Abstract
Candida albicans is a dimorphic fungus that can interconvert between yeast and filamentous forms. Its ability to regulate morphogenesis is strongly correlated with virulence. Tup1, a transcriptional repressor, and the signaling molecule farnesol are both capable of negatively regulating the yeast to filamentous conversion. Based on this overlap in function, we tested the hypothesis that the cellular response to farnesol involves, in part, the activation of Tup1. Tup1 functions with the DNA binding proteins Nrg1 and Rfg1 as a transcription regulator to repress the expression of hypha-specific genes. The tup1/tup1 and nrg1/nrg1 mutants, but not the rfg1/rfg1 mutant, failed to respond to farnesol. Treatment of C. albicans cells with farnesol caused a small but consistent increase in both TUP1 mRNA and protein levels. Importantly, this increase corresponds with the commitment point, beyond which added farnesol no longer blocks germ tube formation, and it correlates with a strong decrease in the expression of two Tup1-regulated hypha-specific genes, HWP1 and RBT1. Tup1 probably plays a direct role in the response to farnesol because farnesol suppresses the haploinsufficient phenotype of a TUP1/tup1 heterozygote. Farnesol did not affect EFG1 (a transcription regulator of filament development), NRG1, or RFG1 mRNA levels, demonstrating specific gene regulation in response to farnesol. Furthermore, the tup1/tup1 and nrg1/nrg1 mutants produced 17- and 19-fold more farnesol, respectively, than the parental strain. These levels of excess farnesol are sufficient to block filamentation in a wild-type strain. Our data are consistent with the role of Tup1 as a crucial component of the response to farnesol in C. albicans.
Candida albicans is the opportunistic fungal pathogen most commonly isolated in humans. C. albicans is part of the normal flora, and it resides in the gastrointestinal and genitourinary tracts, as well as on the skin. However, C. albicans is capable of causing a wide range of diseases, from mild mucosal infections to life-threatening systemic infections termed candidemia (19). Vulnerable patients include those with AIDS and patients undergoing chemotherapy and organ transplantation (19). The annual cost of treating candidiasis in the United States was estimated to be 1 billion dollars, and the mortality rates for patients with candidiasis are 30 to 50%, even with antifungal treatment (28), indicating a need for new antifungal drugs.
The ability of C. albicans to cause disease has been strongly linked to its conversion between two distinct morphological forms, yeast and filamentous. Recently, our research has focused on farnesol, the first quorum-sensing molecule discovered in a eukaryote (17). Farnesol is a virulence factor (35) that is excreted continuously by C. albicans (17), and when it accumulates beyond a threshold level, it blocks the yeast to filament conversion (17). Stationary-phase cultures of C. albicans have accumulated 2 to 4 μM farnesol (17), and the 50% inhibitory concentration value for blocking germ tube formation (GTF) in an N-acetylglucosamine-stimulated assay is ca. 1 to 2 μM (E,E-)farnesol (17, 31, 39), and consequently, these farnesol production levels are physiologically relevant. Other roles described for farnesol include biofilm inhibition (36), protection from oxidative stress (46), and induction of apoptosis in another fungus, Aspergillus nidulans (38). While many phenotypic effects produced by farnesol have been described, little is understood about farnesol's mode of action.
In addition to farnesol, C. albicans yeast and filamentous growth are controlled by an assortment of signaling pathways (4, 13). The yeast-to-filamentous form conversion is activated by many pathways, including components of the CEK1 mitogen-activated protein (MAP) kinase pathway, the Ras/cyclic AMP-dependent pathway, the calcium/calmodulin signaling pathway, the Rim101-independent pathway, and the Chk1 two-component signal transduction pathway. Although each pathway has been implicated in filamentation (10, 26, 37), these pathways show some degree of specialization in that they respond to different environmental inducers. The activation and inhibition of filament development are accomplished largely through changes in gene expression mediated by transcription activators and repressors. Efg1 is a major transcription regulator of filamentous growth and is a central control point for many signaling pathways involved in filamentation (14). Efg1 also regulates the expression of multiple genes, including those involved in virulence (6, 29). Mechanisms have also been identified that block filament development, with Tup1 playing a key role in transcriptional repression (5, 6).
Farnesol is able to block filamentous growth induced by environmental signals for most, and possibly for all, of the signaling pathways activating filament development. These signals include 10% serum, 10 mM l-proline, 2.5 mM N-acetylglucosamine, or the combination of N-acetylglucosamine and l-proline, all at 37°C (17). Thus, farnesol may individually block each of the morphogenic signaling pathways and/or act at a common control point in morphogenesis. Tup1 repression of filament-specific genes is an attractive candidate for a common control point that may be regulated by farnesol (21).
The C. albicans Tup1 protein is a transcription regulator that plays two key roles in the cell: (i) regulation of phase switching, and (ii) inhibition of filamentous growth. Tup1 interacts with the corepressor protein Ssn6 or Tcc1. These complexes function with DNA binding proteins to repress gene expression (23, 42). At least three DNA-binding proteins have been identified that function with Tup1: Nrg1 (homologous to the Saccharomyces cerevisiae Nrg1p protein), Rfg1 (homologous to the S. cerevisiae Rox1p protein), and Mig1 (homologous to the S. cerevisiae Mig1p protein). Homozygous tup1 mutants are unable to grow as yeast and instead remain locked in the filamentous form, in all media tested (5). Deletion of TUP1 results in the up-regulation of approximately one-third of C. albicans genes (32, 33), and these mutants are also avirulent in a murine model of infection. The activation of Tup1 transcription repressor complexes results in the repression of filament-specific gene expression (5, 6, 32, 33).
Here, we tested the hypothesis that the C. albicans response to farnesol involves Tup1. The morphological response to farnesol was tested with wild-type and tup1/tup1, tup1/TUP1, nrg1/nrg1, and rfg1/rfg1 strains to assess the requirement for these genes in the farnesol response. The morphological response and gene expression pattern for MIG1 were not determined because the Mig1 protein does not play a role in the filamentous growth of C. albicans (32). The gene expression patterns of TUP1, NRG1, RFG1, and EFG1, as well as genes under their control, were examined in the presence or absence of farnesol by quantitative Northern and Western analyses. Finally, we compared farnesol production levels in tup1, nrg1, and rfg1 homozygous mutants relative to that in wild-type cells.
MATERIALS AND METHODS
Strains and media.
Candida albicans SC5314 is an independent clinical isolate and the reference strain for the Candida genome sequence (1). C. albicans strains CAF-2 (ura3::imm434/URA3) and CAI-4 (ura3::imm434/ura3::imm434) are derived from SC5314 by gene replacement (16). Strains BCa2-9 (tup1/tup1 in CAI-4 [5]), BCa2-10 (tup1/tup1, frameshift disruption fragment in CAI-4 [5]), DU152 (nrg1/nrg1 in CAI-4 [5]), DU129 (rfg1/rfg1 in CAI-4 [22]), BCa05, which expresses TUP1 ectopically (tup1/tup1, MAL3::p455 in CAI-4 [3, 5]), and BCa2-3 (TUP1/tup1 in CAI-4 [5]) were obtained from Alexander Johnson, University of California, San Francisco, CA. Strain MEN was provided by Richard Cannon, University of Otago, Dunedin, New Zealand.
Resting cells were obtained by growing cells in modified glucose salts biotin media (mGSB) overnight, washing them three times with 50 mM phosphate (pH 6.5), resuspending them in 10 ml of 50 mM phosphate, and storing them at 4°C, to be used within a month.
The defined glucose-salts medium GPP (pH 4.8) contained (per liter of distilled water) glucose, 20 g; l-proline, 1.15 g; NaH2PO4, 3.2 g; KH2PO4, 4 g; MgSO4·7H2O, 0.5g; CuSO4·5H2O, 1 mg; ZnSO4·7H2O, 1 mg; MnCl2, 1 mg; FeSO4, 1 mg; biotin, 20 μg; pyridoxine· HCl, 200 μg; thiamine·HCl, 200 μg. The glucose (20% [wt/vol]) and l-proline (100 mM) were autoclaved separately and added aseptically, as were the filter-sterilized vitamins (27). Modified GPP (mGPP) also contained 2.5 mM N-acetylglucosamine (17). GPP (pH 6.8) contained 3.2 g/liter Na2HPO4 instead of NaH2PO4. For maltose phosphate proline (MPP) medium, filter-sterilized maltose replaced the glucose. Cornmeal agar (Difco, Detroit, MI) was also used. Solid medium included 2% (wt/vol) agar. All media for CAI-4 included uridine at 40 μg/ml.
Microscopy.
Differential interference contrast images were produced with an Olympus BX51 microscope, and colony morphology photographs were made with an Olympus SZX12 microscope.
Quantitative Northern blotting analysis.
To measure mRNA accumulation, SC5314 resting cells were inoculated in mGPP to an optical density at 600 nm of 0.5 to 0.6 and allowed to equilibrate at 37°C for 5 min, whereupon 20 μM farnesol was added to half of the flasks. Cells were grown at 37°C for 0, 20, 40, 60, and 80 min, until the cells were harvested, and total RNA was extracted by the hot phenol method (24). Equal amounts of RNA (15 μg) were resolved on 1.0% agarose-formaldehyde gels, and the RNA was transferred to GeneScreen Plus (NEN Life Science Products, Inc., Boston, MA), using the capillary blot transfer protocol recommended by the manufacturer. The Northern blots were probed with radiolabeled DNA probes. The probe DNA used for synthesis was prepared by PCR using MEN genomic DNA. The probes were labeled with [32P]dCTP (GE Health Sciences, Piscataway, NJ), using an oligo labeling kit, RadPrime DNA labeling system, following the protocol recommended by the manufacturer (Invitrogen, Carlsbad, CA). Northern blots were phosphorimaged using a Storm phosphorimager (Amersham Pharmacia Biotech Inc., Piscataway, NJ) and quantified using ImageQuant software (version 5.0; Molecular Dynamics, Sunnyvale, CA). mRNA abundance measurements were done using a minimum of three independent Northern blots.
Western blotting analysis.
Western blots were prepared as previously described (2), and Tup1 and Act1 proteins were detected with a Supersignal West Pico chemiluminescent substrate, using the manufacturer's protocol (Pierce, Rockford, IL), except that blocking was done with 5% nonfat dried milk. Rabbit polyclonal antibodies against Tup1 were previously described (18). Mouse monoclonal anti-Act1 antibodies and horseradish peroxidase-labeled ant-rabbit immunoglobulin G antibodies were from Amersham Pharmacia Biotech, Inc. (Piscataway, NJ). Horseradish peroxidase-labeled anti-mouse antibody was from Perkin-Elmer (Boston, MA).
Analysis of farnesol levels.
Extracellular farnesol was extracted from cell-free supernatants of cultures grown in mGPP at 30°C and analyzed by gas chromatography-mass spectrometry as described previously (17).
RESULTS
The tup1/tup1 and nrg1/nrg1 mutants lack a morphological response to farnesol, while the rfg1/rfg1 mutant responds to farnesol.
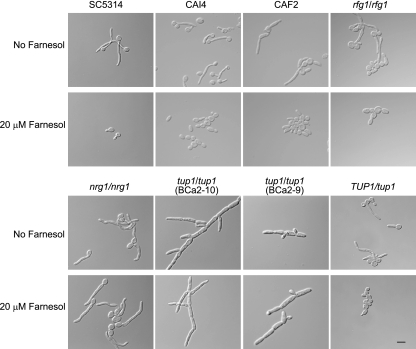
The juxtaposition of farnesol's ability to inhibit differentiation and the role of Tup1 as a transcription repressor for filamentation genes suggest that farnesol could function by activating Tup1 and/or one of its coregulators, Nrg1 or Rfg1. Consequently, we examined the effect of farnesol on the morphology of null mutants lacking TUP1, NRG1, and RFG1. As a control, the wild-type C. albicans SC5314 in filament-inducing media grew as yeast in the presence of 20 μM farnesol and as filaments in media lacking farnesol, demonstrating a positive response to farnesol (Fig. 1). The rfg1/rfg1 mutant responded to 20 μM farnesol in a manner similar to that of SC5314 (Fig. 1). Unlike SC5314 and rfg1/rfg1, the tup1/tup1 and nrg1/nrg1 mutants lacked a detectable response to farnesol and remained filamentous in the presence of 20 μM farnesol (Fig. 1). The filamentous-only cell morphology is the phenotype expected for these known mutants (5, 7, 25, 32, 33). However, in this regard, the tup1/tup1 and nrg1/nrg1 mutants differ from the great majority of filamentous-only mutants recovered from a previous study, 96% of which reverted to a smooth (yeast) colony morphology on yeast malt (YM) agar plates with 50 μM farnesol (20). For the tup1/tup1 mutant, the lack of response to farnesol was specific for the loss of Tup1 because we found that ectopic expression of TUP1 (5) restores the strain's ability to respond to farnesol (data not shown).
FIG. 1.
Response to farnesol by C. albicans under conditions that promote GTF and hyphal growth. SC5314, CAI4, CAF2, rfg1/rfg (DU129), nrg1/nrg1 (DU152), tup1/tup1(BCa2-9), tup1/tup1 (BCa2-10), and TUP1/tup1 (BCa2-3) resting cells were inoculated into mGPP (pH 4.8) medium at 37°C in the presence or absence of 20 μM farnesol, and their cell morphologies were examined at 4 h. Scale bar = 10 μm.
TUP1 mRNA levels increase in the presence of farnesol, while RFG1 and NRG1 mRNA levels were not affected by farnesol.
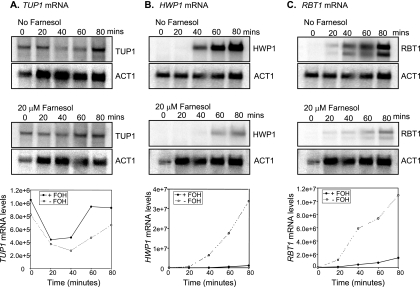
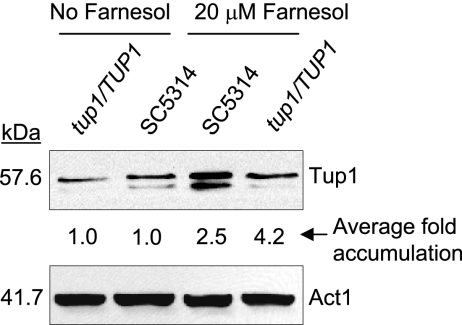
We analyzed the effect of farnesol on TUP1 mRNA levels over time in C. albicans SC5314 cells that had been induced to commence germ tube formation (GTF) by growth at 37°C in mGPP. We previously showed (31) that under these conditions, the first germ tubes appeared at 30 min and the process was complete by 110 min. Furthermore, farnesol no longer blocked GTF when it was added at 60 to 90 min after inoculation (27). Here, our analysis was designed to evaluate changes in TUP1 mRNA just before GTF, when the cells were still responsive to farnesol. Filamentation was induced by transferring resting cells into mGPP (pH 4.8) at 37°C in the presence and absence of 20 μM farnesol, and mRNA levels were determined at 0, 20, 40, 60, and 80 min following induction. In all experiments, the TUP1 mRNA levels decreased over the first 20 min and then increased (Fig. 2A). This pattern is consistent with the single-time-point result of Toyoda et al. (45), who showed that TUP1 mRNA levels increased slightly at 180 min after induction of filamentation. In the presence of farnesol, we found that TUP1 mRNA consistently increased 2.5-fold ± 0.6-fold (n = 4) from 20 to 60 min. Importantly, this is the time period just prior to that at which the cells become committed and are no longer responsive to farnesol (31). In contrast, in the absence of farnesol, there was very little increase (1.4 ± 0.3; n = 4) in TUP1 mRNA levels from 20 to 60 min (Fig. 2A). Thus, farnesol (20 μM) causes a consistent increase in TUP1 mRNA levels during the precise time period when it blocks GTF. This increase of 2.5-fold in TUP1 mRNA corresponded to an increase in SC5314 Tup1 protein levels at 60 min following induction (Fig. 3). Tup1 protein in SC5324 was increased in all three replicate experiments by an average of 2.5-fold.
FIG. 2.
TUP1 mRNA levels increased, while two Tup1-regulated genes, HWP1 and RBT1, were downregulated in the presence of farnesol (FOH). C. albicans SC5314 resting cells were inoculated into mGPP (pH 4.8) medium in the presence or absence of 20 μM farnesol and incubated at 37°C. Cells were then harvested at 0, 20, 40, 60, and 80 min postinoculation. Northern blots were prepared with total RNA from cells incubated in the presence or absence of farnesol. Shown is a phosphorimage of a representative Northern blot probed with radiolabeled TUP1 DNA (A), HWP1 DNA (B), and RBT1 DNA (C) and a plot of average mRNA levels from a minimum of three independent experiments. ACT1 mRNA levels were used as a loading control.
FIG. 3.
Tup1 protein levels are higher in the presence of farnesol. Total protein extracts were prepared from SC5314 and TUP1/tup1 (BCa2-3) resting cells inoculated into mGPP (pH 6.8) medium at 37°C in the presence or absence of 20 μM farnesol and incubated at 37°C for 60 min. The average change (fold) in Tup1 protein accumulation for farnesol-treated cells relative to that of untreated cells is shown. Act1 levels were used as a loading control.
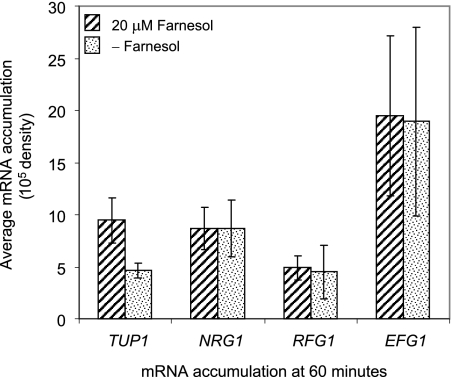
Since Tup1 functions with DNA binding proteins such as Rfg1 and Nrg1, and in C. albicans strain JCM9061 the NRG1 mRNA levels decreased during filamentation (45), we tested the effect of farnesol on the RFG1 and NRG1 mRNA levels during differentiation from yeast to filamentous form. Like TUP1 mRNA, the RFG1 mRNA levels initially decreased and then increased (data not shown). However, unlike TUP1 mRNA, the timing and magnitude of the RFG1 mRNA level changes were similar in the presence and the absence of farnesol (Fig. 4, data not shown). Under the same conditions, the NRG1 mRNA levels did not change during development, and they too were the same in the presence and absence of farnesol (Fig. 4). Thus, we conclude that farnesol does not affect the RFG1 or NRG1 mRNA levels.
FIG. 4.
Farnesol does not affect the expression of RFG1 or NRG1, which encode DNA binding proteins that function with Tup1, or EFG1, which encodes a transcription activator of hypha-specific genes. Quantitative Northern blotting analysis was used to measure the TUP1, NRG1, RFG1, and EFG1 mRNA levels in SC5314 at 60 min after the inoculation of resting cells under conditions that promote GTF in the presence and absence of 20 μM farnesol. The results are averages of three independent experiments.
Expression of the Tup1-regulated filamentation genes HWP1 and RBT1 is inhibited by farnesol.
To determine whether the increased TUP1 expression in the presence of farnesol was biologically significant, we examined the expression of two Tup1-regulated genes, HWP1 and RBT1 (Fig. 2B and C [12]). In the absence of farnesol, the HWP1 and RBT1 transcripts were undetectable at time zero, but they were strongly expressed from 40 to 80 min (Fig. 2B and C). Farnesol delays and dramatically reduces the magnitude of HWP1 and RBT1 mRNA expression (Fig. 2B and C). At 80 min, HWP1 and RBT1 levels were 30- and 7.6-fold lower, respectively, in farnesol-treated cells than in untreated cells. Similar results were observed by Davis-Hanna et al. (11) for HWP1 mRNA at 2 h after treatment with 75 μM farnesol. Thus, there is a strong correlation between elevated TUP1 expression in response to farnesol and the expression of Tup1-regulated genes.
EFG1 mRNA levels remain unaffected by farnesol.
Efg1 is a transcription regulator for genes required for filamentation. EFG1 mRNA levels are downregulated at the initiation of filament development and then increase as filament formation progresses (44). HWP1 and RBT1 are activated by Efg1 during filamentation (6). Therefore, we determined whether farnesol also affected EFG1 mRNA levels (Fig. 4). The EFG1 mRNA levels were high at time zero, decreased to a minimum at 20 min, and then increased steadily throughout the remaining time (data not shown). However, farnesol had no influence on EFG1 mRNA levels, since the timing and magnitude of the changes were similar in the presence and the absence of farnesol (Fig. 4 and data not shown).
Farnesol suppresses the haploinsufficient phenotype of a TUP1/tup1 heterozygote.
Braun and Johnson (5) showed that BCa2-3, a TUP1/tup1 heterozygote, is haploinsufficient in that these cells develop a higher proportion of filaments than the wild-type cells on most media (5). Presumably, these cells do not make enough Tup1 to compensate for the reduced gene copy number. We hypothesized that farnesol might suppress this phenotype because it increases TUP1 expression 2.5-fold in SC5314 and 4.2-fold in TUP1/tup1 (Fig. 3). This increase should restore Tup1 to roughly wild-type levels. To test this hypothesis, we examined the effect of farnesol on C. albicans BCa2-3 on cornmeal agar plus Tween 80, under a coverslip for 25 h at 25°C. Under these conditions, the TUP1/tup1 mutant was more filamentous than the wild-type colonies but less filamentous than the tup1/tup1 mutant (BCa2-10 [4]; see Table 2). As a control, the TUP1/tup1 mutant was shown to respond to farnesol because, although it forms filamentous cells when grown in mGPP medium, the addition of 20 μM farnesol results in growth as yeast (Fig. 1), and Tup1 protein levels were ca. 4.2-fold higher in the TUP1/tup1 mutant treated with farnesol. In contrast to the haploinsufficient phenotype observed with the absence of farnesol, in the presence of farnesol, the TUP1/tup1 mutant looked identical to the wild-type C. albicans (see Table 2). Thus, farnesol suppresses the haploinsufficiency phenotype of the TUP1/tup1 heterozygote.
TABLE 2.
Farnesol suppression of the TUP1/tup1 heterozygotea
| C. albicans strain | Cell morphology at the colony periphery in response to:
|
|
|---|---|---|
| No farnesol | 20 μM farnesol | |
| Wild type (SC5314) | Yeast plus a few filaments | Yeast plus a few filaments |
| TUP1/tup1 (BCa2-3) | Yeast plus filaments | Yeast plus a few filaments |
| tup1/tup1 (BCa10) | Filamentous form | Filamentous form |
Farnesol suppressed the haploinsufficient phenotype of the TUP1/tup1 heterozygote. Cells were plated on a cornmeal agar plus Tween 80 plate, under a coverslip, and grown at 25°C for 25 h. The phenotype for these strains grown on a plate with cornmeal agar plus Tween 80 under a coverslip without farnesol was also previously reported (5).
The tup1/tup1 and nrg1/nrg1 mutants produce excess farnesol.
Jensen et al. (20) tested the farnesol production levels for several filament-only mutants. A subset of these mutants produced levels of farnesol significantly higher than those of the wild-type strains. This overproduction suggests that the ability to respond to farnesol may be linked to the regulation of farnesol production. Here, we tested farnesol production levels in the CAI-4, CAF-2, tup1/tup1, nrg1/nrg1, and rfg1/rfg1 strains. Farnesol production levels were dramatically increased in the tup1/tup1 and nrg1/nrg1 mutants (Table 1), which were unable to respond to farnesol (Fig. 1). The tup1/tup1 and nrg1/nrg1 mutants produced ca. 17- and 19-fold more farnesol, respectively, than did CAF-2 and CAI-4. In contrast, the farnesol-responsive rfg1/rfg1 mutant produced only ca. 2.6-fold more farnesol than the wild-type strains (Table 1). Thus, the two mutants that are unable to respond to farnesol (tup1/tup1 and nrg1/nrg1) produced much higher levels of farnesol than did strains that do respond to farnesol.
TABLE 1.
The tup1/tup1 and nrg1/nrg1 null mutants do not respond to farnesol but overproduce farnesol
| C. albicans strain | Farnesol responsea | Farnesol production (μg/g dry wt of cells) ± SDb | Fold increase in farnesolc |
|---|---|---|---|
| CAI-4 | Positive | 1.6 ± 0.36 | |
| CAF-2 | Positive | 2.0 ± 1.30 | |
| tup1/tup1 (BCa2-10) | Negative | 30.6 ± 6.40 | 17 |
| nrg1/nrg1 (DU152) | Negative | 34.5 ± 12.2 | 19 |
| rfg1/rfg1 (DU129) | Positive | 4.8 ± 2.0 | 2.6 |
Farnesol responses on GPP agar with and without 20 μM farnesol, incubated at 37°C for 48 h. A positive response indicates smooth colony morphology (yeast cells) in the presence of farnesol and rough colony morphology (filamentous cells) without added farnesol. A negative response to farnesol indicates rough colony morphology in the presence and absence of farnesol.
Farnesol production values (μg/g dry weight of cells) ± standard deviation (SD) were the averages of three measurements.
Values are based on fold increases over 1.8, the average value for strains CAI-4 and CAF-2.
tup1/tup1 overproduction of farnesol inhibits SC5314 filamentation.
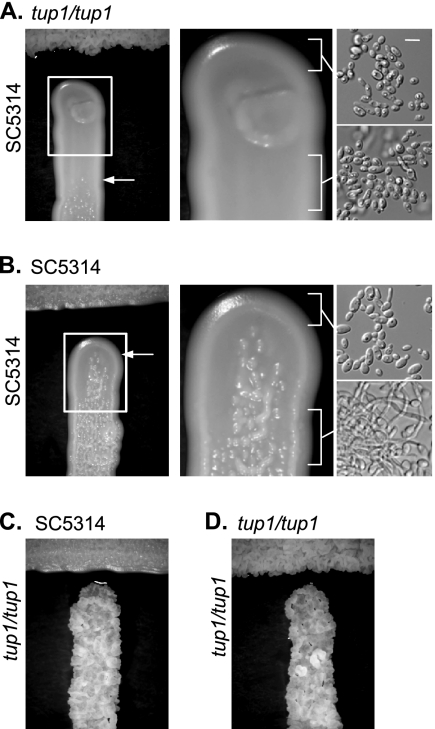
We tested the biological significance of farnesol overproduction by sequentially plating tup1/tup1 and SC5314 next to one another and observing the resultant colony morphologies. When SC5314 was plated and followed 1 day later by another streak with SC5314, a small area of filament inhibition was observed (Fig. 5B). In contrast, when tup1/tup1 was plated first, followed by SC5314, a much larger area of filament inhibition was observed (Fig. 5A). These results are consistent with the tup1/tup1 overproduction of farnesol. As controls, whenever tup1/tup1 was plated second, no filament inhibition was observed (Fig. 5C and D).
FIG. 5.
Overproduction of farnesol by the tup1/tup1 mutant inhibits SC5314 filamentation. Resting cells were grown at 37°C for 24 h on yeast-peptone-dextrose agar plates to allow for farnesol accumulation in the agar (horizontal streak, C. albicans strain SC5314 [B and C]; or tup1/tup1, BCa2-10 [A and D]). Subsequently, either SC5314 (A and B) or tup1/tup1 (C and D) resting cells were plated (vertical streak) and incubated at 37°C for an additional 24 h. The areas above the two arrows (A and B, left panels) are zones of filament inhibition (as evident by smooth morphology) resulting from the farnesol produced by the horizontally streaked strains. Filamentation gives the wrinkled colony morphology seen below the arrows. The pictures in the two white boxes have been magnified ×2.5 so that the colony morphology can be seen more clearly (A and B, center panels). Micrographs of individual cells from the two bracketed regions are shown in the right panels (A and B; scale bar = 10 μm). The cells from the smooth regions are mainly yeast, while there is a much larger proportion of filamentous cells in the wrinkled region.
DISCUSSION
C. albicans responds to farnesol, in part, by changing gene expression (8, 15). We hypothesize that some of these changes are mediated by changes in the activity of the signaling pathways regulating morphogenesis. Here, we show that the tup1/tup1 and nrg1/nrg1 null mutants are strictly filamentous strains and that the cells remain filamentous in the presence of added farnesol (Fig. 1). In these cases, the total farnesol levels are actually much higher than the added farnesol because the mutants themselves produce elevated levels of farnesol (Table 1; see below). Furthermore, Tup1 mRNA and protein levels increased in the presence of farnesol, while mRNA levels of two Tup1-regulated genes, HWP1 and RBT1, decrease (Fig. 2, and 3). Importantly, the timing of this increase (40 to 60 min, Fig. 2) corresponds with the commitment point, beyond which added farnesol no longer blocks GTF (31). Finally, we believe that Tup1 is part of the farnesol response pathway because farnesol suppresses the haploinsufficient phenotype of the TUP1/tup1 strain (Table 2).
Cell synchrony, farnesol concentration, and timing were all important considerations for our experimental design. Previous work examining farnesol-dependent changes in the global transcription profiles of developing biofilms (8, 15) and during resumption of growth following stationary phase (15) were done with mixed cell populations that differed in their ability to respond to farnesol. Furthermore, the effect of adding farnesol on the global gene expression during biofilm formation was determined at a single time point, 24 h after the addition of farnesol (8). This point is significant because such a study could measure only stable long-term farnesol-dependent changes in gene expression. Timing is also important because of the commitment phenomenon. This is the point at which a switch in the environmental stimulus no longer causes the expected switch in morphology (9, 30, 31). It is relevant to farnesol's mode of action because, while farnesol blocks the yeast-to-filament switch, it does not block the elongation of preexisting filaments (31). Thus, for our experiments, we added farnesol at time zero in order to avoid the commitment to filamentous growth, and we harvested cells at 20-min increments to observe changes in transcript levels during the early stages of the farnesol response (31). We also achieved a synchronous cell population by starting with resting cells and inoculating them in mGPP; under these conditions, we routinely got 95 to 100% filamentous cells within 3 to 4 h. Exposing a synchronized cell population to farnesol allowed us to detect subtle and consistent changes in transcript abundance.
Small changes in the expression of a transcription regulator can have profound effects on the genes it regulates. For example, we have shown that nonsense-mediated mRNA decay in S. cerevisiae regulates the accumulation of the mRNA for Adr1, a transcription regulator of the genes responsible for making acetyl-coenzyme A and NADH from nonfermentable substrates. In particular, the respiratory impairment seen with nonsense-mediated mRNA decay mutants is due, in part, to the overexpression of Adr1 (43). The change in ADR1 mRNA levels is small (2.6-fold) but sufficient to affect expression of Adr1-regulated genes. Thus, even though the change in Tup1 expression is relatively small, it can have a profound effect on the expression of the genes it regulates.
Two Tup1 coregulators, encoded by NRG1 and RFG1, were unaffected by farnesol at the mRNA level (Fig. 4). In this regard, it is reasonable that farnesol regulates only one part of the complex, i.e., that farnesol elevates TUP1 mRNA but not NRG1 or RFG1 mRNA. By analogy, for Ca2+ and calmodulin, where only the Ca2+-calmodulin complex is active (40), fungi have calmodulin in excess and regulate the activity of the complex by regulating the availability of cytoplasmic Ca2+ (34).
The tup1/tup1 and nrg1/nrg1 mutants did not respond to farnesol, suggesting that farnesol acts through a pathway requiring Tup1 and Nrg1. The rfg1/rfg1 mutant responded to farnesol, indicating that the genes regulated by Rfg1 are not required for the response to farnesol. Furthermore, the tup1/tup1 and nrg1/nrg1 mutants overproduced farnesol, while the rfg1/rfg1 mutant produced only slightly elevated levels of farnesol (Table 1). This tup1/tup1 mutant overproduction is biologically significant because the excess farnesol produced by the tup1/tup1 mutant inhibits filamentation of the wild-type C. albicans grown on the same plate (Fig. 5). The juxtaposition of farnesol nonresponsive mutants with the overproduction of farnesol implies that a farnesol-Tup1 feedback loop may exist and that Nrg1 may work in concert with Tup1 to negatively regulate farnesol synthesis. This regulation may be direct or indirect. The enzyme responsible for ca. 90% of farnesol synthesis is Dpp3 (35). DPP3 mRNA levels were not significantly elevated in the whole genome profiles of the tup1/tup1 or nrg1/nrg1 mutants (21); however, DPP3 does have a putative Nrg1 binding site in its promoter region.
The increased TUP1 expression we observed for farnesol's blockage of filament development (Fig. 2A) is smaller than that reported for farnesol's blockage of biofilm development, ca. 6.6-fold, as determined with DNA arrays (8). The difference in the response intensity may reflect filament versus biofilm growth conditions, as well as the fact that Cao et al. (8) used one time point 24 h after farnesol addition.
Efg1 is a transcriptional factor that activates hyphal gene expression including that of HWP1 and RBT1 (7). EFG1 mRNA levels are regulated during filamentation, but they were not affected by farnesol, since the timing and magnitude of the changes were similar in the presence and absence of farnesol (Fig. 4). These results are consistent with those of Soto et al. (41), who also found no change in EFG1 mRNA levels at a single time point with added farnesol (41). Together with our results, this suggests that farnesol does not regulate EFG1 mRNA levels, but at this time, we cannot exclude the possibility that posttranslational regulation of Efg1 is affected by farnesol.
Two other farnesol-related findings regarding filamentous growth can be accommodated in a Tup1-dependent model because they are downstream from Tup1. Soto et al. (41) suggested that farnesol acts by causing decreased CPH1 and HST7 mRNA levels. CPH1 is a transcription factor that regulates filamentous growth, and HST7 is a mitogen-activated protein kinase kinase involved in filamentous growth. Both are downregulated by Tup1, and thus, their downregulation by farnesol (41) is consistent with a secondary effect of farnesol on Tup1. Additionally, Chk1, a histidine kinase shown to be required for the farnesol response (26), is also encoded by a Tup1-repressed gene; CHK1 was elevated 6.5-fold in the tup1/tup1 mutant (21). Taken together, these findings indicate that Tup1 is involved in mediating the C. albicans response to farnesol.
Acknowledgments
We thank Alexander Johnson and Richard Cannon for providing us with C. albicans strains and the Tup1 antibody. We also thank Jessica A. Wiles for assisting with the germ tube assays and RNA work.
This work was supported by grants from the National Science Foundation (MCB-0110999), the University of Nebraska Tobacco Settlement Biomedical Research Enhancement Fund, and the Farnesol and Candida albicans Research Fund, University of Nebraska Foundation.
Any opinions, findings, conclusions, or recommendations expressed in this report are ours and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Published ahead of print on 18 April 2008.
REFERENCES
- 1.Arnaud, M. B., M. C. Costanzo, M. S. Skrzypek, G. Binkley, C. Lane, S. R. Miyasato, and G. Sherlock. 2005. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 33D358-D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkin, A. L., N. Altamura, P. Leeds, and M. R. Culbertson. 1995. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Biol. Cell 6611-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backen, A. C., I. D. Broadbent, R. W. Fetherston, J. D. Rosamond, N. F. Schnell, and M. J. Stark. 2000. Evaluation of the CaMAL2 promoter for regulated expression of genes in Candida albicans. Yeast 161121-1129. [DOI] [PubMed] [Google Scholar]
- 4.Berman, J., and P. E. Sudbery. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3918-930. [DOI] [PubMed] [Google Scholar]
- 5.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277105-109. [DOI] [PubMed] [Google Scholar]
- 6.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 15557-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in Candida albicans, is down-regulated during filament induction. EMBO J. 204753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, Y. Y., Y. B. Cao, Z. Xu, K. Ying, Y. Li, Y. Xie, Z. Y. Zhu, W. S. Chen, and Y. Y. Jiang. 2005. cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob. Agents Chemother. 49584-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaffin, W. L., and D. E. Wheeler. 1981. Morphological commitment in Candida albicans. Can. J. Microbiol. 27131-137. [DOI] [PubMed] [Google Scholar]
- 10.Csank, C., K. Schroppel, E. Leberer, D. Harcus, O. Mohamed, S. Meloche, D. Y. Thomas, and M. Whiteway. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 662713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis-Hanna, A., A. E. Piispanen, L. I. Stateva, and D. A. Hogan. 2008. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol. Microbiol. 6747-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Groot, P. W., K. J. Hellingwerf, and F. M. Klis. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20781-796. [DOI] [PubMed] [Google Scholar]
- 13.Dhillon, N. K., S. Sharma, and G. K. Khuller. 2003. Signaling through protein kinases and transcriptional regulators in Candida albicans. Crit. Rev. Microbiol. 29259-275. [DOI] [PubMed] [Google Scholar]
- 14.Eckert, S. E., C. C. Sheth, and F. A. Muhlschlegel. 2007. Regulation of morphogenesis in Candida species, p. 263-291. In C. d'Enfert and B. Hube (ed.), Candida: comparative and functional genomics, 1st ed. Caister Academic Press, Norfolk, United Kingdom.
- 15.Enjalbert, B., and M. Whiteway. 2005. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryot. Cell 41203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornby, J. M., E. C. Jensen, A. D. Lisec, J. J. Tasto, B. Jahnke, R. Shoemaker, P. Dussault, and K. W. Nickerson. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 672982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inglis, D. O., and A. D. Johnson. 2002. Ash1 protein, an asymmetrically localized transcriptional regulator, controls filamentous growth and virulence of Candida albicans. Mol. Cell. Biol. 228669-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis, W. R., and W. J. Martone. 1992. Predominant pathogens in hospital infections. J. Antimicrob. Chemother. 29(Suppl. A)19-24. [DOI] [PubMed] [Google Scholar]
- 20.Jensen, E. C., J. M. Hornby, N. E. Pagliaccetti, C. M. Wolter, K. W. Nickerson, and A. L. Atkin. 2006. Farnesol restores wild-type colony morphology to 96% of Candida albicans colony morphology variants recovered following treatment with mutagens. Genome 49346-353. [DOI] [PubMed] [Google Scholar]
- 21.Kadosh, D., and A. D. Johnson. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 162903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadosh, D., and A. D. Johnson. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 212496-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko, A., T. Umeyama, Y. Utena-Abe, S. Yamagoe, M. Niimi, and Y. Uehara. 2006. Tcc1p, a novel protein containing the tetratricopeptide repeat motif, interacts with Tup1p to regulate morphological transition and virulence in Candida albicans. Eukaryot. Cell 51894-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kebaara, B., T. Nazarenus, R. Taylor, A. Forch, and A. L. Atkin. 2003. The Upf-dependent decay of wild-type PPR1 mRNA depends on its 5′-UTR and first 92 ORF nucleotides. Nucleic Acids Res. 133157-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalaf, R. A., and R. S. Zitomer. 2001. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 1571503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruppa, M., B. P. Krom, N. Chauhan, A. V. Bambach, R. L. Cihlar, and R. A. Calderone. 2004. The two-component signal transduction protein Chk1p regulates quorum sensing in Candida albicans. Eukaryot. Cell 31062-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulkarni, R. K., and K. W. Nickerson. 1981. Nutritional control of dimorphism in Ceratocystis ulmi. Exp. Mycol. 5148-154. [Google Scholar]
- 28.Kullberg, B. J., and S. G. Filler. 2002. Candida and candidiasis. ASM Press, Washington, DC.
- 29.Kumamoto, C. A., and M. D. Vinces. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 71546-1554. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell, L. H., and D. R. Soll. 1979. Commitment to germ tube or bud formation during release from stationary phase in Candida albicans. Exp. Cell Res. 120167-179. [DOI] [PubMed] [Google Scholar]
- 31.Mosel, D. D., R. Dumitru, J. M. Hornby, A. L. Atkin, and K. W. Nickerson. 2005. Farnesol concentrations required to block germ tube formation in Candida albicans in the presence and absence of serum. Appl. Environ. Microbiol. 714938-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42981-993. [DOI] [PubMed] [Google Scholar]
- 33.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 204742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muthukumar, G., A. W. Nickerson, and K. W. Nickerson. 1987. Calmodulin levels in yeasts and filamentous fungi. FEMS Microbiol. Lett. 41253-255. [Google Scholar]
- 35.Navarathna, D. H., J. M. Hornby, N. Krishnan, A. Parkhurst, G. E. Duhamel, and K. W. Nickerson. 2007. Effect of farnesol on a mouse model of systemic candidiasis, determined by use of a DPP3 knockout mutant of Candida albicans. Infect. Immun. 751609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramage, G., S. P. W. Saville, B. L., and J. L. Lopez-Ribot. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 685459-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramon, A. M., A. Porta, and W. A. Fonzi. 1999. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 1817524-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semighini, C. P., J. M. Hornby, R. Dumitru, K. W. Nickerson, and S. D. Harris. 2006. Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol. Microbiol. 59753-764. [DOI] [PubMed] [Google Scholar]
- 39.Shchepin, R., J. M. Hornby, E. Burger, T. Niessen, P. Dussault, and K. W. Nickerson. 2003. Quorum sensing in Candida albicans: probing farnesol's mode of action with 40 natural and synthetic farnesol analogs. Chem. Biol. 10743-750. [DOI] [PubMed] [Google Scholar]
- 40.Soto, T., Y. Ueno, T. Watanabe, T. Mikami, and T. Matsumoto. 2004. Role of Ca2+/calmodulin signaling pathway on morphological development of Candida albicans. Biol. Pharm. Bull. 271281-1284. [DOI] [PubMed] [Google Scholar]
- 41.Soto, T., T. Watanabe, T. Mikami, and T. Matsumoto. 2004. Farnesol, a morphogenetic autoregulatory substance in the dimorphic fungus Candida albicans, inhibits hyphae growth through suppression of a mitogen-activated protein kinase cascade. Biol. Pharm. Bull. 27751-752. [DOI] [PubMed] [Google Scholar]
- 42.Sprague, E. R., M. J. Redd, A. D. Johnson, and C. Wolberger. 2000. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 193016-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, R., B. W. Kebaara, T. Nazarenus, A. Jones, R. Yamanaka, R. Uhrenholdt, J. P. Wendler, and A. L. Atkin. 2005. Gene set coregulated by the Saccharomyces cerevisiae nonsense-mediated mRNA decay pathway. Eukaryot. Cell 42066-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tebarth, B., T. Doedt, S. Krishnamurthy, M. Weide, F. Monterola, A. Dominguez, and J. F. Ernst. 2003. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J. Mol. Biol. 329949-962. [DOI] [PubMed] [Google Scholar]
- 45.Toyoda, M., T. Cho, H. Kaminishi, M. Sudoh, and H. Chibana. 2004. Transcriptional profiling of the early stages of germination in Candida albicans by real-time RT-PCR. FEMS Yeast Res. 5287-296. [DOI] [PubMed] [Google Scholar]
- 46.Westwater, C., E. Balish, and D. A. Schofield. 2005. Candida albicans-conditioned medium protects yeast cells from oxidative stress: a possible link between quorum sensing and oxidative stress resistance. Eukaryot. Cell 41654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]