Abstract
Oxidative stress is recognized as a trigger of different metabolic events in all organisms. Various factors correlated with oxidation, such as the β-oxidation of fatty acids and their enzymatic or nonenzymatic by-products (e.g., precocious sexual inducer factors and lipoperoxides) have been shown to be involved in aflatoxin formation. In the present study, we found that increased levels of reactive oxygen species (ROS) were correlated with increased levels of aflatoxin biosynthesis in Aspergillus parasiticus. To better understand the role of ROS formation in toxin production, we generated a mutant (ΔApyapA) having the ApyapA gene deleted, given that ApyapA orthologs have been shown to be part of the antioxidant response in other fungi. Compared to the wild type, the mutant showed an increased susceptibility to extracellular oxidants, as well as precocious ROS formation and aflatoxin biosynthesis. Genetic complementation of the ΔApyapA mutant restored the timing and quantity of toxin biosynthesis to the levels found in the wild type. The presence of putative AP1 (ApYapA orthologue) binding sites in the promoter region of the regulatory gene aflR further supports the finding that ApYapA plays a role in the regulation of aflatoxin biosynthesis. Overall, our results show that the lack of ApyapA leads to an increase in oxidative stress, premature conidiogenesis, and aflatoxin biosynthesis.
Reactive oxygen species (ROS), such as superoxide anion (O2−), hydrogen peroxide (H2O2), hydroxyl radical (HO), and lipoperoxides (LOOH), which are formed from unsaturated fatty acids and can be produced in the cell during metabolic processes, can be overproduced following the action of oxidative stressors present in the environment (32, 49, 57). To counteract the potentially dangerous accumulation of ROS, cells have evolved strategies (49, 61) based on enzymatic or nonenzymatic systems (28, 45). The main antioxidant enzymes in cells involved in ROS removal are superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GPX). If H2O2 exceeds the cell-scavenging capacity, it can generate highly reactive HO through a Fenton reaction, which initiates the formation of LOOH in the membrane lipids (32).
When ROS accumulation occurs, the oxidant/antioxidant balance is perturbed, which can damage the cell membrane and cell metabolism (free-radical theory of aging) (26). ROS produced at certain time points during the cell's life cycle and at low physiological concentrations play a crucial role in the organism's homeostasis and cell functions. As second messengers, ROS take part in the plant's developmental processes (18, 24, 31) and in the defense mechanisms against pathogens and abiotic stress (5, 24, 52, 62). Similar effects have been shown in mammals, where ROS at proper levels stimulate antioxidant reactions, immune system modulation, and regulation of cell proliferation (3, 4, 55, 59, 65). One of the major objectives of studying the biology of stress is to identify the key factors that control the switch from cytoprotective responses to cell dysfunction following oxidative insult (11).
In fungi, recent studies have evaluated the role played by ROS and fatty-acid metabolism in the differentiation process during growth (1, 8, 9, 38, 42, 63). For example, ROS generated by NADPH oxidase, which are partially controlled by SOD and CAT, play an important role in different aspects of fungal development, such as growth and differentiation (35, 36). In particular, in Neurospora crassa, the start of the transition from conidia to germination is affected by singlet oxygen-generated redox imbalance (38). Oxylipin formation can occur via dioxygenase (DOX) or lipoxygenase (LOX) action and, to a lesser extent, nonenzymatically (24, 58). In Aspergillus nidulans, the presence of the fatty-acid DOXs PpoA, -B, and -C has been reported (63). In Aspergillus, compounds produced by LOXs and DOXs, such as hydroperoxyoctadecadienoic acid (HPODE) and precocious sexual inducer factors, which consist of a mixture of hydroxylated oleic, linoleic, and linolenic acids, have all been shown to stimulate conidiogenesis, and in Aspergillus flavus this occurrence is related to aflatoxin biosynthesis (7, 10, 12, 13). In A. nidulans, ROS can also steer the production of mitospores and meiospores in the regulation of the asexual and sexual phases in development (25, 63). In Aspergillus parasiticus, ROS can control sclerotium formation (14), improving the resistance to adverse environmental conditions. In A. flavus and A. nidulans, the biosynthesis of mycotoxins is closely related to different stages of fungal development, such as conidiogenesis and sclerotium formation (51, 56) in the idiophase, during which an increase in ROS occurs. Other recent studies of different strains of A. parasiticus, some of which are producers of aflatoxin, have demonstrated that oxidative stress is important in aflatoxin production (46).
The efficiency of the cell in maintaining safe levels of ROS mainly depends on the effectiveness of its antioxidant system (29, 49). A quick and effective defensive response depends on the cell's efficient perception of the stress, as well as on the transduction of oxidative signals. In fungi, as well as in animal cells, some transcription factors are able to act as sensors of oxidants in the cell (47, 50). In yeast, it has been shown that oxidative stress-related transcription factors (OSRTFs) (e.g., Yap1, Skn7, Hsf1-2, and Msn2-4) are differentially activated by oxidative stimuli provided by peroxides, diamide, and free-radical generators (45), as well as by antioxidant treatment (34). In particular, Yap1 is a nuclear factor localized in the cytoplasm (where it interacts with the export receptor Crm1) which, under oxidative conditions, migrates to the nucleus, where it binds with responsive elements (TGACTCA). These elements are similar to antioxidant-responsive elements (TGACnnnGC) and promote, together with Skn7 and Hsf1-2, the transcription of many antioxidant-related genes (gst, sod1, sod2, cta1, ctt, trr, and txl [30, 45]). Recently, Saccharomyces cerevisiae has been used as a model for studying the regulation of the response to oxidative stress in A. parasiticus, in particular for investigating the relation between treatment with antioxidant compounds and aflatoxin biosynthesis (34). In A. parasiticus and A. flavus, for some time a correlation has been known to exist among fungal cell oxidative stress, free-radical formation, lipoperoxidation, and aflatoxin biosynthesis (17, 19, 20, 22, 48). Based on the huge quantity of data on fungal development and aflatoxin biosynthesis collected in recent years, the formation of this toxin is considered to be closely related to differentiation and senescence in fungi. However, the extent to which the defense against oxidative stress in the fungal cell plays a role in aflatoxin biosynthesis and the mechanisms underlying this role have not been extensively studied. To this end, we conducted a study of A. parasiticus wild type (WT) and the ΔApyapA null mutant strains and found that the oxidant/antioxidant balance affects aflatoxin biosynthesis and that oxidative stress is one of the main factors involved in the triggering of aflatoxin biosynthesis.
MATERIALS AND METHODS
Fungal strains and culture conditions.
The WT strain used was Aspergillus parasiticus NRRL 2999, a producer of the aflatoxins B1, B2, G1, and G2. ΔApyapA deletion mutants (M) and ApyapA complemented mutants (CM) were generated as described below. The isolates were incubated on potato dextrose agar (PDA; Difco) for 7 days at 30°C, before use. Twenty-five milliliters of potato dextrose broth (PDB; Difco) (which is aflatoxin conducive) in 50-ml Erlenmeyer flasks was inoculated with the WT, Μ, and CM strains, using 0.2 ml of conidial suspension (∼106 conidia) for each flask; incubation was performed at 30°C for different time periods (10, 12, 14, 18, 21, 24, 30, 36, 42, 48, 60, 72, 96, 120, 144, and 168 h). In other experiments, 50 ml of Czapek Dox (CD) broth (Difco) (which is low-conducive for aflatoxins) was inoculated with the WT and M strains; after 4 days of incubation at 30°C, 1 mM cumene hydroperoxide (CH), 0.5 mM menadione (Men), and 1 mM and 10 mM hydrogen peroxide (H2O2) were added to test the strain's sensitivity to oxidant stressors.
Fungal growth and aflatoxin production in culture media.
At each point in time, fungal growth was determined by weighing the mycelium after filtration (Millipore filters; 0.45 μm pore size) and drying it for 48 h at 80°C. To determine the quantity of ROS and LOOH and to perform molecular analyses, the filtered mycelia were lyophilized and weighed.
Aflatoxin production (B1 + B2 + G1 + G2) was analyzed in culture filtrates of the WT, M, and CM strains following extraction with chloroform-methanol (2:1, vol/vol). The extracts were collected, the volume was reduced under a stream of nitrogen, and the quantitative analyses were carried out by high-pressure liquid chromatography, as previously reported (23).
Total hydroperoxides of linoleic acid and percentages of 9- and 13-HODE regioisomers in mycelia.
The WT and Μ strains collected from PDB cultures at different incubation times were homogenized in liquid nitrogen to repress the accidental formation of peroxides. Hydroperoxyoctadecadienoic acids (9- and 13-HPODE) present in the mycelia were analyzed following triple extraction with chloroform-methanol (2:1, vol/vol) in the presence of 100 μg of butylated hydroxytoluene as antioxidant. Peroxides in the extracts were reduced with NaBH4 to obtain hydroxyoctadecadienoic acids (9- and 13-HODE), as previously reported (54). Neither 9-HODE nor 13-HODE was detected in the extracts before reduction. The regioisomers 9-HODE and 13-HODE were analyzed by high-pressure liquid chromatography-atmospheric pressure chemical ionization-mass spectrometry (MS), as reported previously (54). 9-HODE and 13-HODE were identified by comparison with authentic compounds (purchased from Cayman).
Detection and quantification of α-tocopherol.
Analyses of α-tocopherol were performed on lyophilized mycelia at different times, as previously reported (52). Samples were extracted three times in chloroform-methanol (2:1, vol/vol) for 1 h in the presence of 100 μg butylated hydroxytoluene, as antioxidant, and 5 μg heptadecanoic acid (C17:0), as internal standard. The recovered chloroform-methanol mixture was extracted three times in hexane, filtered, vacuum evaporated, and then silylated with trimethyl-silyl-ether (TMS). The TMS derivatives were analyzed by gas chromatography-MS, as described elsewhere (52). Quantitative analyses were performed in single-ion monitoring mode, selecting the ions having m/z values of 502, 277, and 237 for the TMS derivative of α-tocopherol and ions with m/z values of 342 and 327 for the derivative of C17:0. Calibration curves were performed as previously reported; the method was linear (R > 0.99) in the range of 1 to 50 ng α-tocopherol·μl−1.
Levels of anion superoxide and hydrogen peroxide in mycelia.
WT and M lyophilized mycelia (10 mg) collected from PDB cultures at different incubation times (from 10 to 168 h) were homogenized as reported above. O2− and H2O2 accumulation was measured at each point in time. O2− was detected by measuring the reduction of 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) to formazan, according to a published protocol (60), with slight modifications (10). Absorbance at 490 nm was measured in a microplate reader (μQuant; Bio-Tek instruments). XTT levels were expressed as mean absorbance values mg−1 ± standard errors of the means (SEMs).
The quantity of H2O2 was measured using an Amplex Red hydrogen peroxide assay kit (Molecular Probes). Amplex Red, in the presence of horseradish peroxidase enzyme (supplied by the kit), reacts with H2O2 in a 1:1 stoichiometry to produce resorufin, a red fluorescent compound. Samples were incubated for 30 min, and the absorbance at 570 nm was measured using the microplate reader. A standard curve of H2O2 was prepared and used as a reference for quantification.
Activities of SOD, GPX, HPR, and LOX-like enzymes.
The activities of SOD, pH 7.8 and 10.0 (EC 1.15.1.1), GPX (EC 1.11.1.9), hydrogen peroxide reducing enzymes (HPR), and LOX-like enzymes (EC 1.13.11.12) were analyzed in the homogenized mycelia of the WT and Μ strains, as previously described (53).
Zymogram of CAT.
CAT is modified by reacting with ROS, giving rise to more-acidic isoforms (38). CAT conformers, which were extracted from homogenized mycelia of the WT and M strains and collected at different times, were analyzed by zymography. As control, an acidic CAT conformer derived from Aspergillus niger (C1; Sigma-Aldrich) was used, as was the CAT itself, which was oxidized under an O2 stream, which produces a more acidic form (C2). Native minigels (8 to 9 cm and 0.75 nm thick) consisting of 8% polyacrylamide and 0.2% bisacrylamide (Bio-Rad) were loaded with mycelium lysates containing 1 U of putative CAT activity in each lane. Gels were run at 200 V for 2 h 45 min at 4°C on a Miniprotean II Bio-Rad apparatus. Native enzyme was detected as described elsewhere (43).
Cloning and sequencing of ApyapA and Apskn7.
DNA extracted from A. parasiticus NRRL 2999 was amplified in a thermal MasterCycler gradient (Eppendorf) following amplification steps (94°C for 2 min; 35 cycles of 94°C for 30 s, 56°C for 45 s, and 72°C for 1 min; 72°C for 8 min) using the primers Af-yap1 (forward, 5′ TCACACCAGTTCCTCTCATC 3′; reverse, 5′ GCGGAACTTCTCCATAGATT 3′) and Af-skn7 (forward, 5′ TTCCCACTCAACAGATTGAC 3′; reverse, 5′ CATGATTGATCTTTTTCC 3′) designed on the expressed sequence tag sequence of A. flavus and aligned with the yap1 and skn7 gene sequences of S. cerevisiae. Amplification under high-stringency conditions produced a unique band for the yap1-like fragment (720 bp, GenBank accession number DQ104418) and Apskn7 (1,059 bp, GenBank accession number DQ104417). The yap1-like fragment was used for searching for the homologue AfYap1. New primers were designed on the codon start (+1; ApyapA_for, ATGGCCGATTACAATACCCTC) and at the 3′ end of the gene sequences (+1882; ApyapA_rev, TCGAGTTATTTGACCGCGACC) to obtain the complete sequence of ApyapA. This gene was cloned in pGEM-T Easy vector (Promega) and sequenced and aligned with the TBLASTX 2.2.17 in the NCBI website (www.ncbi.nlm.nih.gov/BLAST). The results of BLAST indicated a high homology (amino acid identities of 62% and similarities of 100% [score, 268] with the putative bzip transcription factor [Ap-1] of Aspergillus fumigatus XP_750882.1) of the conceptual amino acid translation of ApyapA with the bzip transcription factor Yap-1 of diverse fungal species. Furthermore, Apskn7 was observed to share a high homology (amino acid identities of 40% and similarities of 57% with the putative response regulator receiver Skn7p of Cochliobolus heterostrophus) with the stress response regulator Skn7 (srrA in A. nidulans) of diverse fungal species. ApYapA (forward, 5′ GTTCTCCATCATCCTCATCC 3′, +1112; reverse, 5′ TGCGGAACTTCTCCATAGAC 3′, +1771) and ApSkn7 (forward, 5′ GGGTACACTACAGGTTCAAA 3′; reverse, 5′ ACGCGTCAAAGCTTCTTAAC 3′) primers were designed and used for the subsequent reverse transcription-PCR (RT-PCR) analysis. The 18S primer pair (forward, 5′ ATGGCCGTTCTTAGTTGGTG 3′; reverse, 5′ GTACAAAGGGGCAGGGACGTA 3′) produced a single fragment of 500 bp (internal standard), whereas the primers chosen for the amplification of Apyap1 and Apskn7 genes produced single fragments of 659 bp and 480 bp, respectively.
Plasmids and transformation.
Two fragments of ApyapA were amplified by PCR from genomic DNA of A. parasiticus NRRL 2999 (WT) with the primer pairs ApyapAXbaI_for (+1112) and ApyapAEcoRI_rev (+1320) and ApyapASphI_for (+1420) and ApyapA_rev (+1772; SalI internal restriction site) and used for the subsequent plasmid transformation. The PCR fragments were eluted from the gel (GelElute extraction kit; Sigma-Aldrich) and cloned in the plasmid pGEM-T (Promega). Plasmid DNA from bacterial colonies containing the resulting pGEM-T::ApyapA constructs was digested with XbaI and EcoRI and with SphI and SalI. The resulting ApyapA fragments were cloned into the vector p3SR2 (8.8 kb) alongside the amdS gene of the acetamide cassette (Fig. 1A). The orientation of the insert was determined by PCR with ApyapA- and AmdS-specific primers. Strain NRRL 2999 of A. parasiticus was transformed with the fragment XbaI-SalI obtained from vector p3SR2::ApyapA (∼9.4 kb), which contains two fragments of the ApyapA gene interrupted by the AmdS cassette (∼4.6 kb) (Fig. 1A). In the A. flavus genome (checked at the website www.aspergillusflavus.org), EcoRI presents two restriction sites ∼1.6 kb downstream and ∼4.0 kb upstream of the AfyapA gene sequence. Thus, considering the high homology of this genome with the A. parasiticus genome (and also the alignment of the A. parasiticus contig present in the NCBI GenBank with the same contig of A. flavus in which the ApyapA homologue is present), the 6.5-kb fragment that originated in the ApyapA knockout mutant could be the result of the sum of the EcoRI downstream fragment (∼1.6 kb), the remaining part of the ApyapA WT gene (∼0.2 kb), the gene fragment used in the deletion construct cloned alongside the amdS resistance cassette (∼0.3 kb), and the amdS fragment (∼4.4 kb) that resulted from EcoRI restriction. The ∼5.5-kb fragment could be the result of ApyapA probe hybridization in a sequence constituted by the 4.0-kb EcoRI ApyapA upstream fragment adjacent to the remaining part of the gene (∼1.1 kb) and to the rest of the deletion construct (∼0.4 kb).
FIG. 1.
DNA gel blot analysis of ApyapA gene replacement mutants and complementation. (A) WT ApyapA locus (ApY); final deletion event with construct containing acetamide resistance cassette (AmdS) and ApyapA gene XhoI-EcoRI (X-E) and SphI-SalI (Sp-Sa) fragments used for transforming WT protoplast; the 5.4-kb BglII-HindIII (B-H) fragment which carries also the hygromycin B resistance cassette (hph) used for complementing ΔApyapA strains. The probe used for the subsequent Southern blot analysis is indicated (p). Genomic DNA was isolated from the wild-type strain NRRL 2999 (WT), the ApyapA complemented mutant (CM), and the gene replacement transformant (M) and digested with EcoRI (E). (B) The blots were hybridized with 1.9-kb ApyapA PCR DIG-labeled probes. (C) PCR amplification of WT, CM, and M strain genomic DNA using AmdS_for and ApyapA_rev primers (expected size of the PCR fragment, ∼2.1 kb) or AmdS_for and AmdS_rev primers (expected size of the PCR fragment, ∼0.7 kb) or Hph_for and Hph_rev primers (expected size of the PCR fragment, ∼0.65 kb) or Hph_for and ApyapA_rev2 primers (expected size of the PCR fragment, ∼0.9 kb). The numbers in the “keys” column indicate the primers used for PCR amplification.
For complementation of the M strain, a 5.4-kb BglII-HindIII fragment resistance cassette was excised from pAN7.1::ApyapA, which carries the hygromycin B resistance selectable marker. The protoplast transformation of the WT and the M strains was performed as described elsewhere (41). As described above, the single ∼4.5-kb fragment present in the CM strain can be generated by the hybridization of the ApyapA probe with a fragment constituted by the ∼1.0-kb EcoRI fragment of the hph resistance cassette, the ∼1.9-kb ApyapA gene sequence (complemented), and the EcoRI ApyapA downstream fragment (∼1.6 kb).
Selection of ApyapA deleted and complemented mutants.
The selection of transformants (strains with deleted ApyapA [M]) was conducted at 30°C on Czapek Dox agar (CDA) containing 30 mM acetamide as the sole nitrogen source; putative transformants were selected, transferred to fresh selective medium, and allowed to sporulate. To obtain homokaryons, single spores were isolated from each selected heterokaryotic transformant and transferred to fresh selective medium. This monoconidial transfer was conducted three times. Finally, 20 homokaryotic progenies were selected and further subcultured to determine the occurrence of abortive transformants. The stability of these transformants was also tested by two additional single-spore transfers on nonselective medium and then again on selective medium and by several mycelial transfers on selective plates. CM (n = 20) strains were selected by testing their resistance to both oxidant stressors and hygromycin B. The protoplasts obtained from conidia of stable Μ strains were transformed with a 5.4-kb BglII-HindIII fragment excised from pAN7.1::ApyapA, which also carries the hygromycin B resistance selectable marker. The protoplasts were plated in CDA in the presence of 1 mM Men and 500 ppm hygromycin B, which at these concentrations completely inhibited the germination of ΔApyapA conidia and their development. The stability of these CM strains was tested by several single-spore transfers on selective medium (Men [1 mM] plus hygromycin B [500 ppm]), as described above for ΔApyapA selection.
For the selection of the M and CM strains, the following criteria were used: (i) Southern blot hybridization with probe (Fig. 1A) obtained by the amplification of the ApyapA WT with the ApyapA primers (see above paragraph) as described above and testing for the presence of the expected bands after Southern analysis of the fungal DNA digested with EcoRI (which does not restrict ApyapA but cuts the amdS sequence once at +4361 and the Hph coding sequence at +2561); the hybridization of the probe is expected to generate a double band in M strain DNA and a single band in the WT and in positive CM strains; (ii) presence/absence of the amdS cassette in the fungal genome, which was tested by PCR with primers AmdS for and AmdS rev; (iii) presence/absence of the Hph cassette in the fungal genome, which was tested by PCR with primers Hph_for (5′ CTTGTATGGAGCAGGAGACC 3′) and Hph_rev (5′ ATTTGTGTACGCCCGACAGC 3′); (iv) PCR amplification of the genomic DNA of the WT, M, and CM strains using AmdS_for and ApyapA_rev primers (expected size of the PCR fragment, ∼2.1 kb); (v) PCR amplification of the genomic DNA of the WT, M, and CM strains using Hph_for and ApyapA_rev2 (5′ GAGGCCTTCTGCAACTCAAG 3′) primers (expected size of the PCR fragment, ∼0.9 kb); (vi) RT-PCR products of cDNA from all of the strains were amplified by using ApyapA_for and ApyapA_rev (in the figures, only one representative strain is shown for the WT, M, and CM strains); and (vii) the ability of the M strain to grow and sporulate similarly to the WT on CDA with or without acetamide and the ability of the CM strains to grow and sporulate similarly to the WT on CDA with or without Men (1 mM) and hygromycin B (500 ppm). Strain development was determined by measuring the growth rate and spore counts of cultures grown on plates of PDA.
aflR, norA, ApyapA, Aphsf2, and Apskn7 semiquantitative RT-PCR analysis.
Total RNA from 100 mg of freeze-dried mycelia was extracted using the Tri-Reagent protocol (Sigma) and was quantified by spectrophotometry, determining the optical density at 260 nm. RNA was treated with RNase-free DNase I and then resuspended in 20 μl of diethyl pyrocarbonate-treated water. RNA was extracted at different points in time (from 10 to 168 h; three tubes for each point in time) from A. parasiticus PDB culture and was used to develop an aflR, norA, ApyapA, Aphsf2, and Apskn7 RT-PCR assay, as previously reported (54). A semiquantitative analysis was conducted by performing RT-PCR amplification under several different conditions, in which the annealing temperature (from 55°C to 65°C), amplification cycles (from 20 to 35), and cDNA quantity (1 to 25 ng) were optimized to obtain a reproducible and reliable amplification curve capable of indicating the best conditions for revealing significant differences in the level of mRNA expression in comparing the various points in time and the different strains. The ratio of gene-target expression to 18S rRNA (used here as an internal standard), which indicates a semiquantitative analysis of mRNA expression, was calculated using quanti-doc, a tool present in the UVI-doc software package.
Southern hybridization.
For Southern blot analysis, 10 μg of genomic DNA from A. parasiticus NRRL 2999 and each M and CM strain was completely digested with EcoRI (10 U) at 37°C for 4 h in the manufacturer's buffer at the recommended concentrations (Fermentas). EcoRI-digested DNA fragments were separated by electrophoresis for 3 h 30 min at 40 V on an 0.8% (wt/vol) agarose gel in 1× Tris-acetate-EDTA buffer. Digoxigenin (DIG)-labeled HindIII cut lambda (λ) (Roche) was used as molecular weight standard. Before blotting, the gel containing the DNA was washed twice with denaturation buffer (0.5 N NaOH and 1.5 M NaCl) for 30 min and twice with neutralization buffer (0.5 M Tris-HCl and 3 M NaCl, pH 7.5) for 15 min. The nucleic acids were transferred to a Hybond-N+ nylon membrane (Roche) using 10× SSC (1.5 M NaCl and 0.15 M sodium citrate) and fixed by UV illumination, in accordance with the Roche method, after overnight blotting. Fluorescent DNA probes were prepared according to the PCR DIG labeling mix method (Roche). The membranes were prehybridized according to the instructions of the manufacturer of the DIG detection kit, at 64°C in DIG Easy buffer (Roche); they were then hybridized for 12 to 16 h in the same buffer containing 250 ng of freshly denatured DIG ApyapA probe at the same temperature.
Statistics.
Data are presented as the mean value (±SEM) of three independent determinations from three separate experiments. In all experiments, data sets were pooled and compared using Student's t test, and the differences were considered significant when the P value was <0.05.
RESULTS
ApyapA deleted and complemented mutant generation.
In yeast, Yap1 modulates the expression of many antioxidant-related genes (2, 16, 45). The expression of the ApyapA gene (Yap1 orthologue) is correlated with responsiveness to oxidative stress (54). Mutants (n = 20) with the ApyapA gene deleted were generated to assess whether ApYapA acts as a sensor of oxidative stress and a modulator of cell antioxidative responses also in A. parasiticus NRRL 2999. ΔApyapA (M), CM, and WT mycelia grown in aflatoxin-conducive medium were analyzed by Southern blotting (Fig. 1B) and PCR analysis (Fig. 1C). As expected, the M strain presented positive hybridization in two fragments of ∼5.5 to 6.5 kb when the ApyapA probe was used (Fig. 1B). This indicates that the ApyapA gene sequence in the M strain was replaced by the deletion cassette that presents an EcoRI restriction site, which is absent in the ApyapA sequence of the WT (Fig. 1A). A unique hybridization signal for a CM strain at a molecular size (∼4.5 kb) lower than that of the WT (∼6.5 kb) is also shown in Fig. 1A. The different size is due to the presence of an EcoRI restriction site in the Hph cassette (Fig. 1A) and indicates (as extensively explained in Materials and Methods) that ApyapA has been correctly reinserted in its locus. The growth of the CM strain (n = 20) treated with 1 mM Men (which severely affected the growth of the M strain) and with hygromycin B (500 ppm) (which was restrictive for the growth of WT and M strains) was not significantly different from that of WT. This confirms the presence of a functioning ApYapA and hygromycin B phosphotransferase in the CM strain. Furthermore, the combination of the primers AmdS_for and ApYapA_rev and of Hph_for and ApyapA_rev2 was positive for PCR amplification (2.1 kb and 0.9 kb in the M and CM strains, respectively) (Fig. 1C). When amdS and hph primers were used, positive amplification (0.7 and 0.65 kb, respectively) appeared only in the M and CM strains, respectively (Fig. 1C).
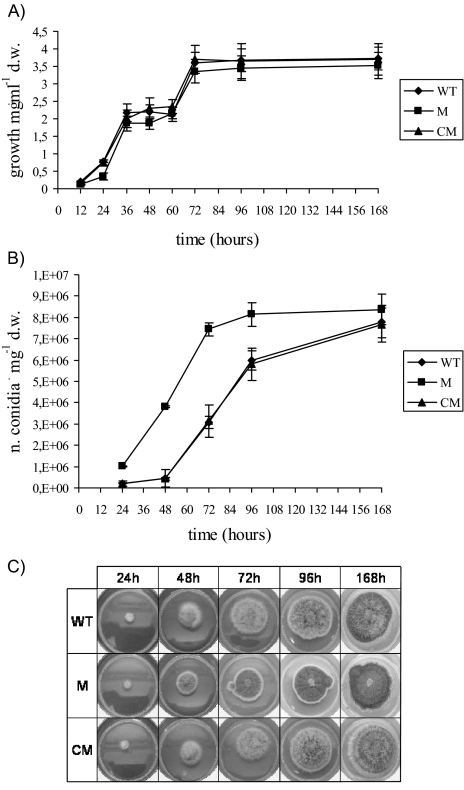
ΔApyapA mutant showed earlier conidium formation than did WT.
The deletion of ApyapA was not directly involved in fungal growth, and yet conidiogenesis was significantly affected. The growth rates of the WT and M strains after 24 h of incubation (Fig. 2A) showed only slight differences. For both, the growth curve presented a biphasic profile: it decreased between 36 and 60 h, whereas the stationary phase was reached 72 h after inoculation. The growth curve of the CM strain did not differ from that of WT (Fig. 2A). With regard to conidiogenesis, the M strain showed a higher number of conidia than did the WT and CM strains, especially between 24 h and 96 h (Fig. 2B and C). However, at 168 h, all three strains had almost the same quantity of conidia. A similar trend has been observed in N. crassa and other fungi, where the undifferentiated vegetative growth is followed by a differentiated status (stimulated by a hyperoxidant condition), in which growth slows down and conidia are formed (1, 2).
FIG. 2.
(A) Mycelial growth (mg [dry weight]·ml−1) of WT, ΔApyapA (M), and ApyapA complemented (CM) strains inoculated in PDB (25 ml) and incubated at 30°C from 12 up to 168 h. (B) Numbers of conidia produced by WT, M, and CM strains cultured under the same experimental conditions. (C) Agar plates (PDA) showing the different timing in conidium formation of WT, M, and CM strains at different time intervals after inoculation (24 to 168 h). The results in panels A and B are the means ± SEMs of three determinations from three separate experiments.
ROS are formed soon after conidium germination and during fungal growth.
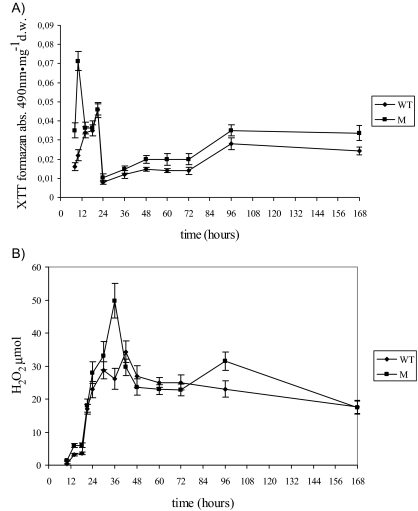
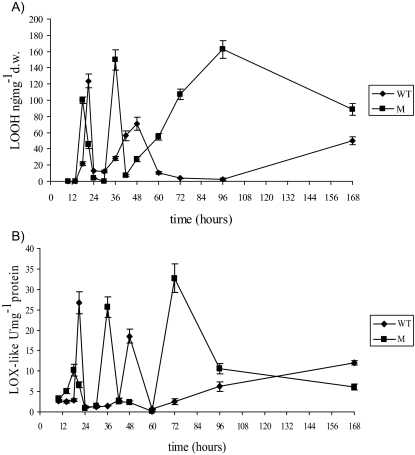
Oxidative stress (i.e., O2− and H2O2 production, the formation of 9- and 13-HODE, and LOX-like activity) was monitored in the mycelia of the WT and M strains between 10 and 168 h (Fig. 3 and 4A and B; Table 1). In both the WT and Μ strains, ROS production occurred quite early (i.e., in the first 24 h of growth), although in the M strain, production occurred earlier and the O2− levels were higher than in the WT strain (Fig. 3A). In the time interval 24 to 60 h, the M strain also had higher levels of H2O2 (about 50 μmol at 36 h, compared to 28 μmol for the WT) and LOOH (about 160 ng·mg−1 at 36 h versus about 80 ng·mg−1 at 48 h in the WT) (Fig. 3B and 4A). After 60 h, for both O2− and H2O2, a slight or nonsignificant difference was observed between the Μ and WT strains, whereas the amount of LOOH was significantly higher for the M strain than for the WT. The trend in LOX-like activity was similar to that for LOOH formation throughout nearly the entire incubation for both the M and the WT strains (Fig. 4A and B).
FIG. 3.
(A) Detection of superoxide anion formation (reported as absorbance of XTT formazan at 490 nm). (B) H2O2 formation (μmol) in mycelia of WT and ΔApyapA (M) strains grown in PDB (25 ml) and incubated at 30°C from 8 to 168 h. The results are the means ± SEMs of three determinations from three separate experiments.
FIG. 4.
(A) 9- and 13-HODE (ng·mg−1 [dry weight]). (B) LOX-like activity measured as diene conjugate formation at 234 nm (U·mg−1 protein) in mycelia of WT and ΔApyapA (M) strains grown in PDB (25 ml) and incubated at 30°C from 10 to 168 h. The results are the means ± SEMs of three determinations from three separate experiments.
TABLE 1.
Relative percentages of 9- and 13-HODE in the mycelia of A. parasiticus WT and ΔApyapA mutant (M) strains
| Time (h) | % HODE (mean ± SEMa) for strain:
|
|||
|---|---|---|---|---|
| WT
|
Μ
|
|||
| 9-HODE | 13-HODE | 9-HODE | 13-HODE | |
| 10 | 3.9 ± 0.5 | 96.1 ± 11.2 | 0.2 ± 0.1 | 99.8 ± 10.2 |
| 14 | 9.1 ± 1.2 | 90.9 ± 11.5 | 46.0 ± 5.2 | 54.0 ± 6.3 |
| 18 | 0.1 ± 0.05 | 99.9 ± 15.2 | 31.0 ± 4.2 | 69.0 ± 7.1 |
| 21 | 3.4 ± 0.4 | 96.6 ± 9.6 | 1.5 ± 0.5 | 98.5 ± 12.2 |
| 24 | 6.3 ± 0.6 | 93.6 ± 8.4 | 0.2 ± 0.1 | 99.8 ± 9.2 |
| 30 | 8.5 ± 0.9 | 91.5 ± 10.2 | 0.1 ± 0.03 | 99.9 ± 8.5 |
| 36 | 10.4 ± 1.1 | 89.6 ± 8.5 | 34.0 ± 4.2 | 66.0 ± 8.5 |
| 42 | 93.9 ± 10.5 | 6.1 ± 1.2 | 87.6 ± 8.2 | 12.4 ± 1.5 |
| 48 | 69.5 ± 6.2 | 30.5 ± 2.1 | 87.2 ± 9.6 | 12.8 ± 1.3 |
| 72 | 36.5 ± 3.9 | 63.5 ± 5.1 | 31.0 ± 3.5 | 69.0 ± 5.2 |
| 96 | 9.80 ± 1.8 | 90.2 ± 10.5 | 23.0 ± 2.6 | 77.0 ± 8.3 |
| 168 | 32.5 ± 4.3 | 67.5 ± 7.5 | 70.0 ± 8.2 | 30.0 ± 5.2 |
The values represent the means ± SEMs of three replicates from three separate experiments.
In fungi, 9- and 13-HPODE can play different physiological roles (63). The 13-HPODE produced by maize LOX inhibits the expression of aflatoxin-related genes in Aspergillus, whereas 9-HPODE stimulates expression (6). To determine whether these two regioisomers were present in A. parasiticus mycelia and played a differential role in regulating aflatoxin biosynthesis, their amount (relative percentage) was analyzed by liquid chromatography-MS (Table 1). At 42 to 48 h, the amount of 9-HODE in both the WT and M strains was significantly higher than that of 13-HODE. However, after 48 h, the amount was higher for 13-HODE.
ΔApyapA inefficiently scavenged ROS in comparison with WT.
In previous studies (53, 54), we showed that fungal cells activated antioxidant systems in response to oxidative stressors and cell aging. In the present study, to demonstrate the role of ApYapA in the control of antioxidant defenses, we carried out a comparison between the WT and M strains, performing experiments with a narrower time interval than that used in the previous studies.
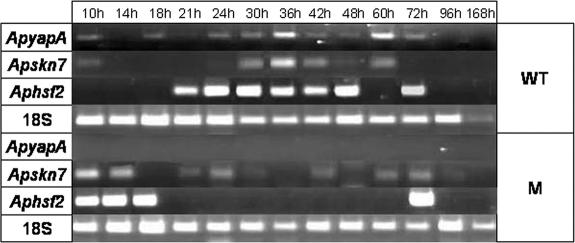
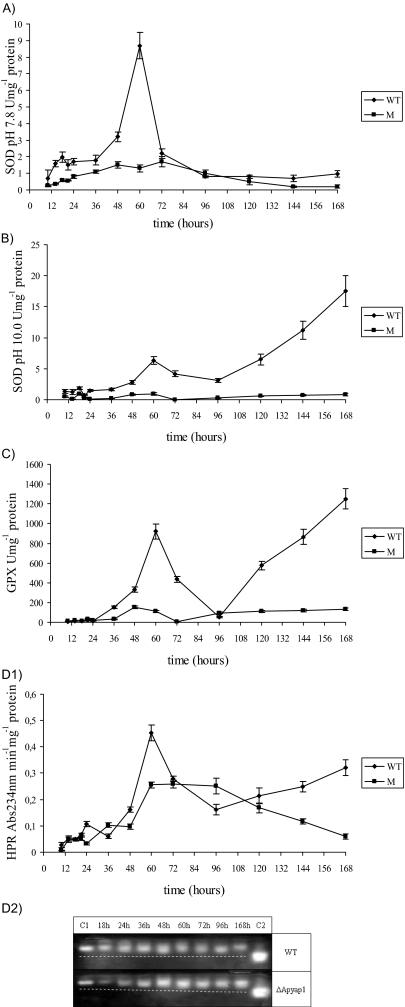
Expression of mRNA was analyzed for three genes (ApyapA, Apskn7, and Aphsf2) whose orthologues are involved in the response to oxidative stress in other fungi (16). In the WT, ApyapA mRNA was expressed from 10 h to 72 h and Apskn7 mRNA was expressed at 10 h and from 30 h to 60 h, whereas Aphsf2 mRNA was expressed from 21 to 48 and 72 h (Fig. 5). In the M strain, at 10 h, there was marked expression of Apskn7 and Aphsf2 mRNA (Fig. 5), which could be the result of an attempt to make up for the lack of ApYapA; in fact, Skn7 can contribute to enhancing CAT gene expression (39), whereas Hsf2 can promote the expression of metallothioneins, which are also involved in ROS scavenging (16). The CM strain showed an amplification profile of ApyapA mRNA that was not significantly different from that of the WT (data not shown). Interestingly, from 0 h to 72 h, the expression of these transcription factors was correlated with antioxidant enzyme activation (SOD, HPR, and GPX) in the fungal cell, and all of the enzyme activities were lower in the M strain than in the WT (Fig. 6A to D1). In the WT, SOD (pH 7.8 and 10.0) and HPR were already activated at 10 h and showed higher levels than those in the M strain. SOD, GPX, and HPR activities peaked at 60 h (Fig. 6A to D1); afterwards, SOD activity at pH 7.8 decreased, whereas SOD at pH 10.0 showed a steep increase beginning at 96 h. SOD activity at basic pH can be peroxisomal or mitochondrial (64). The insufficient response of antioxidant enzymes to the LOOH (very low GPX activity) led to the use of α-tocopherol in the WT mycelia at early points in time. In fact, the amount of α-tocopherol was 12.5 ng·mg−1 at 24 h, followed by a progressive decrease. This compound, which is the most prominent lipophilic antioxidant (66), is thus able to scavenge the excess of LOOH produced at early points in time. In the WT, the marked increase in SOD at pH 10.0 after 96 h could reflect a strategy of the cell to defend mitochondria and peroxisomes from the ROS attack. GPX activity in the WT decreased after 60 h and increased between 96 and 168 h. HPR activity decreased after peaking at 60 h, with a slight increase at 96 h. In general, it was evident that the rate of increase in enzymatic activities was lower from 96 to 168 h than that from 36 to 72 h. In the M strain, between 48 and 72 h, SOD and GPX activity (Fig. 6A to C) and α-tocopherol content (5.8 ng·mg−1 at 24 h) were significantly lower than those in the WT, especially at 60 h, when a difference in HPR activity was also observed (Fig. 6D1). In the M strain, the defective perception of oxidative stress resulted in a less efficient defensive response in the fungal cell.
FIG. 5.
RT-PCR analysis of oxidative stress transcription factor (ApyapA, Apskn7, and Aphsf2) mRNA in A. parasiticus mycelia from WT and M strains, grown in PDB after different periods of incubation. The results are representative of three separate experiments.
FIG. 6.
Antioxidant enzyme SOD, GPX (U·mg−1 protein), and HPR (absorbance at 234 nm min−1·mg−1 protein) activities in mycelia of A. parasiticus WT and ΔApyapA (M) strains grown in PDB at different time intervals of incubation at 30°C. (A and B) SOD activity at pH 7.8 (A) and pH 10.0 (B); (C) GPX activity; (D1) HPR activity; (D2) zymogram analysis of WT and M strain protein extracts obtained from mycelia grown for different time periods at 30°C in PDB and fractionated in a native polyacrylamide gel stained for CAT activity. C1 represents a low-acidic conformer of CAT derived from A. niger, and C2 represents the same CAT oxidized under an O2 stream, giving a more-acidic form. The dashed white line represents the trend (from low- to high-acidic form) of CAT conformers during the different time intervals. The results in panels A to D1 are the means ± SEMs of three determinations from three separate experiments.
The zymograms of the major CAT in the WT and M mycelia are shown in Fig. 6D2. According to other studies (33, 37), CATs are present in fungal mycelia, and their electrophoretic mobility (EM) slightly changes during fungal development and in the presence of oxidative stress in the cell. In our study, one CAT isoform, which was faintly detectable and had a very high molecular weight, did not seem to have been affected by oxidants during fungal growth in either of the strains, whereas the major isoform, which had a higher EM, seemed to have been slightly altered by oxidants. According to a previous study (37), the CAT can be oxidized by ROS and the oxidized form presents a different EM. In our study, in the M strain at 18 h, the EM of the CAT was similar to that of the standard CAT C1; afterwards, the EM increased. In the WT, at 18 h, the CAT had the same EM as that of C1, and at 48 h, a more-acidic form was observed, which could represent a partially oxidized form. At 168 h, the EM of the CAT was very similar to that at 18 h (Fig. 6D2).
ΔApyapA is more sensitive to oxidative stressors than the WT is.
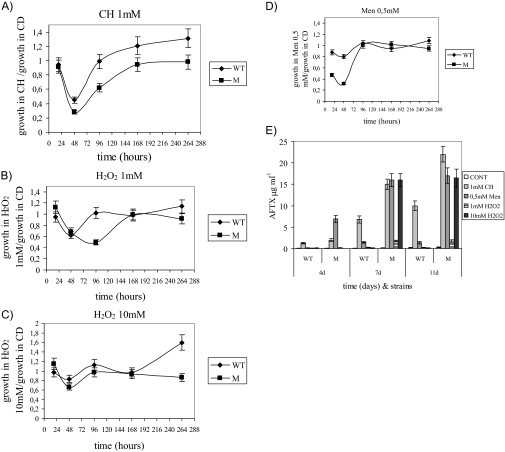
To assess whether the M strain, which lacks an efficient antioxidant defense, was more sensitive than the WT to the presence of oxidants in the environment, several oxidants were added to liquid media. The Μ strain was more susceptible than the WT to all of the oxidants tested, in terms of both growth and aflatoxin biosynthesis. Growth was most influenced by CH (1 mM) and Men (0.5 mM) (Table 2; Fig. 7A to D). Regarding aflatoxin biosynthesis, the M strain was more influenced than the WT at all points in time (4, 7, and 11 days), and the compounds with the greatest stimulating effect were 1 mM CH, 0.5 mM Men, and the highest concentration of hydrogen peroxide (10 mM [Fig. 7E]). These results confirm that the alteration of oxidant perception and the absence of a substantial antioxidant defense are closely related to aflatoxin stimulation.
TABLE 2.
Fungal growth of the WT and ΔApyapA (M) strains in CD and in CD amended with 1 mM CH, 0.5 mM Men, or 1 and 10 mM H2O2
| Strain type and time (h) | Fungal growtha (mg [dry wt] ml−1) in CD amended with:
|
||||
|---|---|---|---|---|---|
| Control | 1 mM CH | 1 mM H2O2 | 10 mM H2O2 | 0.5 mM Men | |
| WT | |||||
| 18 | 2.4 ± 0.3 | 2.3 ± 0.5 | 1.6 ± 0.1 | 1.4 ± 0.1 | 2.1 ± 0.1 |
| 48 | 6.0 ± 0.5 | 2.7 ± 0.4 | 3.7 ± 0.5 | 5.0 ± 0.6 | 4.8 ± 0.2 |
| 96 | 7.2 ± 0.7 | 7.1 ± 0.8 | 7.3 ± 0.5 | 8.1 ± 0.8 | 7.5 ± 0.8 |
| 168 | 7.0 ± 0.8 | 8.5 ± 0.9 | 6.8 ± 0.8 | 8.5 ± 0.8 | 6.6 ± 0.5 |
| 264 | 6.3 ± 1.2 | 7.0 ± 1.5 | 6.1 ± 1.2 | 6.8 ± 0.5 | 5.8 ± 1.0 |
| M | |||||
| 18 | 1.6 ± 0.2 | 1.5 ± 0.2 | 1.8 ± 0.1 | 2.1 ± 0.4 | 0.8 ± 0.1 |
| 48 | 7.0 ± 0.5 | 2.0 ± 0.1 | 4.8 ± 0.2 | 4.6 ± 0.4 | 2.2 ± 0.1 |
| 96 | 9.4 ± 0.8 | 5.9 ± 0.2 | 4.6 ± 0.4 | 9.1 ± 0.4 | 9.4 ± 1.2 |
| 168 | 9.4 ± 1.0 | 8.9 ± 0.5 | 9.4 ± 0.8 | 8.8 ± 0.8 | 9.6 ± 0.7 |
| 264 | 8.3 ± 1.5 | 8.1 ± 0.6 | 7.6 ± 1.0 | 7.1 ± 1.1 | 7.9 ± 1.2 |
The values represent the means ± SEMs of three replicates from three separate experiments.
FIG. 7.
(A to D) Ratios between the fungal growth (mg [dry weight]·ml−1) of WT and ΔApyapA (M) strains in CD amended with CH (1 mM) (A) or 1 (B) or 10 (C) mM H2O2 or Men (0.5 mM) (D) and their growth in CD (control). (E) Aflatoxin (AFTX) production in culture filtrate (μg·ml−1) of WT and M strains grown in media amended with different stressors (CH, 1 mM; Men, 0.5 mM; H2O2, 1 and 10 mM), at different time intervals (4 to 11 days). All the compounds were added after 4 days of incubation at 30°C in CD medium. The results are the means ± SEMs of three determinations from three separate experiments.
ApyapA affects aflatoxin biosynthesis.
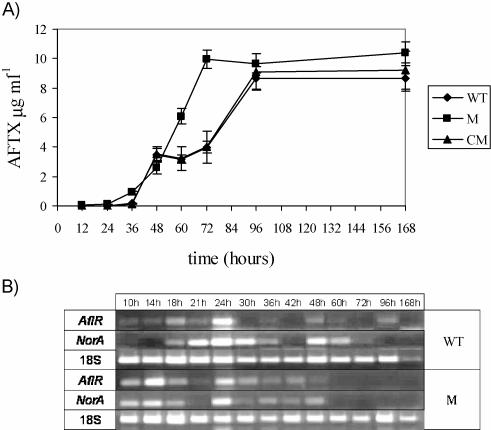
In the WT, between 12 h and 36 h, aflatoxin biosynthesis was lacking or very low, whereas in the M strain it was already observed at 12 h (42 ng·ml−1) (Fig. 8A). Between 48 h and 72 h, the concentration of aflatoxins was significantly higher in the M strain than in the WT. Between 96 and 168 h, although the concentration remained higher for the M strain, the difference was not always significant. In the CM strain, aflatoxin biosynthesis followed the same trend as that in the WT. In the M strain, the expression of aflR and norA mRNA occurred earlier than in the WT (Fig. 8B) and it was highly correlated with the early accumulation of aflatoxin in the medium (Fig. 7E and 8A). In the WT, aflatoxin biosynthesis decreased between 48 and 72 h, soon after fungal vegetative growth began to decline (36 to 60 h) (Fig. 2A). At the same time (36 to 60 h), a higher activity level of antioxidant enzymes was observed (Fig. 6A to D1). At 60 h, all of the enzymes showed a peak in their activities, and at the same time aflatoxin biosynthesis decreased in the WT but not in the M strain, where the enzymatic activities were significantly lower. That there exists an association between the antioxidant/oxidant balance and aflatoxin biosynthesis, probably driven by ApYapA, was also suggested by the results of the in silico analysis of the aflR promoter sequence. This analysis was performed using the N_SITE tool in the Softberry software package, which allowed us to reveal all of the putative regulatory elements present in the promoter region, which were compared with the human N_SITE database. The results obtained showed that the aflR promoter presented diverse regulatory elements, which were similar to those recognized by some OSRTFs in humans, such as AP1 (SiteID, S02349; SiteName, ENKCRE-2, P < 0.05), NF-κΒ (SiteID, S05669; SiteName, NF-κB-E-selectin, P < 0.05), and Rox1 (SiteID, S06246; SiteName, Rox1-HEM13-3, P < 0.01). This suggests that the aflatoxin regulator gene expression could also be affected by oxidative stress.
FIG. 8.
(A) Aflatoxin (AFTX) production in culture filtrate (μg ml−1) of A. parasiticus WT, ΔApyapA (M), and complemented ApyapA (CM) strains inoculated into aflatoxin-conducive medium (PDB). (B) aflR and norA mRNA RT-PCR analysis in A. parasiticus mycelia after different time intervals. The aflatoxin results are the means ± SEMs of three determinations from three separate experiments.
DISCUSSION
In recent years, many authors have found evidence of a close association among oxidative stress, development, differentiation, and secondary metabolism in fungi (1, 2, 27, 54, 68). From these studies it has emerged that the levels of oxidants are finely regulated in fungal cells. In yeast, the maintenance of a favorable redox balance is under the control of Yap1. This factor acts as a sensor of the cell's redox state through a cysteine-rich domain, and it regulates the activation of different antioxidant defense-related genes, such as sod, cat, and gpx (16, 39). In other fungi, such as Candida albicans and Cochliobolus heterostrophus, proteins that are orthologues to Yap1 control the expression of a similar set of genes that are needed to respond to oxidative stress (15, 40).
In A. parasiticus, we found that oxidative stress (i.e., O2−, H2O2, and LOOH formation) occurred soon after conidium germination and during growth. Oxidative perturbation promotes defensive responses such as the expression of OSRTF mRNA, enzymes, and compounds with antioxidant activity. Their modulation leads to metabolic consequences, including an effect on aflatoxin biosynthesis. In A. parasiticus NRRL 2999, ROS production was evident quite early (i.e., 8 to 10 h after inoculation in liquid medium), which is consistent with the findings of a study of N. crassa (1), though different ROS were considered. The production could be due to the emergence of germination tubes which suddenly expose conidia to O2 and/or to the marked increase in metabolic activity. In our study, O2− was formed early on, followed by an increase in H2O2 and LOOH levels after 18 h. LOOH formation can be ascribed both to a LOX-like activity (which also showed a very early activation) and, indirectly, to H2O2 (via a Fenton reaction). In fungi, LOOH could act as a modulator of differentiation events. In other studies of Aspergillus spp. (6, 67), 9-HODE and 13-HODE have been reported to have different physiological roles, although the intracellular detection of regioisomers in relation to aflatoxin biosynthesis has not yet been studied. In our study, the correlation between endogenous levels of 9- and 13-HODE and aflatoxin biosynthesis was not straightforward and needs to be investigated further.
The early accumulation of ROS results in the activation of antioxidant defense mechanisms, through the expression of ApyapA, Apskn7, and Aphsf2 mRNA. In A. parasiticus, it is likely that the putative activation of the above transcription factors organizes intracellular defensive machinery, which includes antioxidant enzymes, such as SODs, HPR, and GPX (which were prevalent at 60 h), and α-tocopherol (at early points in time). In N. crassa, antioxidant activities inhibit cell differentiation, whereas high levels of ROS are required to trigger this process (1). In yeast, the ROS which overwhelmed the endogenous antioxidant system are able to regulate cell aging (66). In our study, in the WT, antioxidant enzyme activity peaked between 48 and 60 h, soon after the increase of oxidants within the cell (36 to 48 h), though a peak in α-tocopherol was present at 24 h. In the same time interval, the hyphal growth rate decreased (36 to 60 h), cell differentiation occurred (as demonstrated by the appearance of conidia [48 h]), and secondary metabolism (36 h) switched on, leading to aflatoxin biosynthesis.
In previous studies (21, 53, 54), we investigated the associations among the expression of OSRTFs, antioxidant responses, and aflatoxin biosynthesis. The results showed that antioxidants inhibited aflatoxin biosynthesis and enhanced ApyapA mRNA expression. In the present study, we describe the role of the Yap1 orthologue, ApYapA, in regulating cell differentiation and aflatoxin biosynthesis following ROS formation and the activation of antioxidant defensive mechanisms. The theoretical translation of the gene sequence of ApyapA shares high similarities with orthologues in A. fumigatus (62% identities and 100% similarities, score of 268), with CHAP1 in C. heterostrophus (53% identities and 79% similarities, score of 128), and with Yap1 in S. cerevisiae (54% identities and 73% similarities, score of 48.5) (see Fig. S1A in the supplemental material).
The presence of regulatory elements in the aflR promoter responsive to AP1 (the human orthologue of Yap1) may suggest that oxidative stress exerts control in the modulation of aflatoxin biosynthesis. Thus, while ApYapA activates antioxidant defenses, it can contribute to the early decrease in aflR and norA mRNA expression. In relation to this, the lack of ApYapA could lead to the enhancement of the expression of aflatoxin-related genes in the mutant strain at the same points in time. In fission yeast, some transcription factors related to sexual differentiation can function both as activators of the expression of specific genes and simultaneously as repressors of the expression of others. In this way, the regulatory options of the cell to express specific gene sets to face different metabolic situations are enhanced (44). Furthermore, the similarity between the A. parasiticus ApyapA and S. cerevisiae yap1 suggests that the gene is involved in modulating the response to oxidative stress. This similarity was also suggested by the presence of 10 well-conserved cysteine residues in the ApyapA AA-deduced sequence, revealed by the Softberry P_SITE analysis. To support this evidence, we used a loss-of-function approach by producing a ΔApyapA mutant of A. parasiticus which strengthens the relationship among oxidative stress, the activation of antioxidant defense mechanisms, cell differentiation, and mycotoxin biosynthesis. In the M strain, ROS production occurred earlier and to a greater extent than in the WT. Consistently, the oxidative signal triggered modest antioxidant defense. The residual antioxidant activity was probably the result of the early enhancement of Apskn7 and Aphsf2 mRNA transcription. In S. cerevisiae, other transcription factors (e.g., Prr1, a homologue of Skn7) are regulated by oxidative stress and, in turn, induce the expression of antioxidant enzyme activities, such as the CAT CTT1 and the SOD SOD1 (39). Moreover, in our study, the CAT zymogram showed that in the M strain the enzyme seems to undergo some oxidation, which is responsible for the slight alteration of the EM during fungal growth. Although the zymogram results did not allow us to reach definitive conclusions concerning the role of the CAT in total HPR activity during oxidative stress, the total HPR activity was less affected than were the activities of other antioxidant enzymes in the M strain; in fact, it has been hypothesized that the oxidation of the tetrapyrrolic ring does not alter CAT activity (37).
Nonetheless, the remaining antioxidant activities were insufficient to scavenge all of the ROS formed, forcing the cell to face a hyperoxidant status and respond by activating earlier cell differentiation and secondary metabolism. In fact, in ΔApyapA aflR and norA, mRNA transcription was enhanced soon after conidium germination, conidiogenesis occurred early and increased starting at 24 h, and aflatoxins were already detectable in culture medium at 12 h. The formation of ROS was slightly delayed in the WT compared to the M strain (delay of 6 to 12 h), which had effects on gene activation, such as a delay in the OSRTF mRNA expression (Apskn7 and Aphsf2) and in the transcription of aflatoxin-related genes (aflR and norA). In relation to this, aflatoxins did not appear before 36 h.
In the M strain, an impaired perception of oxidative stress and a malfunctioning of its signaling led to a defective antioxidant defense response and the consequent stimulation of aflatoxin biosynthesis. This could explain why this strain is more sensitive to different stressors, such as CH and Men, as well as hydrogen peroxide.
Considering our results as whole, similar phases can be envisioned during the growth of A. parasiticus NRRL 2999. Each phase consists of an oxidative burst which triggers the expression of ApyapA, whose product modulates the activation of antioxidant enzymes. Excessive ROS are probably able to trigger aflatoxin biosynthesis, though the underlying mechanism is still unclear. During the growth phase, as soon as the activity of antioxidant enzymes decreases, aflatoxin biosynthesis begins. This hypothesis is supported by our finding that this kind of modulation was not found in ΔApyapA. In this strain, the antioxidant defenses were less effective throughout the time considered.
We demonstrated that oxidative stress, generated either within or outside of the cell, affects aflatoxin formation in A. parasiticus. ApyapA appears to play a significant role in the modulation and maintenance of an appropriate balance between oxidant and antioxidant species and aflatoxin biosynthesis. A complete comprehension of the mechanism by which the carcinogenic aflatoxins are synthesized by the fungus is instrumental in designing appropriate strategies for controlling their production and release into the environment.
Supplementary Material
Footnotes
Published ahead of print on 25 April 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aguirre, J., M. Rios-Momberg, D. Hewitt, and W. Hansberg. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13111-118. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre, J., W. Hansberg, and R. Navarro. 2006. Fungal responses to reactive oxygen species. Med. Mycol. 44(Suppl.)101-107. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, R. S., J. Shi, E. Murad, A. M. Whalen, C. Q. Sun, R. Polavarapu, S. Parthasarathy, J. A. Petros, and J. D. Lambeth. 2001. Hydrogen peroxide mediates the cell growth and transformation caused by mitogenic oxidase Nox1. Proc. Natl. Acad. Sci. USA 985550-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bokoch, G. M., and U. G. Knaus. 2003. NADPH oxidase: not just for leukocytes anymore! Trends Biochem. Sci. 28502-508. [DOI] [PubMed] [Google Scholar]
- 5.Bolwell, G. P., L. V. Bindschedler, K. A. Blee, V. S. Butt, D. R. Davies, S. L. Gardner, C. Gerrish, and F. Minibayeva. 2002. The apoplastic oxidative burst in response to biotic stress in plants: a three-component system. J. Exp. Bot. 531367-1376. [PubMed] [Google Scholar]
- 6.Burow, G. B., T. C. Nesbitt, J. Dunlap, and N. P. Keller. 1997. Seed lipoxygenase products modulate Aspergillus mycotoxin biosynthesis. Mol. Plant-Microbe Interact. 10380-387. [Google Scholar]
- 7.Calvo, A. M., L. L. Hinze, H. W. Gardner, and N. P. Keller. 1999. Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 653668-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo, A. M., H. W. Gardner, and N. P. Keller. 2001. Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans. J. Biol. Chem. 27625766-25774. [DOI] [PubMed] [Google Scholar]
- 9.Calvo, A. M., R. A. Wilson, J. W. Bok, and N. P. Keller. 2002. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66447-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castoria, R., L. Caputo, F. De Curtis, and V. De Cicco. 2003. Resistance of postharvest biocontrol yeast to oxidative stress: a possible new mechanism of action. Phytopathology 93564-572. [DOI] [PubMed] [Google Scholar]
- 11.Ceaser, E. K., D. R. Moellering, S. Shiva, A. Ramachandran, A. Landar, A. Venkartraman, J. Crawford, R. Patel, D. A. Dichinson, E. Ulasova, S. Ji, and V. M. Darley-Usmar. 2004. Mechanisms of signal transduction mediated by oxidated lipids: the role of the electrophile-responsive proteome. Biochem. Soc. Trans. 32151-155. [DOI] [PubMed] [Google Scholar]
- 12.Champe, S. P., P. Rao, and A. Chang. 1987. An endogenous inducer of sexual development in Aspergillus nidulans. J. Gen. Microbiol. 1331383-1387. [DOI] [PubMed] [Google Scholar]
- 13.Champe, S. P., and A. A. el-Zayat. 1989. Isolation of a sexual sporulation hormone from Aspergillus nidulans. J. Bacteriol. 1713982-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, P. K., J. W. Bennett, and P. J. Cotty. 2002. Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia 15341-48. [DOI] [PubMed] [Google Scholar]
- 15.Delaunay, A. D., M. Pflieger, M. Barrault, J. Vinh, and M. Toledano. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111471-481. [DOI] [PubMed] [Google Scholar]
- 16.Estruch, F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24469-486. [DOI] [PubMed] [Google Scholar]
- 17.Fabbri, A. A., C. Fanelli, G. Panfili, S. Passi, and P. Fasella. 1983. Lipoperoxidation and aflatoxin biosynthesis by Aspergillus parasiticus and A. flavus. J. Gen. Microbiol. 1293447-3452. [Google Scholar]
- 18.Fabbri, A. A., C. Fanelli, M. Reverberi, A. Ricelli, E. Camera, S. Urbanelli, A. Rossini, M. Picardo, and M. M. Altamura. 2000. Early physiological and cytological events induced by wounding in potato tuber. J. Exp. Bot. 511267-1275. [PubMed] [Google Scholar]
- 19.Fanelli, C., A. A. Fabbri, E. Finotti, and S. Passi. 1983. Stimulation of aflatoxin biosynthesis by lipophilic epoxides. J. Gen. Microbiol. 1291721-1723. [DOI] [PubMed] [Google Scholar]
- 20.Fanelli, C., A. A. Fabbri, E. Finotti, P. Fasella, and S. Passi. 1984. Free radicals and aflatoxin biosynthesis. Experientia 40191-193. [Google Scholar]
- 21.Fanelli, C., A. A. Fabbri, S. Pieretti, E. Finotti, and S. Passi. 1985. Effect of different antioxidants and free radical scavengers on aflatoxin production. Mycol. Res. 165-69. [DOI] [PubMed] [Google Scholar]
- 22.Fanelli, C., and A. A. Fabbri. 1989. Relationship between lipids and aflatoxin biosynthesis. Mycopathologia 107115-120. [DOI] [PubMed] [Google Scholar]
- 23.Fanelli, C., V. Tasca, A. Ricelli, M. Reverberi, S. Zjalic, E. Finotti, and A. A. Fabbri. 2000. Inhibiting effect of medicinal mushroom Lentinus edodes (Berk.) Sing (Agaricomycetideae) on aflatoxin production by Aspergillus parasiticus Speare. Int. J. Med. Mushrooms 2229-236. [Google Scholar]
- 24.Feussner, I., and C. Wasternack. 2002. The lipoxygenase pathway. Annu. Rev. Plant Biol. 53275-297. [DOI] [PubMed] [Google Scholar]
- 25.Han, K. H., K. Y. Han, J. H. Yu, K. S. Chae, K. Y. Jahng, and D. M. Han. 2001. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol. Microbiol. 41299-309. [DOI] [PubMed] [Google Scholar]
- 26.Harman, D. 1956. Aging, a theory based on free radical and radiation chemistry. J. Gerontol. 11298-300. [DOI] [PubMed] [Google Scholar]
- 27.Hicks, J. K., J. H. Yu, N. P. Keller, and T. H. Adams. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein-dependent signaling pathway. EMBO J. 164916-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh, K., K. I. Tong, and M. Yamamoto. 2004. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 361208-1213. [DOI] [PubMed] [Google Scholar]
- 29.Jayashree, T., and C. Subramanyam. 2000. Oxidative stress as a prerequisite for aflatoxin production by Aspergillus parasiticus. Free Radic. Biol. Med. 29981-985. [DOI] [PubMed] [Google Scholar]
- 30.Jimenez, A., L. Mateos, J. R. Pedrajas, A. Miranda-Vizuete, and J. L. Revuelta. 2007. The txl1(+) gene from Schizosaccharomyces pombe encodes a new thioredoxin-like 1 protein that participates in the antioxidant defence against tert-butyl hydroperoxide. Yeast 24481-490. [DOI] [PubMed] [Google Scholar]
- 31.Jones, A. M. 1994. Surprising signalling in plant cells. Science 263183-184. [DOI] [PubMed] [Google Scholar]
- 32.Kappus, H. 1985. Lipid peroxidation: mechanism, analysis, enzymology and biological relevance, p. 273-310. In H. Sies (ed.), Oxidative stress. Academic Press, London, United Kingdom.
- 33.Kawasaki, L., and J. Aguirre. 2001. Multiple catalase genes are differentially regulated in Aspergillus nidulans. J. Bacteriol. 1831434-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, J. H., B. C. Campbell, J. Yu, N. Mahoney, K. L. Chan, R. J. Molyneux, D. Bhatnagar, and T. E. Cleveland. 2005. Examination of fungal stress response genes using Saccharomyces cerevisiae as a model system: targeting genes affecting aflatoxin biosynthesis by Aspergillus flavus Link. Appl. Microbiol. Biotechnol. 67807-815. [DOI] [PubMed] [Google Scholar]
- 35.Lambeth, J. D. 2004. Nox enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4181-189. [DOI] [PubMed] [Google Scholar]
- 36.Lara-Ortiz, T., H. Riveros-Rosas, and J. Aguirre. 2003. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 501241-1255. [DOI] [PubMed] [Google Scholar]
- 37.Lledias, F., P. Rangle, and W. Hansberg. 1998. Oxidation of catalase by singlet oxygen. J. Biol. Chem. 27310630-10637. [DOI] [PubMed] [Google Scholar]
- 38.Lledias, F., P. Rangle, and W. Hansberg. 1999. Singlet oxygen is part of an hyperoxidant state generated during spore germination. Free Radic. Biol. Med. 261396-1404. [DOI] [PubMed] [Google Scholar]
- 39.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 27416040-16046. [DOI] [PubMed] [Google Scholar]
- 40.Lev, S. R., P. Hadar, S. E. A. Baker, O. C. Yoder, and B. A. Horwitz. 2005. Activation of an AP1-like transcription factor of the maize pathogen Cochliobolus heterostrophus in response to oxidative stress and plant signals. Eukaryot. Cell 4443-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorito, M., C. K. Hayes, A. Di Pietro, and G. E. Harman. 1993. Biolistic transformation of Trichoderma harzianum and Gliocladium virens using plasmid and genomic DNA. Curr. Genet. 24349-356. [DOI] [PubMed] [Google Scholar]
- 42.Malagnac, F., H. Lalucque, G. Lepere, and P. Silar. 2004. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41982-997. [DOI] [PubMed] [Google Scholar]
- 43.Maresca, V., E. Flori, S. Briganti, E. Camera, M. Cario-Andre, A. Taıeb, and M. Picardo. 2006. UVA-induced modification of catalase charge properties in the epidermis is correlated with the skin phototype. J. Investig. Dermatol. 126182-190. [DOI] [PubMed] [Google Scholar]
- 44.Mata, J., A. Wilbrey, and J. Bahler. 2007. Transcription regulatory networks for sexual differentiation in fission yeast. Genome Biol. 8R217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moye-Rowley, W. S. 2003. Regulation of transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryot. Cell 2381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narasaiah, K. W., R. B. Sashidhar, and C. Subramanyam. 2006. Biochemical analysis of oxidative stress in the production of aflatoxin and its precursor intermediates. Mycopathologia 162179-189. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen, T., P. J. Sherratt, and C. B. Pickett. 2003. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 43233-260. [DOI] [PubMed] [Google Scholar]
- 48.Passi, S., C. Fanelli, A. A. Fabbri, E. Finotti, G. Panfili, and M. Nazzarro-Porro. 1985. Effect of halomethanes on aflatoxin induction in cultures of Aspergillus parasiticus. J. Gen. Microbiol. 131687-691. [Google Scholar]
- 49.Passi, S., R. Ricci., E. Aleo, and M. Cocchi. 2005. Oxidative stress, aging and aging-related diseases. Progr. Nutr. 73-22. [Google Scholar]
- 50.Pinkus, R., L. M. Weiner, and V. Daniel. 1996. Role of oxidants and antioxidants in the induction of AP-1, NF-κΒ, and glutathione S-transferase gene expression. J. Biol. Chem. 27113422-13429. [DOI] [PubMed] [Google Scholar]
- 51.Prade, R. A., and W. E. Timberlake. 1993. The Aspergillus nidulans nbrlA regulatory locus consists of overlapping transcription units that are individually required for conidiophore development. EMBO J. 122439-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reverberi, M., M. Picardo, A. Ricelli, E. Camera, C. Fanelli, and A. A. Fabbri. 2001. Oxidative stress, growth factor production and budding in potato tubers during cold storage. Free Radic. Res. 35833-841. [DOI] [PubMed] [Google Scholar]
- 53.Reverberi, M., A. A. Fabbri, S. Zjalic, A. Ricelli, F. Punelli, and C. Fanelli. 2005. Antioxidant enzymes stimulation in Aspergillus parasiticus by Lentinula edodes inhibits aflatoxin production. Appl. Microbiol. Biotechnol. 69207-215. [DOI] [PubMed] [Google Scholar]
- 54.Reverberi, M., S. Zjalic, A. Ricelli, A. A. Fabbri, and C. Fanelli. 2006. Oxidant/antioxidant balance in Aspergillus parasiticus affects aflatoxin biosynthesis. Mycotox. Res. 2239-47. [DOI] [PubMed] [Google Scholar]
- 55.Shackelford, R. E., W. K. Kaufmann, and R. S. Paules. 2000. Oxidative stress and cell cycle check point function. Free Radic. Biol. Med. 281387-1404. [DOI] [PubMed] [Google Scholar]
- 56.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sies, H. 1985. Oxidative stress: introductory remarks, p. 1-8. In H. Sies (ed.), Oxidative stress. Academic Press, London, United Kingdom.
- 58.Spiteller, G. 2001. Peroxidation of linoleic acid and its relation to aging and age dependent diseases. Mech. Ageing Dev. 122617-657. [DOI] [PubMed] [Google Scholar]
- 59.Suh, Y. A., R. S. Arnold, B. Lassegue, J. Shi, X. Xsu, D. Sorescu, A. B. Chung, K. K. Griendling, and J. D. Lambeth. 1999. Cell transformation by the superoxide-generating oxidase Mox1. Nature 40179-82. [DOI] [PubMed] [Google Scholar]
- 60.Sutherland, M. W., and B. A. Learmonth. 1997. The tetrazolium dyes MTS and XTT provide new quantitative assays for superoxide and superoxide dismutase. Free Radic. Res. 27283-289. [DOI] [PubMed] [Google Scholar]
- 61.Talalay, P., A. T. Dinkova-Kostova, and W. D. Holzelaw. 2003. Importance of phase 2 regulation in protection against electrophile and reactive oxygen toxicity and carcinogenesis. Adv. Enzyme Regul. 43121-134. [DOI] [PubMed] [Google Scholar]
- 62.Torres, M. A., J. L. Dangl, and J. D. Jones. 2002. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsitsigiannis, D., and N. P. Keller. 2007. Oxylipins as developmental and host-fungal communication signals. Trends Microbiol. 15109-118. [DOI] [PubMed] [Google Scholar]
- 64.Van Roermund, W. T., M. De Jong, L. Ijlst, J. van Marie, T. B. Dansen, R. J. A. Wanders, and H. R. Waterham. 2004. The peroxisomal lumen in Saccharomyces cerevisiae is alkaline. J. Cell Sci. 1174231-4237. [DOI] [PubMed] [Google Scholar]
- 65.Vignais, P. V. 2002. The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell. Mol. Life Sci. 591428-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilhelm, J., H. Fuksova, Z. Shwippelova, R Vytasek, and A. Pichova. 2006. The effects of reactive oxygen and nitrogen species during yeast replicative ageing. Biofactors 27185-193. [DOI] [PubMed] [Google Scholar]
- 67.Wilson, R. A., H. W. Gardner, and N. P. Keller. 2001. Cultivar-dependent expression of a maize lipoxygenase responsive to seed infesting fungi. Mol. Plant-Microbe Interact. 14980-987. [DOI] [PubMed] [Google Scholar]
- 68.Yu, J., and N. P. Keller. 2005. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43437-458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.