
|---|---|---|---|---|
| >870 nt | <800 nt | |||
| Archaea in total rumen contents | ||||
| Shin et al. (40) | 1 | 104 | 0 | 104 |
| Skillman et al. (42) | 1 | 10 | 0 | 10 |
| Tajima et al. (48) | 1 | 8 | 0 | 23 |
| Whitford et al. (52) | 1 | 41 | 0 | 41 |
| Wright et al. (54) | 3 | 43a | 0 | 405 |
| Wright et al. (55) | 1 | 26 | 0 | 78 |
| Wright et al. (56) | 2 | 28 | 0 | 241 |
| Wright et al. (57) | 1 | 14 | 0 | 104 |
| Yanagita et al. (59) | 1 | 0 | 20 | 20 |
| Protozoan-associated archaea | ||||
| Chagan et al. (3) | 1 | 0 | 8 | 91 |
| Irbis and Ushida (14) | 1 | 0 | 14 | 2,398 |
| Ohene-Adjei et al. (32) | 1 | 0 | 139 | 139 |
| Regensbogenova et al. (37) | 1 | 11 | 9 | 20 |
| Tokura et al. (50) | 1 | 0 | 6 | 69 |
An additional 26 sequences, representing 328 clones, were not included because they were withdrawn from GenBank.
FIG. 1.
Phylogenetic dendrogram of total rumen and rumen protozoan-associated archaea and selected reference sequences. Sequences of >870 nt long (138 reference sequences and 271 cloned sequences) were aligned using the EDIT4 primary sequence editor in ARB (23), and the alignments were checked and corrected to produce a master alignment. All positions were considered for the subsequent analyses. Phylogenetic dendrograms of just the 138 reference sequences and of the reference sequences plus 271 cloned sequences of >870 nt were generated in ARB using the Jukes-Cantor distance correction (17) and the neighbor-joining algorithm (38). These dendrograms were rooted with eight sequences from the Crenarchaeota. These alignments were exported from ARB, and 1,000 bootstrap data sets were generated from each in PAUP (46) using Juke-Cantor distances before 1,000 dendrograms were generated using neighbor joining. Consensus dendrograms were then generated in PAUP. These two consensus dendrograms, with bootstrap values, were used to define the clades referred to in this review. Key bootstrap values from the analysis of the larger data set have been added to the phylogenetic dendrogram illustrated. Sequences of <800 nt long (Table 1) were then inserted into the dendrogram of 271 cloned sequences of >870 nt plus 138 reference sequences using the parsimony insertion tool in ARB. For simplicity, clades are shown as shaded parallelograms, with the total number of sequences used in the dendrogram construction shown in parentheses. Sequences and clades containing sequences that originate from PCR-mediated surveys listed in Table 1 are indicated in boldface text. The scale bar indicates 0.10 inferred nucleotide substitutions per position. Mbb, Methanobrevibacter.
TABLE 2.
Assignment of clones to different clades of Archaea and the number of positive data sets for each cladea
| Clade | Detection in total rumen contents
|
Detection in protozoan-associated archaea
|
||||
|---|---|---|---|---|---|---|
| Mean (%) | Maximum (%) | No. of data sets (n = 12) | Mean (%) | Maximum (%) | No. of data sets (n = 5) | |
| Methanomicrobium | 14.9 | 85.6 | 4 | 20.1 | 80.0 | 3 |
| Methanimicrococcus | 2.4 | 14.6 | 3 | 4.0 | 20.0 | 1 |
| M. gottschalkii | 33.6 | 81.2 | 10 | 42.9 | 98.6 | 3 |
| M. smithii | 0.1 | 0.8 | 1 | 0 | NDc | 0 |
| M. ruminantium | 27.3 | 60.0 | 10 | 12.5 | 62.6 | 1 |
| M. wolinii | 0.1 | 1.0 | 1 | 2.6 | 12.8 | 1 |
| Other Methanobrevibacterb | 0.5 | 5.6 | 1 | 0.6 | 2.9 | 1 |
| Methanosphaera | 3.7 | 26.8 | 5 | 1.9 | 4.0 | 4 |
| Methanobacterium | 0.2 | 2.9 | 1 | 0.4 | 1.1 | 2 |
| RCC | 15.8 | 80.8 | 5 | 15.1 | 74.1 | 2 |
| Qld26 | 1.3 | 8.7 | 3 | 0 | ND | 0 |
| Crenarchaeota | 0.2 | 2.9 | 1 | 0 | ND | 0 |
The 14 studies summarized in this review were carried out by different research groups, with samples from a number of ruminant species and breeds fed different diets in various countries. Different samples were collected, and these were not always treated in the same way. The 16S rRNA genes were amplified using different primer sets, and so divergent amplification biases may have been introduced. Therefore, the studies are not strictly comparable, but taken together, the global data set does offer valuable insight into the identity of the dominant groups of rumen archaea.
Abundant archaea in the rumen.
Based on the analysis of the global data set, the majority (92.3%) of rumen archaea detected in total rumen contents can be placed in three genus-level groups (Table 2). These are Methanobrevibacter (61.6%), Methanomicrobium (14.9%), and a large group of uncultured rumen archaea labeled here as rumen cluster C, or RCC (15.8%). Methanobrevibacter spp. have been considered the dominant methanogens in the rumen (28, 57), and this synthesis supports this hypothesis.
Within the genus Methanobrevibacter, the cloned sequences fall into two major clades. One clade, defined by the species Methanobrevibacter gottschalkii, Methanobrevibacter thaueri, and M. millerae, contains the larger part of the Methanobrevibacter-related clones (a mean of 33.6% of rumen archaea). This group is designated the M. gottschalkii clade. The other major clade, defined by M. ruminantium and M. olleyae, contains 27.3% of rumen archaea, and is designated the M. ruminantium clade. Members of these two clades were found in nearly all of the data sets (Table 2). Methanobrevibacter spp. also appear to be early colonizers of the developing rumen (41). Members of other Methanobrevibacter spp., including M. smithii and Methanobrevibacter wolinii, appear to be rare.
Members of other groups of methanogens, including Methanimicrococcus spp., Methanosphaera spp., and Methanobacterium spp., occurred in fewer data sets or at lower abundances (Table 2). In addition, two groups of uncultured archaea, designated here the Qld26 group, and a clade of Crenarchaeota were also detected. The Qld26 group contains the 16S rRNA gene sequences CSIRO-Qld26 and Ven-04 (55, 57) and nine sequences, mainly associated with large particles in the rumen, detected by Shin et al. (40). The crenarchaeotes were detected rumen in only one study (40), and detection in other samples will be required to show that they were not transients. The physiologies of members of these two groups are not known.
The variation in the sequence composition within the major clades is great enough in some groups to result in almost continual gradation of sequence types with little evidence of subgroups. This is especially apparent within the M. gottschalkii clade. Some of the variation may be due to amplification and sequencing errors, and some may reflect true genetic diversity among strains of ruminal Methanobrevibacter spp. This gradation of 16S rRNA sequence types means that further division based on the available gene sequence evidence is not possible. There is a similar lack of differentiation of some of the other clades into discernable subgroups (e.g., the Methanomicrobium clade, the Methanosphaera clade, the Qld26 clade, the Methanimicrococcus clade, and most of the M. ruminantium clade). The RCC clade, in contrast, does display a considerable variation in sequence types that fall into a number of distinct lineages and may, therefore, represent a number of distinct species and perhaps multiple genera, with greater than 3% difference in sequence between members of different subgroups.
A difference of greater than 3% in the 16S rRNA gene sequences of prokaryotes is often used as an indicator of species-level separation of two strains (45). If the difference is less than 3%, it is not possible to use the 16S rRNA gene sequence comparison to determine whether strains belong to the same or different species. Dighe et al. (8) suggested that 16S rRNA gene sequence differences of >2% correlated with DNA-DNA hybridization similarities of <70% within the genus Methanobrevibacter and so could be used to separate sequences into species. This still relies on the definition of a species as being separated from its relatives by DNA-DNA hybridization values of <70%. In both of the major Methanobrevibacter clades discussed above, the named species have 16S rRNA gene sequence differences of <3%, and the decision to describe multiple species was based largely on differences in the genomes as determined by DNA-DNA hybridization, as well as a limited number of phenotypic differences (29, 36). Whether these can truly be regarded as different species is a matter of interpretation of the meaning of DNA-DNA hybridization results. This means that there may be a large number (>20) of rumen methanogen species within the genus Methanobrevibacter or only a very small number (approximately four). Detailed genome analysis may help reveal which interpretation is correct, although in the end the analysis will depend largely on the definition of a prokaryotic species, especially if the genomes also form a gradation of genotypes (20). It is more significant to determine how many functionally different groups there are within these clades and what the biochemical and ecological differences are.
Comparison with other methods.
RNA-targeted DNA probes have been used to analyze the archaeal community of the rumen using hybridization against extracted RNA (22, 39) or against RNA-containing cells using fluorescence in situ hybridization (FISH) (44, 58). In common with the clone library analyses, these studies found that members of family Methanobacteriaceae (which includes Methanobrevibacter spp., Methanobacterium spp., and Methanosphaera spp.) were the dominant members of the rumen archaeal community (30 to 99% of archaea). Members of the order Methanomicrobiales (which includes Methanomicrobium spp.) were less abundant (0 to 54%), and members of the order Methanosarcinales (which includes Methanimicrococcus) were rare (2 to 3%). Interestingly, significant populations of members of the family Methanococcaceae (8 to 44%) were detected using probes, but this group is wholly absent in gene library-based surveys (Table 2). This suggests that the probes used were not entirely specific or that significant biases occur in library generation. This remains to be resolved. However, members of the order Methanococcales have not been cultured from the rumen (16).
Temporal temperature gradient gel electrophoretic separation of 16S rRNA genes amplified by PCR from total rumen contents of pasture-fed sheep and cows, followed by sequencing prominent bands, confirmed the abundance of members of the M. ruminantium, M. gottschalkii, and RCC clades (31). Members of the Methanimicrococcus clade were also detected in the samples, and after a cultivation-based step, the presence of members of the Methanosphaera clade was also verified. These abundances agree with the global data set. No members of Methanococcales were detected.
Tatsuoka et al. (49) and Denman et al. (7) surveyed the diversity of the methyl-coenzyme M reductase (mcrA) gene in the rumen of cattle. This enzyme and its gene are good markers for the presence of methanogens (10). In both studies of the rumen, mcrA genes from Methanobrevibacter spp. dominated the libraries generated using primers that targeted most mcrA sequence types. Denman et al. (7) also detected a group of mcrA sequences belonging to an unidentified group of archaea also detected in landfill by Luton et al. (24). It is not known if these are derived from one of the uncultured groups of archaea detected in the 16S rRNA libraries. No mcrA genes clearly assignable to the Methanococcales were detected in the two studies on the rumen (7, 49).
The methanogens that have been cultured from the rumen fall into the major clades detected in the global data set, with the exception of Methanosarcina spp. (1, 34) and Methanoculleus olentangyi (16). The isolation of a microorganism from an environment indicates its presence at the time of sampling, but unless abundance is quantified, it is not possible to make statements regarding the numerical significance the organism has in the community from which it was cultured. The 16S rRNA gene-based approach can be expected to detect mainly large populations. At present, the interpretation must be that Methanosarcina spp. and Methanoculleus spp. have been found in samples from the rumen but do not appear to be numerically significant parts of the rumen archaeal community. Methodological biases against their molecular detection could also result in their absence from libraries of PCR-amplified 16S rRNA genes, but the range of different primers used in the different studies suggests that primer bias is unlikely.
Archaeal community structure.
The community compositions uncovered in the different studies varied. This may be attributable to the ruminant host or the diet or to the DNA extraction methods and PCR primers used. Two of the studies were performed using the same DNA extraction methods and the same PCR primers (54, 55). Both assessed archaeal diversity in the rumen of sheep. The rumen archaea of sheep held at the CSIRO Yalanbee Research Station in Western Australia were dominated by members of the M. gottschalkii (75.3%) and M. ruminantium (19.5%) clades. In contrast, sheep in Queensland, Australia, had archaeal populations dominated by members of RCC clade (80.8%) and had only few members of the M. gottschalkii clade (9.0%), no detectable members of the M. ruminantium clade, and some members of the Methanomicrobium clade (7.7%). The differences in community composition could have been caused by differences in diet, environment, health, animal genotype, and animal age (27).
Shin et al. (40) and Tajima et al. (48) reported the dominance of Methanomicrobium spp. among the total rumen archaea of cows (85.6% and 60.9%, respectively) and found that Methanobrevibacter spp. were not detected (40) or not the dominant clade (17.4%) (48). In contrast, Whitford et al. (52) found that Methanosphaera spp. (26.8%) and Methanimicrococcus spp. (14.6%) were abundant in the cows they studied but that Methanobrevibacter spp. were the dominant archaea (58.5%). Wright et al. (56) found that Methanobrevibacter spp. (50.0 to 51.9%) and members of RCC (37.8 to 50.0%) dominated in feedlot cattle in two geographic locations in Canada, while sheep in Australia and Venezuela shared very similar archaeal communities (57). Whether these and other differences and similarities have a methodological basis or are host based or are controlled by diet or animal management choices still remains to be elucidated.
A more detailed analysis of data reported by Wright et al. (54) reveals that diet may have an effect on the composition of the methanogen community. Three groups of sheep at the same site were fed three different diets: pasture, oaten hay based, and lucerne hay based. The abundance of different clades of Archaea varied in these three cohorts (P = 6.8 × 10−6; χ2-test). Diet is known to affect the composition of the bacterial community in the rumen (19, 47), but bacteria can be expected to interact more intimately with the diet since they will be using mainly feed components as their energy substrates. The majority of rumen archaea probably use hydrogen and possibly formate as their energy sources, which are formed during the primary fermentation of feed by the rumen bacteria, fungi, and protozoa. Diet can be expected to have an effect on methanogens due to changes in pH, which can affect methanogen activity (21), and through the presence of toxic compounds that may affect methanogens directly. Feed degradability will affect the rate of passage of digesta (25) and so may select for different species.
Abundant archaea associated with protozoa.
Rumen protozoa are known to harbor ecto- and endosymbiotic methanogens, based on characteristic fluorescence of F350 and F420 and FISH using archaea-specific oligonucleotide probes (9, 51). Based on clone library analyses, the majority (93.8%) of protozoan-associated archaea can be placed in the same three genus-level groups found to dominate the total rumen archaea, the genera Methanobrevibacter and Methanomicrobium and the RCC clade (Table 2). Other genus-level groupings appear to be much less abundant. There were notable variations in the abundance of different groups of protozoan-associated archaea in the five different studies. Members of the M. ruminantium clade were abundant (62.6%) in the study reported by Chagan et al. (3) while Methanomicrobium spp. dominated in the study of Regensbogenova et al. (37), and members of the RCC clade dominated in the study of Ohene-Adjei et al. (32). Sequences known to have originated from protozoa are well interspersed among those from total rumen samples among the Methanomicrobium, M. gottschalkii, and RCC clades. There are not enough sequences to determine if this is also true for the M. ruminantium clade. It is not clear that there are different protozoan-associated and free-living lineages within these clades. All of the studies on rumen-associated archaea are strongly biased because these studies were not made on the total rumen protozoa but on selected protozoa. The abundance of the individual protozoan species and the number of archaeal cells in or on each cell are not taken into account. The aim of these studies was not to describe total rumen protozoan-associated archaea, and so the interpretations made here must be regarded as a very tentative first analysis of abundant protozoan-associated rumen archaea.
The limited amount of information available does not support the idea that there are significant populations of uniquely protozoan-associated archaea in the rumen. Christophersen et al. (4) suggested that in some individual animals, the free-living and protozoan-associated archaea appeared to belong to the same species but that in other individuals they did not. Elimination of protozoa from the rumen of sheep resulted in changes in the archaeal community (30), but it was not clear if this was due to the disappearance of uniquely protozoan-associated methanogens or due to broader changes in rumen function as a result of defaunation that, in turn, affected the total archaeal community structure. The true significance of different clades of protozoan-associated archaea could be determined by careful analysis of the two groups in fractionated rumen samples and by using rRNA targeted FISH methods to observe the localization of cells of different archaeal groups in total rumen samples.
Roles and cooccurrence in the rumen.
Of the 11 clades of Archaea detected (Table 2), 9 consist of hydrogen-utilizing methanogens. These are members of the genera Methanobrevibacter, Methanomicrobium, Methanobacterium, Methanosphaera, and Methanimicrococcus. It is not clear how these manage to coexist in the rumen, but there does appear to be some difference in the abundance of the different groups in different studies, suggesting a host or feed influence. This interpretation assumes no methodological biases. The rumen archaea may also interact with different hydrogen-producing organisms, for example, with protozoa, bacteria, or fungi.
The physiology of members of the Qld26 clade is not known, but based on their close relationship with Methanobrevibacter spp., Methanobacterium spp., and Methanosphaera spp., they are likely to be hydrogen-utilizing methanogens. The physiology of members of the RCC clade and the Crenarchaeota in rumen is not known. The RCC clade corresponds to group C of Kemnitz et al. (18), and relatives have been detected in a number of digestive tract and other anaerobic environments. Uncultured relatives falling into closely allied clades have been detected in soil, on plant roots, in hydrothermal and deep-sea sediments, and in aquifers (18). The RCC clade has been labeled as a group of methanogens (31), but there is little evidence for this. Isolation of members of these groups in pure culture or demonstration of their physiology using molecular ecological techniques is required to define the role of these archaea in the rumen.
Limiting the activity of rumen methanogens in domesticated ruminants may result in gains in animal productivity if the rates and patterns of feed fermentation are not adversely affected. Almost 40 years ago, Czerkawski (5) reviewed the ideas and research in this already active field. Interest in inhibiting rumen methanogens has recently been renewed (26) due to concerns about the amounts of methane generated from domesticated ruminants. This ruminant-derived methane accounts for about one-quarter of all anthropogenic methane emissions and is implicated in human-induced global climate change (58). Knowledge of the ruminal methanogen community is an important part of developing strategies to mitigate rumen methane production. Assuming that the limited data available together constitute a good sample of global ruminant archaeal diversity, it can be concluded that only a few major groups of methanogens need to be targeted by antimethanogen agents in the first instance. It is not clear if elimination of these major groups will allow the less abundant members of other groups to take over the vacated niche. This will depend on the factors that limit or allow the different species to coexist in the rumen. These less abundant archaea may occupy specialist niches, but they may also be relegated to a minor part of the total community through competition. The limited diversity also means that strategies may be widely applicable across different countries. It is not clear whether other groups of methanogens are dominant in less-well-studied ruminant animals or if different feeds will allow otherwise minor groups of methanogens to dominate.
The few studies reported to date and summarized here give an indication of the global rumen archaeal community. This review is meant to act as a call for the development of more information rather than as a definitive summary. Many questions remain to be answered, and some have been briefly raised here. Fifty years after the initial isolation of methanogens from the rumen (1, 33, 43), the census of rumen archaea is beginning to show what groups are present, but this is not yet complete. New questions, such as how the groups apparently coexist and what determines the abundance of the different species, now need to be answered.
Acknowledgments
This work was supported by funding from the Pastoral Greenhouse Gas Research Consortium.
Footnotes
Published ahead of print on 18 April 2008.
REFERENCES
- 1.Beijer, W. H. 1952. Methane fermentation in the rumen of cattle. Nature 170:576-577. [DOI] [PubMed] [Google Scholar]
- 2.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2007. GenBank. Nucleic Acids Res. 35:D21-D25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chagan, I., M. Tokura, J. P. Jouany, and K. Ushida. 1999. Detection of methanogenic archaea associated with rumen ciliate protozoa. J. Gen. Appl. Microbiol. 45:305-308. [DOI] [PubMed] [Google Scholar]
- 4.Christophersen, C. T., A.-D. G. Wright, and P. E. Vercoe. 2004. Examining diversity of free-living methanogens and those associated with protozoa in the rumen. J. Anim. Feed Sci. 13(Suppl. 1):51-54. [Google Scholar]
- 5.Czerkawski, J. W. 1969. Methane production in ruminants and its significance. World Rev. Nutr. Diet. 11:240-282. [DOI] [PubMed] [Google Scholar]
- 6.DeLong, E. F., and N. R. Pace. 2001. Environmental diversity of Bacteria and Archaea. Syst. Biol. 50:470-478. [PubMed] [Google Scholar]
- 7.Denman, S. E., N. W. Tomkins, and C. S. McSweeney. 2007. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol. Ecol. 62:313-322. [DOI] [PubMed] [Google Scholar]
- 8.Dighe, A. S., K. Jangid, J. M. González, V. J. Pidiyar, M. S. Patole, D. R. Ranade, and Y. S. Shouche. 2004. Comparison of 16S rRNA gene sequences of genus Methanobrevibacter. BMC Microbiol. 4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay, B., G. Esteban, K. J. Clarke, A. G. Williams, T. M. Embley, and R. P. Hirt. 1994. Some rumen ciliates have endosymbiotic methanogens. FEMS Microbiol. Lett. 117:157-162. [DOI] [PubMed] [Google Scholar]
- 10.Friedrich, M. W. 2005. Methyl-coenzyme M reductase genes—unique functional markers for methanogenic and anaerobic methane-oxidizing Archaea. Methods Enzymol. 397:428-442. [DOI] [PubMed] [Google Scholar]
- 11.Garrity, G. M., T. G. Lilburn, J. R. Cole, S. H. Harrison, J. Euzéby, and B. J. Tindall. 2007. Taxonomic outline of the Bacteria and Archaea. Part 1. The Archaea, phyla Crenarchaeota and Euryarchaeota. Release 7.7. Michigan State University, Lansing, MI. www.taxonomicoutline.org. Accessed 6 March 2007.
- 12.Hungate, R. E. 1967. Hydrogen as an intermediate in the rumen fermentation. Arch. Microbiol. 59:158-164. [DOI] [PubMed] [Google Scholar]
- 13.Hungate, R. E., W. Smith, T. Bauchop, I. Yu, and J. C. Rabinowitz. 1970. Formate as an intermediate in the bovine rumen fermentation. J. Bacteriol. 102:389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irbis, C., and K. Ushida. 2004. Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J. Gen. Appl. Microbiol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 15.Janssen, P. H. 2007. Growing the recalcitrant. Microbiology 28:127-128. [Google Scholar]
- 16.Joblin, K. N. 2005. Methanogenic archaea, p. 47-53. In H. P. S. Makkar and C. McSweeney (ed.), Methods in gut microbial ecology for ruminants. Springer, Dordrecht, The Netherlands.
- 17.Jukes, T., and C. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 18.Kemnitz, D., S. Kolb, and R. Conrad. 2005. Phenotypic characterization of rice cluster III archaea without prior isolation by applying quantitative polymerase chain reaction to an enrichment culture. Environ. Microbiol. 7:553-565. [DOI] [PubMed] [Google Scholar]
- 19.Kocherginskaya, S. A., R. I. Aminov, and B. A. White. 2001. Analysis of the rumen bacterial diversity under two different diet conditions using denaturing gradient gel electrophoresis, random sequencing, and statistical ecology approaches. Anaerobe 7:119-134. [Google Scholar]
- 20.Konstantinidis, K. T., A. Ramette, and J. M. Tiedje. 2006. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B 361:1929-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lana, R. P., J. B. Russell, and M. E. Van Amburgh. 1998. The role of pH in regulating ruminal methane and ammonia production. J. Anim. Sci. 76:2190-2196. [DOI] [PubMed] [Google Scholar]
- 22.Lin, C., L. Raskin, and D. A. Stahl. 1997. Microbial community structure in gastrointestinal tracts of domestic animals: comparative analyses using rRNA-targeted oligonucleotide probes. FEMS Microbiol. Ecol. 22:281-294. [Google Scholar]
- 23.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 25.Mathison, G. W., E. K. Okine, A. S. Vaage, M. Kaske, and L. P. Milligan. 1995. Current understanding of the contribution of the propulsive activities in the forestomach to the flow of digesta, p. 23-41. In W. von Engelhardt, S. Leonhard-Marek, G. Breves, and D. Giesecke (ed.), Ruminant physiology: digestion, metabolism, growth and reproduction. Ferdinand Enke Verlag, Stuttgart, Germany.
- 26.McAllister, T. A., and C. J. Newbold. 2008. Redirecting rumen fermentation to reduce methanogenesis. Aust. J. Exp. Agric. 48:7-13. [Google Scholar]
- 27.McSweeney, C. S., S. E. Denman, A. D. G. Wright, and Z. Yu. 2007. Application of recent DNA/RNA-based techniques in rumen ecology. Asian-Aust. J. Anim. Sci. 20:283-294. [Google Scholar]
- 28.Miller, T. L. 1995. Ecology of methane production and hydrogen sinks in the rumen, p. 317-331. In W. von Engelhardt, S. Leonhard-Marek, G. Breves, and D. Giesecke (ed.), Ruminant physiology: digestion, metabolism, growth and reproduction. Ferdinand Enke Verlag, Stuttgart, Germany.
- 29.Miller, T. L., and C. Lin. 2002. Description of Methanobrevibacter gottschalkii sp. nov., Methanobrevibacter thaueri sp. nov., Methanobrevibacter woesei sp. nov., and Methanobrevibacter wolinii sp. nov. Int. J. Syst. Evol. Microbiol. 52:819-822. [DOI] [PubMed] [Google Scholar]
- 30.Morgavi, D. P., J. P. Jouany, C. Martin, and M. J. Ranilla. 2006. Archaeal community structure diversity in the rumen of faunated and defaunated sheep. Int. Congr. Ser. 1293:127-130. [Google Scholar]
- 31.Nicholson, M. J., P. N. Evans, and K. N. Joblin. 2007. Analysis of methanogen diversity in the rumen using temporal temperature gradient gel electrophoresis: identification of uncultured methanogens. Microb. Ecol. 54:141-150. [DOI] [PubMed] [Google Scholar]
- 32.Ohene-Adjei, S., R. M. Teather, M. Ivan, and R. J. Forster. 2007. Postinoculation protozoan establishment and association patterns of methanogenic archaea in the ovine rumen. Appl. Environ. Microbiol. 73:4609-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oppermann, R. A., W. O. Nelson, and R. E. Brown. 1957. In vitro studies on methanogenic rumen bacteria. J. Dairy Sci. 40:779-788. [Google Scholar]
- 34.Patterson, J. A., and R. B. Hespell. 1979. Trimethylamine and methylamine as growth substrates for rumen bacteria and Methanosarcina barkeri. Curr. Microbiol. 3:79-83. [Google Scholar]
- 35.Paynter, M. J. B., and R. E. Hungate. 1968. Characterization of Methanobacterium mobilis, sp. n., isolated from the bovine rumen. J. Bacteriol. 95:1943-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rea, S., J. P. Bowman, S. Popovski, C. Pimm, and A. D. G. Wright. 2007. Methanobrevibacter millerae sp. nov. and Methanobrevibacter olleyae sp. nov., methanogens from the ovine and bovine rumen that can utilize formate for growth. Int. J. Syst. Evol. Microbiol. 57:450-456. [DOI] [PubMed] [Google Scholar]
- 37.Regensbogenova, M., P. Pristaš, P. Javorsky, S. Y. Moon-van der Staay, G. W. M. van der Staay, J. H. P. Hackstein, C. J. Newbold, and N. R. McEwan. 2004. Assessment of ciliates in the sheep rumen by DGGE. Lett. Appl. Microbiol. 39:144-147. [DOI] [PubMed] [Google Scholar]
- 38.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 39.Sharp, R., C. J. Ziemer, M. D. Stern, and D. A. Stahl. 1998. Taxon-specific associations between protozoal and methanogen populations in the rumen and a model rumen system. FEMS Microbiol. Ecol. 26:71-78. [Google Scholar]
- 40.Shin, E. C., B. R. Choi, W. J. Lim, S. Y. Hong, C. L. An, K. M. Cho, Y. K. Kim, J. M. An, J. M. Kang, S. S. Lee, H. Kim, and H. D. Yun. 2004. Phylogenetic analysis of archaea in three fractions of cow rumen based on the 16S rDNA sequence. Anaerobe 10:313-319. [DOI] [PubMed] [Google Scholar]
- 41.Skillman, L. C., P. N. Evans, G. E. Naylor, B. Morvan, G. N. Jarvis, and K. N. Joblin. 2004. 16S ribosomal DNA-directed PCR primers for ruminal methanogens and identification of methanogens colonising young lambs. Anaerobe 10:277-285. [DOI] [PubMed] [Google Scholar]
- 42.Skillman, L. C., P. N. Evans, C. Strömpl, and K. N. Joblin. 2006. 16S rDNA directed PCR primers and detection of methanogens in the bovine rumen. Lett. Appl. Microbiol. 42:222-228. [DOI] [PubMed] [Google Scholar]
- 43.Smith, P. H., and R. E. Hungate. 1958. Isolation and characterization of Methanobacterium ruminantium sp. nov. J. Bacteriol. 75:713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soliva, C. R., I. K. Hindrichsen, L. Meile, M. Kreuzer, and A. Machmüller. 2003. Effects of mixtures of lauric and myristic acid on rumen methanogens and methanogenesis in vitro. Lett. Appl. Microbiol. 37:35-39. [DOI] [PubMed] [Google Scholar]
- 45.Stackebrandt, E., and B. M. Göbel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 46.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 47.Tajima, K., R. I. Aminov, T. Nagamine, H. Matsui, H. Nakamura, and Y. Benno. 2001. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tajima, K., T. Nagamine, H. Matsui, M. Nakamura, and R. I. Aminov. 2001. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol. Lett. 200:67-72. [DOI] [PubMed] [Google Scholar]
- 49.Tatsuoka, N., N. Mohammed, M. Mitsumori, K. Hara, M. Kurihara, and H. Itabashi. 2004. Phylogenetic analysis of methyl coenzyme-M reductase detected from the bovine rumen. Lett. Appl. Microbiol. 39:257-260. [DOI] [PubMed] [Google Scholar]
- 50.Tokura, M., I. Chagan, K. Ushida, and Y. Kojima. 1999. Phylogenetic study of methanogens associated with rumen ciliates. Curr. Microbiol. 39:123-128. [DOI] [PubMed] [Google Scholar]
- 51.Vogels, G. D., W. F. Hoppe, and C. K. Stumm. 1980. Association of methanogenic bacteria with rumen ciliates. Appl. Environ. Microbiol. 40:608-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitford, M. F., R. M. Teather, and R. J. Forster. 2001. Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolin, M. J. 1979. The rumen fermentation: a model for microbial interactions in anaerobic ecosystems. Adv. Microb. Ecol. 3:49-77. [Google Scholar]
- 54.Wright, A.-D. G., A. J. Williams, B. Winder, C. T. Christopherson, S. L. Rodgers, and K. D. Smith. 2004. Molecular diversity of rumen methanogens from sheep in Western Australia. Appl. Environ. Microbiol. 70:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright, A.-D. G., A. F. Toovey, and C. L. Pimm. 2006. Molecular identification of methanogenic archaea from sheep in Queensland, Australia, reveal more uncultured novel archaea. Anaerobe 12:134-139. [DOI] [PubMed] [Google Scholar]
- 56.Wright, A.-D. G., C. H. Auckland, and D. H. Lynn. 2007. Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Appl. Environ. Microbiol. 73:4206-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright, A.-D. G., X. Ma, and N. E. Obispo. 29 December 2007. Methanobrevibacter phylotypes are the dominant methanogens in sheep in Venezuela. Microb. Ecol. [Epub ahead of print.] doi: 10.1007/s00248-007-9351-x. [DOI] [PubMed]
- 58.Wuebbles, D. J., and K. Hayhoe. 2002. Atmospheric methane and global change. Earth Sci. Rev. 57:177-210. [Google Scholar]
- 59.Yanagita, K., Y. Kamagata, M. Kawaharasaki, T. Suzuki, Y. Nakamura, and H. Minato. 2000. Phylogenetic analysis of methanogens in sheep rumen ecosystem and detection of Methanomicrobium mobile by fluorescence in situ hybridization. Biosci. Biotechnol. Biochem. 64:1737-1742. [DOI] [PubMed] [Google Scholar]
- 60.Ziemer, C. J., R. Sharp, M. D. Stern, M. A. Cotta, T. R. Whitehead, and D. A. Stahl. 2000. Comparison of microbial populations in model and natural rumens using 16S rRNA-targeted probes. Environ. Microbiol. 2:632-643. [DOI] [PubMed] [Google Scholar]