Abstract
Streptococcus gordonii, an oral commensal organism, is a candidate vector for oral-vaccine development. Previous studies have shown that recombinant S. gordonii expressing heterologous antigens was weakly immunogenic when delivered intranasally. In this study, antigen was specifically targeted to antigen-presenting cells (APC) in order to potentiate antigen-APC interactions and increase the humoral immune response to the antigen. To achieve this goal, a single-chain variable-fragment (scFv) antibody against complement receptor 1 (CR1) was constructed. Anti-CR1 scFv purified from Escherichia coli was able to bind to mouse mixed lymphocytes and bone marrow-derived dendritic cells. The in vivo function of the anti-CR1 scFv protein was assessed by immunizing mice intranasally with soluble scFv and determining the immune response against the hemagglutinin (HA) peptide located on the carboxy terminus of the scFv. The serum anti-HA immunoglobulin G (IgG) immune response was dose dependent; as little as 100 ng of anti-CR1 scFv induced a significant IgG immune response, while such a response was minimal when the animals were given an unrelated scFv. The anti-CR1 scFv was expressed in S. gordonii as a secreted protein, which was functional, as it bound to dendritic cells. Mice orally colonized by the anti-CR1-secreting S. gordonii produced an anti-HA IgG immune response, indicating that such an approach can be used to increase the immune response to antigens produced by this bacterium.
Streptococcus gordonii is a commensal bacterium found in the oral cavities of humans. The organism is considered to be an attractive vector as a live-oral-vaccine vehicle (14, 23). A number of heterologous antigens have been expressed in this organism as either secreted proteins (15, 27) or cell wall-anchored proteins (16, 17, 19, 20, 25, 26). In the murine oral-colonization model, the recombinant S. gordonii was able to establish long-term colonization (16, 20). However, there are difficulties in stimulating a strong protective immune response against recombinant antigens following oral colonization.
Antigen targeting to immune cells has the potential to manipulate the immune system and elicit an immune response more efficiently. Monoclonal immunoglobulin G (IgG) antibodies have long been used as specific targeting vehicles. A number of reports have indicated success in achieving enhanced immune responses using antibodies to complement receptor 1 (CR1) and CR2 (3, 8, 30), Fc receptors (1, 2), and dendritic cell DEC205 receptor (5, 6). However, there are limitations in using intact IgG as a targeting vehicle; these limitations include a weak extravascular transport ability for IgG and difficulties with expressing whole IgG by bacteria. Single-chain variable-fragment (scFv) antibodies, however, offer a number of advantages, e.g., they can be readily produced by bacteria and can be easily engineered genetically as fusion proteins carrying polypeptide antigens. In the context of antigen targeting, scFvs against CR1 and -2 (21, 24), DEC205 (9), CD3 (31), and natural killer NKG2D receptor (29) have been reported with some degree of success.
In this study, we have taken the approach of expressing an scFv antibody against CR1 in S. gordonii to target immune cells. CR1 is a phagocytic receptor expressed by a number of immune cells, including dendritic cells, macrophages, neutrophils, eosinophils, and B cells, as well as erythrocytes. The anti-CR1 scFv was tested for binding to target cells in vitro and used in intranasal immunization in mice.
MATERIALS AND METHODS
Bacteria and growth conditions.
S. gordonii was cultivated in Todd-Hewitt broth containing 0.5% yeast extract at 37°C aerobically without shaking. Kanamycin and tetracycline, when needed, were included in the medium at 250 μg/ml and 10 μg/ml, respectively. Recombinant Escherichia coli was grown aerobically with vigorous shaking at 37°C in Luria Bertani broth (1% tryptone, 0.5% yeast extract, and 1% NaCl [wt/vol]) or Super Broth (1% MOPS [morpholinepropanesulfonic acid], 3% tryptone, 2% yeast extract [wt/vol]) containing either 100 μg/ml of ampicillin or 50 μg/ml of kanamycin. All antibiotics were purchased from Sigma-Aldrich Chemical Co. (Oakville, ON, Canada).
Construction of the anti-CR1 scFv.
The anti-CR1 antibody gene was obtained from the anti-CR1 monoclonal antibody-producing hybridoma HB8592 (American Type Culture Collection, Manassas, VA). The cells were grown in modified Dulbecco's medium supplemented with 5 mM β-mercaptoethanol and 10% fetal calf serum (Sigma-Aldrich). Total RNA was isolated from 1 × 106 hybridoma cells by extraction with the Trizol reagent (Invitrogen Life Technologies, Burlington, ON, Canada). The RNA obtained was dissolved in 40 μl of diethylpyrocarbonate-treated water. cDNA was synthesized from the RNA by reverse transcription using oligo(dT) as the primer and murine leukemia virus reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The variable light-chain (VL) and heavy-chain (VH) antibody fragments were amplified by PCR using mixed primers as described by Barbas et al. (4). The resulting 0.4-kb VL or VH DNA fragments were gel purified and used in overlapping PCR to produce the scFv antibody DNA. The resulting 0.8-kb scFv fragment was ligated into the SfiI sites of the phagemid pComb3X (4). The ligated DNA was transformed into E. coli XL1-Blue. The resulting construct (pCR1) was verified by restriction analysis and DNA sequencing (The John P. Robarts Research Institute DNA Sequencing Facility, London, ON, Canada).
Cloning of the anti-CR1 scFv gene in S. gordonii.
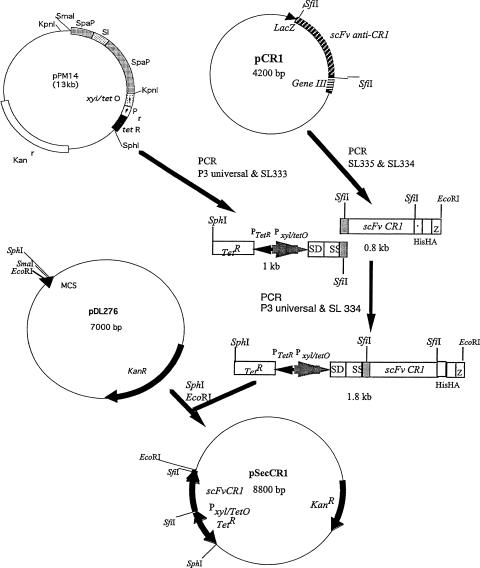
To express the anti-CR1 scFv in S. gordonii, pSecCR1 was constructed. The construction of pSecCR1 is outlined in Fig. 1. Briefly, the DNA coding for the anti-CR1 scFv and the C-terminal histidine and hemagglutinin (HA) tags was obtained by PCR using the primer pair SL334 (CGGAATTCCGTTAAGAAGCGTAGTCCGGAACGTC; the EcoRI site is underlined) and SL335 (GAGGCCCAGGCGGCCGAGCTC). A 1-kb DNA fragment carrying the TetR repressor gene, the tetracycline-inducible xyl/tetO promoter, and the ribosomal binding site and signal sequence of spaP originating from Streptococcus mutans, was amplified from pPM14 (18) using primers P3 Universal (ACGCCAAGCTTGCATGCCTGC; the SphI site is underlined) and SL333 (GAGCTCGGCCGCCTGGGCCTCATCGGCAAAAACCTTTTG). The two fragments were joined together via overlapping PCR, and the resulting 1.8-kb PCR product was cloned into the SphI and EcoRI sites on the E. coli-Streptococcus shuttle vector pDL276, creating pSecCR1. pSecCR1 was transformed into S. gordonii (hppG::Tetr) via natural transformation (13).
FIG. 1.
Construction of pSecCR1. See Materials and Methods for details. SS, signal sequence; SD, ribosomal-binding site; HIS, hexahistidine tag.
Isolation of scFvs from E. coli.
pCR1 was transformed into E. coli TB1 (New England Biolabs, Mississauga, ON, Canada) for the production of scFv. As a control, pK8 (pComb3X carrying scFv recognizing pertussis toxin [S. F. Lee, K. G. Chan, and S. A. Halperin, unpublished data]) was also introduced into E. coli TB1. The recombinant E. coli was grown in 500 ml of Super Broth for 16 h. Cells were harvested by centrifugation (10,000 × g) and resuspended in 5 ml NiCAM wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole) with 1 mM phenylmethylsulfonyl fluoride and sonicated (15 10-s bursts at amplitude 35 separated by 10-s cooling periods; Vibra cell; Sonics and Materials Inc., Danbury, CT). The sonicate was centrifuged at 20,000 × g for 20 min at 4°C. The clear supernatant was passed through a 1.5-ml NiCAM column (Sigma-Aldrich) equilibrated with the wash buffer. Unwanted proteins were removed with 20 ml of wash buffer. The bound scFv proteins were eluted with 5 ml of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole). The protein was concentrated by ultrafiltration using Amicon Ultra-Free-CL centrifugal filter devices (5,000-Da molecular mass cutoff).
scFv isolation from S. gordonii.
Anti-CR1 scFv was isolated from the culture supernatant of an overnight S. gordonii culture (100 ml) using a NiCAM affinity column essentially as described above. Briefly, the culture supernatant was passed through the NiCAM column twice, unwanted proteins were removed by a 20-ml wash, and scFv was eluted with four 500-μl elution buffers. The eluates were used in Western blotting and enzyme-linked immunosorbent assay (ELISA) as described below.
SDS-PAGE and Western immunoblotting.
Protein samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% polyacrylamide gels using the buffer system of Laemmli (12). Proteins were stained with Coomassie blue. The scFv protein concentration was determined by comparison with bovine serum albumin (BSA) standards on the same SDS-PAGE gel using Image J software (National Institutes of Health, Bethesda, MD). For Western immunoblotting, proteins were transferred to nitrocellulose membranes (28) and antigens were detected using an anti-HA monoclonal antibody (1/20,000; Sigma-Aldrich) and goat anti-mouse IgG alkaline phosphatase-conjugated antibody (1/20,000; Sigma-Aldrich).
Isolation of mixed lymphocytes.
Mixed lymphocytes were isolated from blood obtained from BALB/c mice using Ficoll Paque Plus (Amersham Biosciences, Baie d'Urfe, PQ, Canada) according to the manufacturer's instructions. The mixed lymphocytes were used to coat 96-well polystyrene microtiter plates.
Bone marrow-derived dendritic cells.
Dendritic cells were isolated and cultured from the femurs and tibias of BALB/c mice as described previously (7). The cells were harvested on day 6 and used in ELISA and immunofluorescence assays as described below.
Immunization.
The anti-CR1 and -K8 scFvs isolated as described above were dialyzed against phosphate-buffered saline (PBS) and used to immunize BALB/c mice (female; 5 weeks old; n = 4; Charles River Laboratory, St. Constant, Quebec, Canada) using lipopolysaccharide (LPS) (10 μg) from E. coli 6127:B8 (Sigma-Aldrich) as an adjuvant. The mice were immunized intranasally with 0.1 or 5 μg of scFv. One group of mice received LPS as a control. Intranasal immunization was achieved by pipetting 30 μl of prepared antigen dropwise into the nostrils of sedated animals. The animals were immunized on days 1, 23, 33, and 95. Sera and saliva were collected on day 7 prior to immunization and on day 107 using methods described previously (11).
Oral colonization.
The oral colonization of 4-week-old BALB/c mice (female; n = 3) with S. gordonii SecCR1 and S. gordonii SL3 was carried out using methods described previously (16). S. gordonii SL3 is a kanamycin-resistant strain but lacks the anti-CR1 gene (14). To induce the expression of the anti-CR1 scFv, the animals were given intraperitoneal injections of 100 μg tetracycline in 100 μl of PBS on days 1, 8, 16, and 22 (18). The animals were euthanized on day 29, at which time microbiological swabs were obtained from the nasal cavity, oral cavity, and pharynx (14, 16). The swabs were inoculated onto brain heart infusion agar plates containing kanamycin. The colonies obtained were observed to have colony morphology similar to that of S. gordonii and were gram-positive cocci in short chains. Western immunoblot analysis of culture supernatants of selected colonies confirmed the presence of the 33-kDa scFv protein. Saliva was collected 2 days prior to colonization and on day 28. Sera were obtained 1 day prior to colonization and at euthanasia.
ELISA.
ELISAs were used to test the function of scFv in binding to mixed lymphocytes and dendritic cells and detection of HA-specific antibodies in saliva and sera. For the detection of scFv binding, 96-well flat-bottom polystyrene microtiter plates (Corning Inc., Corning, NY) were coated with mixed lymphocytes and dendritic cells (8,000 cells/well), and the cells were lightly fixed with 0.125% gluteraldehyde at 4°C overnight. After overnight incubation, the coated plates were used immediately or stored at −80°C until they were used. The plates were blocked with 1% (wt/vol) BSA in PBS containing 0.1% (wt/vol) Tween 20 (PBST) for 1 h at room temperature, and scFv (200 ng/well) was added. The plates were incubated at 4°C for 16 h and washed with PBST, and the bound scFv was detected with commercial anti-HA monoclonal antibody and goat anti-mouse alkaline phosphatase conjugates.
For the detection of HA-specific antibodies in sera and saliva, microtiter plates were coated with 100 ng/well recombinant cyclophilin 18 (rC18) in ELISA coating buffer at 4°C overnight. The C18 gene originating from Toxoplasma gondii was PCR amplified from pKJ97 (10) using the primer pair SL330 (CCTGGCCCAGGCGGCCGAAAATGCCGGAGTCAGAAAG; the SfiI site is underlined) and SL331 (CCTGGCCGGCCTGGCCCTCCAACAAACCAATGTCCGT; the SfiI site is underlined). The amplified 514-bp PCR fragment encoding the mature C18 was inserted into the SfiI sites on pComb3X. The rC18 protein was isolated as a single 18-kDa protein from E. coli lysate using NiCAM chromatography as described above. rC18 had no homology to the anti-CR1 or K8 scFv but contained the same HA tag, and the protein was large enough to coat the wells easily. Preliminary results showed that consistent ELISA results were obtained compared to the inconsistent results when HA peptide was used. The plates were blocked with 1% BSA in PBST, and serially diluted sera or saliva was added (11). IgG antibodies were detected by the alkaline phosphatase-conjugated goat anti-mouse IgG. IgA antibodies were detected by a biotinylated goat anti-mouse IgA (α-chain specific; 1/20,000; Sigma-Aldrich), followed by an avidin-alkaline phosphatase conjugate (1/20,000; Sigma-Aldrich). The titers of antibodies were expressed as the reciprocal of the dilution that produced an A405 reading 0.05 higher than that of the pooled preimmune samples.
Immunofluorescence.
Bone marrow-derived dendritic cells obtained as described above were used as the CR1-expresssing cells. For a non-CR1-producing control, mouse TC-1 lung epithelial cells (ATCC) were used. TC-1 cells were cultured in RPMI 1640 medium to 90% confluence according to the supplier's instructions. The cells were detached with a 0.25% (wt/vol) trypsin-0.53 mM EDTA solution and centrifuged. One million dendritic cells or TC-1 cells were centrifuged (350 × g; 5 min; 4°C) and washed once in PBS. The cells were resuspended in 0.5 ml of PBS-BSA buffer (PBS plus 1% BSA), and 1 μg of a rat anti-mouse CD16/CD32 (FcγIII/II receptors) monoclonal IgG (BD Biosciences, Mississauga, ON, Canada) was added. The cells were incubated on ice for 5 min to allow the antibody to bind to the FcγIII/II receptors to eliminate background binding in subsequent steps. Following incubation, the cells were centrifuged and resuspended in 0.8 ml PBS-BSA buffer. The cells were divided into four equal aliquots and incubated at 4°C with gentle mixing as follows: aliquot 1 with 200 ng anti-CR1, aliquot 2 with 200 ng K8 scFv, and aliquos 3 and 4 with PBS-BSA buffer alone. After 1 h, the cells were washed in PBS-BSA, anti-HA (1/200) antibody was added to the first three aliquots, and PBS-BSA buffer was added to the fourth aliquot. The cells were further incubated for 1 h. The cells were then washed, and 0.5 μg of a rat anti-mouse IgG-specific fluorescein isothiocyanate-conjugated monoclonal antibody (clone R35-95; BD Biosciences) was added to the first three aliquots. To the fourth aliquot of cells, 0.25 μg of a rat anti-mouse CR1/2-specific fluorescein isothiocyanate-conjugated monoclonal antibody (clone eBIo8D9; eBiosciences, San Diego, CA) was added. The cells were incubated for 30 min at room temperature and washed, and a 10-μl droplet was allowed to dry as a smear on a glass slide in the dark. A drop of emulsion containing 90% glycerol and 10% PBS was applied to prevent rapid quenching of the fluorescence, and a coverslip was placed on top of the emulsion. The cells were viewed with a Leica DM2500 microscope (495-nm excitation; 525-nm emission), and images were captured with a digital camera. The images were converted to black and white and pixel reverted to allow easier viewing using the PhotoShop program.
Statistical analysis.
The results were analyzed with the paired Student's t test, and a P value of <0.05 was considered significant.
Nucleotide sequence accession number.
The GenBank accession number for the anti-CR1 scFv nucleotide sequence is EF694984.
RESULTS
Construction and expression of the anti-CR1 scFv.
The anti-CR1 scFv was constructed from RNA isolated from anti-CR1 monoclonal-antibody-producing hybridoma cells. The construct was verified by restriction analysis and DNA sequencing. The amino acid sequence deduced from the DNA sequence revealed the expected framework and complement-determining regions with the expected cysteine residues in both the VKand VH domains. In addition, the glycine-serine linker and the histidine and HA tags introduced in the cloning were identifiable (Fig. 2).
FIG. 2.
Deduced amino acid sequence of the anti-CR1 scFv. The framework regions (FR), complement-determining regions (CDR; boldface letters), cysteine residues for disulfide bond formation, glycine-serine linker, and hexahistidine and HA tags are indicated.
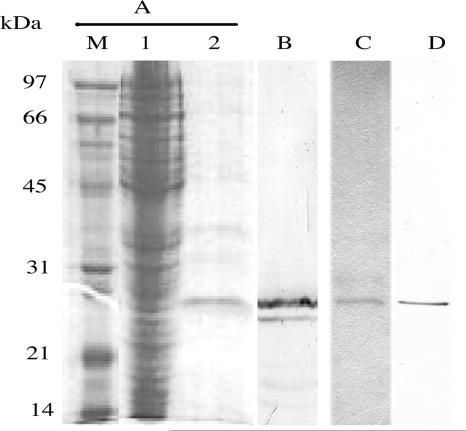
E. coli carrying pCR1 was shown to express a 27-kDa protein recognized by the commercial anti-HA antibody, indicating that scFv was correctly constructed. The 27-kDa protein was subsequently isolated from the E. coli lysate via affinity chromatography on a NiCAM column (Fig. 3). The isolated 27-kDa protein, in addition to a smaller protein, presumably a degradation product, was recognized by the commercial anti-HA antibody. The control K8 scFv was similarly isolated and recognized by the anti-HA antibody (Fig. 3). The amount of isolated anti-CR1 and K8 scFvs was approximately 130 μg/liter of culture.
FIG. 3.
SDS-PAGE and Western immunoblotting of anti-CR1 scFv protein produced by E. coli. (A) Coomassie blue-stained SDS-PAGE gel. Lane M, low-molecular-mass protein markers; lane 1, crude cell lysate; lane 2, anti-CR1 scFv eluted from the NiCAM column. (B) Immunoblot of the isolated anti-CR1 scFv probed with the anti-HA antibody. (C) Coomassie blue-stained K8 scFv isolated by NiCAM chromatography. (D) Immunoblot of the isolated K8 scFv probed with the anti-HA antibody.
Binding of anti-CR1 scFv to lymphocytes and dendritic cells.
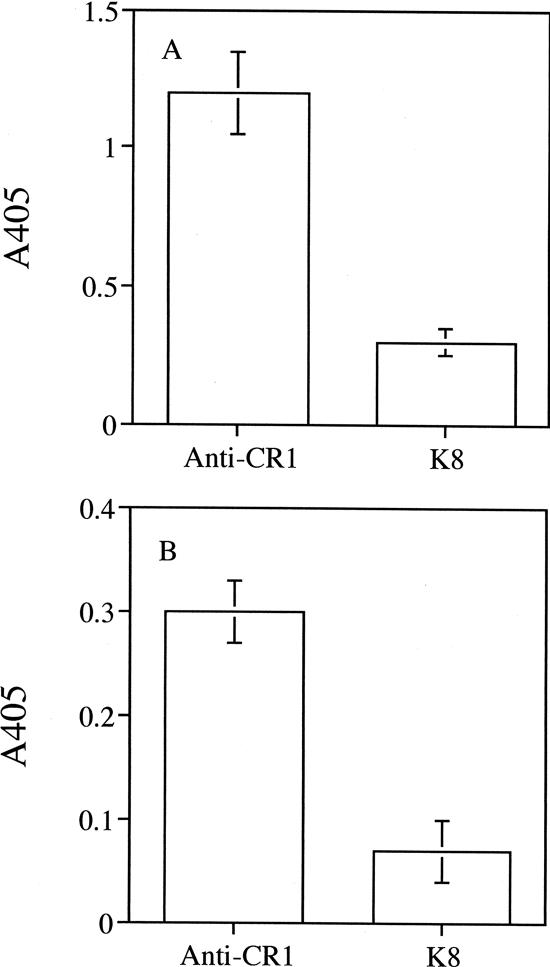
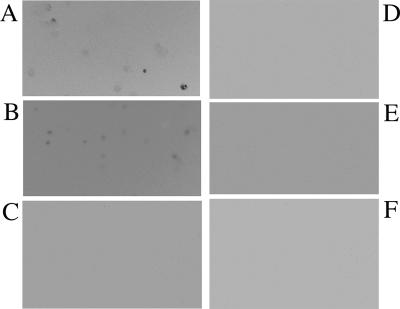
The ability of the isolated anti-CR1 scFv to bind to murine mixed lymphocytes and bone marrow-derived dendritic cells was examined. As shown in Fig. 4, the isolated anti-CR1 scFv bound to the two cell types while the control K8 scFv generated against pertussis toxin did not. The results indicated that the anti-CR1 scFv was functional. Binding was also observed using immunofluorescence microscopy (Fig. 5). The anti-CR1 scFv clearly bound to bone marrow-derived dendritic cells (Fig. 5B), while the K8 scFv did not (Fig. 5C). A commercial anti-mouse CR1 antibody also bound to the dendritic cells (Fig. 5A). As expected, none of the antibodies bound to the control epithelial cells (Fig. 5D to F).
FIG. 4.
Binding of anti-CR1 scFv to mouse mixed lymphocytes (A) and bone marrow-derived dendritic cells (B). The function of the purified scFv protein was determined by its ability to bind to cells expressing the ligand. The bound anti-CR1 scFv protein was detected by the anti-HA antibody. The results shown are means ± standard deviations of triplicate wells.
FIG. 5.
Detection of anti-CR1 scFv binding to mouse bone marrow-derived dendritic cells by immunofluorescence labeling. (A to C) Dendritic cells reacted with a commercial anti-CR1/2 monoclonal antibody, anti-CR1 scFv, and K8 scFv, respectively. (D to F) Mouse TC-1 epithelial cells reacted with a commercial anti-CR1/2 monoclonal antibody, anti-CR1 scFv, and K8 scFv, respectively. See Materials and Methods for details.
Induction of immune response to HA.
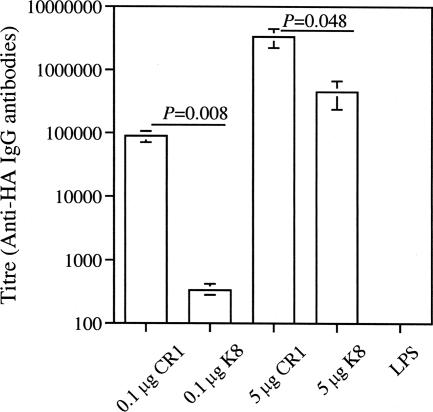
The ability of the anti-CR1 scFv to induce an immune response was tested in a murine immunization experiment. The HA tag present on the carboxy terminus of anti-CR1 served as the antigen. The unrelated K8 scFv, which also contained the same HA tag, was used as a control. The animals were given scFv intranasally with LPS as the adjuvant. As shown in Fig. 6, all groups that received the anti-CR1 scFv showed a strong IgG response, with the strongest response in the group that received 5 μg of protein. The increase in immune response between animals immunized intranasally with 0.1 μg and 5 μg anti-CR1 was statistically significant (P = 0.03). Although an IgG response was also observed in the control group receiving the K8 scFv, the response was weaker. The responses from immunization with 0.1 μg anti-CR1 and 0.1 μg K8 were significantly different (P = 0.008). The response by mice that received 5 μg of anti-CR1 was also higher than in mice that received 5 μg of K8 (P = 0.048). A response was not observed in the group that received only LPS. A salivary IgA response was not observed in any of the groups.
FIG. 6.
Antigen-specific serum IgG response in BALB/c mice following intranasal immunization with the anti-CR1 scFv. CR1, anti-CR1-scFv; K8, antipertussis toxin scFv; LPS, LPS alone. The results shown are means ± standard errors of three or four individual sera.
Expression and function of anti-CR1 in S. gordonii.
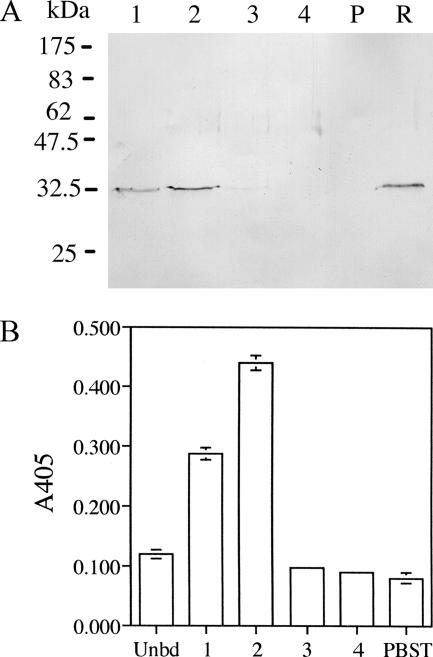
The anti-CR1 scFv gene was subcloned into pDL276 as outlined in Fig. 1. The expression of scFv was under the control of a tetracycline-inducible xyl-tetO promoter. The promoter was shown to be functional in E. coli and S. gordonii (18). E. coli carrying pSecCR1 was shown to express a 33-kDa protein recognized by the commercial anti-HA antibody, indicating the gene had been successfully subcloned (data not shown). pSecCR1 was transformed into a tetracycline-resistant strain of S. gordonii DL-1 (hppG::tet). The transformant secreted a 33-kDa protein recognized by the commercial anti-HA antibody, while such a band was absent in the culture supernatant from the parent strain (Fig. 7A). The 33-kDa protein bound to the NiCAm column and could be eluted with imidazole. The eluted protein was able to bind to mouse dendritic cells in the ELISA (Fig. 7B), indicating that it was functional.
FIG. 7.
Expression and function of anti-CR1 scFv in S. gordonii. (A) Immunoblot of culture supernatant from S. gordonii SecCR1 (lane R) or parent S. gordonii (lane P) concentrated 20-fold by trichloroacetic acid precipitation and anti-CR1 scFv isolated by NiCAM chromatography. Lanes 1 to 4 are fractions from the column. (B) Eluted (bars 1, 2, 3, and 4) and unbound (Unbd) fractions from the NiCAM column were tested for binding to dendritic cells in ELISA. PBST, wells in which PBST was added in place of fractions. The results shown are means ± standard deviations of triplicate wells.
Induction of an immune response following oral colonization with S. gordonii.
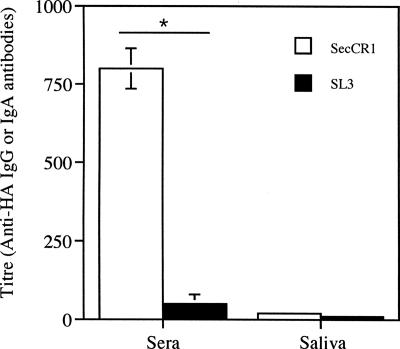
The ability of the anti-CR1-producing S. gordonii to induce an immune response was tested in a mouse oral-colonization study. The results showed that the two groups of mice were colonized to the same degree. In mice given S. gordonii SecCR1, the oral cavity, pharynx, and nasal cavity contained 1,013 ± 200 (mean ± standard error), 115 ± 56, and 20 ± 10 CFU of S. gordonii, respectively. In mice given S. gordonii SL3, there were 950 ± 370, 130 ± 77, and 25 ± 7 CFU of S. gordonii in these sites. Sera from mice colonized with the anti-CR1-producing S. gordonii showed an immune response, while that from mice colonized with the control S. gordonii SL3 did not (Fig. 8). No difference in IgA response was observed in saliva obtained from both groups.
FIG. 8.
Immune response from oral colonization with S. gordonii. Antigen-specific IgG antibody in sera and IgA antibody in saliva from mice colonized with S. gordonii SecCR1 or SL3. The results shown are means ± standard errors of three individual sera or salivas. *, P < 0.05.
DISCUSSION
In the present study, the single-chain recombinant antibody against CR1 was constructed from the cDNA obtained from a hybridoma. The construct was verified by DNA sequencing. The scFv was successfully expressed in E. coli and S. gordonii. The binding assay showed that the scFv was able to bind to dendritic cells and mixed lymphocytes, indicating that the scFv retained its function. The expression of a functional anti-CR1 scFv derived from a different hybridoma, 7G6, was previously reported by Prechl et al. (24), although that scFv remained in the insoluble fraction of E. coli while ours was mostly soluble. To the best of our knowledge, this is the first report of the expression of anti-CR1 scFv in S. gordonii. Oggioni et al. (22) previously described the surface expression of an scFv against the Streptococcus mutans major surface protein antigen P1 in S. gordonii and demonstrated the ability of such an scFv to reduce dental caries in an animal model. These results collectively indicate that functional scFvs can be expressed in E. coli and S. gordonii.
The anti-CR1 scFv isolated from E. coli was able to elicit a very robust antibody response when given intranasally. The antibody response was significantly higher than that from mice immunized with the control K8 scFv, indicating that the anti-CR1 scFv works as postulated and efficiently targets antigen to phagocytic cells, resulting in an increased immune response. These data are in agreement with those observed for the anti-CR1/2 whole IgG molecule coupled to ovalbumin (3) or anthrax lethal toxin (30). However, our result is superior to that reported for the anti-CR1/2 scFv derived from hybridoma 7G6, which failed to elicit a significant antibody response (24) even though the scFv was taken up and the influenza virus peptide carried on the scFv was efficiently presented by antigen-presenting cells in vitro (21). The difference may be due to the use of an LPS adjuvant in our experiment, whereas no adjuvant was used for the 7G6-derived scFv. LPS is known to up-regulate the expression of major histocompatibility complex molecules, CD40 ligand, and cytokines by antigen-presenting cells, leading to full activation of T cells and ultimately a strong immune response. Therefore, we believe that LPS played a role as an adjuvant in facilitating the robust immune response to the HA peptide in our experiment. The adjuvant effect of LPS is likely responsible for the observed immune response to the control K8 scFv, particularly at 5-μg doses.
It is noteworthy that the superior immune response observed in this study was obtained via intranasal immunization. All the studies reported in the literature were performed using the parenteral route. In view of the actual amount of HA administered (0.1 μg of anti-CR1 contains approximately 0.003 μg of HA peptide) and the fact that a large quantity of antigen is usually needed for mucosal immunization, the observed immune response is quite remarkable and further indicates the efficiency of the targeting ability of the anti-CR1 scFv.
The results of the oral-colonization experiment are in agreement with those observed using purified scFv delivered intranasally. We estimated that the amount of anti-CR1 scFv produced by S. gordonii was 17 ng/109 CFU (unpublished data), and in previous colonization studies, we reported that S. gordonii was present to the order of 103 CFU on the oral-nasal pharynx (14). Thus, the amount of scFv produced during colonization at a given time is small (but production is continuous), and yet an immune response was observed, further indicating the efficiency of the anti-CR1 scFv.
It is also interesting that in both scFv protein intranasal-immunization and bacterial-colonization experiments, only a systemic IgG response was observed. Typically, intranasal immunization elicits both a systemic and a mucosal antibody response, and this was certainly the case with other protein antigens (11, 14). On the other hand, our previous results from oral colonization with the recombinant S. gordonii elicited only a weak mucosal IgA response (16). The deviation from these past results suggests that the anti-CR1 scFv may have a unique feature in that it selectively promotes a systemic response, although the mechanism remains unclear.
In summary, an scFv against CR1 was constructed and expressed as a functional protein by both E. coli and S. gordonii. The scFv was able to elicit an enhanced immune response to the HA peptide linked to its C terminus, presumably due to its antigen-targeting ability. An immune response was also observed in a mouse S. gordonii oral-colonization experiment, indicating that the scFv may be used as an antigen-targeting tool in the bacterium.
Acknowledgments
We thank Yi-Jing Li and Annette Morris for their valuable technical assistance. We also thank Carlos Barbas III for providing pComb3X and Keith Joiner for providing pKJ97.
J. B. Knight was an IWK graduate studentship recipient. This study was supported by the Canadian Institutes of Health Research (CIHR).
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Adamova, E., M. C. Walsh, R. R. Gosselin, K. Hale, M. T. Preissler, R. F. Graziano, and E. J. Gosselin. 2005. Enhanced antigen-specific antibody and cytokine responses when targeting antigen to human FcGamma receptor type I using an anti-human Fcγ receptor type I-streptavidine fusion protein in an adjuvant-free system. Immunol. Investig. 34:417-429. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, C. F., and D. M. Mosser. 2002. Cutting edge: biasing immune responses by directing antigen to macrophage Fcγ receptors. J. Immunol. 198:3697-3701. [DOI] [PubMed] [Google Scholar]
- 3.Baiu, D. C., J. Prechl, A. Tchorbanov, H. D. Molina. A. Erdei, A. Sulica, P. J. A. Capel, and W. L. W. Hazenbos. 1999. Modulation of the humoral immune response by antibody-mediated antigen targeting to complement receptors and Fcγ receptors. J. Immun. 162:3125-3130. [PubMed] [Google Scholar]
- 4.Barbas, C. L., D. R. Barton, and J. K. Scott. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 5.Bonifaz, L., D. Bonnyay, K. Mahnke, M. Rivera, M. C. Nussenzwei, and R. M. Steinman. 2002. Efficient targeting of protein antigen to the dendritic cell receptor DEC 205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J. Exp. Med. 196:1627-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifaz, L. C., D. P. Bonnyay, A. Charalambous, D. I. Darguste, S. Fuji, H. Soares, M. K. Brimnes, B. Moltedo, T. M. Moran, and R. M. Steinman. 2004. In vivo targeting of antigens to maturing dendritic cells via the DEC 205 receptor improves T cell vaccination. J. Exp. Med. 199:815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, K. G., M. Mayer, E. M. Davis, S. A. Halperin, T. J. Lin, and S. F. Lee. 2007. The roles of d-alanylation of Streptopcoccus gordonii lipoteichoic acid in innate and adaptive immunity. Infect. Immun. 75:3033-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig, M. L., M. L. Reinagel, E. N. Marti, R. Schlimgen, A. Nardin, and R. P. Taylor. 1999. Infusion of bispecific monoclonal antibody complexes into monkeys provides immunologic protection against later challenge with a model pathogen. Clin. Immunol. 92:170-180. [DOI] [PubMed] [Google Scholar]
- 9.Demangel, C., J. Zhou, A. B. H. Choo, G. Shoebridge, G. M. Halliday, and W. J. Britton. 2005. Single chain antibody fragments for the selective targeting of antigens to dendritic cells. Mol. Immunol. 42:979-985. [DOI] [PubMed] [Google Scholar]
- 10.High, K. P., K. A. Joiner, and R. E. Handschumacher. 1994. Isolation, cDNA sequences, and biochemical characterization of the major cyclosporine-binding proteins of Toxoplasma gondii. J. Biol. Chem. 269:9105-9112. [PubMed] [Google Scholar]
- 11.Knight, J. B., Y. Y. Huang, S. A. Halperin, R. Anderson, A. Morris, A. MacMillan, T. Jones, D. S. Burt, G. Van Nest, and S. F. Lee. 2006. Immunogenicity and protective efficacy of a recombinant filamentous hemagglutinin from Bordetella pertussis. Clin. Exp. Immunol. 144:5543-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Lee C. W., S. F. Lee., and S. A. Halperin. 2004. Expression and immunogenicity of a recombinant diphtheria toxin fragment A in Streptococcus gordonii. Appl. Environ. Microbiol. 70:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, S. F. 2003. Oral colonization and immune responses to Streptococcus gordonii: potential use as a vector to induce antibody against respiratory pathogens. Curr. Opin. Infect. Dis. 16:231-235. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. F., S. A. Halperin, J. B. Knight, and A. Tait. 2002. Purification and immunogenicity of a recombinant Bordetella pertussis S1S3FHA fusion protein expressed by Streptococcus gordonii. Appl. Environ. Microbiol. 68:4253-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, S. F., S. A. Halperin, H. Wang, and A. MacArthur. 2002. Oral colonization and immune responses to Streptococcus gordonii expressing a pertussis toxin S1 fragment in mice. FEMS Microbiol. Lett. 208:175-178. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S. F., R. J. March, S. A. Halperin, G. Faulkner, and L. Gao. 1999. Surface expression of a protective recombinant pertussis toxin S1 subunit fragment in Streptococcus gordonii. Infect. Immun. 67:1511-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallaley, P. P., S. A. Halperin, A. Morris, A. MacMillian, and S. F. Lee. 2006. Expression of pertussis toxin S1 fragment by inducible promoters in oral streptococci and immune responses elicited during oral colonization in mice. Can. J. Microbiol. 52:436-444. [DOI] [PubMed] [Google Scholar]
- 19.Medaglini, D., A. Ciabattini, M. R. Spinosa, T. Maggi, H. Marcotte, M. R. Oggioni, and G. Pozzi. 2001. Immunization with recombinant Streptococcus gordonii expressing tetanus toxin fragment C confers protection from lethal challenge in mice. Vaccine 19:1931-1939. [DOI] [PubMed] [Google Scholar]
- 20.Medaglini, D., G. Pozzi, T. P. King, and V. A. Fischetti. 1995. Mucosal and systemic immune responses to a recombinant protein expressed on the surface of the oral commensal bacterium Streptococcus gordonii after oral colonization. Proc. Natl. Acad. Sci. USA 92:6868-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molnár, E., J. Prechi, A. Isaák, and A. Erdei. 2003. Targeting with scFv: immunomodulation by complement receptor specific constructs. J. Mol. Rec. 16:318-323. [DOI] [PubMed] [Google Scholar]
- 22.Oggioni, M. R., C. Beninati, M. Boccanera, D. Medaglini, M. R. Spinosa, T. Maggi, S. Conti, W. Magliani, F. De Bernardis, G. Teti, A. Cassone, G. Pozzi, and L. Polonelli. 2001. Recombinant Streptococcus gordonii for mucosal delivery of a scFv microbicidal antibody. Int. Rev. Immunol. 20:275-287. [DOI] [PubMed] [Google Scholar]
- 23.Oggioni, M. R., D. Medaglini, T. Maggi, and G. Pozzi. 1999. Engineering the gram positive cell surface for construction of bacterial vaccine vectors. Methods 19:163-173. [DOI] [PubMed] [Google Scholar]
- 24.Prechl, J., A. Tchorbanov, A. Horvath, D. C. Baiu, W. Hazenbos, E. Rajnavolgyi, I. Kurucz, P. J. Capel, and A. Erdei. 1999. Targeting of influenza epitopes to murine CR1/CR2 using single-chain antibodies. Immunopharmacology 42:159-165. [DOI] [PubMed] [Google Scholar]
- 25.Ricci, S., D. Medaglini, C. M. Rush, A. Marcello, S. Peppoloni, R. Manganelli, G. Palu, and G. Pozzi. 2000. Immunogenicity of the B monomer of Escherichia coli heat-labile toxin expressed on the surface of Streptococcus gordonii. Infect. Immun. 68:760-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma, A., H. Nagata, N. Hamada, H. T. Sojar, D. E. Hrudy, H. K. Kuramitsu, and R. J. Genco. 1996. Expression of functional Porphyromonas gingivalis fimbrillin polypeptide domains on the surface of Streptococcus gordonii. Infect. Immun. 62:3933-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma, A., H. T. Sojar, D. E. Hruby, H. K. Kuramitsu, and R. J. Genco. 1997. Secretion of Porphyromonas gingivalis fimbrillin polypeptides by recombinant Streptococcus gordonii. Biochem. Biophys. Res. Commun. 238:313-316. [DOI] [PubMed] [Google Scholar]
- 28.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Von Strandmann, E. P., H. P. Hansen, K. S. Reiners, R. Schnell, P. Borchmann, S. Merkert, V. R. Simhadri, A. Draube, M. Reiser, I. Purr, M. Hallek, and A. Engret. 2006. A novel bispecific protein (ULBP2-BB4) targeting the NKG2D receptor on natural killer (NK) cells and CD138 activates NK cells and has potent antitumor activity against human multiple myeloma in vitro and in vivo. Blood 107:1955-1962. [DOI] [PubMed] [Google Scholar]
- 30.Whipple, E. C., A. H. Ditto, R. S. Shanahan, J. J. Gatesman, S. F. Little, R. P. Taylor, and M. A. Lindorfer. 2007. Low doses of antigen coupled to anti-CR2 mAbs induce rapid and enduring IgG immune responses in mice and cynomolgus monkeys. Mol. Immunol. 44:377-388. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida, S., T. Kobayashi, H. Matsuoka, C. Seki, W. L. Gosnell, S. P. Chang, and A. Ishii. 2002. T cell activation and cytokine production via a bispecific single-chain antibody fragment targeted to blood-stage malaria parasites. Blood 101:2300-2306. [DOI] [PubMed] [Google Scholar]