Abstract
In this study we estimate the seroprevalence of foot-and-mouth disease virus (FMDV) in wildlife from eastern and central Africa. Sera were sourced from between 1994 and 2002 from a rinderpest surveillance program. Our study compared a nonstructural protein enzyme-linked immunosorbent assay (Cedi test) with a virus neutralization test. The study shows that there is only a low seroprevalence of FMDV in sampled nonbuffalo species. The seroprevalence in the Cape buffalo was high for SAT2, lower for SAT1, and lowest for SAT3. As the SAT2 serotype was most prevalent, the Cedi test largely reflected the occurrence of SAT2-positive animals. The results also suggest that SAT2 became dominant around 1998, with a large increase in seroprevalence. The sensitivity and specificity of the Cedi test were estimated by comparison to the combined virus neutralization test results from all three SAT tests. A Bayesian implementation of the Hui-Walter latent class model was used to estimate the test parameters. The model permits estimation in the absence of a gold standard test. The final model, using noninformative priors and assuming conditional independence of test performance, estimated Cedi test sensitivity at 87.7% and specificity at 87.3%. These estimates are similar to those for domestic bovines; they suggest that the Cedi test is a useful tool for screening buffalo for infection with the various serotypes of FMDV.
Foot-and-mouth disease (FMD) is a highly contagious viral disease of even-toed ungulates (Artiodactyla) caused by the single-stranded positive-sense RNA foot-and-mouth disease virus (FMDV; Aphthovirus, Picornaviridae). There are seven distinct serotypes recognized globally, known as O, A, C, SAT1, SAT2, SAT3, and Asia1, of which only Asia1 has not been seen in Africa (25). There is little cross-protection between serotypes, although it has been suggested that there may be problems differentiating exposure in multiply infected or exposed buffalo when using virus neutralization tests (VNT) (21). In the 1900s, serotypes O, A, and C spread from northern Africa and into southern Africa (11), while the SAT viruses spread northwards with sporadic incursions into the Middle East (45). FMD is one of the most important animal diseases and has major impacts on a country's ability to trade in livestock and animal products. In order to facilitate trade between countries and prevent trade barriers, many countries are signatories to the Sanitary and Phytosanitary agreement of the World Trade Organization. To justify any trade barriers, countries are required to demonstrate that trade in particular animals or plants or their products is likely to put them at an unacceptable risk of disease introduction. To satisfy international regulations, these risks need to be assessed as part of a clear and transparent risk analysis as required by the World Organisation for Animal Health (OIE).
Wildlife in Africa, particularly the Cape buffalo (Syncerus caffer), have been identified as natural hosts for the SAT serotypes of FMDV, although they may be infected by all serotypes (20, 22). Also, the strong spatial associations between molecular types from outbreaks in cattle and virus recovered from buffalo suggest that any control strategy for FMD in cattle must address control in buffalo (43). This differs from the general situation in other parts of the world, where control in cattle has tended to result in disappearance of the disease in wildlife populations, and probably reflects the importance of buffalo as a natural reservoir (42). Cattle in many areas of Africa are managed on open rangeland with communal grazing and potential contact with wildlife populations. This wildlife-livestock interface is critical for disease transmission, particularly around common watering points and through contamination of grazing. In southern Africa FMD control has relied upon wildlife fences to keep cattle and buffalo separate, combined with vaccination in buffer zones (40, 43, 45). The planned global eradication of FMDV will require control in buffalo in Africa and a better understanding of the epidemiology in wildlife, particularly in Africa. Serosurveillance will be improved by using the new nonstructural protein (NSP) enzyme-linked immunosorbent assays (ELISAs) now available, which offer the potential to identify animals seropositive for any of the seven serotypes in a single, affordable test (3, 4, 14, 28-30, 34, 35).
This paper describes FMDV serology using the Cedi NSP assay and VNT for the three SAT serotypes. A Bayesian latent class model was used to estimate the parameters (sensitivity and specificity) of the Cedi test and the seroprevalence in the different regions in the absence of a gold standard test for these populations (17).
MATERIALS AND METHODS
Study populations and sampling.
Serum samples were collected from wild ungulates, including buffalo, antelope, and warthogs. These animals were from eastern and central Africa and were originally sampled as part of a wildlife health surveillance project run by the Kenya Wildlife Service, the Pan African Rinderpest Eradication Campaign, and the Programme for the Control of Epizootics from 1994 to 2005, under the auspices of the African Union Inter African Bureau for Animal Resources (IBAR). Serosurveillance for rinderpest virus antibodies in wildlife populations in eastern and central Africa became an important part of the monitoring of virus activity in the closing stages of eradication (27). The wildlife were sampled in and around a large number of mainly unfenced parks and wildlife reserves across eastern and central Africa. Sampled herds were selected partly on the basis of susceptibility and contact with livestock populations. All the buffalo populations sampled had contact with livestock, with the exception of those in Garamba in the Democratic Republic of Congo. Our FMD serological study is based on a subset of sera taken from the bank and sent to the Institute for Animal Health (IAH) Pirbright for screening (Table 1). The selection was made to provide the widest coverage of the sampled zones possible. It focused on the Cape buffalo, a preferred host for FMD virus. The serum samples were obtained from free-ranging wildlife after immobilization by either physical restraint with netting or through remotely injected chemical anesthetic agents (etorphine hydrochloride with a variety of sedative or tranquilizers, e.g., xylazine and acepromazine). Different national teams were used, but all of them were supervised and supported in the field by the wildlife health specialist from the African Union IBAR. A total of 731 sera from 27 species from Kenya, Tanzania, Ethiopia, Sudan, and Chad were screened for FMD virus antibody using a Cedi test FMDV-NS test kit (Cedi Diagnostics B.V.) and by VNT for SAT1, SAT2, and SAT3.
TABLE 1.
Species sampled
| Species | No. sampled |
|---|---|
| Buffalo, Syncerus caffer | 483 |
| African and desert warthog, Phaecocherus africans and P. aethiopicus | 52 |
| White-eared kob, Kobus leucotis | 50 |
| Giraffe, Giraffa camelopardalis | 34 |
| Eland, Taurotragus oryx | 19 |
| Thomson's and Grant's gazelle, Gazella thomsoni and G. grantii | 17 |
| Impala, Aepyceros melampus | 13 |
| Lelwel, Swayne's, and Coke's hartebeest, Alcelaphus buselaphus lelwel, A. buselaphus swaynei, and A. buselaphus buselaphus | 14 |
| Lesser and greater kudu, Tragelaphus imberbis and T. strepsiceros | 9 |
| Topi or tiang, Damaliscus lunatus | 9 |
| Beisa and fringe-eared oryx, Oryx beisa and O. gazella | 7 |
| Cow, Bos indicus | 4 |
| Roan antelope, Hippotragus equinus | 7 |
| Wildebeest, Connochaetes taurinus | 5 |
| Hirola, Beatragus hunteri | 3 |
| Bushbuck, Tragelaphus scriptus | 1 |
| Gerenuk, Litocranius walleri | 1 |
| Mountain nyala, Tragelaphus buxtoni | 1 |
| Sable, Hippotragus niger | 1 |
| Waterbuck, Kobus ellipsyprimnus defassa | 1 |
| Total | 731 |
NSP serology.
The Cedi test FMDV-NS test (Cedi test) is a blocking ELISA that detects antibodies against the nonstructural 3ABC protein of FMDV. It is independent of serotype and may be used to detect exposure in vaccinated animals. In addition, as a blocking ELISA it can be used for all species without the need for a species specific antigen. This test has not been validated for use in wildlife species. The monoclonal antibody (MAb) is based on European viruses (34), so the test can be expected to perform suboptimally for African viruses and the immune response may be as short as 40 days. This means that exposed animals may test negative. Test plates in the kit have been coated with a 3B-specific MAb and incubated with the 3ABC protein, resulting in FMDV-NS antigen bound to the coated MAb. The tests were carried out according to the manufacturer's instructions and as previously reported (36). All samples were tested in duplicate. Plates were read by measuring the optical density (OD) at a wavelength of 450 nm. The ODs of all samples including the controls were calculated and are expressed as the percent inhibition (PI) as follows: PI = 100 − [(OD of test sample)/(mean OD of negative controls)] × 100.
A PI of <50% was considered negative, and a PI of ≥50% was considered positive for recent exposure (within the previous 6 to 12 months) (7, 14). More specifically, a PI value of ≥50% but <70% was considered a weak positive result, and a PI value of ≥70% was considered a strong positive result (36).
VNT.
Titers of neutralizing antibodies of all wildlife sera against FMDV SAT105, SAT2 Eritrea, and SAT309 were measured by microneutralization assay as described in the OIE Manual of Diagnostic Tests and Vaccines (31). Serum samples which were found positive by Cedi test and negative by all three SAT tests were examined for the titers of neutralizing antibodies against O1 Manias, A22 Iraq, C Noville, and Asia Shamir (ISR 3/89). The cutoff for positivity with the VNT was a titer of ≤1:45.
Statistical analysis.
Cedi test and VNT results for each animal were recorded in an Excel spreadsheet (Microsoft Corp.) with species, age, sex, sampling location, and sampling date. An animal which was VNT positive for any SAT serotype was considered VNT SAT positive. The analysis was repeated with a combined VNT for all serotypes but this did not change the results of the parameter estimates, and these data are not included in this paper.
Descriptive statistical analysis was carried out using the R program (http://www.R-project.org). The proportion of samples positive by the Cedi test was estimated for each species as well as by year and by age group for the buffalo samples. The Bayesian latent class model was parameterized using the BRugs package (41) in R, an open access version of WinBugs (38). Convergence of the chains and stability of the estimates was assessed using the Gelman-Rubin statistic (10, 44). The Hui-Walter latent class analysis requires that the buffalo be divided into three distinct subpopulations. This was done geographically using the K-cluster function in Minitab 14 (Minitab Inc.) with the latitude and longitude of the location of each sampled buffalo. Geographical clustering with K-means clustering was used to generate three spatially distinct populations that would have some epidemiological meaning. Randomly allocation to three groups would risk generating three populations with an almost identical prevalence, which would causes poor identifiability for the model.
The no gold standard model.
The sensitivity (Se) of a diagnostic test is the probability of a positive test result conditional on the animal being infected or in this case truly seropositive and can be expressed as Pr(T+ D+). The specificity (Sp) of a diagnostic test is the probability of a negative test result conditional on the animal not being infected or in this case truly seronegative and can be expressed as Pr(T− D−). The basic Hui-Walter latent class model (23) assumes that the test parameters (Se and Sp) are constant across all populations and that the tests are conditionally independent given the true status of the animal, i.e., given an animal's status, knowing the result of the first test does not change the likelihood of a particular result with the second test. We ran several versions of the model, starting first with the simplest three-population two-test model, assuming test conditional independence and uniform priors on test performance. The latent class model can be expressed as follows: Oi Sej,Spj,pi ∼ multinominal(Pri, ni) for populations i = 1 to 3 and tests j = 1, 2, where Oi is the vector of observed counts for test 1 and test 2 for all four possible combinations of the two tests for population i (++, +−, −+, and −−) taken from the two-by-two contingency table and Pri is a vector of probabilities for each count in the table for each population i. For example, the probability of being VNT and Cedi test positive in population i can be written as Pri(++) = Se1 Se2 pi + (1 − Sp1)(1 − Sp2)(1 − pi), where Se1 is the sensitivity of the first test, Sp1 is the specificity of the first test, Se2 is the sensitivity of the second test, Sp2 is the specificity of the second test, and pi is the true seroprevalence in population i (in this model, this will be 1, 2, or 3).
The Bayesian model allows the use of prior information about the test to be included in the estimation process, and this can be thought of as our belief of the distribution of the possible estimates of Se and Sp prior to looking at our new data. This explicitly allows us to state how certain we are about the test and then to combine this with the data from the study to get a posterior belief, which is a weighted estimate based on our prior belief and the data. We then used prior distributions for the Cedi test taken from a review of the Cedi test's performance (6). These were SeCEDI TEST Beta (1.4, 1.2) and SpCEDI TEST Beta (99, 6), which reflect a lack of certainty about the true estimate of the test's Se and a high level of certainty about the estimate of the test's Sp.
As both of the test systems are based around antibody responses, it is likely that there is some conditional dependence in the tests (18). In other words, given that we know the test result from one test, we can know more about what the result of the other test is likely to be, over and above if we knew the infection status. This may lead to biased estimates, as it violates one of the fundamental assumptions of the Hui-Walter model. Therefore, we used a modified version of this model, allowing for conditional dependence between tests, by inclusion of the covariance of test results for disease-positive (covD+) and for disease-negative groups (covD−) (5). The new model can be written as follows: Oi Sej,Spj,pi,covD+,covD− ∼ multinominal(Pri, ni) for populations i = 1 to 3 and tests j = 1, 2, where Oi is the vector of observed counts for test 1 and test 2 cross-tabulated in a two-by-two table (++, +−, −+, and −−) and Pri is a vector of probabilities for each count in the table summing to 1 for each population i. For example, Pr(+ +) = [(Se1 Se2) + covD+]pi + [(1 − Sp1)(1 − Sp2) + (covD−)(1 − pi), and Pr(+ −) = [Se1(Se2 − 1) − covD+]pi + [(1 − Sp1)Sp2 − covD−(1 − pi)].
We had no prior information on the two covariances, so we used uniform priors such that the covariance between the test outcomes for infected animals would satisfy the following criteria: (Se1 − 1)(Se2 − 1) ≤ covD+ ≤ min(Se1;Se2) − (Se1)(Se2), and for the noninfected subpopulation, (Sp1 − 1)(Sp2 − 1) ≤ covD− ≤ min(Sp1;Sp2) − (Sp1)(Sp2) (see reference 15).
Therefore, for instance, a uniform (Se1 − 1)(1 − Se2); min(Se1;Se2) − (Se1)(Se2) prior distribution can be used for covD−.
The three population two-by-two cross-tabulations for each test (see Table 6, below) were used to update the model. For each model, the first 5,000 iterations were discarded as burn-in and the next 20,000 iterations, using two chains, were used to parameterize the model. The convergence of the chains was assessed using the Brooks-Gelman-Rubin diagnostics (10, 44). Goodness of fit was also measured using the deviance information criterion (DIC) which penalizes goodness of fit by “complexity” (37). The 95% credibility intervals are similar to the more widely used confidence intervals but, in contrast to the common misinterpretation of the confidence interval, the credibility interval is the range within which the true population value lies with 95% probability.
TABLE 6.
Parameter estimates from Bayesian formulation of the Hui-Walter latent class model based on 20,000 iterations after convergence model 1a
| Parameter | Mean | SD | 2.5% BCI | 97.5% BCI |
|---|---|---|---|---|
| p coastal | 0.766 | 0.0721 | 0.610 | 0.888 |
| p central | 0.924 | 0.0417 | 0.824 | 0.985 |
| p border | 0.651 | 0.0553 | 0.539 | 0.753 |
| Se Cedi test | 0.877 | 0.0398 | 0.803 | 0.959 |
| Se VNT | 0.952 | 0.0248 | 0.904 | 0.996 |
| Sp Cedi test | 0.873 | 0.0713 | 0.735 | 0.993 |
| Sp VNT | 0.757 | 0.1000 | 0.578 | 0.964 |
Model 1 assumes conditional independence using uniform priors for both tests based on 20,000 iterations (DIC = 57.79). BCI, Bayesian credible interval.
The maps were generated using ArcGISv9.0 (ESRI, Redlands, CA).
RESULTS
Descriptive analysis: nonbuffalo wildlife.
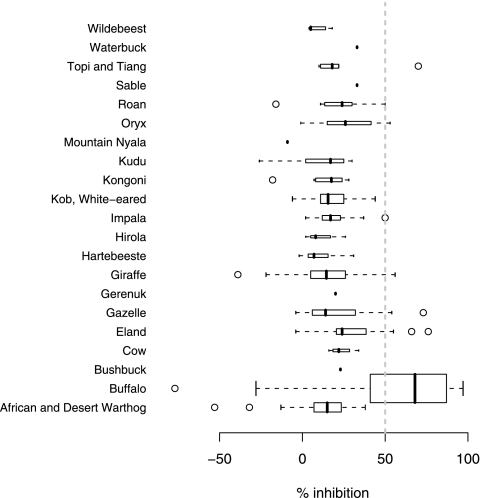
The PIs from the Cedi tests for each species are presented in Fig. 1. There were only small numbers of animals sampled from most species other than buffalo (Table 1); most of the nonbuffalo animals were seronegative on the Cedi test. Of the 248 nonbuffalo wildlife samples tested, 11 were positive by the Cedi test and included seven different species that varied in age and sex. However, not all these were positive by the VNT. All of the Cedi-positive samples were collected in 1999 and 2000 with the exception of the eland, which were sampled in 1996. Nine out of 10 animals (one had no age recorded) were above the age of 3, the other being an approximately 10-month-old gazelle. Only 3/11 had PIs of ≥70%, while 7/11 had very weak positive PI values between 50 and 56%. The warthogs ranged in age from 1 to 10 years, and the kobs ranged from 3 months to 7 years of age. The geographical distribution of the various positive nonbuffalo animals was dispersed across the study area. The results of the VNT also showed a very low seroprevalence, with only 14/248 seropositives in a range of species that included gazelles, giraffe, and impala. A total of 4/22 were positive by both tests: 11/22 were VNT positive Cedi negative, and 7/22 were Cedi positive VNT negative.
FIG. 1.
Box plot of Cedi test results for all species sampled. A PI value of ≥50% indicates a positive test result. ○, outlier.
Descriptive analysis: buffalo.
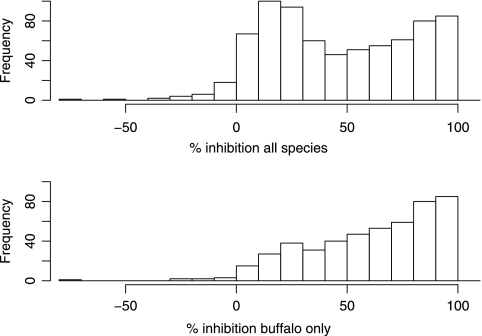
Sampling was carried out between 1994 and 2004. Positive Cedi test samples were seen in buffalo for all 10 years. The PI values for all wildlife and for buffalo only are plotted in Fig. 2. This suggests a bimodal distribution, reflecting the seropositive (mainly buffalo) and seronegative (mainly nonbuffalo animals) populations, with a center around 20% for nonbuffalo animals and a very left-skewed distribution for the buffalo. A total of 327 out of 483 (67.7%) buffalo tested positive for FMDV NSP antibodies with 232 strong positives (≥70) and 95 weak positives (>50 and <70). The proportion of male and female Cedi test-positive buffalo positive for FMDV NSP antibodies was not statistically significantly different (Table 2). The age-stratified and year-stratified summaries of PIs are plotted in Fig. 3A and B. There are very few buffalo calves in the sample, but the data suggest animals seroconvert very early in life, with the age-stratified seropositive proportion increasing to over 60% by 9 months (Table 3). This remains over 60% until the animals are over 15 years old, when there is a slight decline.
FIG. 2.
Histogram of the Cedi test PI for all species and for buffalo only.
TABLE 2.
Cedi test results from male and female buffaloa
| Sex | No. of buffalo
|
Proportion of buffalo that tested positive (95% CI) | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Male | 137 | 56 | 193 | 0.710 (0.646-0.774) |
| Female | 184 | 92 | 276 | 0.667 (0.611-0.722) |
| Total | 321 | 148 | 469 | 0.684 |
The sex of the animal was not recorded for 14 samples, and these were excluded from this analysis. Pearson χ2(1) = 0.9804; P = 0.322.
FIG. 3.
Box plots of Cedi test result versus age group (A) and year of sampling (B). The number of animals sampled in each age category is proportional to the width of the box. ○, outlier.
TABLE 3.
Cedi test results for buffalo, by age categorya
| Age category | No. of buffalo
|
% of buffalo that tested positive | |
|---|---|---|---|
| Positive | Total | ||
| 0-9 mos | 1 | 3 | 33.3 |
| 9-18 mos | 51 | 80 | 63.8 |
| 18 mos-5 yrs | 144 | 201 | 71.6 |
| 5-10 yrs | 79 | 115 | 68.7 |
| 10-15 yrs | 42 | 61 | 68.8 |
| >15 yrs | 7 | 13 | 53.9 |
Ages were not available for 10 animals.
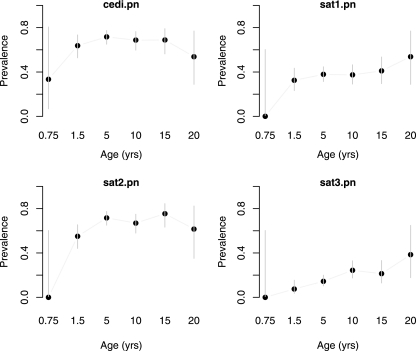
The proportions of seropositive buffalo samples overall by the VNT for SAT serotypes were 37.3% for SAT1, 67.1% for SAT2, and 17.0% for SAT3. The age-stratified seroprevalence for each test is given in Fig. 4. The SAT1 profile suggests early exposure, and then the proportion remains very similar at around 40%. The proportion of SAT2 seropositivity rises to around 70% by 5 years and is stable thereafter. The proportion of SAT3 seropositives is much lower but appears to increase slowly with age. The proportion of Cedi test seropositives largely matches that for SAT2, since this is the predominant serotype. Among the buffalo sampled from eastern Africa, 301 were positive by both Cedi and VNT SAT, 26 were positive by Cedi but negative by VNT SAT, 69 were negative by Cedi but positive by VNT SAT, and 87 were negative by both tests (0.803% agreement; kappa = 0.151; Yules Y = 0.585). The SAT VNT results by serotype, alone and in combination, are shown in Tables 4 and 5.
FIG. 4.
Age-stratified seroprevalence rates based on the Cedi test and VNTs for SAT1, SAT2, and SAT3.
TABLE 4.
SAT VNT seropositive buffalo from 483 sampled
| SAT VNT | No. positive | SAT VNT result | Total no. VNT positive |
|---|---|---|---|
| SAT1 only | 34 | SAT1 | 180 |
| SAT2 only | 165 | SAT2 | 324 |
| SAT3 only | 2 | SAT3 | 82 |
| SAT1/2 | 89 | ||
| SAT1/3 | 10 | ||
| SAT2/3 | 23 | ||
| SAT1/2/3 | 47 |
TABLE 5.
Cross-tabulation of test results for the combined VNT for the three SAT serotypes and the Cedi test ELISA for the three populations of buffaloa
| Populationb and Cedi test result | VNT SAT result
|
|
|---|---|---|
| No. positive | No. negative | |
| 1, Kenya/Tanzania border (4.8093′N, 28.7044′E) | ||
| No. positive | 73 | 12 |
| No. negative | 17 | 15 |
| 2, Central Kenya (4.6801′N, 34.0813′E) | ||
| No. positive | 72 | 2 |
| No. negative | 11 | 4 |
| 3, Coastal Kenya (1.4953′N, 38.3619′E) | ||
| No. positive | 156 | 12 |
| No. negative | 41 | 68 |
Results were generated using K means in Minitab.
The coordinates of the center for each of the three populations are also given.
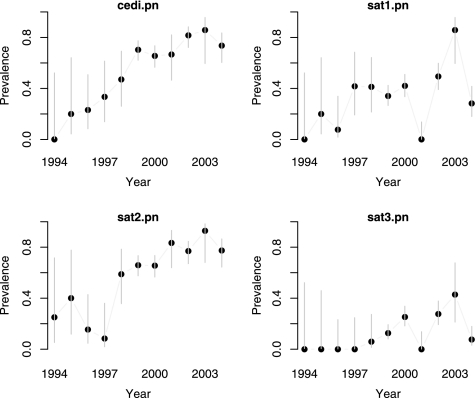
The difference in seroprevalence by year is given in Fig. 5. This gives a variable pattern for SAT1 seroprevalence, with wide fluctuations by year. SAT3 prevalence is low until the 2000s with considerable variation. SAT2 seroprevalence shows more of a pattern with an apparent increase in proportion seropositive by year up to 2000 and then remains high at around 70 to 80%. The Cedi test again seems to largely reflect the SAT2 results, since this is the dominant serotype.
FIG. 5.
Seroprevalence based on the Cedi test and VNTs for SAT1, SAT2, and SAT3 stratified by year of sampling.
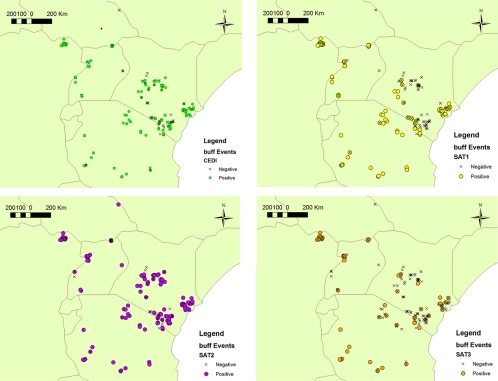
The geographical distributions of the buffalo sampled and their seropositivity for the Cedi test and SAT VNTs are mapped in Fig. 6. There was a small increased odds of seropositivity (odds ratio, 1.15) with large herd size (>50) compared to small herd size (≤50), but this was not statistically significant (P = 0.51).
FIG. 6.
Distribution of seropositive buffalo sampled between 1994 and 2004. Note that colored circles mark positive results (green, Cedi test; yellow, SAT1; purple, SAT2; orange, SAT3), and an X marks samples that were seronegative.
Test parameter estimation in buffalo.
The means and 95% credibility intervals for the three models are summarized in Tables 6, 7, and 8. The sensitivity and specificity of the Cedi test were 0.877 and 0.873 for the model assuming conditional independence and uninformative priors (model 1; i.e., we had no strong beliefs about the test parameters in buffalo from previous studies), compared to 0.872 and 0.942 for the model assuming conditional independence but with informative priors (model 2; i.e., we had strong beliefs about the Sp and less strong beliefs about the Se), and 0.852 and 0.941 for the model including dependence and informative priors (model 3; i.e., we had a strong belief about the Sp and a less strong belief about the Se and thought there may be dependence between tests). The model that included dependence but used uninformative priors was not identifiable. For the three models described, there was good convergence and mixing of the chains based on the Brooks-Gelman-Rubin plots. The fit of each of the models was also compared using the DIC, and there did not appear to be an important difference in fit. The models all estimate the seroprevalence of FMDV to be highest in the central population and lowest in the coastal population. The models also give slightly different estimates of Se and Sp for each test, as might be expected, and the trade-offs of these models are discussed in more detail in the Discussion.
TABLE 7.
Parameter estimates from Bayesian formulation of the Hui-Walter latent class model based on 20,000 iterations after convergence model 2a
| Parameter | Mean | SD | 2.5% BCI | 97.5% BCI |
|---|---|---|---|---|
| p coastal | 0.808 | 0.0551 | 0.687 | 0.902 |
| p central | 0.931 | 0.0376 | 0.844 | 0.989 |
| p border | 0.676 | 0.0469 | 0.582 | 0.764 |
| Se Cedi test | 0.872 | 0.0386 | 0.803 | 0.954 |
| Se VNT | 0.932 | 0.0169 | 0.898 | 0.965 |
| Sp Cedi test | 0.942 | 0.0230 | 0.889 | 0.978 |
| Sp VNT | 0.772 | 0.0975 | 0.593 | 0.966 |
Assumes conditional independence with informative priors for the Cedi test. Se = Beta(1.3,1,2); Sp = Beta(99,6); DIC = 57.019. BCI, Bayesian credible interval.
TABLE 8.
Parameter estimates from Bayesian formulation of the Hui-Walter latent class model based on 20,000 iterations after convergence model 3a
| Parameter | Mean | SD | 2.5% BCI | 97.5% BCI |
|---|---|---|---|---|
| covDn | 0.013 | 0.0182 | −0.0168 | 0.0533 |
| covDp | 0.015 | 0.0168 | −0.0077 | 0.0543 |
| p coastal | 0.833 | 0.0604 | 0.705 | 0.943 |
| p central | 0.948 | 0.0386 | 0.852 | 0.008 |
| p border | 0.692 | 0.0518 | 0.591 | 0.792 |
| Se Cedi test | 0.852 | 0.0426 | 0.773 | 0.942 |
| Se VNT | 0.911 | 0.0266 | 0.852 | 0.956 |
| Sp Cedi test | 0.941 | 0.0232 | 0.888 | 0.978 |
| Sp VNT | 0.753 | 0.1041 | 0.560 | 0.969 |
Model 3 assumptions include dependence between tests with informative priors for the Cedi test based on 20,000 iterations (DIC = 57.64). BCI, Bayesian credible interval.
DISCUSSION
The Cedi FMDV-NS test was used to screen the sera because, as a blocking ELISA, it is species independent and as such should be a good choice for testing wildlife samples from a range of species. In addition, antibodies to the 3ABC protein are considered to be a reliable indicator of infection/exposure regardless of vaccination status and of the serotype of FMDV (36, 39). However, it cannot distinguish between different serotypes.
The seroprevalence estimates in nonbuffalo species were very low based on either the VNT SAT or Cedi test results. However, there does not appear to be very good agreement on individual positive samples between the tests for nonbuffalo. There were insufficient samples to do a more through analysis of the Cedi test in these species, but clearly it would be useful to have better estimates of its performance in a wider range of species.
The Cedi test detected 11 seropositives, but only 4 animals were positive in both test systems. In addition, the distribution of OD readings, although appearing bimodal, did not suggest a clear cutoff. The low seroprevalence by VNT in nonbuffalo species is comparable to the results of a survey done between the years 1989 and 1992 that found only 1 to 2% of 7,970 wild ungulates of 14 different species were positive for FMDV (1). The nonbuffalo wildlife sampled in this study overlapped spatially with the sampled buffalo that were seropositive but appear not to have seroconverted by and large, suggesting some level of resistance if one assumes that they have been exposed through contact with buffalo.
It is interesting that only two warthogs were seropositive by VNT and not by Cedi, although domestic pigs are known to excrete significant quantities of virus at the time of infection (26). The number of samples from other species of wildlife were relatively small, but they suggest, particularly for the warthog species (Phaecocherus african, P. aethiopicus) and kob (Kobus leucotis), that there had been no recent exposures.
These results support previous studies (43) in showing clearly that buffalo (Syncerus caffer) have very high seroprevalences to FMDV by both the Cedi test and by serotype-specific VNTs. In these buffalo populations SAT2 was the most dominant serotype, followed by SAT1 and then SAT3. The data suggest that there has been a marked increase in prevalence of FMDV overall with a shift to SAT2 dominance around 1998. It is interesting that the age-stratified seroprevalence appears to be relatively constant at around 60 to 70% based on the Cedi and SAT2 results across all ages after about 9 months. The reasons for this are not clear, as one might expect that with virus and carrier animals in endemic areas there would be high continuous exposure keeping titers higher. It may reflect the effect of the sampling and spatial components that we have not been able to demonstrate but that reflect a more epidemic pattern, with some populations having lower seroprevalences at certain times but overall giving the appearance of a constant level.
The lack of an increased odds of being seropositive for small versus large herds is evidence for continuous herd-level circulation of SAT2 (and possibly SAT1 and SAT3), as each herd shows a reasonably discrete subpopulation but on occasions aggregation and/or fragmentation of herds occurs, with seasonal or breeding cycles providing opportunities for cross-herd transmission. This is consistent with population-level prevalence across herds in a given ecosystem, which is also high, providing evidence of circulation across the population as well.
Very few SAT3-seropositive animals were only seropositive for SAT3, while a higher proportion were SAT1 seropositive only or SAT2 seropositive only. The majority of samples positive to more than one serotype were SAT2 positive, and this raises the issue of to what extent the SAT results (particularly SAT3) are cross-reactions. Although the SAT1 may be a cross-reaction, interestingly, none of the buffalo sampled in Tchad were SAT1 positive. Most were SAT2 positive, with only two SAT3 positives (21). Without concurrent probangs and virus isolation, it is not possible to separate these and identify how many are cross-reactions and how many are genuine seropositives.
Although there may be some discussion of the precise estimates of sensitivity and specificity of the all of the tests, it is clear that the seroprevalence of FMDV in Cape buffalo in eastern Africa is very high and that animals appear to seroconvert very early in life, probably within the first 1 to 2 years. This is certainly consistent with the situation further south in Africa, where they are the primary wildlife species acting as a reservoir for FMDV in southern Africa (2, 13). There is also some evidence in the data to suggest that the seroprevalence increased through the late 1990s, although SAT2 viruses have been recorded from the region since 1958 (45). However, there were reported outbreaks in Kenya (1994 to 2000), Tanzania (1999 to 2000) and Uganda (1995 to 1999) of SAT1, SAT2, and O (as well as A and C in Kenya) (45). Although buffalo are considered to be the main reservoir of the SAT serotypes, the apparent seroprevalences of SAT1 and SAT3 were low, and based on the NSP test there is little evidence that other serotypes were circulating in the buffalo populations. This may mean that buffalo are not maintaining these serotypes in the region; rather, there may be maintenance in cattle or regular reintroductions from beyond due to cattle movements. This needs to be looked at in more detail, however, with molecular epidemiology tools.
Comparison of the seroprevalence of FMDV in buffalo to the seroprevalence of FMDV in livestock held in similar areas during the time period of the wildlife sampling would be interesting and could possibly shed light on the role that buffalo have in transmission of the disease to domestic livestock. The role of carrier buffalo is a particularly important aspect for control programs, and this needs further study. Dawe et al. (12) demonstrated natural transmission from carrier buffalo to cattle, as well as experimental transmission (13). However, transmission from carrier cattle appears to be much less likely and has not been demonstrated under experimental conditions (16). In cattle it has been seen that although the virus could be isolated up to 57 (19) or 98 (32) days postchallenge from vaccinated and challenged animals, introducing naïve cattle for direct contact with these carrier animals could not transmit the disease.
Data on outbreaks in cattle at the time of sampling were generally not available except from Western Laikipia in Kenya, where there were reports of outbreaks of type O in cattle in October 2002 and samples from the buffalo in the area at the same time were seropositive for SAT2 (19/20), SAT1 (7/20), and SAT3 (2/20). There is little published on non-SAT serotypes in buffalo, but it is unlikely that this type O has come from the buffalo as the reservoir. From reported outbreaks in Saharan Africa (45) it appears that the majority of outbreaks in the southern regions are due to SAT serotypes with only sporadic introductions of O and A. Since this is also the region where most work has been carried out in buffalo, this is reflected in the lack of information of other serotypes in this species.
It is a concern that serum that has been repeatedly freeze-thawed could produce less sensitive test results (36). For the FMD survey, some of the wildlife sera were over 10 years old, as samples were collected from 1994 to 2004. Repeated freeze-thawing also occurred since collection, because originally the samples were tested in eastern and central Africa for rinderpest before being shipped to the United Kingdom to be tested for FMDV. Recent work on sera at Pirbright that examined the consistency in results of antibody screening repeatedly over long storage periods showed remarkable stability in antibody detection by VNT and competitive ELISA (S. Hargrove, personal communication). Despite the lower relative sensitivity of 88% for the 3ABC ELISA, it is a specific and precise test that can be used on populations of animals regardless of species or serotype of FMDV.
The sensitivity and specificity of the 3ABC ELISA (Cedi test) have not previously been estimated for wildlife such as buffalo. Estimates for the sensitivity relative to the VNT results in sheep are reportedly 92%, and for cattle the value is lower at 88%. The relative specificities are higher for both sheep and cattle, between 100% and 99.8%, respectively (36). Recent studies comparing several NSP tests, including the Cedi test, have produced a range of estimates. In cattle the sensitivity ranged from 50.0 to 100.0, depending on how long after infection animals were tested, while the specificity was much more precisely estimated and ranged from 97.2 to 99.0% (6, 33). These estimates formed the basis of the prior distributions used in our latent class models where informative priors were used. These are, of course, based largely on experimental studies (particularly the sensitivity estimates) and on cattle and may in fact not be a very reliable guide to their performance in buffalo. There are also potential problems with using a combined VNT as a second test for an NSP test, because the durations of VNT antibodies and NSP antibodies differ, as does the rate of seroconversion (9). However, the other commercially available kits produced by Svanova, Bommeli, and UBI are not suitable, as they need species-specific conjugates and no wildlife conjugates are available.
The parameter estimates for the Cedi test in buffalo based on the latent class modeling approach suggest that the Cedi test is sensitive and very specific in the buffalo populations of eastern Africa. The specificity for models where an informed prior was used was driven by this prior. Including dependence in the model did not change this. However, using a noninformative prior, which may be more appropriate for this population, reduced the estimated specificity to 87%. The sensitivity on the other hand changed little between models, as the informed prior was very diffuse and added little to the noninformative case, with both producing estimates around 87%. The inclusion of dependence in the model resulted in a slight lowering of the estimate to 85%. The covariances for both the sensitivity and specificity of the two tests were low, indicating that the assumption of independence may be valid, and certainly there is little change in the DIC when it is dropped.
The most appropriate model of the three presented is a matter for discussion. The more conservative approach would be to use model 1, which makes no assumptions about the test parameters in buffalo, as the informative priors used in models 2 and 3 come from cattle studies. For model 3, which included dependence, the test covariances were very small (covsp, 0.01296; covse, 0.01519), and therefore there does not appear to be a problem of dependence between the tests; if we wish to use the priors, model 2 would therefore appear to be a reasonable model.
Although the aim was not to estimate the sensitivity or specificity of the combined VNT for SATs, the estimates gained could be useful for other work. The low estimated specificity of all the models is worrying, given that the VNT is considered the gold standard. However, this must be interpreted in terms of the very different tests and the antibody profiles of neutralizing and NSP antibodies over time postinfection.
Control of FMD in eastern and central Africa is currently unrealistic, as there is a lack of infrastructure to sustain intensive vaccination campaigns and a lack of new vaccines that can produce high and sustained neutralizing antibody titers (24). In addition, Africa will need to also manage FMD in its wildlife or at least in the buffalo populations across the region in order to prevent continuous reintroduction. Contact between free-ranging buffalo in southern Africa is well-recognized (12), and there is some statistical evidence for increased risk to cattle herds in central Africa (8). In southern Africa the problem has been managed through the use of game fencing to keep livestock and wildlife separate (40). However, this is relatively straightforward in southern Africa, where they are at the southern edge of the natural range for the Cape buffalo. In eastern and central Africa, where there are still major buffalo populations mixed with cattle and other livestock, the problem is much more difficult. However, one possibility, as in South Africa, is for the development of FMD-free buffalo herds. Being able to screen them cost-effectively would be a key element in developing such policy. The Cedi test certainly offers a potentially useful tool for screening herds. In the early stages of control the lower specificity will not cause too many problems. However, as seroprevalence and disease decline the predictive value of a positive test will be very poor on an individual basis, though it can still provide useful information at the herd level, particularly if it is combined with a second test or other type of testing, such as probanging.
Acknowledgments
We thank the Director of IBAR for permission to send the samples to Pirbright and to the national authorities for the support in collection of this central bank of sera for the purposes of further study. We also thank Mandy Corteyn (IAH) for lab space and David Paton (IAH) and John Anderson (IAH) for facilitating the work at IAH, Pirbright.
This work is supported by Defra through project SE2918 and SE1122 held by Satya Parida (Institute for Animal Health). Sarah McFarland was funded by DEFRA/SFC through VTRI project VT101 held by M. Woolhouse (University of Edinburgh).
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Anderson, E. C., C. Foggin, M. Atkinson, K. J. Sørensen, R. L. Madekurozva, and J. Nqindi. 1993. The role of wild animals, other than buffalo, in the current epidemiology of foot-and-mouth disease in Zimbabwe. Epidemiol. Infect. 111:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastos, A., C. Boshoff, D. Keet, R. Bengis, and G. Thomson. 2000. Natural transmission of foot-and-mouth disease virus between African buffalo (Syncerus caffer) and impala (Aepyceros melampus) in the Kruger National Park, South Africa. Epidemiol. Infect. 124:591-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, H. G., O. C. Straub, R. Ahl, M. Tesar, and O. Marquardt. 1990. Identification of foot-and-mouth disease virus replication in vaccinated cattle by antibodies to non-structural virus proteins. Vaccine 8:213-216. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, I. E., P. A. de Mello, E. Neitzert, E. Beck, and I. Gomes. 1993. Diagnosis of persistent aphthovirus infection and its differentiation from vaccination response in cattle by use of enzyme-linked immunoelectrotransfer blot analysis with bioengineered nonstructural viral antigens. Am. J. Vet. Res. 54:825-831. [PubMed] [Google Scholar]
- 5.Branscum, A. J., I. A. Gardner, and W. O. Johnson. 2005. Estimation of diagnostic-test sensitivity and specificity through Bayesian modeling. Prev. Vet. Med. 68:145-163. [DOI] [PubMed] [Google Scholar]
- 6.Brocchi, E., I. E. Bergmann, A. Dekker, D. J. Paton, D. J. Sammin, M. Greiner, S. Grazioli, F. De Simone, H. Yadin, and B. Haas. 2006. Comparative evaluation of six ELISAs for the detection of antibodies to the non-structural proteins of foot-and-mouth disease virus. Vaccine 24:6966-6979. [DOI] [PubMed] [Google Scholar]
- 7.Brocchi, E., M. I. De Diego, A. Berlinzani, D. Gamba, and F. De Simone. 1998. Diagnostic potential of Mab-based ELISAs for antibodies to non-structural proteins of foot-and-mouth disease virus to differentiate infection from vaccination. Vet. Q. 20(Suppl. 2):S20-S24. [PubMed] [Google Scholar]
- 8.Bronsvoort, B. M. d. C., C. Nfon, S. M. Hamman, V. N. Tanya, R. P. Kitching, and K. L. Morgan. 2004. Risk factors for herdsman-reported foot-and-mouth disease in the Adamawa Province of Cameroon. Prev. Vet. Med. 66:127-139. [DOI] [PubMed] [Google Scholar]
- 9.Bronsvoort, B. M. d. C., N. Toft, I. E. Bergmann, K.-J. Sørensen, J. Anderson, V. Malirat, V. N. Tanya, and K. L. Morgan. 2006. Evaluation of three 3ABC ELISAs for foot-and-mouth disease non-structural antibodies using latent class analysis. BMC Vet. Res. 2:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks, S. P., and A. Gelman. 1998. Alternative methods for monitoring convergence of iterative simulations. J. Comput. Graph. Stat. 7:434-455. [Google Scholar]
- 11.Brooksby, J. B. 1982. Portraits of viruses: foot-and-mouth disease virus. Intervirology 18:1-23. [DOI] [PubMed] [Google Scholar]
- 12.Dawe, P. S., F. O. Flanagan, R. L. Madekurozwa, K. J. Sørensen, E. C. Anderson, C. M. Foggin, N. P. Ferris, and N. J. Knowles. 1994. Natural transmission of foot-and-mouth disease virus from African buffalo Syncerus caffer to cattle in a wildlife area of Zimbabwe. Vet. Rec. 134:230-232. [DOI] [PubMed] [Google Scholar]
- 13.Dawe, P. S., K. J. Sørensen, N. P. Ferris, I. T. Barnett, R. M. Armstrong, and N. J. Knowles. 1994. Experimental transmission of foot-and-mouth disease virus from carrier African buffalo Syncerus caffer to cattle in Zimbabwe. Vet. Rec. 134:211-215. [DOI] [PubMed] [Google Scholar]
- 14.De Diego, M., E. Brocchi, D. Mackay, and F. De Simone. 1997. The non-structural polyprotein 3ABC of foot-and-mouth disease virus as a diagnostic antigen in ELISA to differentiate infected from vaccinated cattle. Arch. Virol. 142:2021-2033. [DOI] [PubMed] [Google Scholar]
- 15.Dendukuri, N., E. Rahme, P. Belisle, and L. Joseph. 2004. Bayesian sample size determination for prevalence and diagnostic test studies in the absence of a gold standard test. Biometrics 60:388-397. [DOI] [PubMed] [Google Scholar]
- 16.Doel, T. R. 2005. Natural and vaccine induced immunity to FMD, foot and mouth disease virus. Curr. Top. Microbiol. Immunol. 288:103-131. [DOI] [PubMed] [Google Scholar]
- 17.Enoe, C., S. Andersen, V. Sørensen, and P. Willeberg. 2001. Estimation of sensitivity, specificity and predictive values of two serologic tests for the detection of antibodies against Actinobacillus pleuropneumoniae serotype 2 in the absence of a reference test (gold standard). Prev. Vet. Med. 51:227-243. [DOI] [PubMed] [Google Scholar]
- 18.Gardner, I. A., H. Stryhn, P. Lind, and M. T. Collins. 2000. Conditional dependence between tests affects the diagnosis and surveillance of animal diseases. Prev. Vet. Med. 45:107-122. [DOI] [PubMed] [Google Scholar]
- 19.Golde, W. T., J. M. Pacheco, H. Duque, T. Doel, B. Penfold, G. S. Ferman, D. R. Gregg, and L. L. Rodriguez. 2005. Vaccination against foot-and-mouth disease virus confers complete clinical protection in 7 days and partial protection in 4 days: use in emergency outbreak response. Vaccine 23:5775-5782. [DOI] [PubMed] [Google Scholar]
- 20.Hedger, R. S. 1976. Foot-and-mouth disease in wildlife with particular reference to the African buffalo (Syncerus caffer), p. 235-244. In L. A. Page (ed.), Wildlife diseases. Plenum Press, London, England.
- 21.Hedger, R. S., I. T. R. Barnett, D. V. Gradwell, and P. Travassos Dias. 1982. Serological tests for foot-and-mouth disease in bovine serum samples. problems of interpretation. Rev. Sci. Tech. Off. Int. Epizoot. 1:387-393. [DOI] [PubMed] [Google Scholar]
- 22.Hedger, R. S., A. J. Forman, and M. H. Woodford. 1973. Foot-and-mouth disease virus in East African buffalo. Bull. Epizoot. Dis. Afr. 21:99-101. [Google Scholar]
- 23.Hui, S. L., and S. D. Walter. 1980. Estimating the error rates of diagnostic tests. Biometrics 36:167-171. [PubMed] [Google Scholar]
- 24.International Livestock Research Institute. 2006. Global roadmap for improving the tools to control foot-and-mouth disease in endemic settings. International Livestock Research Institute, Nairobi, Kenya.
- 25.Kitching, R. P. 2005. Global epidemiology and prospects for control of foot-and-mouth disease. Curr. Top. Microbiol. Immunol. 288:133-148. [DOI] [PubMed] [Google Scholar]
- 26.Kitching, R. P., and S. Alexandersen. 2002. Clinical variation in foot and mouth disease: pigs. Rev. Sci. Tech. Off. Int. Epizoot. 21:513-518. [DOI] [PubMed] [Google Scholar]
- 27.Kock, R. A., H. M. Wamwayib, P. B. Rossiterc, G. Libeaud, E. Wambwae, J. Okorif, F. S. Shiferawg, and T. D. Mlengeyah. 2006. Re-infection of wildlife populations with rinderpest virus on the periphery of the Somali ecosystem in East Africa. Prev. Vet. Med. 75:63-80. [DOI] [PubMed] [Google Scholar]
- 28.Lubroth, J., and F. Brown. 1995. Identification of native foot-and-mouth disease virus non-structural protein 2C as a serological indicator to differentiate infected from vaccinated livestock. Res. Vet. Sci. 59:70-78. [DOI] [PubMed] [Google Scholar]
- 29.Malirat, V., E. Neitzert, I. E. Bergmann, E. Maradei, and E. Beck. 1998. Detection of cattle exposed to foot-and-mouth disease virus by means of an indirect ELISA test using bioengineered nonstructural polyprotein 3ABC. Vet. Q. 20(Suppl. 2):S24-S26. [DOI] [PubMed] [Google Scholar]
- 30.Neitzert, E., E. Beck, P. Augede-Melo, I. Gomes, and I. E. Bergmann. 1991. Expression of aphthovirus RNA polymerare gene in E. coli and its use together with other bioengineered non-structural antigens in detection of late persistent infection. Virology 184:799-804. [DOI] [PubMed] [Google Scholar]
- 31.OIE. 2005. Manual of diagnostic tests and vaccines for terrestrial animals, 5th ed. OIE, Paris, France.
- 32.Parida, S., J. Anderson, S. J. Cox, P. V. Barnett, and D. J. Paton. 2006. Secretory IgA as an indicator of oro-pharyngeal foot-and-mouth disease virus replication and as a tool for post vaccination surveillance. Vaccine 24:1107-1116. [DOI] [PubMed] [Google Scholar]
- 33.Paton, D. J., K. de Clercq, M. Greiner, A. Dekker, E. Brocchi, I. Bergmann, D. J. Sammin, S. Gubbins, and S. Parida. 2006. Application of non-structural protein antibody tests in substantiating freedom from foot-and-mouth disease virus infection after emergency vaccination of cattle. Vaccine 24:6503-6512. [DOI] [PubMed] [Google Scholar]
- 34.Sørensen, K. J., C. M. Hansen, E. S. Madsen, and K. G. Madsen. 1998. Blocking ELISAs using the FMDV nonstructural proteins 3D, 3AB, and 3ABC produced in the baculovirus expression system. Vet. Q. 20:S17-S20. [PubMed] [Google Scholar]
- 35.Sørensen, K. J., R. L. Madekurozwa, and P. Dawe. 1992. Foot-and-mouth disease: detection of antibodies in cattle sera by blocking ELISA. Vet. Microbiol. 32:253-265. [DOI] [PubMed] [Google Scholar]
- 36.Sørensen, K. J., K. G. Madsen, E. S. Madsen, J. S. Salt, J. Nqindi, and D. K. J. Mackay. 1998. Differentiation of infection from vaccination in foot-and-mouth disease by the detection of antibodies to the non-structural proteins 3D, 3AB and 3ABC in ELISA using antigens expressed in baculovirus. Arch.Virol. 143:1461-1476. [DOI] [PubMed] [Google Scholar]
- 37.Spiegelhalter, D. J., N. G. Best, B. P. Carlin, and A. van der Linde. 2002. Bayesian measures of model complexity and fit (with discussion). J. R. Stat. Soc. B 64:583-639. [Google Scholar]
- 38.Spiegelhalter, D. J., A. Thomas, and N. G. Best. 2003. WinBugs version 1.4 user manual. MRC Biostatistics Unit, Cambridge, United Kingdom.
- 39.Sun, T., P. Lu, and X. Wang. 2004. Localization of infection-related epitopes on the non-structural protein 3ABC of foot-and-mouth disease virus and the application of tandem epitopes. J. Virol. Methods 119:79-86. [DOI] [PubMed] [Google Scholar]
- 40.Sutmoller, P., G. R. Thomson, S. K. Hargreaves, C. M. Foggin, and E. C. Anderson. 2000. The foot-and-mouth disease risk posed by African buffalo within wildlife conservancies to the cattle industry of Zimbabwe. Prev. Vet. Med. 44:43-60. [DOI] [PubMed] [Google Scholar]
- 41.Thomas, A., B. O'Hara, U. Ligges, and S. Sturtz. 2006. Making BUGS open. R News 6:12-17. [Google Scholar]
- 42.Thomson, G. R., and A. D. S. Bastos. 2004. Foot-and-mouth disease, 2nd ed., vol. 2. Oxford University Press, Cape Town, South Africa.
- 43.Thomson, G. R., W. Vosloo, and A. D. S. Bastos. 2003. Foot and mouth disease in wildlife. Virus Res. 91:145-161. [DOI] [PubMed] [Google Scholar]
- 44.Toft, N., G. T. Innocent, G. Gettinby, and S. W. J. Reid. 2007. Assessing the convergence of Markov chain Monte Carlo methods: an example from evaluation of diagnostic tests in absence of a gold standard. Prev. Vet. Med. 79:244-256. [DOI] [PubMed] [Google Scholar]
- 45.Vosloo, W., A. D. S. Bastos, O. Sangare, S. K. Hargreaves, and G. R. Thomson. 2002. Review of the status and control of foot and mouth disease in sub-Saharan Africa. Rev. Sci. Tech. Off. Int. Epizoot. 21:437-449. [DOI] [PubMed] [Google Scholar]