Abstract
To evaluate the fully automated FACSCanto software, we compared lymphocyte subpopulation counts obtained using three-color FACSCalibur-CELLQuest and six-color FACSCanto-FACSCanto software techniques. High correlations were observed between data obtained with these techniques. Our study indicated that FACSCanto clinical software is accurate and sensitive in single-platform lymphocyte immunophenotyping.
Recently, flow cytometry has become the principal technique for the diagnosis and monitoring of cellular immunodeficiencies (3). New instruments, methodologies, and reagents have been developed to improve accuracy, precision, and standardization in lymphocyte subpopulation counts (LSc) (8). Clinical laboratories are now routinely using single-platform assays with a lyse-no-wash methodology, which reduces interlaboratory variability (2) but requires complex analysis with a multiple-gating strategy (1, 4, 5).
The recent availability of new multicolor flow cytometers has allowed the performance of absolute LSc in a single tube, including B lymphocytes and natural killer (NK) cells, with a reduction in cost and time.
In 2004, the six-color flow cytometer FACSCanto provided with FACSCanto and FACSDiva software (BD Biosciences, San Jose, CA) was approved by the FDA for in vitro LSc. Lambert et al. evaluated the performance of this cytometer, using a lyse-no-wash, single-platform technique and a manual-gating analysis. Similarly, Ashman et al. compared the performance of FACSDiva and FACSCanto software using a lyse/wash double-platform technique. They concluded that FACSDiva software was preferable for six-color LSc, as it offered better manual-gating performance.
The aim of our study was to evaluate the performance of the new completely autogating FACSCanto clinical software using the six-color single-tube reagent (TBNK) in a lyse-no-wash and single-platform technique (6-ST). We compared LSc obtained with this approach with those obtained with the three-color multiple-tube technique (3-MT) and FACSCalibur, routinely used in our clinical diagnostic procedures.
LSc were measured in EDTA whole-blood samples of 40 consecutive subjects referred to our laboratory (14 adults, 22 children, and 4 neonates) and 10 healthy adult blood donors. The patients included 17 human immunodeficiency virus infection cases, 6 kidney transplant recipients, and 17 undiagnosed cases. Samples were processed within 8 h of blood draw, using 3-MT in parallel (6) with the following monoclonal antibodies according to the manufacturer's recommendations: CD3-fluorescein isothiocyanate (FITC) (clone SK7), CD4-phycoerythrin (PE) (clone SK3), CD8-PE (clone RPA-T8), CD3/CD16+ CD56 FITC/PE bundle, CD19-PE (clone SJ25C1), and CD45-peridinin chlorophyll protein (PerCP) (clone 2D1) and the 6-ST (TruCount tube) with a TBNK kit that combined CD3-FITC (clone SK7), CD16-PE (clone B73.1), CD56-PE (clone NCAM16.2), CD45-PerCP-Cy5.5 (clone 2D1), CD4-PE-Cy7 (clone SK3), CD19- allophycocyanin (APC) (clone SJ25C1), and CD8-APC-Cy7 (clone SK1).
Full blood counts were performed for all samples using a Sysmex XE 2100 hematology analyzer.
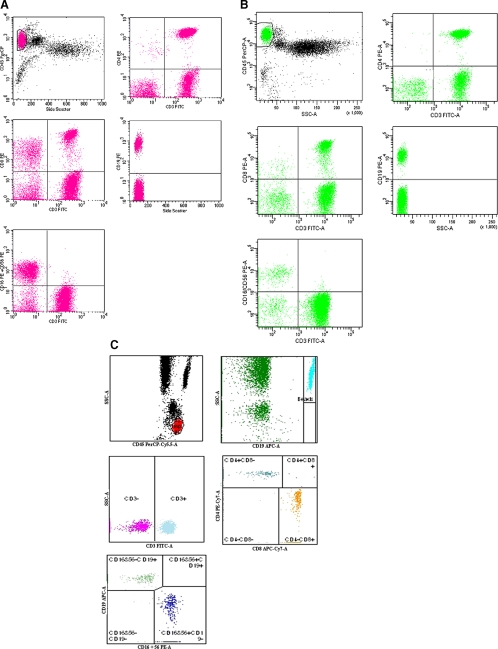
Tubes of 3-MT were acquired both with FACSCalibur (CELLQuest software v. 3.3) (Fig. 1A) and with FACSCanto (FACSDiva software v. 4.0.2) (Fig. 1B). Data analysis employed CD45/side-scatter gating on lymphocytes and then dot plot evaluations of two-antigen coexpression on CD45+ lymphocyte events (CD3/CD4, CD3/CD8, CD19, and CD3/CD16 plus CD56). Each absolute LSc was determined by multiplying the percentages obtained in the analysis by the hematology analyzer lymphocyte counts.
FIG. 1.
Representative lymphocyte subset gating strategies using CellQuest (A), FACSDiva (B), and FACSCanto (C) software.
Samples processed by 6-ST were acquired and analyzed with FACSCanto and FACSCanto clinical software (v. 2.0) (Fig. 1C). This software automatically calculates the absolute LSc using the internal reference beads contained in the TruCount tube. In order to exclude operator variability (7), all the analyses were performed by the same operator.
Correlations of LSc obtained by 3-MT and 6-ST were assessed using the Pearson correlation coefficient and linear regression analysis. The Wilcoxon signed rank test was used to assess differences between the two methods. All statistical analyses were performed with GraphPad Prism 4.0 statistical software.
Instrument calibrations were highly stable over a period of 3 months. The automatic compensation setting included in the FACSCanto software passed in all performances with coefficients of variation always below 20%.
Excellent correlations (r2 > 0.94) were observed between all LSc obtained by 3-MT and 6-ST and between 3-MT performed with CELLQuest and FACSDiva software (r2 > 0.98).
As shown in Table 1, 6-ST produced higher and statistically different counts in all LSc compared to 3-MT.
TABLE 1.
LSc cell counts for 6-ST and 3-MT (single and double platform)
| Cell type | 6-ST FACSCanto cell count (SP)a
|
Pb | 3-MT CELLQuest cell count (DP)a
|
Pc | 3-MT FACSDiva cell count (DP)
|
Pd | |||
|---|---|---|---|---|---|---|---|---|---|
| Median | Range | Median | Range | Median | Range | ||||
| CD3+ | 1,637 | 203-10,475 | <0.0001 | 1,542 | 196-9,512 | >0.05 | 1,585 | 223-9,490 | <0.0001 |
| CD3+/CD4+ | 809 | 63-6,404 | <0.0001 | 757 | 66-5,798 | >0.05 | 737 | 70-5,790 | <0.0001 |
| CD3+/CD8+ | 709 | 127-3,603 | <0.0001 | 621 | 105-3,185 | >0.05 | 602 | 136-3,159 | <0.0001 |
| CD19+ | 305 | 5-2,945 | <0.0001 | 229 | 2-2,119 | <0.05 | 253 | 3-2,067 | <0.0001 |
| CD3−/CD16 + CD56+ | 224 | 24-1,438 | <0.05 | 208 | 15-1,788 | >0.05 | 212 | 8-1,621 | <0.05 |
SP, single platform; DP, double platform.
SP 6-ST FACSCanto versus DP 3-MT CELLQuest.
DP 3-MT CELLQuest versus DP 3-MT FACSDiva.
SP 6-ST FACSCanto versus DP 3-MT FACSDiva.
The 6-ST lymphosums (sums of percentages of CD3+, CD19+, and NK cells) were significantly higher than those obtained with 3-MT (99.5 ± 0.32% versus 94.1 ± 3.7%; P < 0.0001). No difference was found between 3-MT lymphosums obtained with CELLQuest and FACSDiva software (94.1 ± 3.7% versus 93.7 ± 3.7%; P = 0.084). We also recalculated LSc using the 6-ST percentages and the hematology analyzer lymphocyte counts in a double-platform approach (Table 2). Only CD3+/CD4+, CD3+/CD8+, and CD19+ cell counts remained statistically different using 6-ST and 3-MT performed with the FACSCanto flow cytometer. FACSCanto files were also reanalyzed with FACSDiva, and the results were not statistically different from those obtained with the FACSCanto software (data not shown).
TABLE 2.
LSc cell counts for 6-ST and hematology analyzer lymphocyte counts (double platform)
| Cell type | 6-ST FACSCanto cell count
|
Pa | 3-MT CELLQuest cell count
|
Pb | 3-MT FACSDiva cell count
|
Pc | |||
|---|---|---|---|---|---|---|---|---|---|
| Median | Range | Median | Range | Median | Range | ||||
| CD3+ | 1,566 | 219-9,425 | <0.05 | 1,542 | 196-9,512 | >0.05 | 1,585 | 223-9,490 | >0.05 |
| CD3+/CD4+ | 779 | 67-5,759 | <0.005 | 757 | 66-5,798 | >0.05 | 737 | 70-5,790 | <0.005 |
| CD3+/CD8+ | 654 | 137-3,237 | <0.0001 | 621 | 105-3,185 | >0.05 | 602 | 136-3,159 | <0.0001 |
| CD19+ | 285 | 6-2791 | <0.0001 | 229 | 2-2,119 | <0.05 | 253 | 3-2,067 | <0.0001 |
| CD3−/CD16 + CD56+ | 221 | 26-1,466 | <0.05 | 208 | 15-1,788 | >0.05 | 212 | 8-1,621 | >0.05 |
6-ST FACSCanto versus 3-MT CELLQuest.
3-MT CELLQuest versus 3-MT FACSDiva.
6-ST FACSCanto versus 3-MT FACSDiva.
In our study, we evaluated the performance of methodology and instrumentation similar to that used by Lambert et al. but employing the new fully automated FACSCanto clinical software for data analysis, in parallel with our internal reference method. FACSCanto software, although evaluated by Ashman et al. as less accurate than FACSDiva software in LSc, seemed to be as precise and sensitive as the FACSDiva software when we employed it in a lyse-no-wash, single-platform assay using the TBNK reagent. Unlike FACSDiva, the FACSCanto software automatically compensates for all six fluorescences and creates all gates needed to evaluate lymphocyte subsets. Higher LSc obtained with 6-ST could be due to different platform approaches and instrument sensitivities and brighter fluorochromes used in the TBNK reagent. Since we did not find any statistical variation comparing data obtained using only a double-platform technique, we could exclude the possibility that differences were due to the platform approach used. At the same time, a minimally different instrument sensitivity, related only to CD19+ cell counts, was found when 3-MT was performed on either a FACSCalibur or FACSCanto flow cytometer. A unique manual-gating strategy employed by a single operator for both software programs (CELLQuest and FACSDiva) probably contributed to obtaining similar counts. Higher CD4+, CD8+, and CD19+ cell counts found with 6-ST seem to be related to the introduction of brighter fluorochromes in the TBNK reagent, i.e., CD4-PE-Cy7, CD8-APC, and CD19-APC-Cy7, instead of PE-conjugated antibodies in 3-MT. It is important to note that, in spite of these differences in LSc, no modifications in clinical reports were required.
The TBNK reagent, in combination with the FACSCanto software, allows LSc in a shorter time and with a smaller blood sample, a crucial requirement in testing newborns or pediatric patients, as in our setting. The higher lymphosum obtained with 6-ST supports greater accuracy in characterizing a larger number of lymphocytes. Certainly, the use of complex instruments with multicolor analysis in which every fluorochrome has to be accurately compensated for, especially in a lyse-no-wash technique, could be problematic for an inexperienced operator. The implementation of new fully automated software can overcome these problems.
In conclusion, our study indicates that FACSCanto clinical software is accurate, sensitive, and easy to apply for LSc using the TBNK reagent and a single-platform approach in a routine clinical setting.
Footnotes
Published ahead of print on 30 April 2008.
REFERENCES
- 1.Ashman, M., N. Sachdeva, L. Davila, G. Scott, C. Mitchell, L. Cintron, M. Rathore, and D. Asthana. 2007. Influence of 4- and 6-color flow cytometers and acquisition/analysis software on the determination of lymphocyte subsets in HIV infection. Cytometry B 72:380-386. [DOI] [PubMed] [Google Scholar]
- 2.Gratama, J. W., J. Kraan, M. Keeney, V. Granger, and D. Barnett. 2002. Reduction of variation in T-cell subset enumeration among 55 laboratories using single-platform, three or four color flow cytometry based on CD45 and SSC-based gating of lymphocytes. Cytometry 50:92-101. [DOI] [PubMed] [Google Scholar]
- 3.Helbert, M., and J. Breuer. 2000. Monitoring patients with HIV disease. J. Clin. Pathol. 5:266-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacombe, F., F. Durrieu, A. Briais, P. Dumain, F. Belloc, E. Bascans, J. Reiffers, M. R. Boisseau, and P. Bernard. 1997. Flow cytometry CD45 gating for immunophenotyping of acute myeloid leukemia. Leukemia 11:1878-1886. [DOI] [PubMed] [Google Scholar]
- 5.Lambert, C., C. Iobagiu, and C. Genin. 2006. Enumeration of peripheral lymphocyte subsets using 6 vs. 4 color staining: a clinical evaluation of a new flowcytometer. Cytometry B 70:29-38. [DOI] [PubMed] [Google Scholar]
- 6.Schenker, E. L., L. E. Hultin, K. D. Bauer, J. Ferbas, J. B. Margolick, and J. V. Giorgi. 1993. Evaluation of a dual-color flow cytometry immunophenotyping panel in a multicenter quality assurance program. Cytometry 14:307-317. [DOI] [PubMed] [Google Scholar]
- 7.Schnizlein-Bick, C. T., J. Spritzler, C. L. Wilkening, J. K. A. Nicholson, and M. R. G. O'Gorman. 2000. Evaluation of TruCount absolute-count tubes for determining CD4 and CD8 cell number in human immunodeficiency virus-positive adults. Clin. Diagn. Lab. Immunol. 3:336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storie, I., A. Sawle, K. Goodfellow, L. Whitby, V. Granger, R. Y. Ward, J. Peel, T. Smart, J. T. Reilly, and D. Barnet. 2004. Perfect count: a novel approach for the single platform enumeration of absolute CD4+ T-lymphocytes. Cytometry B 57:47-52. [DOI] [PubMed] [Google Scholar]