Abstract
Rhodosporidium toruloides is a heterothallic, bipolar, red yeast that belongs to the Sporidiobolales, an order within a major lineage of basidiomycetes, the Pucciniomycotina. In contrast to other basidiomycetes, considerably less is known about the nature of the mating type (MAT) loci that control sexual reproduction in this lineage. Three genes (RHA1, RHA2, and RHA3) encoding precursors of the MAT A1 pheromone (rhodotorucine A) were previously identified and formed the basis for a genome walking approach that led to the identification of additional MAT genes in complementary mating strains of R. toruloides. Two mating type-specific alleles encoding a p21-activated kinase (PAK; Ste20 homolog) were found between the RHA2 and RHA3 genes, and identification in MAT A2 strains of a gene encoding a presumptive pheromone precursor enabled prediction of the structure of rhodotorucine a. In addition, a putative pheromone receptor gene (STE3 homolog) was identified upstream of RHA1. Analyses of genomic data from two closely related species, Sporobolomyces roseus and Sporidiobolus salmonicolor, identified syntenic regions that contain homologs of all the above-mentioned genes. Notably, six novel pheromone precursor genes were uncovered, which encoded, similarly to the RHA genes, multiple tandem copies of the peptide moiety. This suggests that this structure, which is unique among fungal lipopeptide pheromones, seems to be prevalent in red yeasts. Species comparisons provided evidence for a large, multigenic MAT locus structure in the Sporidiobolales, but no putative homeodomain transcription factor genes (which are present in all basidiomycetous MAT loci characterized thus far) could be found in any of the three species in the vicinity of the MAT genes identified.
Fungal mating type (MAT) loci are specialized genomic regions that determine cell type identity and coordinate the sexual cycle. Their genetic structure has been relatively sparsely sampled in broad phylogenetic terms, but results of published studies have revealed both conserved features and remarkable diversity in ascomycetes and basidiomycetes (8, 13). A common trait of fungal MAT loci is the presence of genes encoding homeodomain or other classes of transcription factors that control the expression of genes that establish cell type identity and activate sexual development. In basidiomycetes, two major types of MAT loci have so far been recognized (9). The tetrapolar mating system of the corn smut Ustilago maydis and the mushrooms Coprinopsis cinerea and Schizophyllum commune is governed by two small (<10-kb) unlinked loci: one encodes homeodomain transcription factors (HD1 and HD2 homologs), and the other encodes lipopeptide pheromones and pheromone receptors (STE3 homologs), which mediate intercellular signaling prior to cell fusion and/or sexual development after mating (9). Each locus may be multiallelic, which can result in up to thousands of compatible mating types in certain tetrapolar species. On the other hand, in the bipolar system of the human pathogen Cryptococcus neoformans and of the barley smut Ustilago hordei the two MAT loci described above for the tetrapolar species are physically linked, forming a single, large (>100-kb) multigene locus, which contains additional genes that may or may not be related to mating (5, 25). In this case there are only two possible combinations of MAT gene alleles, and hence only two compatible mating types exist. A unique feature of C. neoformans is that genes of three components of the pheromone-activated signaling cascade (viz., STE11, STE12, and STE20) are part of the MAT locus and, therefore, mating type specific (25).
Three major lineages are currently recognized in the Basidiomycota (18). One of them, the subphylum Pucciniomycotina, comprises the rust fungi (e.g., the wheat rust Puccinia graminis, whose genome annotation was recently released [http://www.broad.mit.edu/annotation/genome/puccinia_graminis]) and other plant parasites, such as the anther smuts (Microbotryum spp.), as well as many saprobic yeast taxa. The latter include the so-called “red yeasts” in the genera Rhodosporidium and Sporidiobolus, which are classified in the order Sporidiobolales (30). Fungi in this lineage have remained virtually unexplored as to the content and organization of the MAT loci. However, preliminary work with the bipolar anther smut Microbotryum violaceum has shown that chromosomes carrying the MAT loci are sexually size dimorphic and rich in repetitive sequences (20). A recent study based on expressed sequence tag data (39) provided the first clues on their gene content, but the respective genetic organization is yet to be elucidated. The remaining two basidiomycetous subphyla contain the few taxa whose MAT loci have been investigated in detail (9): the Ustilaginomycotina, which includes the majority of smut fungi (e.g., Ustilago spp.) and many other dimorphic plant parasites, and the Agaricomycotina, which includes the mushrooms (e.g., Coprinopsis and Schizophyllum), the jelly fungi, and many yeast taxa (e.g., Cryptococcus). The present study aims to expand our knowledge on the nature and organization of MAT genes to members of the rust lineage, namely, red yeasts in the Sporidiobolales, which may provide invaluable clues on the evolution of MAT loci in basidiomycetes.
As experimental models, saprobic basidiomycetous yeasts have a major advantage over their strictly filamentous or parasitic counterparts because they complete their life cycle on culture medium (11). The vast majority of sexual yeast taxa, such as Rhodosporidium, have dimorphic life cycles: upon fusion of compatible cells, elicited by the mating pheromones, the haploid yeast phase switches to a dikaryotic filamentous phase, during which basidia are eventually produced. In Rhodosporidium basidia arise from teliospores, which are thick-walled, resting cells that form on the dikaryotic hyphae and where karyogamy takes place (11). Germination of basidiospores restores the yeast phase.
All known Rhodosporidium species were isolated as haploid yeasts and have a bipolar mating behavior, i.e., their strains belong to either one of two complementary mating types, designated A1 and A2 or A and a (1, 10). Based on our current understanding of basidiomycete MAT loci, it is hypothesized that the genes encoding the pheromones, the pheromone receptors, and the homeodomain transcription factors are part of a single MAT locus in Rhodosporidium species. Pioneering work by Japanese researchers led by S. Fukui in the late 1980s identified three genes encoding the pheromone precursors in a mating type A1 (or A) strain of R. toruloides (2). These genes were not present in a complementary mating type A2 (or a) strain (3) and have been suggested to be part of the MAT locus (6). Fukui and coworkers (1) had previously characterized the early stages of the mating process in R. toruloides and suggested that A1 cells constitutively produced a pheromone (A factor, later named rhodotorucine A), which induced the production of a factor by A2 cells. The studies of Akada et al. (2, 3) were not continued, and no additional information on the MAT loci of Rhodosporidium species was published since then.
Here we present evidence that the gene encoding a p21-activated kinase (PAK; Ste20-like) possibly involved in pheromone signaling is mating type specific and that, along with the multiple pheromone precursor genes, it belongs to the R. toruloides MAT locus. We also found that this genomic region displays a high level of synteny with homologous genomic regions from the closely related yeast Sporobolomyces roseus (11), whose genome sequence has been recently released (Joint Genome Institute [JGI]; http://genome.jgi-psf.org/Sporo1/Sporo1.home.html). Genomic fragments of a close relative of the latter species, Sporidiobolus salmonicolor (11), are also available and provided an additional source of sequence data for comparison. Analysis of our data on R. toruloides together with that of the S. roseus genome and of the S. salmonicolor Trace Archive provided evidence as to the MAT locus structure of members of the Sporidiobolales and constitutes the second instance of the presence of a PAK kinase gene within a basidiomycete MAT locus.
MATERIALS AND METHODS
Rhodosporidium toruloides strains, culture conditions and mating tests.
The following strains of Rhodosporidium toruloides, obtained from the Portuguese Yeast Culture Collection (CREM; Portugal), were studied in detail: PYCC 4416 (CBS 14; MAT A1), PYCC 5082 (MAT A1), PYCC 4417 (CBS 5745; MAT A2), and PYCC 4661 (MAT A2). Cultures were grown on MYP agar (0.7% malt extract, 0.05% yeast extract, 0.25% soytone-peptone, 1.5% agar) at room temperature. To check sexual compatibility, pairs of 2- to 4-day-old cultures were mixed on an MYP agar plate, incubated at room temperature, and examined microscopically after 1 week for the production of mycelia with clamp connections and teliospores, using phase-contrast optics.
An enlarged set of strains was used to check mating type specificity of pheromone genes by PCR, in addition to the four strains mentioned above, namely, CBS 350, CBS 315, MSD 231, A421, and A415 (MAT A1); CBS 349, PYCC 4786, PYCC 4943, PYCC 5109, PYCC 5081, A412, A401, A413, and A456 (MAT A2). MSD and A strains are environmental isolates obtained by Gadanho et al. (16).
Miscellaneous.
Isolation of genomic DNA was performed essentially as described by Sampaio et al. (29), with a few modifications. After centrifugation, the cell extracts were submitted to digestion with proteinase K (1 mg/ml; 1 h, 37°C) and, subsequently, to phenol (pH 8.0) and chloroform-isoamyl alcohol (24:1) extractions. Nucleic acids were dissolved in 100 μl of distilled water after precipitation.
PCRs were performed in a final volume of 25 μl and contained the following components (unless stated otherwise): 2 mM of MgCl2, 0.25 mM of each of the four deoxynucleoside triphosphates (GE Healthcare), 0.8 μM of primer, 5 μl of template (genomic DNA was diluted 1:750), and 1 U Taq DNA polymerase (Fermentas, Canada). Thermal cycling consisted of a 5-minute denaturation step at 95°C, followed by 35 cycles of denaturation at 94°C for 30 s, 30 s at the annealing temperature (variable), and extension at 72°C (variable time); the annealing temperatures and extension times used in each reaction, as well as the sequences of all the primers, are listed in Table S1 of the supplemental material. A final extension of 7 min at 72°C was subsequently performed.
Amplification products were purified using the GFX PCR DNA and gel purification kit (GE Healthcare) and either cloned into the pMOSBlue vector with the pMOSBlue blunt-ended cloning kit (GE Healthcare) and sequenced or sequenced directly (sequencing was performed by STABVida, Portugal).
Mating type specificity of RHA genes.
The MAT A1 specificities of the RHA1, RHA2, and RHA3 pheromone precursor genes were checked by PCR amplification with gene-specific primers based on the sequences determined by Akada et al. (2) and using genomic DNA from both MAT A1 and MAT A2 strains as template. One forward primer common to the three genes (RHAFw) was used together with one reverse primer specific for each gene (RHA1Rev, RHA2Rev, and RHA3Rev) (see Table S1 in the supplemental material).
Following the identification of the RHA2.A2 gene, mating type specificity was checked in an enlarged set of strains using reverse primers specific for either the RHA2 or the RHA2.A2 coding regions (MC075 and MC071, respectively) (see Table S1 in the supplemental material) and a common forward primer based on a conserved STE20 region in the two mating types (Ste20Dw2A2) (see Table S1 in the supplemental material).
Genome walking.
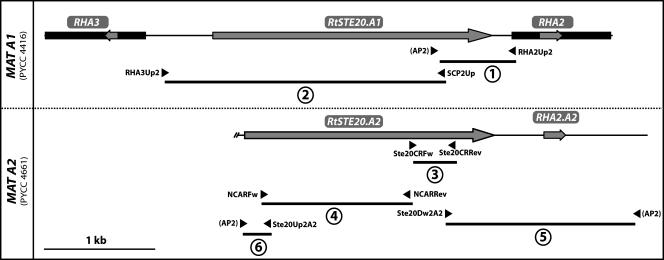
A genome walking approach using the Universal Genome Walker kit (BD Biosciences Clontech) was employed to characterize the MAT loci of R. toruloides. This method consists of preparing a set of genomic DNA libraries each requiring the ligation of adapters to the ends of restriction fragments obtained by complete digestion with a particular enzyme (blunt cutters). These libraries are subsequently screened using combinations of gene-specific and adapter primers (AP1 and AP2). For both MAT A1 strain PYCC 4416 and for MAT A2 strain PYCC 4661, the genomic libraries were prepared using EcoRV, PvuII, and StuI. Each Genome Walker library was used as a template in independent amplification reactions. Primer design and PCR conditions followed the manufacturer's instructions. The fragment upstream of the RHA2 gene (fragment 1; Fig. 1) was obtained from the EcoRV library, whereas the two fragments encompassing part of the MAT A2 STE20 gene (fragments 5 and 6; Fig. 1) were amplified from the StuI MAT A2 library.
FIG. 1.
Structure of the STE20 region of the MAT A1 and MAT A2 loci of R. toruloides. Coding regions are shown as gray arrows, indicating the direction of transcription, and the transcribed regions identified by Akada et al. (2) are shown as black boxes. Genomic DNA fragments 1 to 6 were obtained either by genome walking or by PCR with gene-specific primers. The primers used in each case are indicated by arrowheads (see Table S1 in the supplemental material).
PCR amplification and sequencing of the STE20 alleles.
A fragment encompassing the complete region between the RHA2 and RHA3 genes was obtained by PCR using gene-specific primers RHA3Up2 and SCP2Up (Fig. 1; see also Table S1 in the supplemental material) and genomic DNA from strain PYCC 4416 MAT A1 as a template. The primer pairs Ste20CRFw with Ste20CRRev and NCARFw with NCARRev were based on regions of the MAT A1 STE20 allele that were found to be conserved in the closely related species S. roseus and S. salmonicolor. These primers were used to amplify most of the RtSTE20 gene in the remaining strains under study (MAT A1 and MAT A2). The PCR products were purified and directly sequenced by primer walking.
PCR amplification of a putative MAT A1 pheromone receptor (STE3 homolog).
For the identification of pheromone receptor genes in Rhodosporidium toruloides, the rcb1 gene from Coprinopsis cinereus (accession number Y11082) was used to perform a Tblastx search (4) in the S. roseus genome sequencing project database (JGI; http://genome.jgi-psf.org/Sporo1/Sporo1.home.html). Positive hits were retrieved and used to search the NCBI Trace Archive sequences of S. salmonicolor, yielding an assembled sequence for the SsSTE3 gene. The SsSTE3 and SrSTE3 genes were in turn aligned with other basidiomycete pheromone receptor genes (PDSTE3.3 gene from Pleurotus djamor [AY462110], bbr1 from Schizophyllum commune [U74495], and STE3a from Cryptococcus neoformans var. neoformans [XM_570116]), and the conserved regions were used to design degenerate primers (NCAR3Fw and NCAR4Rev) (see Table S1 in the supplemental material). These degenerate primers were used in PCRs with genomic DNA from R. toruloides but failed to yield an amplification product. A similar reaction using as template genomic DNA from a closely related species (Rhodosporidium kratochvilovae PYCC 4818; MAT A1) (29) produced an amplicon which was directly sequenced and found to encode a partial open reading frame (ORF) of a STE3-like pheromone receptor. The new sequence was used to refine the design of a second set of primers for R. toruloides. These primers (NCAR5STE3Fw and NCAR6STE3Rev) (see Table S1 in the supplemental material) permitted the amplification of part of the STE3 homolog in R. toruloides. The position and orientation of the STE3 homolog relative to the RHA1 gene in R. toruloides were checked by PCR with gene-specific primers (NCAR5STE3Fw and RHA1Rev) (see Table S1 in the supplemental material). PCRs were performed in a final volume of 50 μl and contained the following components: 1 Long PCR buffer, 2 mM of MgCl2, 0.2 mM of each of the four deoxynucleoside triphosphates (GE Healthcare), 1.0% dimethyl sulfoxide, 1.0 μM of primer, 5 μl of template (genomic DNA diluted 100-fold), and 2.5 U Long PCR enzyme mix (Fermentas, Canada). Thermal cycling consisted of a 3-minute denaturation step at 94°C, followed by an initial round of 10 cycles of denaturation at 95°C for 20 s, annealing at 60°C for 30 s, and extension at 68°C for 6 min and a second round of 25 cycles, increasing the extension time 2 s in each cycle. A final extension of 10 min at 68°C was performed. The nucleotide sequences of the ends of the amplified fragment were obtained by direct sequencing using the same primers.
In silico analyses.
Primers were designed using Primer Premier 5.0 software (Premier Biosoft International). The assembly of DNA sequences was performed using SeqMan II software (v.6.1) from DNASTAR. Gene models were constructed by combining nucleotide homology searches in the NCBI database, using Blastn and tBlastx (4) and in silico predictions of putative introns and coding sequences using AUGUSTUS software (http://augustus.gobics.de/submission) (34). Blastp searches in NCBI were used to check for conserved domains within the deduced amino acid sequences and to assign putative gene functions.
The genomic DNA sequences of S. roseus (IAM 13481) showing a high degree of identity to the STE20 homolog of R. toruloides PYCC 4416 (MAT A1) were retrieved from the first release of the S. roseus genome sequencing project by JGI (http://genome.jgi-psf.org/Sporo1/Sporo1.home.html; scaffold 9, contig 11). Annotation of the 40-kb region encompassing the STE20 gene was completed using Tblastx. The NCBI Trace Archive sequences of S. salmonicolor (IAM 12258) contained sequences with a high percent sequence identity to that of the STE20 gene of S. roseus. The relevant S. salmonicolor fragments were assembled stepwise from the STE20 region. Following homology searches using the SsRHA2 gene, the SsRHA1 and SsRHA3 genes of S. salmonicolor were also retrieved and assembled (the Trace Archive sequences used are listed in Table S2 of the supplemental material). Nucleotide and amino acid sequences were aligned using ClustalW (37) and T-Coffee (28), respectively.
Tblastx was used to search the entire genome of S. roseus for homologs of the homeodomain transcription factors (HD1 and HD2).
Nucleotide sequence accession numbers.
The accession numbers for the novel R. toruloides DNA sequences are as follows: EU386160 (RtSTE20.A1, PYCC 4416, fragments 1 and 2) (Fig. 1); EU386161 (RtSTE20.A2 and RHA2.A2, PYCC 4661, fragments 3 to 6) (Fig. 1); EU401861 and EU401862 (RtSTE20.A1, PYCC 5082, and RtSTE20.A2, PYCC 4417, respectively; region corresponding to the fragments 3 and 4) (Fig. 1); EU401863 (RtSTE3.A1, PYCC 4416, partial sequence).
RESULTS
MAT status of R. toruloides strains.
To characterize genomic regions involved in mating in R. toruloides, the four strains to be studied (two of each mating type) were tested for their ability to mate and form hyphae with clamp connections and teliospores, characteristic of the sexual cycle in this yeast (10). All four strains were fertile, MAT A1 isolates mated only with MAT A2 isolates, and none of the strains were self-fertile. The mating results were in accord with the presence of the pheromone precursor genes previously described by Fukui and coworkers (2) (RHA1, RHA2, and RHA3), since they were detected by PCR with gene-specific primers only in strains that mated as MAT A1 in crossing experiments.
Genomic regions in the vicinity of the pheromone precursor genes in R. toruloides MAT A1.
Two of the pheromone precursor genes previously identified (RHA2 and RHA3) were found to be located close to each other on opposite DNA strands (2) (Fig. 1), while the location of the third gene (RHA1) in relation to the former remained to be established. Since in basidiomycetes the genes encoding pheromone precursors have so far been found almost invariably inside the MAT loci, we used the sequences of the three RHA genes as anchors for a genome walking approach to explore the adjacent regions, presumed to be part of the MAT A1 locus in this yeast.
To this end, a genome walking approach was designed that resulted in the amplification of a 0.7-kb genomic DNA fragment upstream of the RHA2 gene (Fig. 1, fragment 1). Sequencing of this fragment revealed two notable features: first, a region of 204 bp located immediately upstream of the RHA2 gene was found to be identical to the corresponding region upstream of the RHA3 gene sequenced by Akada et al. (2); second, a region of approximately 200 bp at the 5′ end of the fragment turned out to encode a peptide with strong similarity to the catalytic domain of PAK kinases (19). To investigate whether this was part of a PAK kinase gene, the entire intervening genomic region between the RHA2 and RHA3 genes was amplified and sequenced by primer walking (Fig. 1, fragments 1 and 2). This region was indeed found to contain a putative PAK kinase gene (named RtSTE20), homologous to genes found in, for example, Ustilago maydis, Cryptococcus neoformans, and Pneumocystis carinii (32, 33, 38). In silico analysis predicted the presence of at least four introns in this gene.
The RtSTE20 gene is mating type specific in R. toruloides MAT A1.
Since the RtSTE20 gene is located between two mating type-specific genes, we anticipated that it would be part of the MAT A1 locus. In this case, its sequence would be expected to exhibit mating type-specific polymorphisms. To investigate this, the RtSTE20 gene was amplified and sequenced almost completely in the four strains under study (the regions sequenced, approximately 1.7 kb, correspond to fragments 3 and 4 depicted in Fig. 1). The sequences were indeed found to be almost identical between alleles obtained from strains of the same mating type (eight differences between the two MAT A1 strains and three differences between the two MAT A2 strains). However, sequences obtained for the opposite mating types had approximately 80 single nucleotide differences, which would lead to nine amino acid changes in the conserved domains of the RtSte20 protein (see Fig. S1 in the supplemental material). The large majority of the nucleotide differences between the two alleles were, however, located in less-conserved regions of the RtSTE20 gene. This observation supports the conclusion that RtSTE20 is a mating type-specific gene in R. toruloides.
A putative pheromone precursor gene in R. toruloides MAT A2.
To explore the organization of the MAT A2 locus of R. toruloides, the region around the RtSTE20 gene in MAT A2 strain PYCC 4661 was analyzed by genome walking. A fragment of 2 kb downstream of the STE20 gene was amplified and sequenced (Fig. 1, fragment 5). A completely dissimilar sequence was found downstream of the RtSTE20 gene in the MAT A2 strain when compared to the same region in a MAT A1 strain (encoding the RHA2 gene). To ascertain if this region also encoded a pheromone precursor, the sequence was examined for the presence of an open reading frame exhibiting characteristics in keeping with the general features of previously characterized pheromone precursor genes (6, 7). Indeed, a region was found potentially encoding two identical copies of a 13-amino-acid peptide and exhibiting at the C terminus a sequence of four amino acids (CAAX motif) very similar to that of the MAT A1 pheromone precursors (Fig. 2 and 3). The most likely initiation codon is preceded by two CT-rich regions (Fig. 2), one of which is identical to that found upstream of the transcription initiation site of the RHA (MAT A1) genes (2). Therefore, we postulate that this novel gene, named RHA2.A2, encodes a precursor of the peptide moiety of the R. toruloides MAT A2 pheromone (rhodotorucine a).
FIG. 2.
Nucleotide sequence of the genomic region encoding the RHA2.A2 gene, showing the predicted pheromone precursor sequence. CT-rich regions possibly involved in transcription initiation are underlined. The two repeats proposed to represent the peptide moiety of the mature pheromone are shadowed. The sequences resembling the CAAX prenylation motif are enclosed in boxes.
FIG. 3.
Alignment of the predicted pheromone precursor peptides from the three sporidiobolaceous species, as deduced from the sequence of the respective genes: RHA (R. toruloides), SsRHA (S. salmonicolor), and SrRHA (S. roseus). Sequences representing the mature pheromones are shadowed, and the sequences resembling the CAAX prenylation motif are underlined in the C terminus of each precursor.
Mating type specificity of RtSTE20/RHA regions.
In order to check linkage between mating behavior and the presence of the newly identified RHA2.A2/STE20 region, a specific primer based on the RHA2.A2 coding region was used in a MAT A2 diagnostic PCR in combination with a RtSTE20 forward primer. A total of 14 additional strains (five MAT A1 and nine MAT A2) whose mating behavior was previously checked were tested for the presence of the putative MAT A2 locus sequences. As expected, an amplicon was obtained only for MAT A2 strains (results not shown). Conversely, when the same RtSTE20 primer was used in combination with a primer based on the RHA2 (MAT A1) coding sequence in a PCR assay with the same strain set, only MAT A1 strains yielded an amplification product (results not shown). These results strongly suggest that this region is indeed part of the MAT locus of R. toruloides. Notably, one R. toruloides isolate (A399) (16), which was not included in the previous strain set because it exhibited ambiguous mating behavior (apparent sexual compatibility with several MAT A1 and MAT A2 strains), tested positive in both MAT A1 and MAT A2 diagnostic PCR assays, although amplification of MAT A1 sequences was weaker than observed for MAT A1 strains.
Direct evidence of linkage of the characterized genomic region in R. toruloides to MAT by genetic analysis of F1 progeny was not possible since basidiospores form only upon germination of teliospores embedded in the agar medium and are thus not easily subjected to micromanipulation.
Comparison with syntenic genomic regions from related red yeasts.
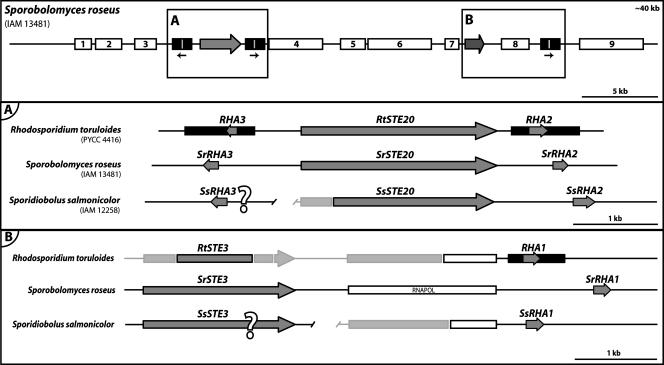
A search in the recently released genome data of Sporobolomyces roseus for genes with a high percent sequence similarity with the catalytic domain of RtSTE20 identified a gene likely to represent a RtSTE20 homolog. Notably, this gene (named henceforth SrSTE20) is also flanked by two divergently transcribed putative pheromone precursor genes, which were named SrRHA2 and SrRHA3, based on synteny with the RHA2 and RHA3 genes from R. toruloides. Unlike the latter, the two genes from S. roseus are completely identical not only in the coding sequence but also in the promoter and terminator regions. Furthermore, a third identical gene (named SrRHA1) was found downstream of a gene probably encoding a 30- to 40-kDa subunit of RNA polymerase III (Pol III). Notably, the 3′ end of the coding sequence of the R. toruloides homolog of this Pol III subunit was found upstream of the RHA1 gene in the genomic DNA fragment sequenced by Akada et al. (2).
For Sporidiobolus salmonicolor, genomic sequences are available as Trace Archives (short unassembled genomic DNA fragments). A search of this database revealed the presence of a gene very similar to the STE20 genes of S. roseus and R. toruloides. This gene (SsSTE20) was used as an anchor to assemble a contiguous fragment that extended to approximately 2 kb downstream of its 3′ end, where a putative pheromone precursor gene (named SsRHA2) was similarly found (Fig. 4). The sequence of the SsRHA2 gene was used in new searches in the Trace Archives and led to the identification of two additional genes potentially encoding pheromone precursors. While for one of the genes (SsRHA3) the longest possible contiguous fragment did not provide any clue as to its location, the second gene (SsRHA1) was found downstream of a partial ORF encoding the 30- to 40-kDa subunit of RNA polymerase III (Fig. 4). The SsRHA1 and SsRHA3 coding sequences are completely identical.
FIG. 4.
Structure of a putative MAT locus in S. roseus and comparison with homologous genomic regions in R. toruloides and S. salmonicolor. The top panel shows the gene content and organization in a 40-kb region around the putative pheromone precursor (box A) and pheromone receptor (box B) genes in S. roseus. Additional genes identified in this region are depicted as white boxes and are listed in Table S3 of the supplemental material. The lower panels show the gene content and organization of the regions around the pheromone precursor (A) and the pheromone receptor (B) homologs in the three species. Unknown sequences are shown in light gray. The relative position and orientation of the genes marked “?” are unknown. All the remaining features are represented as in Fig. 1.
In S. roseus a long contiguous sequence around the SrSTE20 gene is available. Therefore, in order to find out whether more MAT-related genes could be found in the vicinity of this SrSTE20 core, a genomic region of approximately 40 kb was scrutinized for the presence of ORFs. Notably, a STE3-like gene (17), potentially encoding a pheromone receptor, was found close to the SrRHA1 gene (Fig. 4, upper panel). Many other genes, apparently unrelated to mating but exhibiting a high percent sequence similarity with genes identified in other organisms, were also found in this region and are indicated in Fig. 4 and listed in Table S3 of the supplemental material. These results suggest that the entire region between the pheromone receptor core and the SrSTE20 core may be part of a large MAT locus in this yeast. A STE3-like gene was also identified in S. salmonicolor, but the incompleteness of the genome sequence data did not allow any inference about its location with respect to the other S. salmonicolor genes identified.
A pheromone receptor gene in Rhodosporidium.
The two STE3-like genes identified in S. roseus and S. salmonicolor share regions of considerable sequence identity. Using a first pair of degenerate primers based on these regions, we were unable to amplify a STE3 fragment from R. toruloides. However, it was possible to obtain an amplicon when genomic DNA from Rhodosporidium kratochvilovae, a species closely related to R. toruloides (29), was used as a template in the PCR. Sequencing of this fragment and its alignment with the above-mentioned STE3-like genes (results not shown) allowed the design of another set of primers that led to the amplification of the corresponding STE3 fragment from R. toruloides MAT A1 strain PYCC 4416, as confirmed by direct sequencing. Amplification of the latter fragment was possible only in MAT A1 strains, as expected for a mating type-specific pheromone receptor gene.
PCR with RtSTE3 and RHA1 gene-specific primers (see Table S1 of the supplemental material) yielded an amplification product of approximately 5 kb, consistent with a gene organization identical to that found in S. roseus. Sequencing of the ends of the amplified fragment confirmed the suspected synteny, including the presence of the gene encoding the RNA Pol III subunit between the STE3 and RHA1 genes (Fig. 4B).
Search for putative HD homologs in S. roseus.
We found a gene encoding a putative HD1 homolog in the S. roseus genome (scaffold 7, contig 11). Blastp searches in fungal protein databases using part of the sequence of the encoded protein retrieve most of the basidiomycetous HD1 proteins previously characterized. No HD2 candidates with such characteristics were found, although additional genes potentially encoding homeodomain proteins were identified in the S. roseus genome.
The gene encoding the putative HD1 homolog was recently annotated in the S. roseus genome, but at this stage of the assembly of the sequence data the distance to the STE20/RHA region cannot be precisely determined. However, even if they lie on the same chromosome, the distance of the SrSTE20 core to this HD1 candidate would be necessarily in excess of 600 kb, as judged from the positions of the genes within the scaffolds; the shortest distance to one of the ends of the respective scaffolds is 213 kb (STE20 scaffold) and 410 kb (HD1 scaffold), respectively. The region around this putative HD1-like gene was carefully screened for the presence of a HD2 homolog, since in most basidiomycetous MAT loci the HD1 and HD2 genes are closely linked and divergently transcribed. However, no HD2 candidate gene was found, even when the homology searches using tBlastx were extended to the entire genome. Nevertheless, it should be noted that the presence of introns may considerably complicate this kind of search based on a short conserved region, as for example in the case of the Sxi2a gene (HD2 homolog) of Cryptococcus neoformans (21).
DISCUSSION
In this paper we present the first insight into the structure of the MAT locus of a group of fungi that has remained unexplored in this respect. While the basis of the work consisted of the identification of novel genomic regions in Rhodosporidium toruloides, we have also analyzed syntenic genomic regions in two additional species belonging to the Sporidiobolales, namely, Sporobolomyces roseus and Sporidiobolus salmonicolor.
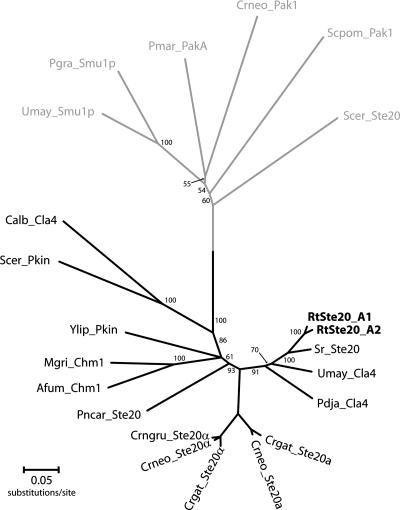
We found that two of the R. toruloides pheromone precursor genes previously characterized by Akada et al. (2) flanked a gene potentially encoding a kinase of the PAK family, whose homologs were also present in the S. roseus and S. salmonicolor genomes. These three PAK kinase orthologs encode proteins that possess the two functional domains common to the entire PAK kinase family, namely, a highly conserved C-terminal catalytic domain and a CRIB domain (cdc42/ras interaction binding domain) (see Fig. S1 in the supplemental material). In addition, they have a third domain (PH-pleckstrin homology) (see Fig. S1 in the supplemental material) that is present in a subset of the PAK family members, such as the Ste20 kinases of Cryptococcus neoformans and C. gattii (38) and the Cla4 kinase of Ustilago maydis (26). The former proteins were shown to be involved in mating or required for the onset of the filamentous phase and are present in the MAT loci of the two Cryptococcus sibling species. However, the MAT-specific alleles of STE20 have diverged considerably more in C. neoformans than in R. toruloides (approximately 70% versus 90% sequence identity) (Fig. 5). It is therefore unlikely that the two R. toruloides proteins exhibit functional dissimilarities comparable to those found for the C. neoformans MAT-specific kinases (38).
FIG. 5.
Phylogenetic tree showing the relationships between PAK kinases from different fungi, based on alignment of the respective protein sequences. Kinases lacking the PH domain are located in the gray branch of the tree. The sequences used and respective accession numbers are listed in Table S4 of the supplemental material, and the alignment of the three conserved domains in a subset of the proteins is shown in Fig. S1 of the supplemental material. The evolutionary distances were computed using Protdist (JTT matrix-based method) (12). Neighbor-joining (12) and MEGA4 (36) were used to construct and visualize the phylogenetic tree. Numbers on branches are bootstrap values inferred from 10,000 replicates (values below 50% are not shown).
The rhodotorucine A (RHA) genes were the first pheromone precursor genes whose structure was determined in basidiomycetes (2). The same authors (23, 24) had previously determined the chemical structure of the mature pheromone as a lipopeptide (farnesyl-undecapetide), in which the C-terminal cysteine of the peptide moiety was shown to be covalently linked to a farnesyl group through a thioether bond. That was the first demonstration of this type of posttranslational modification, which was subsequently found in many other fungal pheromones, such as the a factor of Saccharomyces cerevisiae, as well as in other eukaryotic proteins (6, 7).
Like other prenylated proteins, the predicted rhodotorucine A precursors contain a C-terminal CAAX motif (2), where A stands for an aliphatic amino acid and X for any amino acid (often an alanine in fungal pheromones) (7). The C-terminal motif of the rhodotorucine A precursor is actually CTVA, where threonine does not conform to the general pattern of fungal lipopeptide pheromones, which have either valine, isoleucine, or leucine as the middle amino acids (7, 27). However, recently Schirawski et al. (31) predicted six pheromone precursors in the smut Sporisorium reilianum, four of which have a terminal CTIA motif. The rhodotorucine A precursors are, nevertheless, unique among fungal lipopeptide pheromones, since they contain multiple tandem copies of the peptide moiety (2, 7). Akada et al. (2) suggested that those multiple repeats could have originated by an unequal crossing-over mechanism. The same authors (2) also affirmed that the production of multiple peptides per mole of precursor (as well as the presence of multiple RHA genes) may increase the production of the mating pheromone per cell and that this may be a prerequisite for the poorly diffusible small lipophilic peptides to function effectively. In fact, the mating reaction in R. toruloides appears to be triggered by the production of rhodotorucine A by MAT A1 (or A) cells, according to the chain of events suggested by Abe et al. (1). The other unusual trait of the rhodotorucine A precursors is that CAAX motifs are present between the repeats, where they are followed by a lysine residue, which has been proposed as a signal for proteolytic processing (2, 6).
Our findings provide interesting new insights into this topic. On one hand the putative rhodotorucine a precursor predicted from the RHA2.A2 gene sequence appears to have also a CAAX C-terminal motif (CTIA instead of CTVA) but only two repeats of the peptide moiety, which are apparently preceded by a much longer N terminus tail and separated by a longer peptide spacer (Fig. 3). These differences suggest that the pheromone precursor genes of the two opposite mating types of R. toruloides could be processed by different mechanisms. On the other hand our analyses of the S. roseus and S. salmonicolor genome data revealed the presence of three putative pheromone precursor genes which resemble those of R. toruloides MAT A1 (Fig. 3). The presence of three pheromone precursor genes in the three species examined, each encoding three to six copies of the mature peptide moiety, suggests that they have in common the production of abundant pheromone as an important feature of the mating process. This could mean that compatible partner recognition occurs mainly or exclusively through pheromone-receptor interactions. This is in contrast with tetrapolar species, like U. maydis, where mate recognition requires a second level of compatibility determined by the multiallelic mating type b locus, which encodes the bE and bW homeodomain transcription factors.
One striking difference between the RHA genes of R. toruloides and those found in the other two species is that the C-terminal motifs of the latter appear not to correspond to CAAX boxes. However, a lysine residue is present in the C terminus of the S. roseus and S. salmonicolor precursors immediately after a putative CAAX box and may signal a proteolytic processing site that could produce a CAAX-bearing intermediate. In all three sporidiobolaceous species, the internal CAAX boxes (as well as the putative terminal CAAX motifs of the S. roseus and S. salmonicolor precursors) have the consensus sequence C(T/I)VS, where the terminal amino acid is a serine instead of alanine (Fig. 3). However, the former amino acid is also compatible with a potential farnesylation of the terminal cysteine of the peptide moiety (7).
In the bipolar basidiomycetes Cryptococcus neoformans and Ustilago hordei, the MAT loci are large and multigenic (100 kb and 500 kb, respectively) and encompass both a pheromone-receptor gene region and a region encoding homeodomain transcription factors (5, 25). We did not come across candidate homeodomain transcription factor genes upon inspection of a 40-kb genomic region surrounding the pheromone precursor and pheromone receptor genes in S. roseus, but a very likely HD1 candidate was found at a distant location in the genome. This could mean that the MAT locus extends beyond the region inspected in S. roseus or that the homeodomain transcription factors are not linked to the MAT locus in this species, a situation corresponding to the Aon MAT status proposed by Fraser et al. (15) as one of the possible modes of transition from tetrapolar to bipolar mating systems. The other instance of a bipolar basidiomycete whose MAT locus structure has been elucidated, the mushroom Coprinellus disseminatus, is an example of the reverse situation with respect to MAT locus content, since in this case mating type identity is conferred by a short locus containing one or two pairs of closely linked homeodomain transcription factor genes but the pheromone and pheromone receptor genes are not mating type specific (9, 22). However, it should be kept in mind that although S. roseus is the only species in the Sporidiobolales for which the entire genome is available for scrutiny, it has been deemed an asexual species and therefore its MAT gene organization cannot be unequivocally linked to a functional sexual cycle. On the other hand, the sequenced S. roseus strain may well correspond to a haploid mating type of a yet-unrecognized sexual species, a situation frequently encountered among basidiomycetous yeasts (11). In favor of the latter hypothesis is the fact that none of the putative S. roseus MAT genes examined showed any sign of being degenerated or nonfunctional. On the other hand, S. salmonicolor is a heterothallic species, and the sequenced strain (IAM 12258, CBS 483) has a known mating status (MAT A2) (35).
Putative pheromone receptor genes (STE3 homologs) with a high degree of sequence identity were found in R. toruloides MAT A1 strains and in the other two sporidiobolaceous species. In R. toruloides and S. roseus the genes appear to be located upstream of the RHA1 gene, with the same gene unrelated to mating (a subunit of RNA Pol III) in between. Therefore, also in this region, located in S. roseus at a distance of approximately 10 kb from the SrSTE20 core, a strict synteny occurs between the three species (Fig. 4). Since the pheromone receptor gene remains to be identified in MAT A2 strains of R. toruloides, it is so far unknown whether the MAT A2 locus is also syntenic in this region. The strong sequence similarity observed between the Ste3 proteins and even between the putative mature pheromone regions in the three species suggests that the three MAT loci depicted in Fig. 3 represent the “same mating type,” analogous to MAT A1 in R. toruloides. The limited amount of information available for the opposite mating type (restricted to the RtSTE20.A2 and RHA2.A2 genes in R. toruloides) precludes any conclusion with respect to extended MAT synteny conservation between the two mating types.
In conclusion, our results strongly suggest that the studied group of bipolar basidiomycetes possess extended multigenic MAT loci, containing both mating-related and unrelated genes. This gene content resembles that of the MAT locus of the distantly related bipolar Cryptococcus species (14). However, the apparent high level of synteny observed between the three sporidiobolaceous species is in sharp contrast with the highly variable MAT gene order and orientation observed in the Cryptococcus species. Future studies will be aimed at a more complete definition of the Sporidiobolales MAT loci, with special emphasis on the identification of additional MAT gene candidates, determination of the full extension of the MAT loci, and investigation of the role of homeodomain transcription factors in sexual reproduction. The recent release of genomic data from two other fungi in the Pucciniomycotina (Puccinia graminis and Microbotryum violaceum) (39) and the future completion of the Rhodosporidium babjevae genome (an ongoing sequencing project at the JGI) will also contribute to placing our findings in a broader context and help to retrace the evolution of the MAT loci in this basidiomycete subphylum.
ADDENDUM IN PROOF
The strain sequenced by the JGI (IAM 13481), labeled as Sporobolomyces roseus, does not actually belong to that species, as was recently shown by Valério et al. (Int. J. Syst. Evol. Microbiol. 58:736-741, 2008). It corresponds to an unnamed taxon with an unknown sexual status (see Fig. 2 of the above-mentioned paper).
Supplementary Material
Acknowledgments
The work was partially supported by project POCTI/BME/44322/2002 and Ph.D. grant SFRH/BD/29580/2006 (M.C.) from the Portuguese Foundation for Science and Technology.
We thank Kenneth Wolfe (Smurfit Institute of Genetics, University of Dublin) for providing access to the Sporobolomyces roseus genome data before it was publicly available and José Paulo Sampaio (CREM, Universidade Nova de Lisboa) and coworkers for the R. toruloides strains.
Footnotes
Published ahead of print on 11 April 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Abe, K., I. Kusaka, and S. Fukui. 1975. Morphological change in the early stages of the mating process of Rhodosporidium toruloides. J. Bacteriol. 122710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akada, R., K. Minomi, J. Kai, I. Yamashita, T. Miyakawa, and S. Fukui. 1989. Multiple genes coding for precursors of rhodotorucine A, a farnesyl peptide mating pheromone of the basidiomycetous yeast Rhodosporidium toruloides. Mol. Cell. Biol. 93491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akada, R., J. Kai, I. Yamashita, T. Miyakawa, and S. Fukui. 1989. Genomic organization of multiple genes coding for rhodotorucine-a, a lipopeptide mating pheromone of the basidiomycetous yeast Rhodosporidium toruloides. Arch. Microbiol. 152484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakkeren, G., G. Jiang, R. L. Warren, Y. Butterfield, H. Shin, R. Chiu, R. Linning, J. Schein, N. Lee, G. Hu, D. M. Kupfer, Y. Tang, B. A. Roe, S. Jones, M. Marra, and J. W. Kronstad. 2006. Mating factor linkage and genome evolution in basidiomycetous pathogens of cereals. Fungal Genet. Biol. 43655-666. [DOI] [PubMed] [Google Scholar]
- 6.Bölker, M., and R. Kahmann. 1993. Sexual pheromones and mating responses in fungi. Plant Cell 51461-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldwell, G. A., F. Naider, and J. M. Becker. 1995. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol. Rev. 59406-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casselton, L. A. 2002. Mate recognition in fungi. Heredity 88142-147. [DOI] [PubMed] [Google Scholar]
- 9.Casselton, L. A., and M. P. Challen. 2006. The mating type genes of the basidiomycetes, p. 357-374. In U. Kües and R. Fischer (ed.), The Mycota, vol. I. Growth, differentiation and sexuality, 2nd ed. Springer, Berlin, Germany. [Google Scholar]
- 10.Fell, J. W., and A. Statzell-Tallman. 1998. Rhodosporidium Banno, p. 678-692. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, a taxonomic study, 4th ed. Elsevier, Amsterdam, The Netherlands.
- 11.Fell, J. W., T. Boekhout, Á. Fonseca, and J. P. Sampaio. 2001. Basidiomycetous yeasts, p. 3-35. In D. J. McLaughlin, E. G. McLaughlin, and P. A. Lemke (ed.), The Mycota, vol. VIIB. Systematics and evolution. Springer, Berlin, Germany. [Google Scholar]
- 12.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. Department of Genetics, University of Washington, Seattle.
- 13.Fraser, J. A., and J. Heitman. 2004. Evolution of fungal sex chromosomes. Mol. Microbiol. 51299-306. [DOI] [PubMed] [Google Scholar]
- 14.Fraser, J. A., S. Diezmann, R. L. Subaran, A. Allen, K. B. Lengeler, F. S. Dietrich, and J. Heitman. 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2e384. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, J. A., Y. Hsueh, K. M. Findley, and J. Heitman. 2007. Evolution of the mating-type locus: the basidiomycetes, p 19-34. In J. Heitman, J. W. Kronstad, J. W. Taylor and L. A. Casselton (ed.). Sex in fungi: molecular determination and evolutionary implications. ASM Press, Washington, DC.
- 16.Gadanho, M., D. Libkind, and J. P. Sampaio. 2006. Yeast diversity in the extreme acidic environments of the Iberian pyrite belt. Microb. Ecol. 52552-563. [DOI] [PubMed] [Google Scholar]
- 17.Hagen, D. C., G. McCaffrey, and G. F. Sprague, Jr. 1986. Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor: gene sequence and implications for the structure of the presumed receptor. Proc. Natl. Acad. Sci. USA 831418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hibbett, D. S., M. Binder, J. F. Bischoff, M. Blackwell, P. F. Cannon, O. E. Eriksson, S. Huhndorf, T. James, P. M. Kirk, R. Lücking, H. Thorsten Lumbsch, F. Lutzoni, P. B. Matheny, D. J. McLaughlin, M. J. Powell, S. Redhead, C. L. Schoch, J. W. Spatafora, J. A. Stalpers, R. Vilgalys, M. C. Aime, A. Aptroot, R. Bauer, D. Begerow, G. L. Benny, L. A. Castlebury, P. W. Crous, Y. C. Dai, W. Gams, D. M. Geiser, G. W. Griffith, C. Gueidan, D. L. Hawksworth, G. Hestmark, K. Hosaka, R. A. Humber, K. D. Hyde, J. E. Ironside, U. Kõljalg, C. P. Kurtzman, K. H. Larsson, R. Lichtwardt, J. Longcore, J. Miadlikowska, A. Miller, J. M. Moncalvo, S. Mozley-Standridge, F. Oberwinkler, E. Parmasto, V. Reeb, J. D. Rogers, C. Roux, L. Ryvarden, J. P. Sampaio, A. Schüssler, J. Sugiyama, R. G. Thorn, L. Tibell, W. A. Untereiner, C. Walker, Z. Wang, A. Weir, M. Weiss, M. M. White, K. Winka, Y. J. Yao, and N. Zhang. 2007. A higher-level phylogenetic classification of the fungi. Mycol. Res. 111509-547. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann, C., M. Shepelev, and J. Chernoff. 2004. The genetics of Pak. J. Cell Sci. 1174343-4354. [DOI] [PubMed] [Google Scholar]
- 20.Hood, M. E., J. Antonovics, and B. Koskella. 2004. Shared forces of sex chromosome evolution in haploid-mating and diploid-mating organisms: Microbotryum violaceum and other model organisms. Genetics 168141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hull, C. M., M. J. Boily, and J. Heitman. 2005. Sex-specific homeodomain proteins Sxi1α and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot. Cell 4526-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James, T. Y., P. Srivilai, U. Kües, and R. Vilgalys. 2006. Evolution of the bipolar mating system of the mushroom Coprinellus disseminatus from its tetrapolar ancestors involves loss of mating-type-specific pheromone receptor function. Genetics 1721877-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamiya, Y., A. Sakurai, S. Tamura, N. Takahashi, K. Abe, E. Tsuchiya, and S. Fukui. 1978. Isolation of rhodotorucine A, a peptidyl factor inducing the mating tube formation in Rhodosporidium toruloides. Agric. Biol. Chem. 431239-1243. [Google Scholar]
- 24.Kamiya, Y., A. Sakurai, S. Tamura, N. Takahashi, K. Abe, E. Tsuchiya, S. Fukui, C. Kitada, and M. Fujino. 1978. Structure of rhodotorucine A, a novel lipopeptide inducing mating tube formation in Rhodosporidium toruloides. Biochem. Biophys. Res. Commun. 831077-1083. [DOI] [PubMed] [Google Scholar]
- 25.Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich, and J. Heitman. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leveleki, L., M. Mahlert, B. Sandrock, and M. Bölker. 2004. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol. Microbiol. 54396-406. [DOI] [PubMed] [Google Scholar]
- 27.McClelland, C. M., J. Fu, G. L. Woodlee, T. S. Seymour, and B. L. Wickes. 2002. Isolation and characterization of the Cryptococcus neoformans MATa pheromone gene. Genetics 160935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirot, O., E. O'Toole, and C. Notredame. 2003. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 313503-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sampaio, J. P., M. Gadanho, S. Santos, F. L. Duarte, C. Pais, A. Fonseca, and J. W. Fell. 2001. Polyphasic taxonomy of the basidiomycetous yeast genus Rhodosporidium: Rhodosporidium kratochvilovae and related anamorphic species. Int. J. Syst. Evol. Microbiol. 51687-697. [DOI] [PubMed] [Google Scholar]
- 30.Sampaio, J. P., M. Gadanho, R. Bauer, and M. Weiß. 2003. Taxonomic studies in the Microbotryomycetidae: Leucosporidium golubevii sp. nov. and the new orders Leucosporidiales and Sporidiobolales. Mycol. Prog. 253-68. [Google Scholar]
- 31.Schirawski, J., B. Heinze, M. Wagenknecht, and R. Kahmann. 2005. Mating type loci of Sporisorium reilianum: novel pattern with three a and multiple b specificities. Eukaryot. Cell 41317-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, D. G., M. D. Garcia-Pedrajas, W. Hong, Z. Yu, S. E. Gold, and M. H. Perlin. 2004. An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot. Cell 3180-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smulian, A. G., T. Sesterhenn, R. Tanaka, and M. T. Cushion. 2001. The STE3 pheromone receptor gene of Pneumocystis carinii is surrounded by a cluster of signal transduction genes. Genetics 157991-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanke, M., R. Steinkamp, S. Waack, and B. Morgenstern. 2004. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 32(Web Server issue)W309-W312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Statzell-Tallman, A., and J. W. Fell. 1998. Sporidiobolus Nyland, p. 693-699. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, a taxonomic study, 4th ed. Elsevier, Amsterdam, The Netherlands.
- 36.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, P., C. B. Nichols, K. B. Lengeler, M. E. Cardenas, G. M. Cox, J. R. Perfect, and J. Heitman. 2002. Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryot. Cell 1257-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yockteng, R., S. Marthey, H. Chiapello, A. Gendrault, M. E. Hood, F. Rodolphe, B. Devier, P. Wincker, C. Dossat, and T. Giraud. 2007. Expressed sequences tags of the anther smut fungus, Microbotryum violaceum, identify mating and pathogenicity genes. BMC Genomics 10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.