Abstract
Nutrient-sensing kinases play important roles for the yeast Saccharomyces cerevisiae to adapt to new nutrient conditions when the nutrient status changes. Our previous global gene expression analysis revealed that the Pho85 kinase, one of the yeast nutrient-sensing kinases, is involved in the changes in gene expression profiles when yeast cells undergo a diauxic shift. We also found that the stationary phase-specific genes SNZ1 and SNO1, whch share a common promoter, are not properly induced when Pho85 is absent. To examine the role of the kinase in SNZ1/SNO1 regulation, we analyzed their expression during the growth of various yeast mutants, including those affecting Pho85 function or lacking the Pho4 transcription factor, an in vivo substrate of Pho85, and tested Pho4 binding by chromatin immunoprecipitation. Pho4 exhibits temporal binding to the SNZ1/SNO1 promoter to down-regulate the promoter activity, and a Δpho4 mutation advances the timing of SNZ1/SNO1 expression. SNZ2, another member of the SNZ/SNO family, is expressed at an earlier growth stage than SNZ1, and Pho4 does not affect this timing, although Pho85 is required for SNZ2 expression. Thus, Pho4 appears to regulate the different timing of the expression of the SNZ/SNO family members. Pho4 binding to the SNZ1/SNO1 promoter is accompanied by alterations in chromatin structure, and Rpd3 histone deacetylase is required for the proper timing of SNZ1/SNO1 expression, while Asf1 histone chaperone is indispensable for their expression. These results imply that Pho4 plays positive and negative roles in transcriptional regulation, with both cases involving structural changes in its target chromatin.
The budding yeast Saccharomyces cerevisiae changes its gene expression profiles upon alterations in nutrient status to adapt to new conditions. Nutrient-sensing kinases play important roles in this adaptation process. Cyclic AMP-dependent kinase and Snf1 regulate gene expression, cell growth, and carbohydrate metabolism in response to glucose availability, Tor kinases regulate protein synthesis and autophagy responding to nitrogen availability, and Pho85 kinase regulates the PHO system responding to environmental phosphate (Pi) (4, 34). The Pho4 transcription factor activates the transcription of the PHO genes involved in the PHO system in response to Pi limitation in the medium (20). Under high-Pi conditions where the yeast cells do not need to express the PHO genes, the Pho85 kinase-Pho80 cyclin complex phosphorylates Pho4 to facilitate its exclusion from the nucleus, resulting in repression of the PHO genes (7, 8). When the Pi concentration in the medium becomes low, Pho85-Pho80 is inactivated by Pho81, a Cdk inhibitor, resulting in the accumulation of Pho4 in the nucleus to enable activation of the PHO genes. Pho85 also facilitates the degradation of Gcn4, a transcription factor that activates genes involved in amino acid metabolism under amino acid-starvation conditions, when sufficient amounts of the nutrients are available (2, 13). Based on a global gene expression analysis, we previously reported that Pho85 kinase was involved in the changes in gene expression responding to a diauxic shift (18). When yeast cells undergo a diauxic shift, they shift their metabolism from fermentation to respiration, and accordingly, the genes involved in mitochondrial function, gluconeogenesis, and storage carbohydrate synthesis are induced, while those involved in glycolysis are repressed (5). Pho85 function is required for yeast cells to carry out these changes properly (18). Thus, Pho85 functions in responding to alterations in the environmental nutrient conditions that are more general than those previously considered (18).
SNZ1 was discovered as a gene expressed in the stationary phase, and SNO1, which is adjacent to SNZ1, is regulated coordinately with the neighboring gene through a common promoter (3, 21). In the absence of Pho85, these genes were not induced properly at the late growth stage (18). When we looked at the SNZ1/SNO1 promoter sequence, we found a putative Pho4-binding sequence (CACGTT) at −380 (taking the A of ATG of SNZ1 as +1) in addition to three possible Gcn4-binding sites. Gcn4 is required for the activation of SNZ1/SNO1 under amino acid depletion conditions (17). The involvement of Pho85 in the alterations of gene expression accompanying a diauxic shift and the presence of a prospective Pho4-binding site in the SNZ1 promoter prompted us to test whether Pho85-Pho4 is involved in the regulation of SNZ1. We analyzed in detail the effects of various mutations that affect the functions of Pho85 and Pho4 and the chromatin structure on SNZ/SNO expression and timing during yeast growth. Here we report that Pho4 binds to the SNZ1/SNO1 promoter in a Pi-dependent manner to repress SNZ1/SNO1 to ensure the appropriate timing of their expression. We also demonstrate that Pho4 binding is accompanied by alterations in the chromatin structure at the SNZ1 promoter and that the Rpd3 histone deacetylase (HDAC) is involved in the regulation of the timing of SNZ1/SNO1 expression.
MATERIALS AND METHODS
Strains and media.
The yeast strains used in this work are listed in Table S1 in the supplemental material. To disrupt the PHO81 locus, the BglII (+472)-XhoI (+2164) fragment, in which the XbaI (+1194)-to-BamHI (+1764) region had been replaced by a URA3 fragment, was used, and successful disruption was confirmed by PCR using MN289/MN290 primers. MFY409 (Δcyc8::URA3) was constructed by using pJS22 plasmid (26). Escherichia coli strain DH5α was used as a host for plasmids. Media for E. coli growth and rich medium (YPAD) for yeast were prepared as described previously (24, 25). Low-Pi medium was prepared by using a yeast nitrogen base without Pi (Q-Biogene) instead of the normal yeast nitrogen base in synthetic dextrose (SD) medium and was supplemented with 0.2 μM sodium Pi. For the incubation under low-Pi conditions, yeast cells were grown in YPAD medium for 6 or 24 h, collected by centrifugation, and resuspended in the same volume of low-Pi medium followed by incubation for an additional 6 h. For incubation under high-Pi conditions for 30 h, sodium Pi buffer was added after 24 h to reach a final concentration of 2 mM. To analyze the effect of 3-aminotriazole (3-AT), yeast cells grown to mid-log phase (A600 = 0.5) were collected, resuspended in SD medium lacking histidine and supplemented with 100 mM 3-AT, and incubated at 30°C for 1 h before proceeding to RNA or chromatin isolation.
DNA manipulation.
Standard E. coli and yeast genetic methods and DNA manipulations were as described previously (24, 25). The primers used in this work are listed in Table S2 in the supplemental material. To construct the PHO4-tagged MFY376 strain, a fragment containing PHO4 tagged with His6 and Flag3 tags was amplified using primers Pho4-Flag-F and -R and the pUG6H3Flag plasmid (10) as the template, followed by transformation. Successful replacement and production of tagged Pho4 were confirmed by PCR and an immunoblot analysis with an anti-Flag antibody, respectively (data not shown). To construct the yeast strain with a deletion of ASF1, HDA1, RPD3, or SIN3, the adaptamer-mediated PCR method was employed to prepare the DNA fragments for transformation (22). The target fragments for each locus were amplified with the appropriate primer pairs (listed in Table S2 in the supplemental material) to place the forward tag sequence (GGAATTCCAGCTGACCACC) and the reverse tag sequence (GATCCCCGGGAATTGCCATG) at the 5′ and 3′ termini, respectively. Two partially overlapping fragments of the URA3 gene from Kluyveromyces lactis (KlURA3) were synthesized by PCR with primer set MN464 and MN345 and primer set MN344 and MN465 to place the forward tag and the reverse tag at the 5′ and 3′ termini, respectively. The target and the KlURA3 fragments with the corresponding tags were combined and annealed through the tag and were then subjected to PCR to prepare fusion fragments with the 5′ and 3′ portions of KlURA3, respectively. The two PCR fragments were combined and used to transform yeast. Successful disruption was confirmed by PCR. A reporter plasmid bearing the SNZ1 promoter and lacZ was constructed as follows. The promoter fragment (−970 to +30) was cloned by PCR using the MN449/MN450 primer pair to incorporate BamHI and BglII sites at the 5′ and 3′ termini, respectively, and was incorporated into the pMF811 plasmid (19). To mutagenize the putative Pho4-binding sequence in the SNZ1 promoter, a QuikChange II site-directed mutagenesis kit (Stratagene), the MN459/MN460 primer pair, and the SNZ1 promoter fragment were used. Successful mutagenesis was confirmed by DNA sequencing. To replace the chromosomal SNZ1 locus with the mutant SNZ1 lacking the putative Pho4-binding site, the DNA fragments for transformation were prepared by the adaptamer-mediated PCR method (22) as described above. The resulting Ura-positive transformants were transferred to medium containing 5-fluoroorotic acid to select Ura-negative colonies. Loss of the KlURA3 marker and successful replacement were confirmed by PCR and DNA sequencing, respectively.
Chromatin immunoprecipitation (ChIP) was carried out essentially as described previously (9). Briefly, yeast cells producing Flag-tagged Pho4 were cultivated in high- or low-Pi medium at 30°C to an A600 of 1.0 to 1.2, and the proteins were then cross-linked to DNA by adding formaldehyde. For time course experiments, cells were grown in YPAD medium at 30°C at an initial A600 of 0.05, and at 12, 18, 24, and 36 h, portions of the culture were removed and the proteins were cross-linked. After an incubation at 4°C for 12 h, the cells were disrupted and centrifuged to obtain crude extracts. An anti-Flag monoclonal antibody (Sigma) was added to the extracts to precipitate the cross-linked material, and then the cross-links were reversed by incubation at 65°C. The precipitated DNA was analyzed by PCR using gene-specific primers (MN1023/MN1025, MN1186/MN1187, MN1178/MN1179, and MN972/MN973 for SNZ1, SNZ2, SNO2, and PHO5, respectively).
Analytical methods.
RNA was isolated from yeast cells grown in YPAD medium for 12, 18, 24, and 36 h by use of a RiboPure yeast RNA extraction kit (Ambion). Northern analysis was carried out as described previously (18). The β-galactosidase assay was described previously (19). Chromatin was isolated from yeast cells grown in YPAD medium at 30°C for 24 to 27 h, as described previously (27), and was digested with micrococcal nuclease (MNase) (0.1 or 0.2 units/μl) in a 200-μl reaction mixture at 37°C for 10 min. The digested chromatin was then treated with RNase, digested with proteinase K, extracted with phenol-chloroform, and precipitated with ethanol. For high-resolution mapping by primer extension, a TCGACTTTCCGGACATTGTACTGTGGGT primer covering −495 to −461 (taking the A of ATG of the SNZ1 open reading frame [ORF] as +1) was end labeled with 32P. To analyze the chromatin structure of the fully activated SNZ1 promoter, 3-AT was added to the W303 culture at an A600 of 1.0, and the culture was incubated at 30°C for 1 h before the chromatin preparation. The chromatin and the purified DNA samples were digested with MNase and purified as described above. The digested DNA was then mixed with the labeled primer and subjected to a primer extension reaction as described previously (16). The products were separated on a 6% polyacrylamide-50% urea denaturing gel, and the gel was exposed to X-ray film. The autoradiogram was scanned with an Epson ES-10000G scanner, and the TIFF image thus generated was analyzed using MultiGauge software (Fujifilm, Japan).
RESULTS
Pho85 is required for SNZ/SNO gene expression.
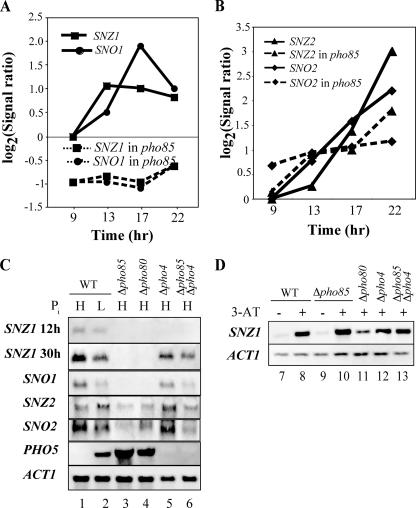
In the course of our previous microarray study (18), we noticed that SNZ1 and SNO1 were not properly induced during the late growth stage when Pho85 was absent (Fig. 1A). The expression of SNZ2 and SNO2, the other members of the SNZ/SNO family, was also affected by a Δpho85 mutation, with a lesser effect on SNZ2 (Fig. 1B). SNZ1 has a prospective Pho4-binding site at −380 (CACGTT), whereas SNZ2 and SNO2 lack the binding sequences in their promoters, but two are present in the SNO2 ORF at +271 and +492 (CTGCAC). In the absence of Pho85, Pho4 accumulates in the nucleus to activate the PHO genes regardless of the Pi condition (7, 8). Under this condition, if Pho4 binds to the SNZ1 promoter, then the observed defect in the induction of SNZ1/SNO1 in a Δpho85 mutant suggests repression by Pho4, which is generally considered to be a transcriptional activator. Therefore, we first used Northern analysis to examine whether the expression of the SNZ and SNO genes required Pho85. We detected SNZ1 expression in the wild-type (wt) cells grown for 30 h under high-Pi conditions (Fig. 1C, lane 1). When the Pi concentration in the medium was low, SNZ1 expression was reduced to some extent (lane 2), suggesting Pi-dependent expression of SNZ1. In the cells grown for 12 h, that is, to the mid-log phase, SNZ1 expression was barely detectable under both sets of Pi conditions (lanes 1 and 2). In the absence of Pho85, SNZ1 expression was diminished (lane 3), which coincided with the results of the GeneChip analysis (Fig. 1A). Similarly, in the absence of Pho80, a cyclin partner of Pho85 kinase for the phosphorylation of Pho4 in vivo, SNZ1 expression was also diminished (lane 4). The introduction of a Δpho4 mutation to the Δpho85 mutant restored the SNZ1 expression, but not to the wt level (lane 6), while a Δpho4 mutation alone did not affect SNZ1 expression significantly (lane 5). Similarly, SNO1 expression was diminished in the absence of either Pho85 or Pho80 (lanes 3 and 4) but was not affected significantly by the absence of Pho4 (lane 5). A Δpho4 Δpho85 double mutation, however, failed to restore SNO1 expression to the wt level (lane 6). SNZ2 and SNO2 also required Pho85 for their expression (Fig. 1C, lane 3) but were not as dependent on Pho80 as SNZ1 and SNO1 (lane 4). A loss of Pho4 did not appear to affect SNZ2/SNO2 expression (lane 5), and again the introduction of a Δpho4 mutation to the Δpho85 mutant could restore their expression to some extent but not to the wt level (lane 6). PHO5 is a typical PHO gene and is shown as a control that is activated by Pho4 under low-Pi conditions and inactivated by Pho85-Pho80 under high-Pi conditions, as demonstrated in Fig. 1C.
FIG. 1.
Requirement of Pho85 for SNZ/SNO gene expression. (A) Time course of SNZ1/SNO1 expression as determined by GeneChip analysis (based on data from reference 18). The extents of SNZ1 (squares) and SNO1 (circles) expression are shown as log2(signal ratio) values. Solid and broken lines designate expression in the wt and a Δpho85 mutant, respectively. (B) Time course of SNZ2/SNO2 expression by GeneChip analysis (18). SNZ2 (triangles) and SNO2 (diamonds) expression levels in the wt and a Δpho85 mutant are shown as described for panel A. (C) Northern analysis of SNZ1, SNO1, SNZ2, and SNO2 expression in the wt and various pho mutants and under high (H)- or low (L)-Pi conditions. Total RNA was extracted from the yeast cells (designated at the top of the panel) grown for 12 (SNZ1 only) or 30 h and was subjected to Northern analysis as described in Materials and Methods, and then the blot was hybridized with the respective digoxigenin-labeled probe. To analyze PHO5 expression, yeast cells were incubated in high- or low-Pi medium for 5 h before RNA isolation. ACT1 was detected with RNA prepared from a 12-h culture. (D) Effect of 3-AT on SNZ1 expression. Yeast strains, as designated at the top of the panel, were grown to mid-log phase, resuspended in SD medium lacking histidine and with (+) or without (−) 100 mM 3-AT, and then incubated for 1 h before RNA isolation.
These results revealed that Pho85 is required for SNZ1/SNO1 expression at a late growth stage (30 h) and suggested that Pho4 inhibits SNZ1/SNO1 expression in the absence of either member of the Pho85-Pho80 complex. Notably, the fact that SNZ1 expression failed to restore the wt level in a Δpho85 Δpho4 double mutant suggests that Pho85 may have a different function than the inactivation of Pho4 in the regulation of SNZ1. Pho85 is also required for the expression of the other SNZ/SNO genes tested, but their dependence on Pho80 or Pho4 is different from that of SNZ1.
SNZ1 expression is responsive to amino acid starvation (17). To determine whether Pho85 is involved in this process, we analyzed SNZ1 expression in the presence of 3-AT in the wt and various pho mutants (Fig. 1D). SNZ1 was induced regardless of the presence or absence of Pho85 when the cells were treated with 3-AT (Fig. 1D, lanes 8 and 10), and a Δpho4 mutation did not affect the induction (lanes 12 and 13). These results indicated that Pho85 is not required for SNZ1 expression in response to amino acid starvation and suggested that the Pho85 requirement is elicited specifically in the cells at the late growth stage. However, Pho85 does not appear to have a general effect on the genes whose expression is induced specifically in the late growth phase, since the loss of Pho85 does not cause reduced expression of SPG1 and NGR1, which are both induced at the stationary phase (12, 18).
Pho4 binds to the SNZ1 promoter to inhibit SNZ1 expression.
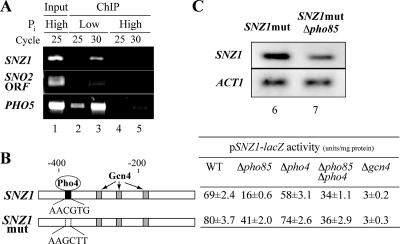
To study whether Pho4 is directly involved in the transcriptional regulation of SNZ1, we analyzed in vivo Pho4 binding to the SNZ1 promoter by ChIP from cells grown under either high- or low-Pi conditions. Pho4 is known to bind to the PHO5 promoter in a Pi-dependent fashion, as confirmed by our ChIP experiment (Fig. 2A, lanes 2 to 5). The SNZ1 promoter fragment was also enriched in the chromatin fragments prepared from cells grown under low-Pi conditions (Fig. 2A, lane 3), indicating that Pho4 binds to the SNZ1 promoter in vivo in a Pi-dependent manner. We also analyzed the in vivo binding of Pho4 to the SNZ2/SNO2 region. The SNZ2 promoter fragment (the intergenic region of SNZ2/SNO2) was not enriched (data not shown), whereas Pho4 bound weakly to the SNO2 ORF under low-Pi conditions (Fig. 2A, lane 3).
FIG. 2.
Pho4 binding to the SNZ1 promoter and involvement of the Pho4-binding site in transcriptional regulation of SNZ1. (A) In vivo Pho4 binding to the SNZ1 promoter and the SNO2 ORF demonstrated by ChIP followed by gene-specific PCR. Total DNA in the extract (Input) and the immunoprecipitated chromatin fragment prepared from the wt (MFY376) cells grown under low- or high-Pi conditions were subjected to PCR with either the SNZ1 or PHO5 promoter-specific or SNO2 ORF-specific primers, and the DNA was amplified by 25 or 30 cycles of the reaction. (B) A schematic representation of SNZ1 and its mutant (SNZ1mut) promoters, showing the Pho4-binding site (black box) with the wt (AACGTG) or mutant (AAGCTT) sequences and the three Gcn4-binding sites (shaded boxes). The activities of the wt and SNZ1mut promoters, represented by β-galactosidase activity data, are shown on the right side. The wt and various mutant strains harboring the reporter plasmid were grown in high-Pi medium for 30 h prior to the assay of the reporter activity. The values represent averages and standard errors of the results of three independent assays. (C) Northern analysis of chromosomal SNZ1 expression in the SNZ1mut and SNZ1mut Δpho85 double mutant. Total RNA was isolated from cells grown for 30 h in YPAD medium and was subjected to Northern analysis as described in Materials and Methods. ACT1 is a loading control.
Next we examined whether the prospective Pho4-binding site in the SNZ1 promoter was functioning in the transcriptional regulation of SNZ1 by two methods, a reporter assay and Northern analysis. After growth for 30 h, the wt promoter fused to lacZ showed a reduced level of activity in a Δpho85 mutant (about 23% of the wt level); activity was restored to about 50% of the wt level by introducing a Δpho4 mutation (Fig. 2B). A Δpho4 mutation alone did not affect significantly the activity level (Fig. 2B). A deletion of Gcn4 almost abolished the promoter activity, indicating that Gcn4 is required for SNZ1 activation. When the sequence of the prospective Pho4-binding site (AACGTG) was altered to AAGCTT (SNZ1mut), the promoter activity in a Δpho85 mutant was reduced to about 50% of the wt level, and this was barely affected by the introduction of a Δpho4 mutation to the mutant (Fig. 2B). This activity level was almost equal to that of the wt promoter observed in a Δpho85 Δpho4 double mutant, suggesting that this binding site is required for Pho4 to exert its inhibitory function. This notion was further strengthened by Northern analysis of the expression of the chromosomal SNZ1mut in the wt (MFY365) cells or in Δpho85 mutant (MFY366) cells in which the wt SNZ1 promoter sequence had been replaced by the mutant promoter sequence lacking the Pho4-binding site (Fig. 2C). SNZ1mut was expressed in the absence of Pho85, but not fully (Fig. 2C, lane 7), indicating that Pho4 binding is required for SNZ1 repression in the absence of Pho85. Taken together, these results suggested that Pho4, when activated in the absence of Pho85 and bound to the SNZ1 promoter, functions to inhibit SNZ1 expression, while Gcn4 plays a major role in the activation of SNZ1. Since the mRNA level and the promoter activity of SNZ1 were not restored to the wt level in the double mutant or with the SNZ1mut promoter, Pho85 seems to exert its effect through an additional, unknown mechanism to regulate SNZ1 expression.
Pho4 functions to delay the timing of SNZ1 expression.
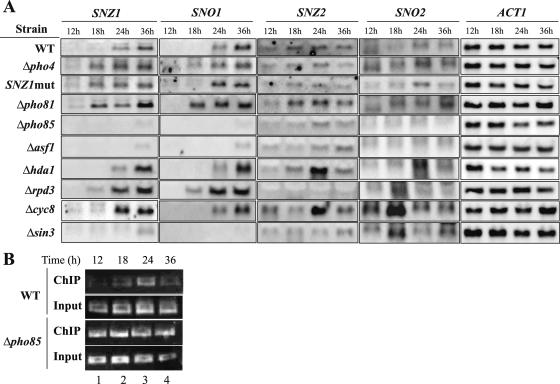
We next assessed the biological relevance of the inhibition of SNZ1 by Pho4. One idea is that Pho4 may prevent the untimely expression of SNZ1, since SNZ1 is expressed specifically in the late growth (postdiauxic) stage to stationary phase (21). If this hypothesis is correct, then in the absence of Pho4, SNZ1 could be transcribed in an earlier growth stage. To test this idea, we analyzed periodical SNZ1 expression from 12 h to 36 h in the wt, Δpho4, SNZ1mut, Δpho81, and Δpho85 strains (Fig. 3A, five rows from the top). In the wt cells, SNZ1 expression became apparent at 24 h and was maximal at 36 h (Fig. 3A, left panel). On the other hand, when Pho4 was absent, SNZ1 expression was detected at 18 h and increased along with the incubation period (24 and 36 h). A similar SNZ1 expression profile was observed for the SNZ1mut strain. In a Δpho81 mutant in which the low-Pi signal was not transmitted to inactivate Pho85, and in which Pho4 was therefore inactive, advanced expression of SNZ1 was also observed. In the absence of Pho85, SNZ1 expression was not detected throughout the incubation period. Taken together, these results suggested that SNZ1 expression at the late growth stage is regulated by Pho85-Pho4, depending on the Pi conditions. SNO1 expression is coregulated with SNZ1 (21), as observed with the wt panel (Fig. 3A). The expression of SNO1 was affected by these mutations in a manner similar to that seen with SNZ1, except that the absence of Pho4 slightly advanced the timing whereas that of the Pho4 binding site (SNZ1mut) did not (Fig. 3A, second panel from the left). Although it is a member of the SNZ/SNO family, SNZ2 is expressed at the prediauxic stage (21). Its expression in the wt became detectable at 12 h, peaked at 18 and 24 h, and decreased at 36 h. We observed a similar profile in the mutant strains, except for Δpho85, in which SNZ2 expression was reduced (Fig. 3A, middle panel). The loss of Pho4 appears to have little effect on SNZ2 expression in the time course experiment compared to the data shown in Fig. 1C. These results indicate that Pho4 does not play a major role in the regulation of the timing of SNZ2 expression. SNO2 is considered to be coregulated with SNZ2 (21), but our results showed that the timing of expression of the two genes was different, in that SNO2 peaked at 24 h with a barely detectable level at 18 h in the wt and SNZ1mut (Fig. 3A, second panel from the right). The absence of Pho4 appeared to increase the SNO2 expression level, whereas that of Pho81 shifted the peak to 36 h. The absence of Pho85 abolished SNO2 expression. These results revealed that SNZ2 and SNO2 are not coregulated and that Pho4 appears to inhibit SNO2 expression but that expression of SNZ2 is not inhibited to an appreciable extent (Fig. 1C and 3A). A control experiment with the ACT1 probe revealed no significant differences in the amounts of loaded RNA (Fig. 3A, right panel). Thus, the Pi-dependent Pho4 binding appears to delay both SNZ1 and SNO1 expression until the appropriate growth stage, probably with less effect on the latter, and to inhibit SNO2 expression.
FIG. 3.
(A) Northern analysis of the time course of SNZ1, SNO1, SNZ2, and SNO2 gene expression in the wt and in various mutants affecting the PHO system or chromatin structure. Total RNA was isolated from cells grown in YPAD medium at 12, 18, 24, and 36 h and was subjected to Northern analysis as described in Materials and Methods. ACT1 is shown as a loading control. (B) Temporal Pho4 binding to the SNZ1 promoter in the wt (MFY376) and Δpho85 (MFY377) strains as demonstrated by ChIP and PCR. Total DNA in the extract (Input) and the immunoprecipitated chromatin fragments prepared from the wt and mutant cells at the designated times were subjected to PCR with the SNZ1 promoter-specific primer and were amplified by 30 cycles of the reaction.
To examine whether Pho4 binding to the SNZ1 promoter is temporary or constitutive, we carried out a ChIP experiment with cell extracts prepared from cells grown for 12, 18, 24, and 36 h in YPAD medium. In the wt cells, an enrichment of the Pho4-bound fragment was observed at 24 h, and weak enrichment was observed at 18 and 36 h (Fig. 3B, lanes 2 to 4), indicating that Pho4 binding occurs at the late growth stage. In Δpho85 cells in which Pho4 is localized in the nucleus constitutively, Pho4 bound to the promoter throughout the growth stages (Fig. 3B).
Chromatin structure affects SNZ1 expression and its timing.
How can Pho4 exert an inhibitory function by binding to the SNZ1 promoter? When Pho4 activates PHO5, Pho4 binding triggers alterations in the nucleosome positioning at the PHO5 promoter and the Asf1 histone chaperone removes histones from the promoter region, thereby opening the TATA box (1, 32). We considered the possibility that similar chromatin alterations caused by Pho4 binding may lead to transcriptional repression of SNZ1. To examine this possibility, we first analyzed the effects of mutations that affect chromatin structure (Δasf1, Δhda1, Δrpd3, and Δsin3) and general repression (Δcyc8) on the timing of SNZ/SNO gene expression. A proteome analysis revealed a physical interaction between Pho4 and Sin3 forming the HDAC complex with Rpd3 (6). Repression by the Cyc8-Tup1 corepressor reportedly involves histone deacetylation (30), and the corepressor can interact with Rpd3 in vitro (33). Asf1 counteracts silencing by chromatin, and its absence diminished SNZ1, SNO1, and SNO2 expression and significantly reduced SNZ2 expression (Fig. 3A), indicating that Asf1 activity is required for SNZ/SNO expression. HDAC usually functions to repress transcription by stabilizing chromatin. The loss of Rpd3 HDAC advanced the timing of SNZ1/SNO1 expression, whereas that of Hda1, another yeast HDAC, did not appear to affect the timing but increased the expression levels of the two genes at 36 h (Fig. 3A). This observation suggests the possibility that a specific HDAC activity is required to regulate the timing of SNZ1/SNO1 expression. The absence of Cyc8 appeared to increase SNZ1 expression at 24 h and to advance the timing slightly (Fig. 3A) but did not appear to affect SNO1 expression (Fig. 3A). These results suggested the possibility that Cyc8 may also be involved in SNZ1 repression at the late growth stage. Although Sin3 forms a complex with Rpd3, the effect of a Δsin3 mutation was completely different from that of Δrpd3, i.e., a decrease in SNZ1/SNO1 expression, as seen with Δasf1 (Fig. 3A). These mutations showed different effects on SNZ2/SNO2. The absence of Hda1 or Cyc8 made SNZ2 expression peak at 24 h, whereas that of Rpd3 or Sin3 caused a decrease in SNZ2 expression (Fig. 3A). With respect to SNO2, the absence of Hda1 increased its expression at 24 h whereas that of Rpd3, Cyc8, or Sin3 advanced its peak to 18 h (Fig. 3A). These results indicated that the chromatin structure is involved in the regulation of the SNZ/SNO genes by altering the timing and probably the expression levels of these genes, with different effects on the individual members.
Pho4 alters chromatin structure in the SNZ1 promoter region.
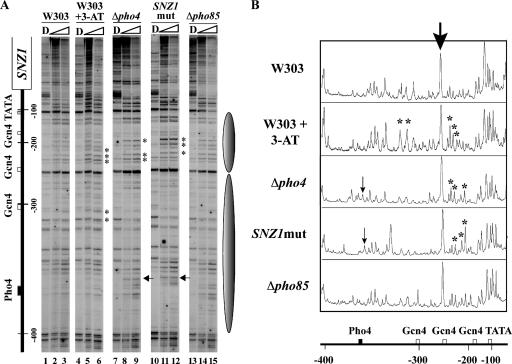
We next analyzed whether Pho4 binding affects the chromatin structure at the SNZ1 promoter region by limited digestion of isolated nuclei with MNase followed by high-resolution mapping of MNase cleavage sites by primer extension analysis with a primer covering −495 to −461 (taking the A of ATG of the SNZ1 ORF as +1) (Fig. 4). In naked DNA as well as in the chromatin samples in the tested strains, MNase cleaved strongly at around −400, −255, and −105 in the SNZ1 promoter (Fig. 4A), and the spaces between each pair of bands were about 145 bp in length, which could accommodate a single nucleosome. Thus, we supposed that nucleosomes might be positioned in these promoter regions (Fig. 4A). Since the cells from which the chromatin was isolated were grown for 24 to 27 h in YPAD medium, the SNZ1 promoter in the wt should have been partially activated (Fig. 3A). When the SNZ1 promoter was fully activated by the addition of 3-AT, the two regions encompassing the upstream Gcn4-binding site (at −305) and that sandwiched between the other two Gcn4-binding sites (at −255 and −180) became more susceptible to MNase digestion in the chromatin samples (Fig. 4A, lanes 5 and 6) compared to those in the wt (lanes 2 and 3) and Δpho85 (lanes 14 and 15) strains. Similarly, in the chromatin samples prepared from Δpho4 and SNZ1mut strains in which Pho4 was absent from the cell and Pho4 failed to bind to the promoter because of the loss of the binding site, respectively, the region between the two downstream Gcn4-binding sites (at −255 and −180) showed increased accessibility (Fig. 4A, lanes 8, 9, 11, and 12). These observations were confirmed by densitometric analysis of the gel image (Fig. 4B). Thus, in the absence of Pho4 binding, the chromatin structure of these regions became unstable, allowing proteins more access to the SNZ1 promoter.
FIG. 4.
A high-resolution analysis of chromatin structure in the SNZ1 promoter region. (A) Chromatin DNA was isolated from the strains designated at the top of the panel, digested with MNase (0.1 or 0.2 units/μl) as described in Materials and Methods, and subjected to a primer extension reaction using a 32P-labeled primer (−495 to −461). A schematic representation of the SNZ1 promoter region, showing the TATA box and the Pho4- and Gcn4-binding sites, is on the left side of the panel. Numerals designate nucleotide distance, taking the A of ATG of the SNZ1 ORF as +1. D, purified DNA digested with MNase. The arrows indicate enhanced MNase cleavage signals near the Pho4-binding site observed in the chromatin samples from Δpho4 and SNZ1mut strains. Asterisks are for those observed between the two downstream Gcn4-binding sites (at −255 and −180) and near the upstream Gcn4 site at −305 compared to those in the wt and Δpho85 strains. Possible nucleosome positions are designated by gray ellipses on the right-hand side of the panel. (B) A densitometric analysis of the gel images. The lanes containing the chromatin samples digested with 0.1 U/μl MNase were analyzed (lanes 2, 5, 8, 11, and 14 in Fig. 4A). The intensity of the bands was normalized with reference to the strongly cleaved band at −255 (bold arrow). The asterisks and arrows designate the peaks of the corresponding bands in panel A. A schematic representation of the SNZ1 promoter is shown at the bottom.
In the absence of either Pho4 (Δpho4) or the Pho4-binding site (SNZ1mut), MNase cleaved a site adjacent to the Pho4-binding site (Fig. 4A and 4B), which was not observed in the wt or Δpho85 strain (Fig. 4A and 4B), indicating that the accessibility of MNase to this site is Pho4 dependent. This result revealed in vivo binding of Pho4 to the SNZ1 promoter, which is in good agreement with ChIP analysis data for Pho4 binding (Fig. 2A).
DISCUSSION
SNZ1 and SNO1 are expressed specifically in the postdiauxic to stationary phases (21). In this work, we have shown that Pho4 functions to repress or down-regulate the transcription of SNZ1 so that SNZ1 is appropriately expressed in this late growth stage. Three lines of evidence support this conclusion. (i) Northern analysis and a reporter assay demonstrated that Pho4 inhibits SNZ1 expression in the absence of Pho85, which allows Pho4 to become active (Fig. 1 and 2). (ii) The ChIP analysis revealed that Pho4 binds to the SNZ1 promoter in vivo in a Pi-dependent and transient fashion, and this binding is required for repression/downregulation of SNZ1 (Fig. 2). (iii) The absence of Pho4 in the cell, Pho4 binding to the SNZ1 promoter lacking the Pho4 binding site (SNZ1mut), or Pho81, which transmits low-Pi signal, advances the timing of SNZ1 expression (Fig. 3A). SNO1 is coregulated with SNZ1 (3, 21), and its timing appears to be regulated similarly by Pho4 but in a weaker manner (Fig. 3A). We can imagine that a destabilized nucleosome in the absence of Pho4 could affect the bidirectional transcription of SNZ1 and SNO1 to different extents. SNZ2 and SNO2, other members of the SNZ/SNO family, are expressed at an earlier growth stage than SNZ1/SNO1, that is, at the diauxic to postdiauxic stage (21). Pho4 does not appear to affect the timing of SNZ2 expression, although it may affect the expression level to some extent (Fig. 3A). On the other hand, Pho4 binding within the SNO2 ORF may play an inhibitory role in SNO2 transcription and thus regulate its timing of expression (Fig. 2A and Fig. 3A). These observations showed that Pho4 binding is likely to contribute to the different timings of expression of the genes of the SNZ/SNO family, although we are still unable to exclude the possibility that the effect of Pho4 is indirect.
Pho85 is required for the expression of all of the SNZ/SNO genes tested (Fig. 1 and 3). The observations that the mRNA level and the promoter activity of SNZ1 are not restored to the wt level in a Δpho4 Δpho85 double mutant and with the SNZ1mut promoter suggest that Pho85 is required to counteract an as-yet-unknown repressor other than Pho4 or to activate an unknown activator. This notion is more significant for SNO1, SNZ2, and SNO2, since expression of those genes was barely recovered in the double mutant, suggesting that Pho85 regulates those genes mainly through a different pathway than via Pho4. SNZ1 and SNO1 are required for yeast growth when the intracellular vitamin B6 level is low, suggesting an involvement of these genes in the biosynthesis of the vitamin (23). SNZ2 does not appear to be required for the vitamin B6 pathway (23). On the other hand, SNZ2 and SNO2 are induced by thiamine depletion whereas SNZ1 and SNO1 are not (23). Although physical interactions among the Snz and Sno proteins have been reported (23), at present, we cannot provide a clear explanation of why the timing of the expression of these SNZ/SNO genes is differently regulated and how the difference is related to apparently distinct functions of the members of the SNZ/SNO gene family.
Since the binding of Pho4 to the SNZ1 promoter is dependent on the Pi condition (Fig. 2A), although not as strongly as that of PHO5, the repression of SNZ1 by Pho4 should be regulated by the Pi condition. In the late growth stage, the amount of Pi in the medium is usually limited, resulting in derepression of some PHO genes; specifically, PHO84 and PHO5, which show a quick response to Pi limitation (31), are induced at 22 h of incubation, whereas the other PHO genes are still repressed (5, 18). Since the amount of nuclear Pho4 increases as the environmental Pi concentration decreases, this different response probably reflects the different affinities of binding of Pho4 to the specific promoters. Pho4 appears to bind transiently to the SNZ1 promoter, peaking at 24 h (Fig. 3B), and we assume that this binding responds to the Pi limitation to prevent the inappropriate or untimely expression of SNZ1. Advanced SNZ1 expression in a Δpho81 mutant, in which the low-Pi signal is not transmitted to inactivate Pho85-Pho80, supports the idea of SNZ1 repression by Pi-responsive Pho4 binding (Fig. 3A). Upon entering the postdiauxic to stationary phase, an activator of SNZ1, possibly Gcn4, is induced sufficiently to overcome the repression by Pho4. Gcn4 is likely to play a major role in the activation process, since, in the absence of Gcn4, SNZ1 promoter activity at 30 h was drastically decreased (Fig. 2B). We can imagine the possibility that Gcn4 may destabilize Pho4 binding, possibly by nucleosome replacement. In the absence of Pho85, too much Pho4 accumulates in the nucleus for the activator to relieve repression. Although the Pho85-Pcl5 Cdk-cyclin complex phosphorylates Gcn4 to trigger its degradation (2, 13), PCL5 expression is drastically reduced in the postdiauxic stage (19), making the complex much less active, and therefore Gcn4 becomes much more stable, thus leading to the activation of the transcription factor at this growth stage. When fully activated by amino acid starvation (treatment with 3-AT), Gcn4 can overcome the repression of SNZ1 caused by the presence of excess Pho4 in a Δpho85 mutant (Fig. 1D).
Our conclusion implies that Pho4 functions as both an activator and a repressor. Rap1 and Abf1 are also known to possess the two opposite functions. Rap1 activates genes encoding glycolytic enzymes and ribosomal proteins and maintains the silent chromatin structure at telomeres and the silent mating type locus (15, 28). Repression by Abf1 also involves the silent chromatin structure (14, 36). In the case of Pho4, activation (derepression) of PHO5 requires alterations in nucleosome positioning at its promoter, which involves Pho4 binding and its activation domain (32). The Asf1 histone chaperone displaces histones for PHO5 activation by Pho4 (1, 32). Our high-resolution mapping results indicated that a loss of Pho4 binding increases the accessibility of MNase to the SNZ1 promoter region (Fig. 4). Since the Pho4-binding site at −380 may exist at the edge of a nucleosome (Fig. 4A), we suppose that Pho4 can stabilize an array of nucleosomes positioned in the SNZ1 promoter, leading to the repression of SNZ1. In the absence of Pho4 binding, the nucleosome array becomes unstable, making Gcn4 more easily accessible to its binding sites, as represented by the increased MNase cleavage bands around −255 and −180. This scenario can account for the advanced activation of SNZ1 in the Δpho4 and SNZ1mut strains (Fig. 3A). The loss of Asf1 abolished SNZ1/SNO1 expression (Fig. 3A), indicating that Asf1 activity is required in the activation process of SNZ1/SNO1, probably by removing histones from the promoter region. Since Asf1 activity is also required for SNZ2 expression, its participation does not appear to be specific to Pho4-involved regulation but is instead rather general.
The idea of involvement of chromatin structure in the transcriptional regulation of SNZ1 is also supported by a microarray analysis demonstrating that histone H4 depletion activates SNZ1/SNO1 (35). A proteome analysis revealed a physical interaction between Pho4 and Hht1 comprising histone H3 (11), and with Sin3 forming the HDAC complex with Rpd3 (6), suggesting the direct involvement of Pho4 in transcriptional repression through interactions with the nucleosome. In fact, we have shown that the absence of Rpd3, but not Hda1, advances the timing of SNZ1/SNO1 expression (Fig. 3A), suggesting the possibility that the Rpd3-Sin3 HDAC complex is recruited to the SNZ1 promoter through an interaction with Pho4. The absence of Sin3, however, abolished SNZ1/SNO1 expression, which argues against this model. Since Sin3 functions as a scaffold protein interacting with a variety of factors to form heterogeneous HDAC complexes regulating global gene expression (29), we suppose that the loss of Sin3 causes a global effect that leads to rather indirect repression of SNZ1/SNO1. SNZ2 expression is altered by the deletion of ASF1, RPD3, HDA1, CYC8, or SIN3, suggesting that the chromatin structure is also involved in SNZ2 regulation. However, their effects on the expression level or timing are different from the results seen with SNZ1. This implies that a complex interplay of factors, including Pho4 and those that remodel chromatin structure, may be responsible for the different timing of expression among the SNZ/SNO gene family members.
The frequent appearance of the Pho4-binding sequence (CACGTG/T and CTGCAC) in the intergenic regions of the yeast genome (ca. 2,800) suggests that Pho4 may be involved more generally in either positive or negative transcriptional regulation than is currently considered to be the case. A Pho4-binding analysis conducted on a genome-wide scale by ChIP-on-chip analysis, combined with a global analysis of the Pho4 effect on gene expression, should yield more insights into the broad functions of this transcription factor.
Supplementary Material
Acknowledgments
We are grateful to Y. Katou and K. Shirahige for help in the construction of yeast strains and ChIP experiments.
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology (M.N., M.S., and A.T.).
Footnotes
Published ahead of print on 11 April 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14657-666. [DOI] [PubMed] [Google Scholar]
- 2.Bömeke, K., R. Pries, V. Korte, E. Scholz, B. Herzog, F. Schulze, and G. H. Braus. 2006. Yeast Gcn4p stabilization is initiated by the dissociation of the nuclear Pho85p/Pcl5p complex. Mol. Biol. Cell 172952-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, E. L., E. K. Fuge, P. A. Padilla, and M. Werner-Washburne. 1996. A stationary-phase gene in Saccharomyces cerevisiae is a member of a novel, highly conserved gene family. J. Bacteriol. 1786865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll, A. S., and E. K. O'Shea. 2002. Pho85 and signaling environmental conditions. Trends Biochem. Sci. 2787-93. [DOI] [PubMed] [Google Scholar]
- 5.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278680-686. [DOI] [PubMed] [Google Scholar]
- 6.Graumann, J., L. A. Dunipace, J. H. Seol, W. H. McDonald, J. R. Yates III, B. J. Wold, and R. J. Deshaies. 2004. Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol. Cell. Proteomics 3226-237. [DOI] [PubMed] [Google Scholar]
- 7.Kaffman, A., N. M. Rank, E. M. O'Neill, L. S. Huang, and E. K. O'Shea. 1998. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396482-486. [DOI] [PubMed] [Google Scholar]
- 8.Kaffman, A., N. M. Rank, and E. K. O'Shea. 1998. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 122673-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katou, Y., K. Kaneshiro, H. Aburatani, and K. Shirahige. 2006. Genomic approach for the understanding of dynamic aspect of chromosome behavior. Methods Enzymol. 409389-410. [DOI] [PubMed] [Google Scholar]
- 10.Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 4241078-1083. [DOI] [PubMed] [Google Scholar]
- 11.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440637-643. [DOI] [PubMed] [Google Scholar]
- 12.Martinez, M. J., S. Roy, A. B. Archuletta, P. D. Wentzell, S. S. Anna-Arriola, A. L. Rodriguez, A. D. Aragon, G. A. Quinones, C. Allen, and M. Werner-Washburne. 2004. Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: gene expression and identification of novel essential genes. Mol. Biol. Cell 155295-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meimoun, A., T. Holtzman, Z. Weissman, H. J. McBride, D. J. Stillman, G. R. Fink, and D. Kornitzer. 2000. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol. Biol. Cell 11915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyake, T., J. Reese, C. M. Loch, D. T. Auble, and R. Li. 2004. Genome-wide analysis of ARS (autonomously replicating sequence) binding factor 1 (Abf1p)-mediated transcriptional regulation in Saccharomyces cerevisiae. J. Biol. Chem. 27934865-34872. [DOI] [PubMed] [Google Scholar]
- 15.Moretti, P., and D. Shore. 2001. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol. 218082-8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morohashi, N., Y. Yamamoto, S. Kuwana, W. Morita, H. Shindo, A. P. Mitchell, and M. Shimizu. 2006. Effect of sequence-directed nucleosome disruption on cell-type specific repression by α2/Mcm1 in the yeast genome. Eukaryot. Cell 51925-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 214347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishizawa, M., Y. Katou, K. Shirahige, and A. Toh-e. 2004. Yeast Pho85 kinase is required for proper gene expression during the diauxic shift. Yeast 21903-918. [DOI] [PubMed] [Google Scholar]
- 19.Nishizawa, M., M. Tanabe, N. Yabuki, K. Kitada, and A. Toh-e. 2001. Pho85 kinase, a yeast cyclin-dependent kinase, regulates the expression of UGP1 encoding UDP-glucose pyrophosphorylase. Yeast 18239-249. [DOI] [PubMed] [Google Scholar]
- 20.Oshima, Y. 1997. The phosphate system in Saccharomyces cerevisiae. Genes Genet. Syst. 72323-334. [DOI] [PubMed] [Google Scholar]
- 21.Padilla, P. A., E. K. Fuge, M. E. Crawford, A. Errett, and M. Werner-Washburne. 1998. The highly conserved, coregulated SNO and SNZ gene families in Saccharomyces cerevisiae respond to nutrient limitation. J. Bacteriol. 1805718-5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid, R. J. D., M. Lisby, and R. Rothstein. 2002. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol. 350258-277. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez-Navarro, S., B. Llorente, M. T. Rodríguez-Manzaneque, A. Ramne, G. Uber, D. Marchesan, B. Dujon, E. Herrero, P. Sunnerhagen, and J. E. Perez-Ortin. 2002. Functional analysis of yeast gene families involved in metabolism of vitamins B1 and B6. Yeast 191261-1276. [DOI] [PubMed] [Google Scholar]
- 24.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. A laboratory course manual. Cold Harbor Spring Laboratory Press, Cold Spring Harbor, NY.
- 25.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Schultz, J., and M. Carlson. 1987. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol. Cell. Biol. 73637-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu, M., S. Y. Roth, C. Szent-Gyorgyi, and R. T. Simpson. 1991. Nucleosomes are positioned with base pair precision adjacent to the α2 operator in Saccharomyces cerevisiae. EMBO J. 103033-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shore, D. 1994. RAP1: a protean regulator in yeast. Trends Genet. 10408-412. [DOI] [PubMed] [Google Scholar]
- 29.Silverstein, R. A., and K. Ekwall. 2005. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 471-17. [DOI] [PubMed] [Google Scholar]
- 30.Smith, R. L., and A. D. Johnson. 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25325-330. [DOI] [PubMed] [Google Scholar]
- 31.Springer, M., D. D. Wykoff, N. Miller, and E. K. O'Shea. 2003. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 1E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svaren, J., and W. Horz. 1997. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 2293-97. [DOI] [PubMed] [Google Scholar]
- 33.Waterborg, J. H. 2000. Steady-state levels of histone acetylation in Saccharomyces cerevisiae. J. Biol. Chem. 27513007-13011. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, W. A., and P. J. Roach. 2002. Nutrient-regulated protein kinases in budding yeast. Cell 111155-158. [DOI] [PubMed] [Google Scholar]
- 35.Wyrick, J. J., F. C. Holstege, E. G. Jennings, H. C. Causton, D. Shore, M. Grunstein, E. S. Lander, and R. A. Young. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402418-421. [DOI] [PubMed] [Google Scholar]
- 36.Zou, Y., Q. Yu, and X. Bi. 2006. Asymmetric positioning of nucleosomes and directional establishment of transcriptionally silent chromatin by Saccharomyces cerevisiae silencers. Mol. Cell. Biol. 267806-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.