Abstract
The lack of an experimentally amenable sexual genetic system in Aspergillus fumigatus is a major limitation in the study of the organism's pathogenesis. A recent comparative genome analysis revealed evidence for potential sexuality in A. fumigatus. Homologs of mating type genes as well as other genes of the “sexual machinery” have been identified in anamorphic A. fumigatus. The mat1-2 gene encodes a homolog of MatA, an HMG box mating transcriptional factor (MatHMG) that regulates sexual development in fertile Aspergillus nidulans. In this study, the functionalities of A. fumigatus mat1-2 and the Mat1-2 protein were determined by interspecies gene exchange between sterile A. fumigatus and fertile A. nidulans. Ectopically integrated A. fumigatus mat1-2 (driven by its own promoter) was not functional in a sterile A. nidulans ΔmatA strain, and no sexual development was observed. In contrast, the A. fumigatus mat1-2 open reading frame driven by the A. nidulans matA promoter and integrated by homologous gene replacement at the matA locus was functional and conferred full fertility. This is the first report showing that cross species mating type gene exchange between closely related Ascomycetes did not function in sexual development. This is also the first report demonstrating that a MatHMG protein from an asexual species is fully functional, with viable ascospore differentiation, in a fertile homothallic species. The expression of mat1-2 was assessed in A. fumigatus and A. nidulans. Our data suggest that mat1-2 may not be properly regulated to allow sexuality in A. fumigatus. This study provides new insights about A. fumigatus asexuality and also suggests the possibility for the development of an experimentally amenable sexual cycle.
Mating type genes (mat loci) have been characterized in a number of homothallic, heterothallic, and asexual filamentous Ascomycetes. In fertile fungi, the mat locus functions as a master regulatory locus controlling sexual reproduction (8-10, 21). Aspergillus (Emericella) nidulans is a homothallic, self-fertile ascomycete. It has both sexual and asexual reproductive strategies and provides a robust genetic system for the study of eukaryotic development and cell biology (6, 35). Sexual reproduction in A. nidulans is a complex multistep process that involves coordinated differentiation of three tissue types: Hülle cells, ascogenous hyphae, and cleistothecium wall. Hülle cells function as nurse cells for fruiting body development in A. nidulans, which forms mature cleistothecia that are hard and highly pigmented (6). However, Hülle cells are not required for sexual reproduction per se and, Hülle cell production varies among A. nidulans strains. Other fertile Aspergillus species lack Hülle cells and produce cleistothecia that are soft and lightly pigmented (6). Sexual conjugation is believed to occur within the foci of Hülle cells, where mating and fertilization between cells that are equivalents of male and female gametangia take place. Upon fertilization, parental nuclei divide synchronously and ascogenous hyphae proliferate within the female organ. Pairs of nuclei are ultimately segregated into dikaryotic cells where karyogamy, meiosis, and two additional mitoses lead to the formation of asci filled with eight binucleate ascospores. Vegetative hyphae of female origin grow in a circular fashion surrounding the fertile ascogenous tissue and eventually form the hard cleistothecial wall (3, 4, 6, 48). As in other fertile Ascomycetes, the mating type regulatory genes matA (HMG box) and matB (alpha box) are required for mating and completion of sexual development, with matA being required to initiate fruiting body development (matA, accession number AY328028 and locus identification number [ID] AN4734; matB, accession number AY399600 and locus ID AN2755) (B. L. Miller and K. Y. Miller, unpublished data).
Aspergillus fumigatus is an opportunistic human pathogen and major cause of life-threatening invasive aspergillosis in immunodeficient individuals, with an overall mortality rate of about 50%. The species has been classified among the “imperfect fungi” (17). No sexual cycle has been observed to date. The lack of a sexual cycle significantly limits the application of genetic analyses to the study of mechanisms involved in pathogenesis. However, a high degree of genetic diversity among clinical and environmental isolates suggests existing latent, recently lost, or rare sexual activity within populations of A. fumigatus. (11, 13, 15, 32, 36, 37). Comparative genome analysis provides further evidence for sexual ability in A. fumigatus. The homologs of mating type genes (mat1-1 and mat1-2), as well as all key components of the core meiotic program, pheromone signaling, and genes involved in fruiting body development have been identified in A. fumigatus (12, 45). Two complementary A. fumigatus mating types with heterothallic structures at the mat locus are equally distributed in nature (13). The expression of putative mating type genes and other sex-related genes have been detected at low abundance during mycelial growth (29). However, neither pheromone nor pheromone receptor genes are expressed in a mating type-specific pattern. It is possible that the lack of A. fumigatus sexuality could be a result of a mutation in one of the key genes of the “sexual machinery” that blocks sexual development. A study of the mating type genes in sterile A. fumigatus is essential for understanding the basis for asexuality and may provide important insights for potential reconstruction or induction of a viable sexual cycle. The availability of an experimentally amenable sexual state in A. fumigatus would have a major impact upon the study of the pathogenesis and biology of this species.
The A. fumigatus mat1-2 gene encodes a homolog of the A. nidulans MatA, an HMG box mating type transcriptional factor (MatHMG) that is a key regulator of sexual development in fertile A. nidulans (B. L. Miller and K. Y. Miller, unpublished data). This mating type-specific, high-mobility-group, DNA-binding domain is highly conserved among the Ascomycetes. The main goal of this study was to use the A. nidulans genetic system to address the hypothesis that the A. fumigatus mat1-2 gene and encoded Mat1-2 protein carry functional mating type information and can regulate sexual development in A. nidulans. Our data demonstrate that the A. fumigatus mat1-2 gene is not properly expressed or developmentally regulated and, consequently, cannot support the sexual cycle in an A. nidulans ΔmatA mutant. By contrast, A. fumigatus Mat1-2 protein expressed under control of the A. nidulans matA promoter is functional and can drive sexual reproduction in A. nidulans. Although mat1-2 may not be properly expressed in A. fumigatus, our results provide evidence for the potential ability of A. fumigatus to undergo sexual reproduction and the possibility of constructing compatible mating partners by manipulation of mat loci.
MATERIALS AND METHODS
Strains, growth conditions, and genetic manipulations of A. nidulans.
A. nidulans strains used in this study are listed in Table 1. Appropriately supplemented media were prepared as described previously by Pontecorvo et al. (35), Kafer (19), and Vallim et al. (43). Standard A. nidulans culture conditions and genetic techniques were used as described previously by Pontecorvo et al. (35). DNA and RNA isolation, standard molecular manipulations, and Southern blot analysis were performed as described previously by Miller et al. (26, 27, 43). DNA-mediated transformation of A. nidulans was performed according to the protocols of Miller et al. (27) and Yelton et al. (50).
TABLE 1.
Aspergillus strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| FGSCA26 | biA1 | Fungal Genetics Stock Center |
| FGSCA237 | pabaA2 yA2; trpC801 | Fungal Genetics Stock Center |
| UI401 | pyrG89 ya2 biA1; argB2 ΔmatA::argB; riboB2 | B. L. Miller and K. Y. Miller |
| UI404 | pyrG89 biA1 pabaA1; argB2 ΔmatA::argB | B. L. Miller and K. Y. Miller |
| UI414 | pyrG89:pWP1:pyrG biA1 pabaA1; argB2 ΔmatA::argB | This study |
| UI412 | pyrG89:pWP3:pyrG biA1 pabaA1; argB2 ΔmatA::argB | This study |
| UI421 | biA1 pabaA1; matA::Afmat1-2 ORF | This study |
| UI422 | yA2 biA1 pabaA1; matA::Afmat1-2 ORF | This study |
| UI423 | biA1 pabaA1; matA::Afmat1-2 ORF | This study |
| Af293.139 | pyrG1 | G. S. May (MD Anderson) |
Database analyses of the A. fumigatus genome.
The matA locus encoding the HMG box domain mating type protein has been analyzed in A. nidulans (B. L. Miller and K. Y. Miller, unpublished data). To identify the homolog of the A. nidulans matA gene in A. fumigatus genome, the translated open reading frame (ORF) of the A. nidulans matA gene (locus ID AN4734.3; accession number AY32808) was used in a BLASTP search of the TIGR A. fumigatus database (www.tigr.org) and in comparative analysis of three aspergilli genomes (12). A. nidulans genome sequences are available from the Broad Institute (www.broad.mit.edu/annotation/genome/aspergillus_nidulans/home.html). The locus ID for A. fumigatus mat1-2 is Afu3g06170.
Plasmid construction.
Plasmid pWP1 was constructed using the pCR8-TOPO vector, TOPO TA cloning kit for entry into the Gateway technology and AccuPrime Pfx polymerase (Invitrogen) (41). Genomic DNA from the Aspergillus fumigatus pyrG1 strain Af293.139 was extracted according to standard protocol (50). The A. fumigatus mat1-2 gene (Afu3g06170), including the ORF, the 1.2-kb upstream flank, and the 1.7-kb downstream flank, was amplified from the genomic DNA with primers AfmatAF6 and AfmatARR4 (Table 2). The PCR product was cloned into the pCR8-TOPO vector. The resulting plasmid was subsequently subjected to a recombination reaction with pAN7, a destination plasmid containing the Gateway RfC reading frame cassette and the A. nidulans pyrG selectable marker cloned into pDK101 (20). The final vector created upon recombination contained both the A. fumigatus mat1-2 gene and the A. nidulans pyrG marker. The mat1-2 gene was sequenced to confirm the correct sequence and to determine its orientation in pWP1.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| AnMatAR11 | TGC CGT ATG CTA CCT GAG |
| AnMatAF11 | TGG GAG TGT ATC AGC TTC ATG |
| AnpyrG1 | GAA TTC GAT ACC TGT GGA AAG |
| AnpyrG2 | TGA TCA GTG CTT GTC TAC CAG |
| AnmatAF33 | CCG ACA GCA TCA CCG AGC TCC |
| AnmatAR29 | GGT GTG CGC AGA ACA CGC AGA |
| AnbenAF2 | GAT GTT CGA CCC CAA GAA CA |
| AnbenAR2 | CTT GAA GAG CTC CTG GAT GG |
| AfmatA(ATG) | ATG GCT ACA GTC CCA ATC GCC |
| AfmatA(TAG) | CTA GAA GCA ATC AGA GAT AAA ATC |
| Afmat1-2F4 | ATG TGA CCG ACA TGA TCG GCC AGG |
| AfmatAF6 | GCA AGC CTG ACA GCA GAG CAC |
| Afmat1-2R5 | TGT CTT GAC AGC TTC ACC GTG |
| AfmatARR4 | CCT TCT ACC TAC GTC GGG ACG CTT |
| AfbenAF1 | GGC CTC AAG ATG TCC TCG ACC |
| AfbenAR1 | CTC CTC GCC GTA CTC CTC CT |
The destination vector pmatA An/Af ORF swap containing the A. fumigatus mat1-2 ORF under the A. nidulans matA promoter and terminator was created by using the pCR4-TOPO and pCR8-TOPO vectors, the TOPO TA cloning kit for entry into the Gateway technology (Invitrogen), and Phusion high-fidelity PCR master mix (New England Biolabs). Genomic sequences containing the A. nidulans matA ORF flanked by 1-kb upstream and 1.8-kb downstream regulatory regions were amplified from FGSCA26 (wild-type [WT]) genomic DNA with primers AnmatAF11 and AnmataR11 (Table 2). The PCR product was cloned into the pCR4-TOPO vector (41). Subsequently, 990 bp of the 1,054-bp coding region was removed by ClaI and ApaI digestion. Blunt ends were generated with Klenow fragment, and the vector was ligated with the Gateway RfA reading frame cassette. The A. fumigatus mat1-2 coding region beginning with ATG and ending with TAG was amplified from the genomic DNA of Af293.139 strain with primers AfmatA (ATG) and AfmatA (TAG) and cloned into pCR8 vector. Both pCR8 and pCR4 constructs were subjected to recombination reaction, resulting in the formation of a destination vector that contained the A. fumigatus mat1-2 ORF incorporated at the ClaI/ApaI site and flanked by A. nidulans 1-kb promoter and 1.8-kb terminator sequences. The region of transgene between priming sites of AnmatAF11 and AnmatAR11 was sequenced to confirm in-frame cloning and correct sequence. Both plasmids, pWP1 and pmatA An/Af ORF swap, were propagated using TOP10 Escherichia coli strain (Invitrogen). Standard methods for the manipulation of E. coli cells and DNA were performed as described previously (38).
A. fumigatus mat1-2 complementation assay.
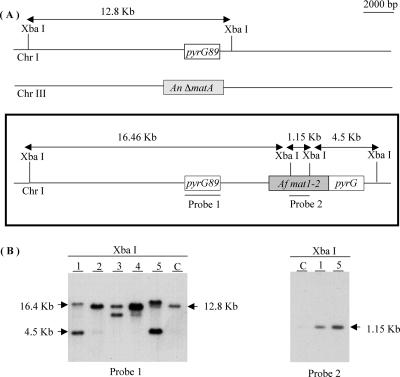
The UI404 matA deletion strain was made by replacing 300 bp of the matA coding region between two ClaI sites with the A. nidulans argB selectable marker (B. L. Miller and K. Y. Miller, unpublished data). DNA-mediated transformation of the A. nidulans UI404 matA deletion strain with pWP1 plasmid containing the A. fumigatus mat1-2 gene and A. nidulans pyrG as a selectable marker was performed to determine whether the A. fumigatus mat1-2 gene can function as a mating type regulator in the A. nidulans sexual cycle. Transformants were selected based on pyrG prototrophy and screened by Southern analysis for ectopic integration of A. fumigatus mat1-2 at the pyrG89 locus. Genomic DNA of isolated transformants was digested with the XbaI restriction enzyme, and Southern blot analysis with two different probes was performed (Fig. 1). Transformants having the disrupted endogenous A. nidulans matA gene and an ectopically integrated copy of A. fumigatus mat1-2 were further analyzed for the ability to undergo sexual reproduction (Fig. 2).
FIG. 1.
Ectopic integration of A. fumigatus mat1-2. (A) Schematic representation of the targeted genomic region of recipient strain UI404 on Chr I. The A. nidulans ΔmatA allele on Chr III is also shown. Genetic organization of transformants created upon ectopic integration of A. fumigatus mat1-2 at the pyrG89 locus is shown in the box. Positions of restriction enzymes, predicted fragment size, and probes used to verify ectopic integration are indicated. (B) Southern blot analysis was performed to confirm A. fumigatus mat1-2 integration. Genomic DNA from A. nidulans WT and five transformants (T1 to T5) was digested with XbaI and probed with pyrG-specific probe 1. Transformants T1 and T5 were additionally verified with A. fumigatus mat1-2-specific probe 2. C, control.
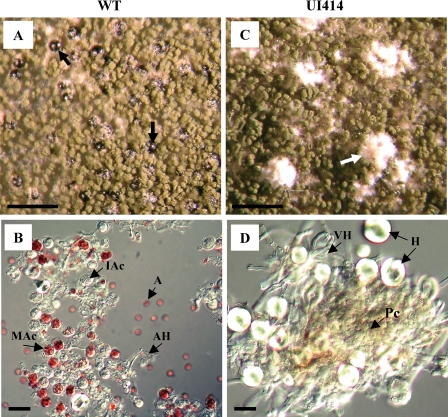
FIG. 2.
Phenotype of the A. nidulans transformant UI412 with resident ΔmatA and ectopic A. fumigatus mat1-2 at 6 days postinduction of the sexual cycle. (A) Dark purple-pigmented WT cleistothecia (black arrows) and mature green conidiophores. (B) Contents of broken WT cleistothecium at 6 days postinduction. A, mature ascospores; AH, ascogenous hyphae; IAc, immature transparent ascus; MAc, mature red-pigmented ascus with ascospores. (C) Clusters of white Hülle cells (white arrow) in UI414 at 6 days postinduction. (D) UI414 protocleistothecium within a cluster of Hülle cells under higher magnification. H, individual Hülle cells; VH, vegetative hyphae; Pc, protocleistothecium. Magnification, 700 μm (A and C) and 20 μm (B and D).
A. fumigatus mat1-2 ORF swap experiment.
The argB-disrupted A. nidulans matA ORF was removed from the A. nidulans UI404 strain genome and replaced with the A. fumigatus mat1-2 ORF to determine the Mat1-2 protein functionality in sexual development. The UI404 strain was transformed with the pmatA An/Af ORF construct. A linear fragment with the transgene was generated by digestion with NotI and SpeI restriction enzymes to promote gene replacement at the matA locus via homologous sequences flanking the A. fumigatus mat1-2 ORF. DNA fragments were recovered from agarose gel using a Geneclean Turbo kit DNA purification kit (Bio 101 Systems). The recipient strain UI404 was cotransformed with the linear fragment of A. fumigatus mat1-2 ORF flanked by A. nidulans matA regions (2 μg/μl) and the circular ppyrG plasmid (6 μg/μl) that contains the A. nidulans pyrG fragment cloned into pDK101. The total amount of DNA used to transform protoplasts was experimentally determined to optimize single-copy integration and limit multiple tandem integrations. Transformants generated by the integration of pyrG plasmid at the pyrG89 locus and the integration of the NotI/SpeI linear fragment by double crossover at the matA locus were isolated based on argB auxotrophy. Transformants were crossed with FGSCA237 to obtain homokaryotic single-spore isolates of primary transformants carrying the A. fumigatus mat1-2 ORF and argB prototrophy, which is essential for successful sexual development. Genomic DNA was extracted from single-ascospore isolates, and Southern blot analysis was performed to discriminate single-copy gene replacement from multiple tandem or ectopic integrations. The presence of a single copy of the A. fumigatus mat1-2 ORF at the A. nidulans matA locus was confirmed by two different digests using SalI and ApLI restriction enzymes (Fig. 3). Three progeny were isolated and designated UI421, UI422, and UI423.
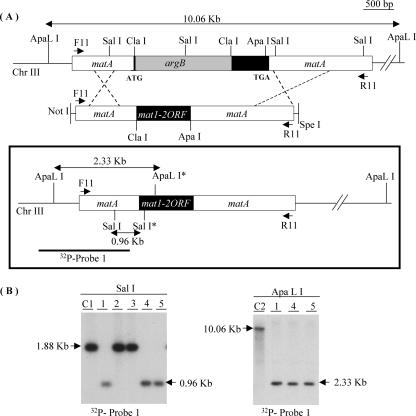
FIG. 3.
ORF swap strategy. (A) Schematic representation of the WT A. nidulans matA genomic region on Chr III. A. nidulans matA disruption allele and construct used to introduce the A. fumigatus mat1-2 ORF by homologous recombination. The argB selectable marker is incorporated between the Cla1 sites of the A. nidulans matA ORF (black solid box). Position of primers (F11 and R11) used to create the ORF swap construct are indicated. The mat1-2 ORF was cloned between Cla1 and Apa1 as described in Materials and Methods. The flanking regions with A. nidulans matA homology for double-crossover gene replacement are indicated as white boxes. Structure of the A. nidulans matA::mat1-2 genomic region created upon homologous recombination (bold frame) is shown within the box. Enzymes used for Southern blot analysis are indicated. Sal1* and ApaL1* are present exclusively in A. fumigatus mat1-2. The restriction enzymes and probe used to confirm gene replacement are marked. (B) Results of Southern blot analysis. Sal1 digestion differentiates between A. nidulans matA ORF and A. fumigatus mat1-2 ORF. ApaL1 digestion was used to determined the number of integrated mat1-2 copies. C1, control, WT matA in FGSCA26; C2, control, disrupted ΔmatA in UI404.
Induction of sexual development and fruiting body analysis.
A total of 104 conidia per plate were spread onto 1.2% agar plates containing rich medium (YGMTV [yeast extract, glucose, MgSO4, trace elements, and vitamins]). Cultures were grown at 37°C, and plates were sealed 24 h after inoculation to promote sexual development. Cleistothecial tissue and Hülle cells were subjected to microscopy observations from 4 to 24 days (43). The efficiency of fruiting body formation was calculated based on the percentage of protocleistothecia that developed into mature cleistothecia. Protocleistothecia in Aspergillus nidulans are defined as female coiled hyphal elements found within clusters of Hülle cells. Successful mating and fertilization lead to cleistothecium development. Three plates of each strain with developmental cultures were examined under a dissecting scope, and the average number of protocleistothecia and number of cleistothecia/cm2 were determined. Ten individual cleistothecia of the WT and each of the ORF swap strains were collected for size measurements. The diameter of each fruiting body was measured using a standardized microscope ruler. The production and viability of ascospores were assessed by cleaning 20 individual cleistothecia on 3% agar plates and crushing them in 1.5-ml Eppendorf tubes containing 0.1% Tween 80. Aliquots of ascospores were loaded onto a hemacytometer chamber, and the total number of ascospores per cleistothecium was obtained. The ascospore viability was determined as the percentage of ascospores that germinated on solid minimal medium out of the total number of spores that were plated.
Light microscopy.
Photomicrographs of developmental cultures on petri dishes were taken by using a Nikon Coolpix 5400 camera and a Zeiss SV8 stereomicroscope. Fruiting bodies were suspended in 15 μl of sterile water on a glass slide and crushed under a coverslip. Fruiting body tissue was examined by differential interference contrast optics using a Zeiss Axioplan. Photographs were taken with either a Nikon Coolpix 5400 or a Photometrics CoolSnap ES camera and MetaVue software (Universal Imaging Corp.).
Comparative quantitative PCR (QTPCR) analysis.
Developmental tissues of FGSCA26 (WT), UI422, UI412, UI414, and Af293.139 were collected 6 days postinduction. Strains FGSCA26 and UI412 were analyzed for the expression of matA at resident and ectopic loci. Strains UI422 and UI414 were analyzed for A. fumigatus mat1-2 expression in A. nidulans, at either resident or ectopic locus. Additionally, the natural expression of mat1-2 in Af293.139 was assessed. Total RNA was extracted, treated with Turbo DNase (Ambion), and reverse transcribed from oligo(dT) primers by using SuperScript First-Strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen). Transcript levels were quantitated using either the threshold cycle (ΔΔCT) method or a relative standard curve. SYBR green sequence detection was performed using the StepOne real-time PCR system (Applied Biosystems). To monitor the expression of matA in the reference sample FGSCA26 (matA at its resident locus) and in UI412 (matA integrated ectopically at the pyrG locus), the primers AnmatAF33 and AnmatAR29 were used (Table 2). To detect A. fumigatus mat1-2 expression in A. nidulans and A. fumigatus, the primers Afmat1-2F4 and Afmat1-2R5 were used. β-tubulin (benA) was used as the endogenous control to normalize expression of the mat gene in both A. nidulans and A. fumigatus. AnbenAF2 and AnbenAR2 primers were used to amplify benA in all A. nidulans strains. AfbenF1 and AfbenAR1 primers were used to amplify the A. fumigatus benA homolog. Validation experiments of target and control genes for the comparative ΔΔCT method were performed according to the instructions of Applied Biosystems (1). SYBR green exhibits some sequence-specific DNA binding with higher preference for AT-rich sequences (52). To avoid fluorescence differences resulting from different DNA sequences between matA and mat1-2, we analyzed both genes separately. For a valid ΔΔCT method calculation, the efficiency of the target amplification and the efficiency of reference amplification must be approximately equal. The direct comparison of A. fumigatus mat1-2 expressions in A. fumigatus and A. nidulans was possible and valid since amplification efficiencies of endogenous control genes, A. nidulans benA and A. fumigatus benA, were nearly equal. Relative quantitation, by the ΔΔCT method, is expressed as a difference in target gene expression with respect to an endogenous control in different samples. Each cDNA sample was assayed in triplicate, and RNAs were obtained from three separate biological samples.
RESULTS
The A. fumigatus mat1-2 gene cannot complement an A. nidulans ΔmatA mutant.
The A. nidulans genetic system was employed to investigate the potential functionality of A. fumigatus mat1-2 in sexual development. All A. nidulans ΔmatA strains are sterile and form only undifferentiated protocleistothecia within the foci of Hülle cells. Fruiting bodies (cleistothecia), ascogenous hyphae, and ascospores are not observed (K. Y. Miller and B. L. Miller, unpublished data) (Fig. 2C and D). (This ΔmatA phenotype differs from that described previously by Paoletti et al. [30], where fruiting bodies are formed at reduced numbers and these lack ascospores.) Therefore, matA is not needed for Hülle cell formation and acts downstream of key initiators of sexual development. A. fumigatus mat1-2 encodes a structural homolog of the A. nidulans MatA HMG box mating type protein. A. nidulans MatA and A. fumigatus Mat1-2 display 53% identity and 70% similarity at the amino acid level. The HMG box of A. fumigatus Mat1-2 is the region of highest homology to A. nidulans MatA with 74% identity. Outside the HMG box, the N termini of the two proteins share 69% identity, while there is only limited identity (33%) in the C termini. At the DNA level, the HMG box has 75% sequence similarity, but there is no significant sequence similarity outside this domain. A. fumigatus mat1-2 has previously been identified and annotated (11, 12, 32). The A. fumigatus mating type homolog containing the HMG domain was previously identified by Poggeler (32). A comparison of mat loci in fertile A. nidulans and sterile A. fumigatus revealed extensive synteny (12). The function of the putative protein encoded by A. fumigatus mat1-2 is unknown. The sexual reproduction of A. fumigatus has been proposed but never observed in nature or the laboratory. To determine whether A. fumigatus mat1-2 can function in A. nidulans as a mating type regulator, we performed a complementation assay by using an A. nidulans ΔmatA strain.
We first tested whether A. fumigatus mat1-2 can confer mating function and drive sexual reproduction in A. nidulans. The sterile ΔmatA UI404 strain was transformed with the pWP1 plasmid containing the A. fumigatus mat1-2 gene (ORF plus the 1-kb promoter region and the 1.8-kb transcription termination region) and a selectable marker, A. nidulans pyrG. Due to a lack of extensive DNA sequence homology between A. nidulans matA and A. fumigatus mat1-2 outside the HMG box domain, recovered transformants were screened for single-copy ectopic integration of A. fumigatus mat1-2 at A. nidulans pyrG (Fig. 1). Two transformants, T1 and T5, were analyzed in detail. These transformants were induced to undergo sexual development. Six days after induction, the control strain FGSCA26 had differentiated mature fruiting bodies: cleistothecia filled with ascospores. Neither T1 nor T5 had differentiated fruiting bodies by day 6, but formed only foci of Hülle cells. No sexual development was observed beyond this developmental block, which is characteristic of the UI404 matA deletion strain (Fig. 2). Therefore, the A. fumigatus mat1-2 gene did not complement the A. nidulans matA deletion. By contrast, ectopically integrated A. nidulans matA (the ORF plus the 1-kb promoter region and the1.8-kb transcription termination region) complemented UI404 and fully restored fruiting body development.
Construction of an ORF swap strain by replacement of the endogenous A. nidulans matA ORF with the A. fumigatus mat1-2 ORF.
We performed an ORF swap experiment to determine whether the Mat1-2 protein was functional in sexual development. A transcriptional fusion gene was created in which the expression of the A. fumigatus mat1-2 ORF is driven by the A. nidulans matA promoter, and potential posttranscriptional regulation is under control of A. nidulans matA 5′ and 3′ untranscribed regions. This construct, with the A. fumigatus mat1-2 ORF flanked by 5′ and 3′ sequences of A. nidulans matA, was used to replace the endogenous A. nidulans ΔmatA allele by homologous recombination at the matA locus (Fig. 3A). Transformants created upon gene replacement at the matA deletion were argB2 auxotrophs. Transformant T4 was crossed to FGSCA237 to restore argB prototrophy and ensure that the ORF swap transformant was homokaryotic. Three argB+ progeny, P1, P4, and P5, were isolated, and the presence of a single copy of the A. fumigatus mat1-2 ORF at the A. nidulans matA locus was confirmed by Southern blot analysis (Fig. 3B). Digestion with SalI was designed specifically to distinguish A. nidulans matA from the FGSCA237 parent and the A. fumigatus mat1-2 ORF swap parent in the selected progeny. A. fumigatus mat1-2 has a unique SalI (SalI*) restriction site in the coding region, whereas A. nidulans matA lacks this site. The 1.88-kb SalI fragment corresponds to WT A. nidulans matA, while the presence of the A. fumigatus mat1-2 ORF was distinguished by a 0.96-kb fragment (Fig. 3A and B). ApaL1 digestion was designed to confirm the presence of a single copy of the A. fumigatus mat1-2 ORF at the resident A. nidulans matA locus. The A. fumigatus mat1-2 ORF has a unique ApaLI (ApaLI*) restriction site. The 10.06-kb ApaLI fragment represents ΔmatA control of the UI404 recipient strain. Single 2.33-kb ApaLI fragments in P1, P4, and P5 indicate a single copy of the A. fumigatus mat1-2 ORF (Fig. 3B). Two or more tandem integrations would be indicated by an additional fragment of 3.5 kb.
The A. fumigatus Mat1-2 protein supports differentiation of atypical hypertrophic cleistothecia in A. nidulans.
ORF swap strains UI421 (P1), UI422 (P4), UI423 (P5), and WT FGSCA26 were induced to undergo sexual development, and developmental tissue was examined 10 days postinduction. All ORF swap strains developed extremely large fruiting bodies that were ∼300 times bigger per volume than those of the WT. The abundance of ORF swap cleistothecia was significantly lower than the value for the WT. In the WT, 100% of the protocleistothecia developed into mature cleistothecia by 10 days postinduction. By contrast, ORF swap strains had abundant undeveloped protocleistothecia within clusters of Hülle cells. Only 1.2% of all protocleistothecia underwent successful mating and fertilization which resulted in fruiting body formation (Table 3; Fig. 4).
TABLE 3.
Size and efficiency of cleistothecia development at 10 days postinductiona
| Strain | Average size C (μm) | Average no./cm2
|
% Fertilized P | |
|---|---|---|---|---|
| P | C | |||
| FGSCA26 | 145 | 0 | 866 | 100 |
| UI421 | 650 | 213.5 | 2.6 | 1.2 |
P, protocleistothecia; C, cleistothecia.
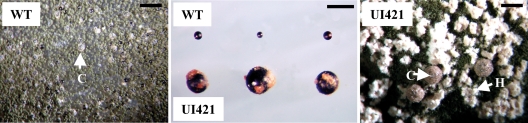
FIG. 4.
Developmental competence and fruiting body size in WT and ORF swap (UI421). Sexual development was induced, and the size and efficiency of cleistothecial development were determined as described in Materials and Methods. The middle panel illustrates the difference in cleistothecial size between WT (top of the panel) and ORF swap strain UI421 (bottom of the panel). C, cleistothecia; H, clusters of Hülle cells and protocleistothecia. Magnification bar, 700 μm.
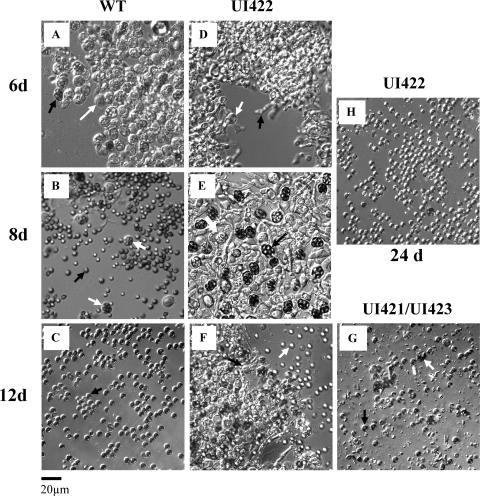
Developmental time points (6, 8, 12, and 24 days postinduction) were selected based upon different stages of Aspergillus nidulans sexual development. Four separate sexual cycle inductions were analyzed. Samples of fruiting body tissue from each time point were examined microscopically. By 5 days postinduction, the WT and all ORF swap strains differentiated foci of Hülle cells that represent the earliest identifiable stage of sexual development. Within the foci of Hülle cells, mating and fertilization occur to trigger fruiting body development. After 6 days, the WT strain differentiated small pink cleistothecia, within which all ascogenous hyphae had differentiated into distinct asci with transparent or red-pigmented ascospores (Fig. 5A) In contrast, UI421, UI422, and UI423 at 6 days had differentiated significantly larger transparent cleistothecia containing a mass of proliferating ascogenous hyphae but no distinct asci (Fig. 5D). Sexual development of all ORF swap strains was apparently delayed at this time. After 8 days postinduction, the WT strain had abundant, almost fully mature, purple cleistothecia that were filled with ascospores. However, not all ascospores were released from asci at that time (Fig. 5B). Cleistothecia formed by the ORF swap strains were less abundant but much bigger than those formed by the WT as described previously. Large cleistothecia were dark purple pigmented and filled with asci at various developmental stages, with little or no differentiated ascospores at that time (Fig. 5E). Small amounts of debris, apparently coming from lysis of undeveloped asci and ascospores, were present in all ORF swap strains. After 12 days of sexual development, cleistothecia of the WT had reached full maturity. All ascospores were released from the asci and were ready for discharge (Fig. 5C). Similarly, development in UI421 and UI423 was completed. Ascospores were released from asci, but a large number of asci failed to develop, leading to the accumulation of debris (Fig. 5G). UI422 was not yet fully mature at 12 days and had large amounts of ascogenous hyphae and asci at different stages of development, and small numbers of mature released ascospores were present (Fig. 5F). UI422 continued to develop up to 24 days postinduction, at which time fruiting bodies were full of released ascospores and small amounts of ascospores enclosed in asci (Fig. 5H). All ORF swap strains represent different progeny from a cross between the original transformant and FGSC237 as described in Materials and Methods. The influence of genetic backgrounds of the parental stains may explain an observed variation in sexual morphogenesis among ORF swap progeny.
FIG. 5.
Sexual development in WT and ORF swap strains (UI421, UI422, and UI423). Cleistothecia were isolated, and the internal contents were examined at different time points after induction (see Materials and Methods). (A) WT at 6 days (6d) postinduction. Young ascus with immature ascospores (white arrow), and mature ascus with pigmented ascospores (black arrow). (B) WT at 8 days postinduction. Released mature ascospores (black arrow) and a few intact asci (white arrow). (C) WT at 12 days (12d) postinduction. Released mature ascospores (black arrow). (D) UI422 at 6 days postinduction. Young ascus (white arrow) and ascogenous hyphae (black arrow). (E) UI422 at 8 days postinduction. Immature ascus (white arrow) and mature ascus with ascospores (black arrow). (F) UI422 at 12 days postinduction. Released mature ascospores (white arrow) and young ascus (black arrow). (G) The phenotype of UI421 and UI423 up to 8 days postinduction is the same as UI422; however, at 12 days postinduction, development is completed and debris from undeveloped asci (white arrow) and ascospores (black arrow) are indicated. (H) UI422 at 24 days (24d) postinduction and released mature ascospores. Magnification bar, 20 μm.
A. fumigatus Mat1-2 protein confers full fertility in A. nidulans.
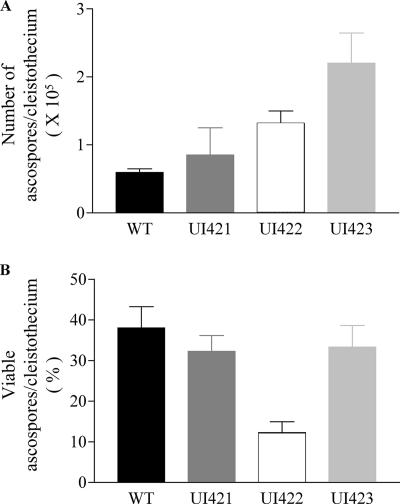
The total number and viability of ascospores were examined to confirm fertility of the ORF swap strains. Twenty mature cleistothecia from the WT and each ORF swap strain were examined. Because of differences in temporal development between WT and ORF swap strains, all cleistothecia were collected at the same developmental stage, when they released ascospores. The average number of ascospores per cleistothecium in the WT was 0.59 × 105. The number of ascospores in UI421 was similar to that in the WT (average, 0.85 × 105). By contrast UI422 and UI423 differentiated significantly larger amounts of ascospores: 3.1 × 105 and 2.2 × 105, respectively, which represents a four- to fivefold increase over the value for the WT (Fig. 6A). Ascospore viability in the WT was an average of 37%, and similar viabilities were observed for UI421 and UI423, whereas UI422 ascospore viability was about twofold lower (Fig. 6B).
FIG. 6.
Ascospore production and viability in WT and ORF swap strains (UI421, UI422, and UI423). Illustrated are the means and standard errors (error bars) of the means for 20 separate cleistothecia from each strain. Statistical significance was determined by the one-way analysis of variance (P < 0.05).
A. fumigatus Mat1-2 protein supports outcrossing in A. nidulans.
WT A. nidulans strains (matA) are readily able to outcross. Crossed cleistothecia differentiate ascospore progeny with meiotic recombination frequencies consistent with established recombination units found in the A. nidulans genetic linkage map. To examine whether A. fumigatus Mat1-2 can also function in an outcross, we crossed the yellow conidial strain FGSCA237 and the green conidial strain UI421. Large cleistothecia were picked from a region of well-balanced and stabile vegetative heterokaryon. Crossed cleistothecia were initially selected based upon conidial color of the progeny. Progeny from each cleistothecium were then analyzed for recombination frequency between markers on dropout medium. Recombination frequencies between the linked markers yA (Chr 1R) and biA (Chr 1R) and the unlinked markers yA and trpC (Chr VIII) were analyzed for 100 progeny. For the linked markers, progeny were ∼93% parental-to-7% recombinant, which is consistent with the established genetic linkage of seven recombination units. The segregation of unlinked markers was ∼50% parental-to-50% recombinant as predicted. Therefore, the A. fumigatus Mat1-2 protein supports self-fertility, cross-fertility, and meiotic recombination.
A. fumigatus mat1-2 gene expression in A. nidulans is significantly higher than the natural transcript levels in A. fumigatus.
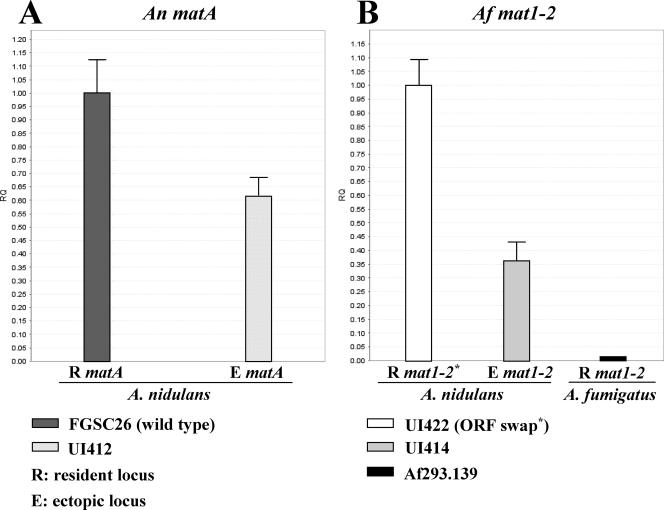
Functionalities of the A. fumigatus mat1-2 gene and A. fumigatus mat1-2 ORF were correlated with the level of transcript expressed under conditions favorable for sexual development. The expression of the A. nidulans matA gene in A. nidulans and the A. fumigatus mat1-2 gene in both A. nidulans and A. fumigatus were analyzed using comparative QTPCR. Developmental tissues of four A. nidulans strains, FGSCA26 (WT), UI412, UI414, and UI422, and A. fumigatus strain Af293.139 were collected 6 days postinduction and RNA prepared as described in Materials and Methods. Genomic position appears to play an important role in proper developmental expression of mating type genes (2, 34, 46, 47). Therefore, to analyze ectopic mat1-2 expression in UI414, we included two reference control strains, FGSCA26, expressing A. nidulans matA at its resident locus (Chr III), and UI412, expressing A. nidulans matA at the ectopic site (Chr I). The expression of the ectopic copy of A. nidulans matA (ORF plus 1 kb upstream and 1.8 kb downstream) was ∼60% of A. nidulans matA expression at its resident locus (Fig. 7A). This level of expression was sufficient to confer fruiting body development in A. nidulans. To correlate expression and functionality of A. fumigatus mat1-2, we analyzed two A. nidulans strains expressing the mat1-2 transcript: UI422 (mat1-2 ORF driven by the A. nidulans matA promoter at the A. nidulans matA resident locus) and UI414 (A. fumigatus mat1-2 gene with ORF driven by the native A. fumigatus promoter at the ectopic locus). Relative expression levels in FGSCA26 and UI422 are comparable because the matA promoter at its resident locus drives transcription in both strains. The A. fumigatus mat1-2 gene (ORF plus 1.2 kb upstream and 1.7 kb downstream) expressed by its own promoter was ∼36% of the transcript level expressed from the A. nidulans matA promoter in the ORF swap strain UI422 (Fig. 7B). While expression in reference strain UI422 is fully capable of driving sexual reproduction in A. nidulans, the level of expression driven by the native A. fumigatus promoter is not sufficient to support fruiting body formation in A. nidulans. Therefore, the A. fumigatus promoter responds to upstream A. nidulans regulators, but this response is not as robust as that of the A. nidulans promoter. Native mat1-2 transcript levels in A. fumigatus strain Af293.139 were evaluated with reference to expression in A. nidulans. Significantly, mat1-2 expression from its resident locus in A. fumigatus was only about 1.1% relative to that in UI422 (Fig. 7.B). These data suggest that there may exist a critical threshold level of mating type gene expression that is required to initiate fruiting body formation. Similar threshold specificities have been observed for other A. nidulans developmental transcription factors (28). If the above suggestion is the case, the level of mat1-2 expression in A. nidulans (UI414) and A. fumigatus (Af293.129) would be below this threshold. It is also significant that a comparison of UI414 and Af293.139 suggests that the A. fumigatus mat1-2 promoter is not responding to an important upstream regulator(s) in A. fumigatus or that these critical regulators are missing/nonfunctional in A. fumigatus.
FIG. 7.
Comparative expression (QTPCR) of A. nidulans matA and A. fumigatus mat1-2. (A) Comparative expression of A. nidulans matA at its resident locus (FGSCA26 [WT]) and ectopically integrated at pyrG (UI412). (B) Comparative expression of the A. fumigatus mat1-2 ORF at the resident A. nidulans matA locus, driven by the A. nidulans matA promoter (UI422), ectopically integrated A. fumigatus mat1-2 at pyrG driven by its own promoter (UI414), and native A. fumigatus mat1-2 expression in the A. fumigatus strain Af293.139. Comparative QTPCR was performed using A. nidulans benA or A. fumigatus benA as a reference. Relative quantitation is the normalized difference in target gene expression (A. nidulans matA or A. fumigatus mat1-2) in different samples. Error bars indicate standard deviations.
DISCUSSION
In this study, we investigated the function of the mat1-2 mating type gene and its encoded protein to elucidate possible reasons for asexuality in the opportunistic human pathogen Aspergillus fumigatus. Mating ability and sexual development in filamentous Ascomycetes are controlled by two alternate forms of the mat locus, one encoding an HMG box protein and the other encoding an alpha box protein (8-10). The discovery of mat1-1 (alpha box) and mat1-2 (HMG box) A. fumigatus isolates that express putative mating type genes suggests a potential for sexual compatibility and fertility or already existing, but cryptic, sexual activity (29, 45). However, the physical presence and expression of mat loci, as reported for many asexual species, is not sufficient to confer sexuality. The mat genes may carry mutations rendering them nonfunctional or redirecting their regulatory functions to another biological process not related to the sexual cycle. Alternatively, other genes of the “sexual machinery” may have been lost. To clarify the basis for A. fumigatus asexuality, it is important that the functionality of proteins encoded by mating type genes is determined (1a, 11). Therefore, the main goal of this study was to determine functionality of the A. fumigatus mat1-2 gene and the Mat1-2 protein by using the well-characterized genetic system of fertile homothallic A. nidulans. We used two approaches to separately analyze gene function and protein function.
We first determined the functionality of A. fumigatus mat1-2 by interspecies gene exchange between sterile heterothallic-like A. fumigatus and fertile homothallic A. nidulans. Interspecies mating type gene exchanges and heterologous expression have previously been used to understand the role and function of the mat locus in homothallic, heterothallic, and sterile Ascomycetes. Mating type gene exchanges between members of the Sordariaceae (homothallic Sordaria macrospora and heterothallic Podospora anserina and Neurospora crassa) revealed cross-species functionality. Mating type genes were interchangeable between these species with respect to mating and fruiting body development but were not interchangeable with regard to postfertilization functions, such as meiosis and ascospore formation (2, 34). Similar results were obtained when mating type genes from asexual Alternaria alternata were ectopically integrated and expressed in fertile heterothallic Cochliobolus heterostrophus (1a). Turgeon et al. reported that the mating type gene MAT-2 from asexual Bipolaris sacchari was able to support full fertility with normal ascospore differentiation in a ΔMAT C. heterostrophus strain (40, 42). Therefore, in all reported cases, mating type function is conserved among both closely and distantly related species, even though gene structure was highly divergent. Turgeon et al. concluded that the reason for B. sacchari asexuality was not due to a defective mat gene but to another function required for sexual reproduction (40, 42). By contrast, our data demonstrate that A. fumigatus mat1-2 introduced ectopically into an A. nidulans ΔmatA strain was not properly expressed and, consequently, did not function to support either cleistothecia or ascospore development. To our knowledge, this is the first report showing that mating type gene exchange between closely related asexual and fertile Ascomycetes did not function in any aspect of sexual development. In line with observations of Turgeon et al., our data suggest that A. fumigatus asexuality might be caused by defective regulation of mat1-2 expression. The lack of proper A. fumigatus mat1-2 expression in A. nidulans might be due to an A. fumigatus mat1-2 promoter that does not respond appropriately to upstream regulators so that the level of expression is less than the threshold required to drive sexual reproduction. Other possible reasons for reduced expression could be position effect or subtle differences in mechanisms that regulate the expression of mating type genes in homothallic fertile A. nidulans and heterothallic sterile A. fumigatus. Our data further provide evidence that mat1-2, as it exists in the genome of the sequenced clinical isolate Afu293, is expressed at such a low level that it may not be sufficient to drive sexual reproduction. The expression of mat1-2 at its resident locus in A. fumigatus represents only 1.1% of the level of mat1-2 expression required to confer fertile fruiting body differentiation in A. nidulans. This level of expression in A. fumigatus is also significantly less than mat1-2 expression driven by its native promoter in A. nidulans. Therefore, the absence of a critical upstream regulator may also explain a deficiency in mat1-2 expression and asexuality in A. fumigatus. Many other A. fumigatus isolates may be similar to the sequenced strain Af293 used in this study, and this phenomenon may be widespread throughout the worldwide A. fumigatus population. Paoletti et al. also observed very low expression levels of mating type genes in A. fumigatus environmental and clinical strains by using a reverse transcription-PCR/gel analysis approach (29). Detectable expression of both mating type genes required 35 cycles, a cycle threshold similar to what we observed for strain Af293.139 in our QTPCR analysis. In two independent studies, 50 and 70 A. fumigatus strains of clinical and environmental origin, respectively, were subjected to pair-wise matings in all possible combinations and on various mycological media promoting fungal mating. No signs of mating or sexual development were observed for any combination of strains (G. S. May and J. Kwon-Chung, personal communication). These studies support the argument that although there are two extant A. fumigatus mating type idiomorphs, the mat genes are not responding to key upstream regulators or key transcriptional factors for initiating sexual reproduction are missing. It remains possible that undetected strains that do express mat1-2 at functional levels may still exist in the current world population. Because A. fumigatus sexual reproduction has not been observed in nature or in the laboratory, these strains may be quite rare.
Because the mat1-2 gene was nonfunctional in A. nidulans, we were prompted to determine whether the Mat1-2 protein was functional by using the ORF swap approach. We created a gene replacement strain in which the A. nidulans matA ORF was effectively replaced with the A. fumigatus mat1-2 ORF. The expression of the A. fumigatus mat1-2 ORF was therefore driven by the A. nidulans matA promoter at the resident matA locus on Chr III. All ORF swap transformants were fertile, although sexual development was delayed and cleistothecium morphogenesis and ascosporogenesis were altered. Previous studies have shown that heterologous expression of functional mating type proteins conferred the differentiation of fruiting bodies that were indistinguishable from the WT in size and shape but had no, or only few, asci and ascospores (1a, 2, 34, 40, 42). By contrast, we observed the very unique formation of atypical hypertrophic fruiting bodies when the sexual cycle was driven by A. fumigatus Mat1-2 when it was expressed under control of the A. nidulans matA promoter. These atypical cleistothecia were filled with proliferating ascogenous hyphae and asci at various stages of maturation. The ascogenous tissue did not completely fill the entire volume of the cleistothecium. Hence, the cleistothecium was very soft and fragile. The A. fumigatus Mat1-2 protein has the highly conserved mating type HMG box DNA binding domain. Therefore, it probably binds to the same sets of target genes as A. nidulans MatA does and supports full fertility. However, limited homology at the N and C termini of the two proteins could influence interactions with other transcription factors and affect the transcript abundance of target genes. These differences would, in turn, alter sexual cycle progression and fruiting body development. For example, ascospore differentiation and meiosis in Saccharomyces cerevisiae diploids are induced in the presence of the mating type a1/alpha2 heterodimer which represses the expression of RME1 (repressor of meiosis) (5). The formation of a MatAHMG/MatB alpha box (MatBalpha) mating type heterodimer has not yet been demonstrated for A. nidulans. However, Jacobsen et al. have shown that the alpha box and HMG box mating type proteins of homothallic Sordaria macrospora are able to form heterodimers (18). Our results suggest that the A. fumigatus Mat1-2 HMG box protein may not interact with the A. nidulans MatB alpha box mating factor to form a functional heterodimer. Alternatively, the protein-protein interaction may not be sufficiently stable. In addition to self-fertility, A. fumigatus Mat1-2 supports cross fertility and meiotic recombination when mated with a WT (matA) strain. It was previously shown that the Saccharomyces pombe mating type genes are required for both mating and meiosis and also influence meiotic recombination (25, 44). It has also been shown that HMG box proteins bind to, and potentially stabilize, the four-way (Holliday) DNA junction, an intermediate in homologous genetic recombination (49). In Ascomycetes, the expression of genes encoding proteins required for meiotic homologous recombination could be regulated by MatHMG transcription factors, including A. fumigatus Mat1-2, when expressed in A. nidulans.
Fruiting body development in Ascomycetes is a complex process of cellular differentiation that is under polygenic control and requires special intrinsic signals and environmental conditions (31). Sexual morphogenesis in A. nidulans has been described previously; however, the molecular mechanisms controlling coordinated development of ascogenous hyphae and cleistothecium wall remain largely unknown (6). It has previously been demonstrated that the HMG box mating type transcription factor is necessary for fruiting body development in homothallic as well as in heterothallic fungi. MatHMG is required for the expression of different sets of target genes directly involved in male and female fertility, fruiting body morphogenesis, fruiting body abundance, and ascospore formation (7, 8, 21, 33). Sexual cycle genes include not only those encoding pheromones, pheromone receptors, and components of signal transduction pathways but also enzymes involved in cell wall biogenesis and metabolism (31). The delay in sexual development of the ORF swap strains may be due to unfavorable metabolic conditions created by reduced activities of critical enzymes when regulated by A. fumigatus Mat1-2. One of the essential enzymes being expressed at the onset of sexual development is endo-1-3-beta-glucanase. This enzyme is required to degrade glucan as a carbon source for fruiting body formation. Reduced endo-1-3-beta-glucanase expression could result in lowered carbon source availability and cause a delay in development. The delay in cleistothecia and ascospore development in the ORF swap strains might also be due to altered regulation of genes encoding components of signal transduction cascades that control expression of enzymes involved in cell wall biogenesis. These would also be important determinants of cleistothecium size and efficiency of development. Two well-characterized signaling pathways, the cyclic AMP-dependent protein kinases protein kinase A and mitogen-activated protein kinase, are known to regulate fruiting body formation in response to external stimuli. In particular, mating type proteins are known to regulate genes of the pheromone-mediated mitogen-activated protein kinase signaling pathway. N. crassa strains with a Galpha subunit mutation (Δgna-1) form fewer protoperithecia with no ascospores (22). Fewer cleistothecia and ascospores were reported for A. nidulans gprA and gprB mutants that encode putative G protein-coupled pheromone receptors (39). Therefore, reduced efficiency of cleistothecia formation in ORF swap strains could be explained by the altered expression of genes encoding important components of pheromone signaling, including G proteins, pheromones, and pheromone receptors, when regulated by the A. fumigatus Mat1-2 transcription factor.
In conclusion, our results confirm that the A. fumigatus Mat1-2 protein is not only a structural but also a functional homolog of the A. nidulans MatA HMG box mating type transcription factor and alone is able to regulate sexual development in A. nidulans. Additionally, A. fumigatus Mat1-2, like A. nidulans MatA, supports self-fertility, cross-fertility, and normal meiotic recombination frequency when expressed in the A. nidulans genetic background under control of the A. nidulans matA promoter. By contrast, the A. fumigatus mat1-2 gene in A. nidulans is not expressed at levels sufficient to confer biological function for sexual reproduction. Therefore, we infer that improper developmental regulation of mat1-2 could be a reason for asexuality in A. fumigatus. This may result from differences in mat1-2 promoter function or from key initiators of sexual development acting upstream of mat1-2 that are missing or not functional. Several lines of evidence demonstrate both functional conservation of the mat loci and distinctive plasticity in reproductive life style. Within the Ascomycetes, it has been possible with some success to change the reproductive mode by using intraspecies manipulation of mating type loci in Gibberella zeae and interspecies mating type gene exchanges among Cochliobolus sp. (23, 51). Intergenus mating type gene exchanges between asexual Bipolaris sacchari and heterothallic Cochliobolus heterostrophus, between homothallic Sordaria macrospora and heterothallic Podospora anserina, and between heterothallic Neurospora crassa and heterothallic Podospora anserina are also functional (2, 34, 40). Furthermore, in separate studies, Magee and Magee (24) and Hull et al. (16) were able to alter the mating-type like (MTL) locus in the asexual pathogen Candida albicans to create compatible mating partners able to mate in vitro. Hull et al. also showed evidence of mating in vivo within a mammalian host during infection (16). Overall, these data suggest that in most species, conservation of mating type function allows intraspecies, interspecies, and intergenus gene exchanges and supports alternate reproductive life styles. Further studies using a combination of genetic manipulations of mat loci and interspecies gene exchanges between sterile A. fumigatus and fertile Aspergilli, such as A. nidulans, Emericella heterothallica, and Neosartorya fischeri, would be a next step in clarifying the expression of A. fumigatus mating type genes and their potential roles in A. fumigatus asexuality. These investigations should help elucidate whether A. fumigatus is truly asexual or has a cryptic sexual activity; moreover, they may reveal ways to manipulate mat loci to reconstruct compatible mating partners able to undergo a successful sexual cycle.
Acknowledgments
We thank G. Arrizabalaga for valuable discussions.
This work was supported by grants NSF IBN/DCB 0318801 and NIH/COBRE P20RR15587 to B.L.M.
Footnotes
Published ahead of print on 1 February 2008.
REFERENCES
- 1.Applied Biosystems. 2004. Guide to performing relative quantitation of gene expression using real-time quantitative PCR. Applied Biosystems, Foster City, CA.
- 1a.Arie, T., I. Kaneko, T. Yoshida, M. Noguchi, Y. Nomura, and I. Yamaguchi. 2000. Mating-type genes from asexual phytopathogenic Ascomycetes Fusarium oxysporum and Alternaria alternata. Mol. Plant-Microbe Interact. 131330-1339. [DOI] [PubMed]
- 2.Arnaise, S., D. Zickler, and N. L. Glass. 1993. Heterologous expression of mating-type genes in filamentous fungi. Proc. Natl. Acad. Sci. USA 906616-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin, C. R. 1955. Ascocarps of Aspergillus and Penicillium. Mycologia 47669-687. [Google Scholar]
- 4.Bruggeman, J., A. J. Debets, K. Swart, and R. F. Hoekstra. 2003. Male and female roles in crosses of Aspergillus nidulans as revealed by vegetatively incompatible parents. Fungal Genet. Biol. 39136-141. [DOI] [PubMed] [Google Scholar]
- 5.Casselton, L. A. 2002. Mate recognition in fungi. Heredity 88142-147. [DOI] [PubMed] [Google Scholar]
- 6.Champe, S. P., D. L. Nagle, and L. N. Yager. 1994. Sexual sporulation. Prog. Ind. Microbiol. 29429-454. [PubMed] [Google Scholar]
- 7.Coppin, E., S. Arnaise, V. Contamine, and M. Picard. 1993. Deletion of the mating-type sequences in Podospora anserina abolishes mating without affecting vegetative functions and sexual differentiation. Mol. Gen. Genet. 241409-414. [DOI] [PubMed] [Google Scholar]
- 8.Coppin, E., R. Debuchy, S. Arnaise, and M. Picard. 1997. Mating types and sexual development in filamentous Ascomycetes. Microbiol. Mol. Biol. Rev. 61411-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debuchy, R., and B. G. Turgeon. 2006. Mating-type structure, evolution, and function in Euascomycetes, p. 293-323. In U. Kües and R. Fisher (ed.), The mycota I growth, differentiation and sexuality, 2nd ed. Springer-Verlag, Berlin, Germany.
- 10.Dyer, P. S. 2007. Sexual reproduction and significance of MAT in the Aspergilli, p. 123-142. In J. Heitman, J. W. Kronstad, J. W. Taylor, and L. A. Casselton (ed.), Sex in fungi: molecular determination and evolutionary implications, 1st ed. American Society for Microbiology, Washington, DC.
- 11.Dyer, P. S., and M. Paoletti. 2005. Reproduction in Aspergillus fumigatus: sexuality in a supposedly asexual species? Med. Mycol. 43(Suppl. 1)S7-S14. [DOI] [PubMed] [Google Scholar]
- 12.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 4381105-1115. [DOI] [PubMed] [Google Scholar]
- 13.Gow, N. A. 2005. Fungal genomics: forensic evidence of sexual activity. Curr. Biol. 15R509-R511. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, B., S. E. Eckert, S. Krappmann, and G. H. Braus. 2001. Sexual diploids of Aspergillus nidulans do not form by random fusion of nuclei in the heterokaryon. Genetics 157141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hope, W. W., M. J. Kruhlak, C. A. Lyman, R. Petraitiene, V. Petraitis, A. Francesconi, M. Kasai, D. Mickiene, T. Sein, J. Peter, A. M. Kelaher, J. E. Hughes, M. P. Cotton, C. J. Cotten, J. Bacher, S. Tripathi, L. Bermudez, T. K. Maugel, P. M. Zerfas, J. R. Wingard, G. L. Drusano, and T. J. Walsh. 2007. Pathogenesis of Aspergillus fumigatus and the kinetics of galactomannan in an in vitro model of early invasive pulmonary aspergillosis: implications for antifungal therapy. J. Infect. Dis. 195455-466. [DOI] [PubMed] [Google Scholar]
- 16.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289307-310. [DOI] [PubMed] [Google Scholar]
- 17.Ibrahim-Granet, O., B. Philippe, H. Boleti, E. Boisvieux-Ulrich, D. Grenet, M. Stern, and J. P. Latge. 2003. Phagocytosis and intracellular fate of Aspergillus fumigatus conidia in alveolar macrophages. Infect. Immun. 71891-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobsen, S., M. Wittig, and S. Poggeler. 2002. Interaction between mating-type proteins from the homothallic fungus Sordaria macrospora. Curr. Genet. 41150-158. [DOI] [PubMed] [Google Scholar]
- 19.Kafer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1933-131. [DOI] [PubMed] [Google Scholar]
- 20.Kovalic, D., J. H. Kwak, and B. Weisblum. 1991. General method for direct cloning of DNA fragments generated by the polymerase chain reaction. Nucleic Acids Res. 194560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kronstad, J. W., and C. Staben. 1997. Mating type in filamentous fungi. Annu. Rev. Genet. 31245-276. [DOI] [PubMed] [Google Scholar]
- 22.Krystofova, S., and K. A. Borkovich. 2006. The predicted G-protein-coupled receptor GPR-1 is required for female sexual development in the multicellular fungus Neurospora crassa. Eukaryot. Cell 51503-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, J., T. Lee, Y. W. Lee, S. H. Yun, and B. G. Turgeon. 2003. Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol. Microbiol. 50145-152. [DOI] [PubMed] [Google Scholar]
- 24.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289310-313. [DOI] [PubMed] [Google Scholar]
- 25.Meade, J. H., and H. Gutz. 1978. Influence of the mat1-M allele on meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 88235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, B. L., K. Y. Miller, K. A. Roberti, and W. E. Timberlake. 1987. Position-dependent and -independent mechanisms regulate cell-specific expression of the SpoC1 gene cluster of Aspergillus nidulans. Mol. Cell. Biol. 7427-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, B. L., K. Y. Miller, and W. E. Timberlake. 1985. Direct and indirect gene replacements in Aspergillus nidulans. Mol. Cell. Biol. 51714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, K. Y., J. Wu, and B. L. Miller. 1992. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 61770-1782. [DOI] [PubMed] [Google Scholar]
- 29.Paoletti, M., C. Rydholm, E. U. Schwier, M. J. Anderson, G. Szakacs, F. Lutzoni, J. P. Debeaupuis, J. P. Latge, D. W. Denning, and P. S. Dyer. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 151242-1248. [DOI] [PubMed] [Google Scholar]
- 30.Paoletti, M., F. A. Seymour, M. J. Alcocer, N. Kaur, A. M. Calvo, D. B. Archer, and P. S. Dyer. 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 171384-1389. [DOI] [PubMed] [Google Scholar]
- 31.Poggeler, S., M. Nowrousian, and U. Kuck. 2006. Fruiting-body development in Ascomycetes, p. 325-356. In U. Kües and R. Fisher (ed.), The mycota I growth, differentiation and sexuality, 2nd ed. Springer, Berlin, Germany.
- 32.Poggeler, S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr. Genet. 42153-160. [DOI] [PubMed] [Google Scholar]
- 33.Poggeler, S., M. Nowrousian, C. Ringelberg, J. J. Loros, J. C. Dunlap, and U. Kuck. 2006. Microarray and real-time PCR analyses reveal mating type-dependent gene expression in a homothallic fungus. Mol. Genet. Genomics 275492-503. [DOI] [PubMed] [Google Scholar]
- 34.Poggeler, S., S. Risch, U. Kuck, and H. D. Osiewacz. 1997. Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics 147567-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. Mackdonald, and A. W. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5141-238. [DOI] [PubMed] [Google Scholar]
- 36.Rydholm, C., P. S. Dyer, and F. Lutzoni. 2007. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot. Cell 6868-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rydholm, C., G. Szakacs, and F. Lutzoni. 2006. Low genetic variation and no detectable population structure in aspergillus fumigatus compared to closely related Neosartorya species. Eukaryot. Cell 5650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Seo, J. A., K. H. Han, and J. H. Yu. 2004. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 531611-1623. [DOI] [PubMed] [Google Scholar]
- 40.Sharon, A., K. Yamaguchi, S. Christiansen, B. A. Horwitz, O. C. Yoder, and B. G. Turgeon. 1996. An asexual fungus has the potential for sexual development. Mol. Gen. Genet. 25160-68. [DOI] [PubMed] [Google Scholar]
- 41.Shuman, S. 1994. Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. J. Biol. Chem. 26932678-32684. [PubMed] [Google Scholar]
- 42.Turgeon, B. G., A. Sharon, S. Wirsel, K. Yamaguchi, S. K. Christiansen, and O. C. Yoder. 1995. Structure and function of mating type genes in Cochliobolus spp. and asexual fungi. Can. J. Bot. 73778-783. [Google Scholar]
- 43.Vallim, M. A., K. Y. Miller, and B. L. Miller. 2000. Aspergillus SteA (sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 36290-301. [DOI] [PubMed] [Google Scholar]
- 44.Van Heeckeren, W. J., D. R. Dorris, and K. Struhl. 1998. The mating-type proteins of fission yeast induce meiosis by directly activating mei3 transcription. Mol. Cell. Biol. 187317-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varga, J. 2003. Mating type gene homologues in Aspergillus fumigatus. Microbiology 149816-819. [DOI] [PubMed] [Google Scholar]
- 46.Wirsel, S., B. Horwitz, K. Yamaguchi, O. C. Yoder, and B. G. Turgeon. 1998. Single mating type-specific genes and their 3′ UTRs control mating and fertility in Cochliobolus heterostrophus. Mol. Gen. Genet. 259272-281. [DOI] [PubMed] [Google Scholar]
- 47.Wirsel, S., B. G. Turgeon, and O. C. Yoder. 1996. Deletion of the Cochliobolus heterostrophus mating-type (MAT) locus promotes the function of MAT transgenes. Curr. Genet. 29241-249. [PubMed] [Google Scholar]
- 48.Wu, J., and B. L. Miller. 1997. Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol. Cell. Biol. 176191-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xin, H., S. Taudte, N. R. Kallenbach, M. P. Limbach, and R. S. Zitomer. 2000. DNA binding by single HMG box model proteins. Nucleic Acids Res. 284044-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yelton, M. M., J. E. Hamer, E. R. de Souza, E. J. Mullaney, and W. E. Timberlake. 1983. Developmental regulation of the Aspergillus nidulans trpC gene. Proc. Natl. Acad. Sci. USA 807576-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yun, S. H., M. L. Berbee, O. C. Yoder, and B. G. Turgeon. 1999. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. USA 965592-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zipper, H., H. Brunner, J. Bernhagen, and F. Vitzthum. 2004. Investigations on DNA intercalation and surface binding by SYBR green I, its structure determination and methodological implications. Nucleic Acids Res. 32e103. [DOI] [PMC free article] [PubMed] [Google Scholar]