Abstract
Ca2+-mediated signaling events in eukaryotic cells are initiated by Ca2+ channels located in the plasma membranes and endomembranes. Cch1, a high-affinity Ca2+ channel in the plasma membranes of Cryptococcus neoformans and other fungi, plays a role in many different cellular processes, but the mechanisms that regulate Cch1 are not well understood. A Ras recruitment two-hybrid screen was used to identify protein partners of Cch1 as a means of identifying possible mechanisms of channel regulation. Here, we show that Cch1 specifically associates with a cytoplasmic protein known as elongation factor 3 (EF3). The robust interaction between the cytosolic C terminus of the Cch1 protein and EF3 shown here was confirmed by demonstrating that Cch1 could coimmunoprecipitate with EF3 in yeast lysates. To examine the effects of EF3 on Cch1 behavior, we altered the EF3 gene function by constructing a C. neoformans antisense EF3 repression strain. Our results show that the repression of EF3 led to the mislocalization of Cch1, suggesting a role for EF3 in targeting Cch1 to the plasma membrane of C. neoformans. Consistent with this notion, the antisense EF3 repression strain displayed a growth defect under conditions of limited extracellular Ca2+. Collectively, these results suggest that EF3 and Cch1 are functionally coupled and that EF3 has a function apart from its role in the protein translation cycle.
Ca2+-mediated signaling events are central to diverse cellular processes in eukaryotic cells (6). Fungal pathogens, including Cryptococcus neoformans, depend on a Ca2+ signaling pathway for their virulence, their ability to cope with the host environment, and their tolerance to many stresses including stress mediated by triazoles, a class of antifungal drugs (4, 8, 16, 33, 34, 37, 42, 44, 48, 50). In other fungi, cell cycle regulation, pheromone arrest, and morphogenesis are among the physiological processes that are regulated by Ca2+-mediating signaling pathways (5, 17, 20, 27, 32, 40).
A rise in cytosolic Ca2+ levels is required to initiate and propagate signaling, and these transient changes in Ca2+ concentration are initiated by Ca2+-permeable channels that allow Ca2+ ions to enter the cytoplasmic space along its steep electrochemical potential gradient (6). C. neoformans and other fungi possess a high-affinity Ca2+-permeable channel (Cch1) protein in their plasma membranes. Despite the similarity between the overall topology of Cch1 and that of other voltage-gated channel proteins (i.e., the four repeats of the six transmembrane-spanning regions), Cch1 lacks the predicted voltage sensor signature motif that is the hallmark of all voltage-gated channels (37, 38). This finding suggests that Cch1 may be gated/regulated by a mechanism other than voltage and thus may not operate like the classic voltage-gated Ca2+ channels in higher eukaryotes. A glycine residue within the cytoplasmic linker between S2 and S3 of Cch1 was identified as essential for Cch1 activity because a missense mutation that caused a Gly1265-to-Glu substitution resulted in a complete loss of function (31); however, it is not clear whether this residue contributes to the gating mechanism of Cch1. It is unlikely that Gly1265 plays any role in the subcellular localization of Cch1 or in the stability of the Cch1 protein (31).
It has been suggested that changes in secretory Ca2+ levels in Saccharomyces cerevisiae can promote the influx of Ca2+ via Cch1 and its subunit Mid1 (38); however, a direct demonstration of the mechanism that leads to Cch1 activity remains elusive. Cch1-mediated Ca2+ uptake requires Mid1, and accordingly their association is supported by in vivo co-IP experiments (11). It is not known how Mid1 contributes to or promotes Cch1 channel activity and, interestingly, Mid1 does not have any structural homologues in higher eukaryotes (30). Certain cellular stress conditions like membrane perturbations and endoplasmic reticulum (ER) stress have been shown to promote the influx of Ca2+ by Cch1-Mid1 in S. cerevisiae (10, 11). It was postulated that the activation of Cch1-Mid1 during ER stress was promoted by mitogen-activated protein kinase 1 (Mpk1) signaling, and its activation appeared to be independent of the unfolded protein response (11, 46). Consistent with these findings, fluconazole treatment of Candida glabrata infection resulted in a Cch1-Mid1-mediated Ca2+ uptake, which was essential for survival during prolonged fluconazole exposure, suggesting that Cch1-Mid1 constitutes a cell survival pathway (33).
Herein we sought to identify proteins that partner with Cch1 as a means to further explore Cch1 regulation. This was achieved by using the Sos-Ras recruitment yeast two-hybrid screen, which allows the bait and the target to interact in the cytosol instead of the nucleus (12, 28). We found that Cch1 associates specifically with the cytoplasmic protein elongation factor 3 (EF3). It has been demonstrated that EF3 performs an essential function during the protein translation cycles in S. cerevisiae (14, 18, 49), Candida albicans (21), and Pneumocystis carinii (53). In C. neoformans, EF3 has been found to possess many of the same structural features as that of EF3 from S. cerevisiae, suggesting a similar role in protein translation (7). Interestingly, EF3 is unique to fungi because the mammalian protein translation process does not require EF3. Apart from the function of EF3 in translation, additional roles for EF3 in C. neoformans or other fungi are currently unknown. However, in higher eukaryotes, some elongation factors have been shown to function in areas unrelated to protein translation (1, 9, 39, 43). We found that the cytosolic C termini of the Cch1 and EF3 proteins form a robust interaction, which we confirmed by demonstrating that Cch1 could coimmunoprecipitate with EF3 in yeast lysates. Our results show that the repression of EF3 mRNA resulted in the mislocalization of Cch1, suggesting a role for EF3 in targeting Cch1 to the plasma membrane of C. neoformans. Further support for this notion was demonstrated by the significant growth defect of the C. neoformans antisense EF3 repression strain under conditions of limited extracellular Ca2+. Because low-affinity Ca2+ channels or transporters could not mediate the uptake of Ca2+ under these conditions, Cch1 would become essential for survival, consistent with the growth defect of the cch1Δ null mutant strain in low-Ca2+ medium (37). Taken together, our results suggest that EF3 and Cch1 are functionally related and that EF3 maintains a function apart from its role in the protein translation cycle.
MATERIALS AND METHODS
Fungal strains and media.
The strains used in this study are listed in Table 1. All the strains were recovered from 15% glycerol stocks stored at −80°C prior to use in these experiments. Strains were maintained on YPD (1% yeast extract, 2% peptone, and 2% dextrose) medium or synthetic (SD) medium lacking leucine and/or uracil. The C. neoformans H99 and the GAL7-antisense EF3 strain were grown in YP (1% yeast extract, 2% peptone) medium supplemented with either glucose or galactose for the repression and induction of antisense EF3, respectively. Where indicated, a cell-impermeable version of 1,2-bis(2-aminophenoxy)ethane-N,N,N,N,-tetraacetic acid, cesium salt (BAPTA; Invitrogen Molecular Probes) was added at a final concentration of 1 mM (52). Tunicamycin (Sigma-Aldrich) was dissolved in dimethyl sulfoxide. Unless otherwise noted, cells of C. neoformans and Saccharomyces cerevisiae were cultured in YPD medium at 30°C for 24 h.
TABLE 1.
Strains used in this study
| Strain | Genotype | Parent strain or background | Source or reference |
|---|---|---|---|
| H99 | Wild type MATα | 47 | |
| AGCn16 | cch1::URA5 MATα | H99 | 37 |
| AGCn96 | Antisense GAL7-EF3 URA5 MATα | H99 | This study |
| AGSc29 | Wild type S. cerevisiae; MATaade2-1 can1-100 his3-11 leu2-3 trp1-1ura3 | W303 | 25 |
| AGSc27 | S. cerevisiae; cdc25-2 MATaura3 leu2 trp1lys2 his200 ade101 GAL+ | 28 |
CytoTrap yeast two-hybrid assay.
The open reading frame of the cytosolic portion of the C terminus of CCH1 from C. neoformans (CCH1-C; 1,018 bp) was cloned into a pSos vector (Stratagene) to create the bait, human Ras exchange factor (hSos) fusion protein. The forward primer was 5′-CCCCGTCGACCC GCCTTACCGGCGTA TTCGAAGTGCGA-3′, and the reverse primer was 5′-CCCGCGGCC GCTCAACCC TGTTCATTTT CAATGCCTTC-3′. A C. neoformans cDNA library was cloned into the pMyr vector accordingly (using a Cytotrap XR library construction kit) and expressed as a target fusion protein with a myristoylation sequence that anchored the fusion protein to the plasma membrane. Both the bait and the target constructs were coexpressed in the yeast cdc25-2 temperature-sensitive strain. Candidate colonies expressing interacting proteins were screened by plating yeast cells on galactose medium lacking leucine and uracil at the restrictive temperature of 37°C. pMyr cDNA plasmid DNA were isolated from positive yeast cultures and recovered through the transformation of Escherichia coli, and inserts were sequenced. A BLASTX sequence similarity search was performed against a nonredundant database at NCBI to identify the potential positive candidates.
Coimmunoprecipitation (co-IP) and Western blot analysis.
For co-IP experiments, the yeast epitope-tagged shuttle vector pESC-LEU (Stratagene, La Jolla, CA) was used. These vectors contain the GAL1 and GAL10 yeast promoters in an opposing orientation so that two genes can be coexpressed and protein-protein interactions can be analyzed by immunoprecipitation analysis. The FLAG epitope is located downstream of the GAL10 promoter and the c-myc epitope is located downstream of the GAL 1 promoter. The open reading frame of the positive candidate, EF3, was PCR amplified from cDNA with the EF3 forward primer 5′TTT GCGGCCGCTATGGCTCCT GCTGCTACCGCT GCTGCCTCCTCTGGCA-3′ and the reverse primer 5′-CCCACTAGTCCAAGCTC TTCATCACTGAAGACTTCCTCTCCTCG-3′. The EF3 gene was subcloned into the Xho1 and NheI sites of the pESC-LEU vector in front of the FLAG epitope tag to generate a C-terminal FLAG-tagged EF3. The CCH1-C DNA fragment was PCR amplified using the CCH1-C forward primer 5′-CCCGAATTCACA TGGAAC AGAGGTTGATT-3′ and reverse primer 5′-CCCGTCGACATCA ACCCTGTTCATTTTGA-3′. The CCH1-C DNA fragment was subcloned into the XhoI and NheI sites of the same vector described above in front of the c-myc epitope to create a C-terminal Myc-tagged CCH1-C fragment (pESC-Cch1-C-myc). The pESC-CCH1-C-EF3 vector was transformed into the S. cerevisiae cdc25-2 strain (Table 1, strain AGSc29). Cells were cultured in SD medium minus leucine overnight and induced for 14 h in galactose.
Western blot analysis and co-IP were performed as explained below. Briefly, 10 ml of cells at mid-logarithmic growth phase were collected and washed in cold H2O. Pellets were resuspended in 300 μl of modified extraction buffer (150 mM NaCl, 50 mM Tris-Cl [pH 7.5], 50 mM NaF, 5 mM EDTA, 0.1% NP-40) with a protease inhibitor cocktail (Sigma) and lysed using a Vortexer with 0.4-ml glass beads per sample. Lysates were centrifuged at 2,000 rpm for 5 min, and the supernatants (whole-cell extracts) were precleared with 20 μl of protein G Sepharose beads for 2 h (Pierce Biotechnology). Immunoprecipitation was performed by incubating the precleared extract with 1 μl of mouse FLAG (DDDDK) tag antibody (Abcam, Cambridge MA) and 20 μl of protein G beads for 2 h. Beads were washed five times with 1 ml of extraction buffer, and samples were finally resuspended in 10 μl of 2× sodium dodecyl sulfate (SDS) sample buffer before they were loaded onto a 10% SDS-polyacrylamide gel. Western blotting for Myc-tagged proteins was performed by using a mouse monoclonal 9E10 antibody as the primary antibody (1:5,000 dilution) followed by detection with rabbit polyclonal antibody to mouse immunoglobulin G (IgG) (product H&L [horseradish peroxidase]; Abcam).
Antisense repression assay of EF3.
The open reading frame of EF3 was inserted into the JMM170 vector containing the C. neoformans galactose-inducible GAL7 promoter (26) at the NotI site, using the forward primer 5′-TTTGCGGCCGC TATGGCTCCTGC TGCTACCGCTG CTGCCTCCTC TGGCA-3′ and the reverse primer 5′-TTTGCGGCC GCTTCAGATCTTATC GTCGTCATCCTTGTAATC CAT-3′ . The resulting construct (in the sense orientation) was introduced into the JEC43 strains by biolistic transformation (19, 23). The JEC43 strain was also transformed with the GAL7-JMM170 construct, in which the orientation of EF3 was reversed to create the antisense EF3 strain (19, 23). Transformants were verified by colony PCR amplification.
Northern blot analysis.
Total RNA was isolated from the C. neoformans wild-type strain and from the antisense EF3 repression strain. Cells were grown for 24 h in YPD medium at 30°C. Equal amounts of cells were pelleted, washed with distilled water, and resuspended in YP medium with 2% galactose for 3 h to induce the EF3 antisense transcript. Cells were washed with sterile water, and subsequently, 2% glucose was added to chase out the EF3 antisense transcript. Cells were collected at 2 h, 4 h, 6 h, and 14 h after the addition of 2% glucose and centrifuged in a tabletop centrifuge. The pellet was lysed by vortexing with 2-mm glass beads, and RNA extraction was performed using an RNeasy mini-kit (Qiagen). For Northern blot hybridization studies, 7.5 μg of total RNA from cells collected from the above-described cultures was fractionated by electrophoresis in a formaldehyde-agarose gel (1.2% [wt/vol]) and blotted onto Hybond membranes. Blots were prehybridized at 65°C for 4 h in a high-stringency sodium phosphate buffer (0.5 M [pH 7.2]) with 7% SDS, and 1 mM EDTA. The EF3 transcript was detected by using [32P]dCTP (1 × 107 cpm/ml; 550-bp DNA probe against EF3). The probe was prepared with a forward (5′-ATGGCTCCTGCTGCTACC-3′ ) and reverse (5′-ATCAACTGCTGGAGAATCTC-3′ ) oligonucleotide. Northern blot hybridization was carried out at 65°C for 14 h. After blots were hybridized, they were washed in a buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% SDS) solution for 20 min at 65°C and then washed again in a 0.5× SSC buffer for 10 min at 65°C. Blots were exposed on scientific autoradiographic imaging film (Kodak) for 14 h.
Fluorescence microscopy.
Cells were grown to mid-log phase in YPD medium with 2% glucose overnight, washed with distilled water, and resuspended in YP medium plus galactose for 3 h at 30°C to induce the EF3 antisense transcript. Cells were fixed in 3% formaldehyde for 1 h at 30°C and washed twice, and spheroplasts were obtained by digesting cell walls with 40 mg/ml of lysing enzyme from Trichoderma harzianum (Amersham) in 1 M sorbitol, 10 mM sodium citrate (pH 5.8) for 3 h at 30°C. Cells were then washed in 1 M sorbitol and 10 mM sodium citrate buffer and diluted in phosphate-buffered saline (PBS) and dried on microscope slides. Cells were then incubated with a peptide antibody raised against Cch1 (1:500 dilution) (Antibodies Incorporated, Davis, CA) plus 1 mg/ml of bovine serum albumin (Sigma) at 4°C overnight. Cells were subsequently washed extensively in PBS and then incubated for 1 h with 1:1,000 diluted fluorescein isothiocyanate (FITC)-conjugated secondary antibody (Abcam, Cambridge, MA). Cells were subsequently washed three times with PBS. Immunofluorescence was examined with a Leica DMR series fluorescence microscope equipped with a Chroma 86013 filter set (Chroma Technology, Rockingham, VT) and CoolSNAP-HQ software (Roper Scientific, Tucson, AZ). The fluorescence label was visualized by using filters S484/15 for excitation and S517/30 for emission. All images were pseudocolored with Metamorph software (Universal Imaging, Downingtown, PA).
Peptide antibody of Cch1.
Peptide antibodies were raised against two short cytosolic regions of the Cch1 protein, (i) DGRDIWGDPN and (ii) SDDAHYRRDSKP (Antibodies Inc., Davis CA).
Disc diffusion halo assays and spot assays.
The antisense EF3 repression strain and a wild-type strain were cultured in YPD medium overnight at 30°C. Cells were pelleted, washed twice with sterilized water, and resuspended in YP medium. Disk diffusion assays were carried out by resuspending cells (2 × 107) in 0.7% top agar in YP medium supplemented with glucose or galactose, and this was poured onto YP medium plus glucose or YP medium plus galactose. The concentrations of tunicamycin used were 2, 8, 16, and 32 μg. Plates were incubated at 30°C for 2 days before they were imaged.
For sensitivity spot assays, the antisense EF3 repression strain and a wild-type strain were cultured in YPD medium overnight at 30°C. Cells were pelleted, washed twice with sterilized water, and resuspended in YP medium. Serially diluted cells (105, 104, 103, 102) were added to YP medium plus glucose or YP medium plus galactose supplemented with 1 mM cell-impermeant BAPTA.
RESULTS
The Ras recruitment two-hybrid method identifies EF3 as an associating protein of Cch1.
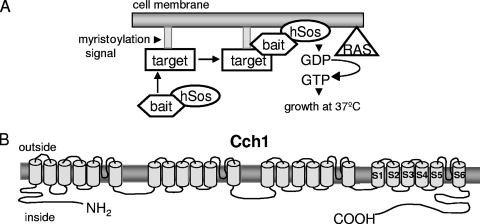
To elucidate the mechanism(s) of Cch1 regulation, we sought to identify proteins that associate specifically with Cch1, using a modified yeast two-hybrid screen. We used the Sos and Ras recruitment two-hybrid strategy because the interaction between the bait and the cDNA library takes place in the cytosol instead of the nucleus, and since Cch1 is a plasma membrane Ca2+ channel, we reasoned that this approach was more likely to find proteins that were physiologically relevant (12, 28). This two-hybrid screen uses the Ras pathway in S. cerevisiae in that when it is localized to the plasma membrane, the Ras guanyl nucleotide exchange factor Cdc25 stimulates GDP/GTP exchange on Ras (12, 28). This process activates downstream signaling events that promote cell growth. The screen is performed with a yeast mutant strain carrying a cdc25-2 allele that is not viable at 37°C; however the human Ras exchange factor (hSos) complements this mutation and restores growth at 37°C when it is targeted to the plasma membrane (Fig. 1A). The translocation of hSos from the cytosol to the plasma membrane is dependent on the bait-target interaction. The bait is fused to a C-terminally truncated hSos, which is active but unable to target the plasma membrane, and the target is anchored to the plasma membrane via a myristoylation signal. The physical interaction between bait and target recruits hSos to the plasma membrane, activates Ras signaling, and promotes growth of the cdc25-2 strain at 37°C (Fig. 1A).
FIG. 1.
(A) Schematic representation outlining the strategy in the Sos-Ras recruitment two-hybrid assay. This two-hybrid screen exploits the Ras pathway in S. cerevisiae by using a cdc25-2 mutant strain that is unable to grow at 37°C. The bait is fused to a C-terminally truncated hSos, which is active but unable to target to the plasma membrane, and the target is anchored to the plasma membrane via a myristoylation signal. A physical interaction between bait and target recruits hSos to the plasma membrane, activates Ras signaling, and promotes growth of the cdc25-2 strain at 37°C. (B) Predicted topology of Cch1 showing the cytoplasmic C-terminal portion (1,018 amino acids) expressed in yeast cells and tested in the interaction assay.
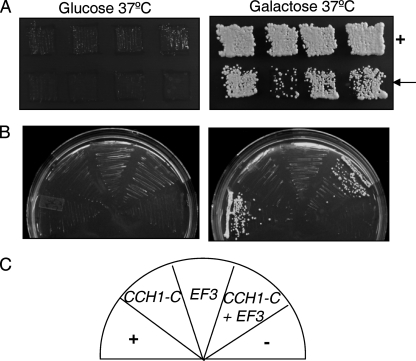
Since much of the Cch1 protein spans the plasma membrane, the regions of Cch1 most accessible to cytosolic proteins are likely to be the N- and C-terminal cytosolic portions of Cch1 (Fig. 1B) (2). In addition, these regions are often found to have a regulatory function in many different ion channels (13, 54). Thus, a Sos fusion protein of the cytosolic portion of the C terminus of Cch1 (CCH1-C) from C. neoformans was used as bait to screen a myristoylated, galactose-inducible cDNA library (the target) of C. neoformans (Fig. 1B). The S. cerevisiae temperature-sensitive (cdc25-2) strain expressing the cDNA library and CCH1-C were grown on glucose or on galactose plates lacking leucine and uracil. Approximately 5 ×105 colonies were screened, and the colonies that grew only in the presence of galactose and at the restrictive temperature of 37°C, similar to the positive control, were identified as putative candidate colonies (Fig. 2A). Upon sequencing the plasmids harboring the candidate cDNAs, we found that one encoded EF3. We were able to confirm that Cch1-C interacted with EF3, since we found that the yeast strain harboring plasmids expressing Cch1-C and full-length EF3 grew at 37°C only in the presence of galactose, suggesting that the interaction was dependent on the expression of EF3 (Fig. 2B).
FIG. 2.
Cch1 forms a robust interaction with EF3. (A) A Sos- and Ras-recruitment two-hybrid assay was used to identify potential interactors of Cch1. The bait (the cytosolic portion of the C terminus of CCH1) and target (a cDNA library from Cryptococcus neoformans) constructs were coexpressed in a temperature-sensitive (cdc25-2) strain of Saccharomyces cerevisiae. Candidate colonies expressing interacting proteins were identified by growth on galactose medium lacking leucine and uracil at the restrictive temperature of 37°C (arrow). +, positive control. (B and C) EF3 was identified as a candidate target protein. Full-length EF3 was subcloned and expressed in the cdc25-2 strain to confirm the interaction with Cch1. −, negative control.
EF3 coimmunoprecipitates with Cch1 in yeast lysates.
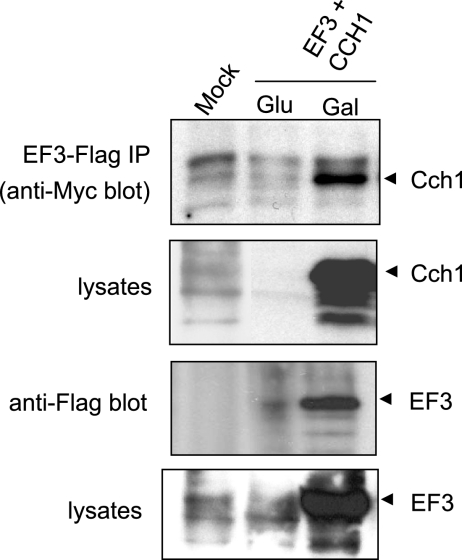
Given the interaction between Cch1-C and EF3, we asked whether Cch1-C from C. neoformans could coimmunoprecipitate with EF3 from C. neoformans. A Myc-epitope-tagged Cch1-C and a FLAG-epitope-tagged EF3, both under the control of a galactose-inducible promoter, were cotransformed into yeast. EF3 was immunoprecipitated from yeast lysates using anti-FLAG antibodies. Protein blots of the IP (IPs) were probed with anti-Myc antibodies to detect Cch1-C. We successfully detected Cch1-C in the anti-FLAG IPs and in lysates from cells that were exposed to galactose, where the expression of Cch1-C was specifically induced (Fig. 3, upper two panels). Accordingly, Cch1-C was not detected in IPs or lysates from cells exposed to glucose, where the expression of Cch1-C was repressed, or in mock experiments (Fig. 3, upper two panels). Furthermore, EF3 was also detected when the same protein blots were probed with anti-FLAG antibodies (Fig. 3, lower two panels). In contrast, EF3 was not detected in IPs or lysates from cells grown in glucose or in similar mock experiments. Moreover, the EF3-Cch1 complex was not detected in IPs from lysates of cells expressing EF3 alone (data not shown). Taken together, the data suggest that Cch1 formed a complex with EF3 in S. cerevisiae. The interaction between EF3 and full-length Cch1 could not be confirmed because the full-length gene encoding the Cch1 protein cannot be propagated in E. coli and therefore was not expressed in yeast (Liu et al., unpublished data).
FIG. 3.
EF3 coimmunoprecipitates with Cch1 in yeast cell lysates. Yeast cells were transformed with Myc-tagged Cch1 and FLAG-tagged EF3, both under the control of a galactose-inducible promoter. Yeast cell lysates were isolated from cells grown in YP medium supplemented with galactose or glucose. The Cch1-EF3 complex was immunoprecipitated via a FLAG tag on EF3, using anti-FLAG antibodies. Immune complexes were immunoblotted and probed with anti-Myc antibodies to detect Cch1 and probed with anti-FLAG antibodies to detect EF3.
The predicted structural features of C. neoformans EF3 and the creation of an EF3 antisense repression strain.
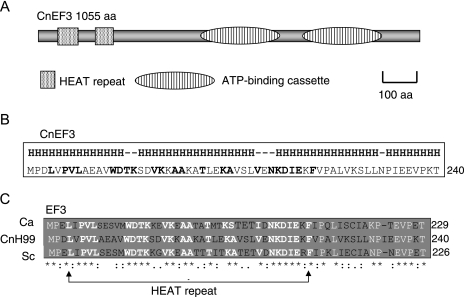
EF3 has previously been identified and characterized in C. neoformans (CnEF3) (7), as well as in Candida albicans (CaEF3) (15, 21) and S. cerevisiae (ScEF3) (2, 14, 18, 49, 51). The predicted structural features of CnEF3 include two ATP-binding regions that form ATP-binding cassettes near the C terminus (NCBI, protein family [Pfam] database) (Fig. 4A). These ATP-binding sites are also present in ScEF3 and CaEF3 (2, 14, 15, 18, 21). The ATPase activity of EF3 has been previously demonstrated, and it is thought to provide an essential function in the protein translation cycles of several fungi (14, 15, 18). Surprisingly, EF3 is unique among translational factors in fungi because mammalian translational systems do not require EF3. ScEF3 belongs to a family of proteins containing HEAT repeats, which have been defined as an alpha helical hairpin that is tandemly repeated to form a superhelical structure with hydrophobic cores (3, 36). The predicted analysis of the secondary structure of CnEF3 (SCRATCH Protein Predictor server, http://scratch.proteomics.ics.uci.edu/; PSIPRED protein structure prediction sever, http://bioinf.cs.ucl.ac.uk/psipred/) revealed that CnEF3 is a highly helical protein (Fig. 4B). This is consistent with the predicted high-helical content reported for other HEAT repeat-containing proteins (3, 35, 36). A comparison of the amino acid sequences of EF3 from C. neoformans with those of C. albicans and S. cerevisiae revealed a high degree of conservation within the HEAT repeats (Fig. 4C). Collectively, the evidence suggests that CnEF3 shares structural features that are common to EF3 from other fungi (Fig. 2C). Interestingly, HEAT repeats appear to mediate protein-protein interactions that are involved in cytoskeletal organization, vacuolar transport, and nucleocytoplasmic transport (3, 36, 45).
FIG. 4.
Schematic diagram representing the predicted structure of EF3 in Cryptococcus neoformans. (A) The structure of CnEF3 consists of two ATP-binding cassettes with ATPase activity and two HEAT repeats near the N terminus. (B) Proteins with HEAT domains such as EF3 have a very high helical content (≥50%). The amino acid sequence corresponds to the second HEAT domain in CnEF3 (where His the helical portion of protein; boldface amino acids represent conserved amino acids in the HEAT repeat). (C) HEAT repeats are highly conserved among fungi. (Ca, Candida albicans; Sc, Saccharomyces cerevisiae; CnH99, Cryptococcus neoformans var. grubii).
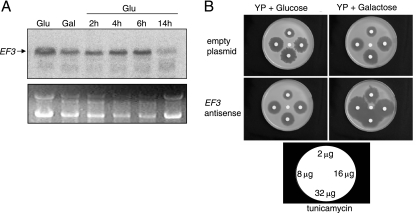
Although other elongation factors in eukaryotes have been shown to function in areas unrelated to protein translation, additional roles for EF3 have not been reported. To begin to elucidate the significance of the Cch1-EF3 complex, we examined cells of C. neoformans that lacked a functional EF3 protein. However because EF3 appears to be an essential protein, a galactose-inducible EF3 antisense repression strain was constructed (26). To confirm that the EF3 antisense strain did not result in any detectable EF3 native transcript, Northern blot analysis was performed, and the EF3 transcript was detected by an EF3 double-stranded DNA probe (550 bp) (Fig. 5A). Since the EF3 probe was made with a portion of the coding region of the EF3 cDNA sequence and the antisense strain was made by using the full-length antisense EF3, we found that this probe could bind to both the sense and antisense sequences of EF3, which resulted in cross-reactivity between the antisense and native transcripts (26). Thus, to confirm the antisense repression of the EF3 native transcript in this strain, it was necessary to chase out the galactose-induced antisense EF3 transcript. This was achieved by first inducing EF3 antisense with galactose for 3 h and subsequently repressing the EF3 antisense by the addition of glucose. We found that at 14 h after the addition of glucose, native EF3 transcript was significantly reduced in the Northern blots probed with a 32P-labeled DNA fragment of EF3, suggesting that the native mRNA of EF3 was destroyed by the antisense transcript (Fig. 5A).
FIG. 5.
Antisense repression of EF3. A. Northern blot analysis demonstrated decreased levels of native EF3 transcript in an EF3 antisense strain. RNA was isolated from a strain of Cryptococcus neoformans expressing a galactose-inducible antisense construct of EF3. Cells were grown in YP medium plus glucose (Glu) overnight, resuspended in galactose (Gal) for 3 h to induce the antisense repression of EF3 transcript, and subsequently resuspended in glucose. Cells grown in glucose for 2, 4, 6 and 14 h were harvested and RNA was isolated, separated by electrophoresis in a formaldehyde gel, blotted and probed with a 32P labeled double stranded DNA fragment of EF3. The transcript of EF3 was significantly reduced in cells grown in glucose for 14 h. rRNA was used as a loading control. (B) The antisense repression of EF3 potentiates the growth inhibition caused by tunicamycin, an ER-stressing agent. A strain of Cryptococcus neoformans expressing a galactose-inducible EF3 antisense construct was grown in YP medium plus glucose overnight, and subsequently, 2 × 107 cells were resuspended in top agar poured onto YP solid medium supplemented with either glucose or galactose. Disks containing the indicated concentrations of tunicamycin were added to the solidified medium, and a disk containing dimethyl sulfoxide was added to the center as a solvent control. Cells were incubated for 48 h at 30°C.
Because EF3 has been shown to mediate protein translation, we examined the effects of antisense EF3 in cells exposed to an ER-stressing agent. Disk diffusion halo assays were performed in the presence of tunicamycin, an agent that disrupts the glycosylation of newly synthesized proteins and promotes ER stress (10, 11, 33). The EF3 antisense strain displayed significantly larger and clear halos surrounding the disks spotted with tunicamycin when the strain was grown in the presence of galactose, conditions that induced the antisense transcript (Fig. 5B, lower panel). In the presence of glucose, the EF3 antisense strain resulted in halos similar to those observed with a wild-type strain (Fig. 5B, upper panel). This suggested that the repression of EF3 potentiated the growth inhibition caused by tunicamycin (Fig. 5B). Collectively, the hypersensitivity of the EF3 antisense strain to tunicamycin and the reduction in EF3 native transcript observed with Northern blot analysis suggest that the antisense mRNA led to the repression of the EF3 native transcript.
Repression of the EF3 transcript leads to the mislocalization of Cch1.
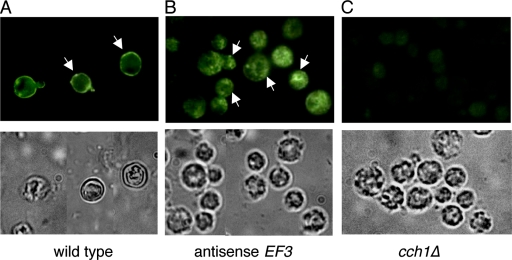
Based on the apparent role of some elongation factors in protein trafficking, we questioned whether EF3 could be affecting the localization of Cch1 to the plasma membrane. To test this notion, cells from a wild-type strain and an EF3 antisense strain were fixed, and spheroplasts were produced by digesting the cell walls. Cells were incubated with a primary peptide antibody raised against Cch1, followed by an FITC-conjugated secondary antibody. Fluorescence microscopy revealed a wild-type strain of cryptococcal cells with a fluorescent ring surrounding the cell, indicative of Cch1's localization to the plasma membrane (Fig. 6A), consistent with previous reports that have shown that Cch1 localizes to the plasma membrane in S. cerevisiae (11). Remarkably, cells from the EF3 antisense strain showed significant intracellular fluorescence with very little cell surface fluorescence, indicating that Cch1 was mislocalized under conditions that induced the repression of EF3 mRNA (Fig. 6B). The peptide antibody raised against Cch1 appeared to be specific for Cch1, since no detectable fluorescence could be observed in the cch1Δ null mutant strain (Fig. 6C). Also, fluorescence could not be detected in cells incubated with the FITC-conjugated secondary antibody alone (data not shown).
FIG. 6.
The antisense repression of EF3 leads to the mislocalization of Cch1 in Cryptococcus neoformans. Cells from an EF3 antisense repression strain were grown in YP medium plus galactose, and harvested and fixed, and spheroplasts were generated by enzyme digestion of cell walls. Cells were incubated with a primary peptide antibody raised against Cch1, followed by the addition of a FITC-conjugated secondary antibody. (A) Fluorescence microscopy revealed a fluorescent ring surrounding a wild-type cell, consistent with Cch1's localization to the plasma membrane. (B) The fluorescence pattern of Cch1 was significantly altered in the EF3 antisense repression strain. (C) No detectable fluorescence was observed for a cch1Δ null mutant strain incubated with the Cch1 peptide antibody. The corresponding bright field images of cells is shown in the lower panels. Magnification, ×100.
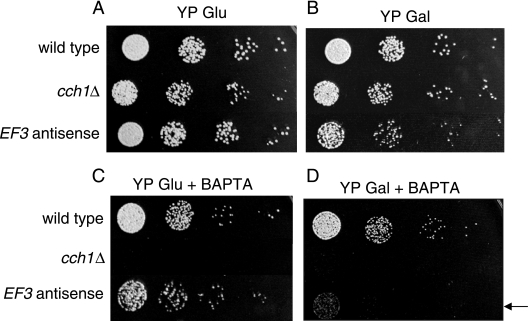
The mislocalization of Cch1 by the repression of EF3 suggests that the ability of Cch1 to mediate the influx of Ca2+ could be compromised. We have previously established that under conditions of limited extracellular Ca2+, Cch1 was the only high-affinity channel that could mediate Ca2+ uptake (37). Accordingly, we would predict that the EF3 antisense strain may be unable to grow under conditions of limited extracellular Ca2+, where Ca2+ uptake would be dependent solely on Cch1. To test this hypothesis, the effects of EF3 repression on cell growth were tested. Serially diluted cells from a galactose-inducible EF3 repression strain were used for spot assays. In the presence of YPD medium with glucose, where the free extracellular Ca2+ concentration was ∼140 μM, the EF3 antisense strain grew at a rate comparable to that of the wild-type strain (Fig. 7A). In contrast, the EF3 antisense strain showed a severe growth defect under induction conditions (YPD medium plus galactose supplemented with BAPTA, a cell-impermeant Ca2+ chelator), where the free Ca2+ concentration was significantly limiting (∼100 nM) (Fig. 7B). The growth sensitivity of the EF3 antisense strain was similar to the growth defect of the cch1Δ null mutant when it was exposed to low Ca2+ medium, as shown previously (37) and repeated here (Fig. 7C and D). The similarities in the growth sensitivity phenotypes and the mislocalization of Cch1 by the repression of EF3 would suggest that EF3 and Cch1 are functionally coupled.
FIG. 7.
The antisense repression of EF3 promotes a significant growth defect in low-Ca2+ medium. Serially diluted cells from three Cryptococcus neoformans strains, the wild type, the cch1Δ strain, and the EF3 antisense strain were added to YP medium plus glucose (A), YP medium plus galactose (B), YP medium plus glucose supplemented with cell-impermeant BAPTA (1 mM) (C), and YP medium plus galactose plus BAPTA (D). The antisense repression of EF3 led to a significant growth defect in cells grown in YP medium plus galactose in the presence of BAPTA (arrow). Strains were grown in YP medium plus glucose overnight and subsequently washed in sterile water, and serially diluted cells were resuspended in YP medium plus glucose or YP medium plus galactose before cells were spotted onto plates.
DISCUSSION
Ca2+-mediated signaling events promote many different cellular responses in pathogenic fungi, including growth at the host temperature and tolerance to the azole class of antifungals (4, 8, 16, 22, 33, 34, 37, 42, 44, 48, 50). At the top of the Ca2+ signaling cascade is Cch1, a high-affinity Ca2+ channel that mediates the influx of Ca2+ across the plasma membrane and is essential for survival in low-Ca2+ environments (24, 37, 38). Despite Cch1's primary role in promoting the extracellular influx of Ca2+ to initiate downstream Ca2+-dependent signaling events, details about the regulation of Cch1 remain unresolved.
We sought to explore the regulatory mechanisms of Cch1 from C. neoformans by identifying proteins in C. neoformans that associate specifically with Cch1. We purposely used the hSos-Ras recruitment two-hybrid strategy, since the interaction between Cch1 and its putative target would take place in the cytosol instead of the nucleus (12, 28). This approach would most likely enrich for targets that were physiologically relevant to a plasma membrane-bound protein like Cch1. This same approach has successfully identified protein partners of ligand-gated ion channels and other integral proteins (54). We found a robust interaction between Cch1 and EF3, and this interaction was confirmed by demonstrating that Cch1 could coimmunoprecipitate with EF3. Unfortunately, we were unable to test the interaction between EF3 and full-length Cch1, since subcloning and propagating CCH1 of C. neoformans in E. coli has not been possible so far. Alternative subcloning strategies for this seemingly toxic gene are currently being pursued (Liu et al., unpublished data). However, collectively, our results suggest that the interaction between EF3 and Cch1 is specific. The association of EF3 and Cch1 was demonstrated by the two-hybrid method and by the detection of the EF3-Cch1-1 immune complex in yeast lysates. In addition, the effects of the antisense repression of EF3 on the localization of Cch1 in C. neoformans and the growth inhibition of the C. neoformans EF3 repression strain in low-Ca2+ medium supports a functional relationship between EF3 and Cch1.
EF3 performs a vital role in protein translation, where it dictates the binding affinities of the ternary complex and the deacylated tRNA to the ribosomal A site and E site, respectively (2, 7). In doing so, EF3 appears to maintain a balance between the protein translation rate and amino acid fidelity (2, 7). Because EF3 is an essential protein, antisense repression under the control of an inducible promoter provided a convenient means with which to alter the EF3 gene function (26). The EF3 antisense phenotype was observed only under conditions that induced the antisense transcript, suggesting that the native EF3 mRNA was destroyed by the antisense transcript. Apart from its role in protein translation, additional functional roles for EF3 in C. neoformans or other fungi are not known. The findings reported here suggest that EF3 could indeed have alternative functions in cells.
The mislocalization of Cch1 under conditions of EF3 repression is noteworthy and suggests that EF3 is involved in regulating the plasma membrane localization of Cch1 by directly targeting Cch1 to the plasma membrane and maintaining the steady-state distribution of Cch1 channels and/or by regulating the internalization and recycling of Cch1. The role of elongation factors in protein trafficking is not without precedent; a recent report demonstrating that elongation factor 1 alpha (eEF1A) is an interaction partner of the M4 muscarinic acetylcholine receptors showed that eEF1A specifically regulates the cell surface density of the receptors by directly trafficking these proteins to the plasma membrane (39). In addition, the involvement of eEF1A in nontranslational activities is further supported by the presence of eEF1A isoforms in regions that lack translation (39). eEF1A was also found to colocalize with the alpha 2 subunit of glycine receptors, which are members of a family of ligand-gated ion channels, and to redistribute to the cytoskeletal microtubules (9). These findings support the notion that similar to eEF1A, EF3 may also have noncanonical functions that might involve protein targeting and/or protein internalization and recycling.
Further support for the potential role of EF3 in protein targeting might be attributed to the presence of the HEAT repeats, which are believed to function as protein-protein interaction domains. For example, in yeast, the plasma membrane localization of Tor2 was dependent on the HEAT repeats within Tor2 (35). Whether the HEAT repeats in EF3 dictate its ability to orchestrate the localization of Cch1 to the plasma membrane of C. neoformans remains to be seen.
The effect of EF3 repression on the localization of Cch1 suggests a probable defect in the ability of Cch1 to mediate the influx of Ca2+ across the plasma membrane. This notion is supported by the growth inhibition of the EF3 antisense repression strain in low-Ca2+ medium. Under these conditions, low-affinity Ca2+ channels or transporters could not mediate Ca2+ influx, and thus, Cch1 would become essential for survival, consistent with the inability of the cch1Δ null mutant to grow on low-Ca2+ medium. Thus, survival of C. neoformans under conditions of limited extracellular Ca2+ is dependent on the proper targeting of Cch1 to the plasma membrane.
Acknowledgments
We thank O. Cagnag and E. Blumwald for providing assistance in acquiring fluorescence images of Cryptococcus neoformans strains. We thank J. Bautos for technical assistance and members of the laboratory for critical discussions and reading of the manuscript.
This study was supported by a National Institutes of Health grant awarded to A.G.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Amiri, A., F. Noei, S. Jeganathan, G. Kuklarni, D. E. Pinke, and J. M. Lee. 2006. eEF1A2 activates Akt and stimulates Akt-dependent actin remodeling, invasion and migration. Oncogene 163027-3040. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, C. B. F., T. Becker, M. Blau, M. Anand, M. Halic, B. Balar, T. Mielke, T. Boesen, J. S. Pedersen, C. M. T. Spahn, T. G. Kinzy, G. R. Andersen, and R. Beckmann. 2006. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature 443663-668. [DOI] [PubMed] [Google Scholar]
- 3.Andrade, M. A., C. Petosa, S. I. O'Donoghue, and P. Bork. 2001. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 3091-18. [DOI] [PubMed] [Google Scholar]
- 4.Batanghari, J. W., and W. E. Goldman. 1997. Calcium dependence and binding in cultures of Histoplasma capsulatum. Infect. Immun. 655257-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bencina, M., M. Legisa, and N. D. Reed. 2005. Cross-talk between cAMP and calcium signaling in Aspergillus niger. Mol. Microbiol. 56268-281. [DOI] [PubMed] [Google Scholar]
- 6.Berridge, M. J., M. D. Bootman, and H. L. Roderick. 2003. Calcium signaling dynamics, homeotstasis and remodeling. Nat. Rev. Mol. Cell Biol. 4517-529. [DOI] [PubMed] [Google Scholar]
- 7.Blakely, G., J. Hekman, K. Chakraburtty, and P. R. Williamson. 2001. Evolutionary divergence of an elongation factor 3 from Cryptococcus neoformans. J. Bacteriol. 1832241-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blankenship, J. R., F. L. Wormley, M. K. Boyce, W. A. Schell, S. G. Filler, J. R. Perfect, and J. Heitman. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot. Cell 3422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bluem, R., E. Schmidt, C. Corvey, M. Karas, A. Schlicksupp, J. Kirsch, and J. Kuhse. 2007. Components of the translational machinery are associated with juvenile glycine receptors and are redistributed to the cytoskeleton upon aging and synaptic activity. J. Biol. Chem. 28237783-37793. [DOI] [PubMed] [Google Scholar]
- 10.Bonilla, M., K. K. Nastase, and K. W. Cunningham. 2002. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 212343-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonilla, M., and K. W. Cunningham. 2003. Mitogen-activated protein kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Cell. Biol. 144296-4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Border, Y. C., S. Katz, and A. Aronheim. 1998. The Ras recruitment system, a novel approach to the study of protein-protein interactions. Curr. Biol. 81121-1130. [DOI] [PubMed] [Google Scholar]
- 13.Catterall, W. A., J. T. Hulme, X. Jiang, and W. P. Few. 2006. Regulation of sodium and calcium channels by signaling complexes. J. Recept. Signal Transduct. Res. 26577-598. [DOI] [PubMed] [Google Scholar]
- 14.Chakraburtty, K., and F. J. Triana-Alonso. 1998. Yeast elongation factor 3: structure and function. J. Biol. Chem. 379831-840. [DOI] [PubMed] [Google Scholar]
- 15.Colthurst, D. R., B. S. Schauder, M. V. Hayes, and M. F. Tuite. 1992. Elongation factor 3 (EF3) from Candida albicans shows both structural and functional similarity to EF-3 from Saccharomyces cerevisiae. Mol. Microbiol. 61025-1033. [DOI] [PubMed] [Google Scholar]
- 16.Cruz, M. C., A. L. Goldstein, J. R. Blankenship, M. Del Poeta, D. Davis, M. E. Cardenas, J. R. Perfect, J. H. McCusker, and J. Heitman. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 4546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cyert, M. S. 2001. Genetic analysis of calmodulin and its targets in Saccharomyces cerevisiae. Annu. Rev. Genet. 35647-672. [DOI] [PubMed] [Google Scholar]
- 18.Dasmahapatra, B., and K. Chakraburtty. 1981. Protein synthesis in yeast. I. Purification and properties of elongation factor 3 from Saccharomyces cerevisiae. J. Biol. Chem. 2569999-10004. [PubMed] [Google Scholar]
- 19.Davidson, R. C., M. C. Cruz, R. A. L. Sia, B. Allen, J. A. Alspaugh, and J. Heitman. 1999. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 2938-48. [DOI] [PubMed] [Google Scholar]
- 20.Dayton, J. S., M. Sumi, N. N. Nanthakumar, and A. R. Means. 1997. Expression of a constitutively active Ca2+/calmodulin-dependent kinase in Aspergillus nidulans spores prevents germination and entry into the cell cycle. J. Bio. Chem. 2723223-3230. [DOI] [PubMed] [Google Scholar]
- 21.Di Domenico, B. J., J. Lupisella, M. Sandbaken, and K. Chakraburtty. 1992. Isolation and sequence analysis of the gene encoding translation elongation factor 3 from Candida albicans. Yeast 8337-352. [DOI] [PubMed] [Google Scholar]
- 22.Edlind, T., Smith, L. H. Karl, S. Katiyar, and J. Nickels. 2002. Antifungal activity in S. cerevisiae is modulated by calcium signaling. Mol. Microbiol. 46257-268. [DOI] [PubMed] [Google Scholar]
- 23.Edman, J. C., and K. J. Kwon-Chung. 1990. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol. Cell. Biol. 104538-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer, M. N., N. Schnell, J. Chattaway, P. Davies, G. Dixon, and D. Saunders. 1997. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 419259-262. [DOI] [PubMed] [Google Scholar]
- 25.Gelli, A. 2002. Rst1 and Rst2 are required or the a/α diploid cell type in yeast. Mol. Microbiol. 46844-854. [DOI] [PubMed] [Google Scholar]
- 26.Gorlach, J. M., H. C. McDade, J. R. Perfect, and G. M. Cox. 2002. Antisense repression in Cryptococcus neoformans as a laboratory tool and potential antifungal strategy. Microbiology 148213-219. [DOI] [PubMed] [Google Scholar]
- 27.Hallen, H. E., and F. Trail. 2008. The L-type calcium ion channel Cch1 affects ascospore discharge and mycelial growth in the filamentous fungus Gibberella zeae (Anamorph Fusarium graminearum). Eukaryot. Cell 7415-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, W., S. L. Wang, G. Lozano, and B. de Crombrugghe. 2001. cDNA library screening using the SOS recruitment system. BioTechniques 3094-100. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Iida, H., H. Nakamura, T. Ono, M. S. Okumura, and Y. Anraku. 1994. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein is required for Ca2+ influx and mating. Mol. Cell. Biol. 148259-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iida, K., T. Jinfeng, T. Tada, A. Saka, M. Tamai, H. Izumi-Nakaseko, S. Adachi-Akahane, and H. Iida. 2007. Essential, completely conserved glycine residue in the domain III S2-S3 linker of voltage-gated calcium channel alpha1 subunits in yeast and mammals. J. Biol. Chem. 28225659-25667. [DOI] [PubMed] [Google Scholar]
- 32.Joseph, J. D., and A. R. Means. 2000. Identification and characterization of two Ca2+/CaM-dependent protein kinases required for normal nuclear division in Aspergillus nidulans. J. Biol. Chem. 27538230-38238. [DOI] [PubMed] [Google Scholar]
- 33.Kaur, R., I. Castano, and B. Cormack. 2004. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 481600-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraus, P. R., C. B. Nichols, and J. Heitman. 2005. Calcium- and calcineurin-independent roles for calmodulin in Cryptococcus neoformans morphogenesis and high temperature growth. Eukaryot. Cell 61079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunz, J., U. Schneider, I. Howald, A. Schmidt, and M. N. Hall. 2000. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 27537011-37020. [DOI] [PubMed] [Google Scholar]
- 36.Li, W., L. C. Serpell, W. J. Carter, D. C. Rubinsztein, and J. A. Huntington. 2005. Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J. Biol. Chem. 28115916-15922. [DOI] [PubMed] [Google Scholar]
- 37.Liu, M., P. Du, G. Heinrich, G. M. Cox, and A. Gelli. 2006. Cch1 mediates calcium entry in Cryptococcus neoformans and is essential in low-calcium environments. Eukaryot. Cell 51788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Locke, E. G., M. Bonilla, L. Liang, Y. Takita, and K. W. Cunningham. 2000. A homolog of voltage-gated Ca2+ channels is stimulated by depletion of secretory Ca2+ in yeast. Mol. Cell. Biol. 206686-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McClatchy, D. B., G. Fang, and A. I. Levey. 2006. Elongation factor 1A family regulates the recycling of the M4 muscarinic acetylcholine receptor. Neurochem. Res. 31975-988. [DOI] [PubMed] [Google Scholar]
- 40.Muller, E. M., E. G. Locke, and K. W. Cunningham. 2001. Differential regulation of two Ca2+ influx systems by pheromone signaling in Saccharomyces cerevisiae. Genetics 1591527-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Odom, A., S. Muir, E. Lim, D. L. Toffaletti, J. Perfect, and J. Heitman. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 102576-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olarewaju, O., P. A. Ortiz, W. Q. Chowdhury, I. Chatterjee, and T. G. Kinzy. 2004. The translation elongation factor eEF1B plays a role in the oxidative stress response pathway. RNA Biol. 189-94. [DOI] [PubMed] [Google Scholar]
- 44.Onyewu, C., N. A. Afshari, and J. Heitman. 2006. Calcineurin promotes infection of the cornea by Candida albicans and can be targeted to enhance fluconazole therapy. Antimicrob. Agents Chemother. 113963-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, J. H., L. Aravind, E. C. Wolff, J. Kaevel, Y. S. Kim, and M. H. Park. 2006. Molecular cloning, expression and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc. Natl. Acad. Sci. USA 10351-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13349-355. [DOI] [PubMed] [Google Scholar]
- 47.Perfect, J. R., S. D. Lang, and D. T. Durack. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 62177-194. [PMC free article] [PubMed] [Google Scholar]
- 48.Sanglard, D., F. Ischer, O. Marchetti, J. Entenza, and J. Bille. 2003. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 4959-976. [DOI] [PubMed] [Google Scholar]
- 49.Sarthy, A. V., T. McGonigal, J. O. Capobianco, T. H. Holzman, K. A. Walter, D. A. Egan, and R. C. Goldman. 1997. High-level overexpression of yeast elongation factor 3 and detailed kinetic analysis using a coupled spectrophotometric assay. Anal. Biochem. 254288-290. [DOI] [PubMed] [Google Scholar]
- 50.Sebahati, T. S., J. T. Engle, and W. E. Goldman. 2000. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science 2901368-1372. [DOI] [PubMed] [Google Scholar]
- 51.Triana-Alonso, F., J. K. Chakraburtty, and K. H. Nierhaus. 1995. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol Chem. 27020473-20478. [DOI] [PubMed] [Google Scholar]
- 52.Tsien, R. Y. 1980. New calcium indicators and buffers with high selectivity against magnesium and protons: design, synthesis and properties of prototype structures. Biochemistry 192396-2404. [DOI] [PubMed] [Google Scholar]
- 53.Ypma-Wong, M. F., W. Fonzi, and A. Sypherd. 1992. Fungus-specific translation elongation factor 3 gene present in Pneumocystis carinii. Infect. Immun. 604140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yusifov, T., N. Savalli, C. S. Gandhi, M. Ottolia, and R. Olcese. 2008. The RCK2 domain of the human BKCa channel is a calcium sensor. Proc. Natl. Acad. Sci. USA 105376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]