Abstract
To better understand the role of histone lysine acetylation in transcription in Plasmodium falciparum, we sought to attenuate histone acetyltransferase (HAT) activity using anacardic acid (AA). We showed that AA reversibly and noncompetitively inhibited the HAT activity of recombinant PfGCN5. To a lesser extent, AA inhibited the PfGCN5 activity in parasite nuclear extracts but did not affect histone deacetylase activity. AA blocked the growth of both chloroquine-sensitive and -resistant strains, with a 50% inhibitory concentration of ∼30 μM. Treatment of the parasites with 20 μM of AA for 12 h had no obvious effect on parasite growth or gross morphology but induced hypoacetylation of histone H3 at K9 and K14, but not H4 at K5, K8, K12, and K16, suggesting inhibition of the PfGCN5 HAT. Microarray analysis showed that this AA treatment resulted in twofold or greater change in the expression of 271 (∼5%) parasite genes in late trophozoites, among which 207 genes were downregulated. Cluster analysis of gene expression indicated that AA mostly downregulated active genes, and this gene pool significantly overlapped with that enriched for H3K9 acetylation. We further demonstrated by chromatin immunoprecipitation and real-time PCR that AA treatment reduced acetylation near the putative promoters of a set of downregulated genes. This study suggests that the parasiticidal effect of AA is at least partially associated with its inhibition of PfGCN5 HAT, resulting in the disturbance of the transcription program in the parasites.
Chromatin is the physiological template for cellular processes such as DNA replication, repair, and transcription in eukaryotes. Histones, the major protein components of chromatin, not only play essential roles in packaging nuclear DNA but also serve as key regulators of genomic function (26). Histone tails are subjected to a plethora of posttranslational modifications including acetylation, methylation, phosphorylation, ubiquitylation, and sumoylation, which altogether constitute the “histone code” (7, 35, 50). These modifications affect the physical architecture of chromatin (10), generate binding platforms recognized by other proteins (19), and are important elements of epigenetic gene regulation.
The most extensively studied histone modification event is lysine acetylation, regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Based on a number of conserved structure motifs, HATs are divided into GNAT (GCN5-related N-acetyltransferase), MYST (MOZ, YBF2/SAS3, and TIP60), and p300/CBP (CREB-binding protein) families (48). These HAT families use different catalytic mechanisms: whereas p300 catalyzes a double-displacement reaction (53), the GNAT family members GCN5 and PCAF (p300/CBP associated factor) are involved in the ordered binding and release of substrates and products (52). Despite extensive pharmaceutical efforts, only a few HAT inhibitors are known to date (54). Lys-coenzyme A (Lys-CoA) and H3-CoA-20 are potent, selective inhibitors of the HATs p300/CBP and PCAF, respectively (38), but their impermeability to cells limits their uses in vivo. Along this line, a series of Lys-CoA derivatives with enhanced cell permeability have been synthesized, and some of them possess potent inhibitor activities against p300 both in vitro and in vivo (57). Toward the development of synthetic HAT inhibitors, Biel et al. designed γ-butyrolactone derivatives as GCN5 inhibitors (8). High-throughput screening also led to the identification of isothiazolones as inhibitors of PCAF and p300, which have antiproliferative activity against tumor cell lines (49). A Saccharomyces cerevisiae-based assay system has been used to screen natural and synthetic compounds and determine their specificity for GCN5 (41, 46). Recently, chemicals from natural medicinal plants such as anacardic acid (AA), garcinol, and curcumin have been shown to be potent inhibitors of p300 and PCAF (2-4). Among them, curcumin and garcinol are able to induce histone hypoacetylation in cancer cells, leading to apoptosis (2, 4, 36). In addition, garcinol and AA have served as excellent backbones from which a series of analogues have been synthesized and tested for activities inhibitory to HATs. Specifically, most of the 28 synthesized AA analogues could inhibit HAT activity in vitro and were cytotoxic to a wide range of cancer cells (23), whereas one of the compounds was a p300 activator (3). Remarkably, modifications of garcinol have led to the identification of a p300-specific inhibitor which is relatively nontoxic to human cells but able to repress human immunodeficiency virus replication (42). The fact that most eukaryotes contain multiple HAT family members and their use of distinct catalytic mechanisms offer a good opportunity for designing specific inhibitors.
Drug-resistant malaria parasites are an escalating problem for malaria control, and there is an urgent need for novel chemotherapy. The completion of the Plasmodium falciparum genome sequence has provided insights into the biological pathways of pathogenesis and offered a great opportunity for identifying effective drug targets (29). The ∼2.3-megabase genome of the P. falciparum genome, containing ∼5,640 genes, is distributed among 14 chromosomes. Like those of other eukaryotes, the malaria parasite genome is packed with histones into chromatin (12, 43) and encodes a battery of enzymes that catalyze the dynamic addition and removal of covalent modifications of histones during development (14, 24, 43). The malaria parasite has PfGCN5 and a MYST family HAT homologue (PfMYST) but lacks a p300/CBP homologue (24). Histone acetylation in the parasite has been shown to regulate the monoallelic expression of the var genes, which mediates the antigenic switching and virulence of the parasite (21, 27, 55, 56). A recent study revealed that variegated expression of genes involved in erythrocyte invasion in different parasite clones may be also under epigenetic control (13), suggesting that an epigenetic mechanism is a conserved theme for genome-wide transcription regulation in this parasite. Furthermore, the H3 acetylation mediated by PfGCN5 appears to play a key role in gene activation in the parasite (16, 24). Consequently, disturbance of histone acetylation has negative impacts on parasite development and malaria parasite HATs are viable targets for therapy. For instance, the polyphenolic compound curcumin inhibits malaria parasite growth both in vitro and in vivo (45, 47), and this antiparasitic effect is partially due to downregulation of PfGCN5 HAT activity (15).
To further explore HAT enzymes as potential targets for controlling malaria, we assessed the effect of AA, an inhibitor for both p300 and PCAF HAT families, on PfGCN5 using in vitro and in vivo assays. AA, also known as 6-pentadecanylsalicylic acid, is found in plants of the family Anacardiceae. Cashew nut shell liquid is a rich natural source, containing approximately 70% AA. We show here that AA inhibits PfGCN5 HAT activity and P. falciparum growth in culture. Consistent with the role of PfGCN5 in regulating global gene expression, AA treatment inhibits PGCN5 activity, resulting in histone hypoacetylation and downregulation of a panel of developmentally regulated genes in the parasite.
MATERIALS AND METHODS
Inhibition of recombinant PfGCN5 by AA.
A 50 mM stock solution of AA (C15:0) (Axxora, San Diego, CA) was made in dimethyl sulfoxide (DMSO). Its effect on the HAT activity of the recombinant PfGCN5 HAT domain was studied using an established HAT filter assay (15). Briefly, 1 μg (6.25 μM) of recombinant PfH3 and 30 ng (60 nM) of the recombinant PfGCN5 HAT domain were incubated in a HAT buffer in the absence (DMSO control) or presence of AA (10 to 60 μM) at 30°C for 10 min. The reaction mixtures were further incubated with 0.1 μCi of [3H]acetyl-CoA (Amersham, Piscataway, NJ) for 10 min, which was determined to be in the linear range of the reaction (data not shown). The reaction mixtures were blotted onto P-81 filters (Whatman, Florham Park, NJ), washed with 50 mM NaHCO3-Na2CO3 buffer (pH 9.2), and dried, and the radioactive counts were determined with a Beckman Coulter liquid scintillation counter (LSC). To study the inhibition kinetics of AA, the filter assays were performed similarly using a constant amount of recombinant PfH3 (10 μM) and increasing concentrations (6.5 to 1,080 nM) of [3H]Ac-CoA in the absence or presence of 10 and 15 μM of AA. The results were plotted using Kaleidograph software, version 3.5 (Synergy Software, Reading, PA). To determine whether the inhibitory effect of AA on PfGCN5 is reversible, HAT reaction mixtures were assembled with DMSO control or 10, 20, or 40 μM of AA. Whereas half of the reaction mixture was used directly for the HAT assay, the other half was dialyzed extensively for 30 min in a cold HAT buffer using a microdialyzer (Cole-Parmer, Vernon Hills, IL). Afterwards, the reaction was continued as described above, and radioactivity was measured by LSC. The assay was performed in triplicate, and the HAT activity was expressed as a percentage of the DMSO control, which was set as 100%. The relative HAT activities for the DMSO control and different concentrations of AA were compared using Tukey's t test.
Parasite culture and drug assays.
In vitro drug assays were performed using chloroquine (CQ)-sensitive P. falciparum strains 3D7 and D10 and CQ-resistant strains Dd2 and 7G8. Parasites were cultured in human O+ erythrocytes at 5% hematocrit in RPMI 1640 supplemented with 25 mM HEPES, pH 7.5, 25 mM sodium bicarbonate, 50 mg/liter hypoxanthine, 0.5% Albumax II, and 40 μg/ml gentamicin sulfate. Cultures were maintained at 37°C in a gas mixture of 5% CO2 and 3% O2. Synchronization was carried out by two 5% sorbitol treatments of ring stage parasites (37). The inhibitory effect of AA on parasite growth was determined using a standard [3H]hypoxanthine labeling method with serial dilutions of AA at final concentrations of 5 to 100 μM (15, 20). Inhibitory concentration (IC) values were calculated by linear regression analysis using Kaleidograph. Statistical analysis of IC values between different parasite strains was performed using Tukey's t test. The long-term effect of a sub-50% IC (IC50) of AA (5 or 20 μM) on the growth of 3D7 and 7G8 strains was evaluated by daily examination of parasite morphology and parasitemias for 5 days at starting parasitemias of 0.5% and 5% hematocrit. To test whether AA treatment affected particular stages of the asexual parasites, synchronized 12-h ring stage parasites treated with or without 20 μM AA were monitored for development. The proportion of each stage was determined from 300 live parasites at 24, 36, 48, and 60 h.
AA inhibition of HAT and HDAC activity in parasites.
To determine the effect of AA on PfGCN5 in cultured parasites, nuclear extracts were prepared from parasites treated with 0, 30, or 50 μM of AA for 12 and 18 h (15). HAT filter assays were performed using 1 μg (6.25 μM) of PfH3, 5 μl of nuclear extract from each treatment, and 0.1 μCi of [3H]Ac-CoA as described above. HDAC activity in cultured parasites was measured using a Histone Deacetylase kit (Millipore, Billerica, MA). Briefly, biotin-conjugated H4 peptide was radiolabeled by using PCAF and [3H]Ac-CoA and purified by streptavidin-conjugated agarose beads. In a 200-μl reaction mixture, 10 μl of each nuclear extract prepared as described above was incubated with 40,000 cpm of 3H-acetylated H4 peptide at room temperature for 24 h. After the addition of 50 μl of quenching solution, 100 μl of supernatant was used for counting with the LSC. Additionally, to test the effect of AA on HDAC in vitro, 10 μl of control nuclear extract and 40,000 cpm of 3H-acetylated H4 peptide were incubated with 10, 20, and 30 μM of AA for 24 h and HDAC activity was measured with the LSC. The HDAC inhibitor sodium butyrate (200 mM) was included as a positive control. The differences in HAT and HDAC activity between control and treated parasites were analyzed by analysis of variance (ANOVA).
Effects of AA on histone modifications.
To measure the effect of AA on covalent modifications of histone lysines in P. falciparum, histones were extracted from control 3D7 parasites and those treated with 20 μM of AA for 12 h (43). Equal amounts of histones were separated by 18% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed in Western blots with a panel of antibodies specific for histone acetylation and methylation on different lysine residues (Millipore). The specificity of these antibodies has been previously described (14, 43). To determine whether AA treatments resulted in change of PfGCN5 expression, 100 μg of protein extracts from parasites treated with 0 and 20 μM of AA for 12 h was separated by 4 to 12% NuPAGE gel (Invitrogen, Carlsbad, CA) and the PfGCN5 protein levels were estimated by immunoblotting with anti-PfGCN5 antibodies (25).
Microarray analysis.
To evaluate the effect of AA on parasite global gene expression, 3 × 109 18-h-old synchronized trophozoites were treated for 12 h with 0, 5, 10, or 20 μM of AA. Microarray analysis was performed using spotted arrays consisting of 7,462 70-mer oligonucleotides (9). RNA was isolated from control and treated parasites using TRIzol reagent and treated with RNase-free DNase I (Promega, Madison, WI) to remove contaminating genomic DNA. Fluorescence labeling with dUTP-Cy3 or dUTP-Cy5 was performed by reverse transcription using 30 μg of total RNA with both oligo(dT)12-20 and random hexamers according to the manufacturer's instructions (Amersham). Hybridization to microarray slides and data transformation were performed as described previously (16, 28). Data were further filtered to remove spots with standard deviations of >0.5 from three independent experiments. The 48-h expression profiles of the genes (9) whose expression levels had undergone twofold or greater changes after treatment with 20 μM of AA were selected for cluster analysis using the Cluster Analysis and Visualization software (22).
Real-time RT-PCR.
To validate the results from the microarray analysis, synchronized 3D7 parasites at 18 h were treated with AA (0, 5, 10, and 20 μM) for 12 h and harvested for total RNA extraction. cDNA was synthesized from 5 μg of total RNA using SuperScript III reverse transcriptase (RT) (Invitrogen) with the oligo(dT)12-17 primer. A total of 17 genes from three groups whose expression levels were either reduced, elevated, or unchanged after AA treatment were selected for real-time RT-PCR confirmation using specific primers (see Table S1 in the supplemental material). Amplification efficiency of individual genes was normalized to that of the reference gene seryl-tRNA synthetase. Relative expression levels of tested genes were determined using the 2−ΔΔCT method with ΔΔCT = (CT of AA-treated target gene − CT of control target gene) − (CT of AA-treated reference gene − CT of control reference gene) (16), where CT is the threshold cycle.
ChIP assays.
To determine whether the effect of the AA on gene expression was the result of changed histone acetylation status in the promoter regions, aliquots of the parasites for real-time RT-PCR analysis were also used for chromatin immunoprecipitation (ChIP) analysis with antibodies against acetylated H3K9 (anti-H3K9ac). ChIP was performed on equal numbers of control and AA-treated parasites. Real-time PCR was performed on the 17 selected genes using primers designed around 500 bp upstream of putative translation start codons (see Table S1 in the supplemental material). A 200-bp fragment of the PF07_0047 coding region, whose H3K9ac level from ChIP analysis did not experience change after curcumin or AA treatment, was used as an internal reference (15). The relative enrichment in treated parasites was calculated using the 2−ΔΔCT method with PF07_0047 as the internal control. To determine whether the changes in histone acetylation observed after AA treatment might be due to changes in nucleosomal occupancy at these genes, ChIP analysis was performed using anti-histone H4 antibody. After treatments with AA for 12 h, equal numbers of parasites were used for ChIP with the anti-H4 antibody and real-time PCR was done using DNA from the immunoprecipitate and input genomic DNA. The relative enrichment of H4 in treated parasites was calculated using the 2−ΔΔCT method: ΔΔCT = (CT-treated − CT-input) − (CT-control − CT-input). Finally, the relative enrichment values (representing of the relative levels of histone occupancy at the putative promoters of the selected genes) were used to normalize the H3K9ac enrichment values.
Microarray data accession number.
The microarray data were posted at http://www.ncbi.nlm.nih.gov/projects/geo/ with an accession number of GSE11763.
RESULTS
AA inhibits recombinant PfGCN5.
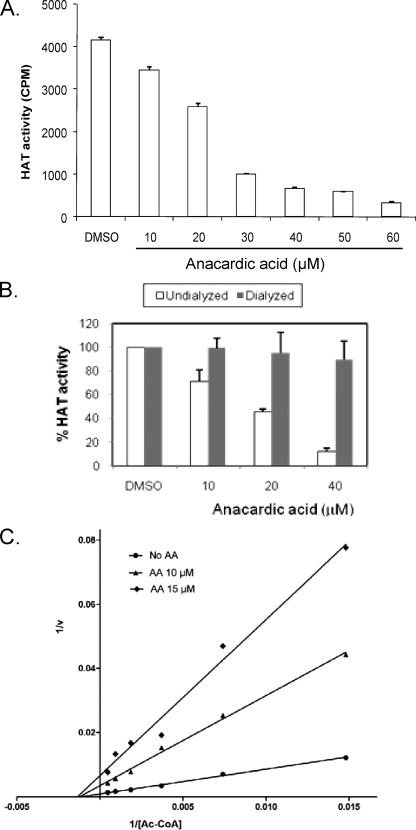
AA was found to be a noncompetitive inhibitor of the HATs p300 and PCAF (3). To determine whether AA inhibits PfGCN5, a GNAT family HAT related to PCAF, we performed filter HAT assays using the recombinant PfGCN5 HAT domain (24). Compared with the solvent DMSO control, AA inhibited the HAT activity of recombinant PfGCN5 with an IC50 of 26.6 μM (Fig. 1A). To determine whether inhibition of PfGCN5 by AA was reversible, dialysis was performed on preassembled HAT reaction mixtures. The results showed that inhibition of PfGCN5 HAT activity by AA was reversible and that the PfGCN5 HAT activity was fully restored after removal of AA by dialysis (Fig. 1B). To determine the nature of the inhibition, we analyzed the kinetics of PfGCN5 inhibition by AA. The double-reciprocal plot showed that, in the presence of AA, Km changed little, whereas Vmax of the reaction decreased (Fig. 1C). This indicated that AA acted as a noncompetitive inhibitor for PfGCN5, consistent with its mechanism of inhibition of other HAT members (3).
FIG. 1.
Effect of AA on recombinant PfGCN5 HAT activity. (A) Filter HAT assays with the recombinant PfGCN5 HAT domain and PfH3 in the absence or presence of different concentrations of AA. CPM, counts per minute. (B) Reversibility of AA's inhibition of recombinant PfGCN5. Filter HAT assays were assembled, and the reaction mixtures were either dialyzed or undialyzed before proceeding with the addition of [3H]Ac-CoA. The HAT activity of reactions using AA was compared with that from DMSO controls, which was set as 100%. Results represent means and standard deviations from three replicates. (C) Lineweaver-Burk plot showing the inhibition kinetics of AA for PfGCN5, as measured with a constant amount of PfH3 and increasing amounts of [3H]Ac-CoA.
AA inhibits parasite growth in culture.
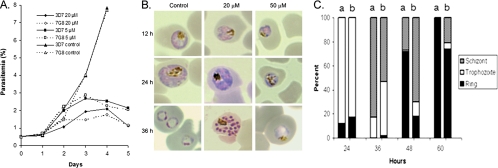
To study the effect of AA on the cultured malaria parasite, we first tested the effect of a range of AA concentrations on synchronized ring stage P. falciparum 3D7 and found that 100 μM of AA completely blocked parasite growth. Using the [3H]hypoxanthine incorporation assay, we determined the IC50s for four parasite strains to be 30 to 34 μM and IC90s to be 54 to 57 μM (Table 1). No significant difference in sensitivity between the strains tested was found (Tukey's t test, P > 0.05). Treatments with sub-IC50 concentrations of AA (5 and 20 μM) inhibited parasite growth to different degrees, and the growth curves for 3D7 and 7G8 strains were similar (Fig. 2A). After two asexual erythrocytic cycles, parasitemias in the controls had increased from 0.5 to almost 8%, whereas parasitemias in AA-treated parasites only reached less than 2.5%. At 20 μM of AA, parasite development and morphology appeared normal at 12 h, but growth retardation began to show at 24 h and became increasingly obvious at later time points, resulting in a less synchronous culture (Fig. 2B and C). We did not observe accumulation of a particular developmental stage, suggesting that AA's action was not stage specific. In comparison, 50 μM of AA blocked parasite growth and gross morphological abnormalities such as chromatin condensation and vacuole formation were evident even at 24 h after treatment (Fig. 2B).
TABLE 1.
Sensitivity of four P. falciparum strains to AA in vitroa
| Parasite strain | IC50 (μM) | IC90 (μM) |
|---|---|---|
| 3D7 | 30.41 ± 4.54 | 54.74 ± 5.31 |
| D10 | 32.13 ± 1.41 | 57.83 ± 1.57 |
| 7G8 | 34.76 ± 0.57 | 60.75 ± 3.22 |
| Dd2 | 31.06 ± 5.68 | 55.91 ± 6.73 |
Values are means ± standard deviations from three replicates. Values in each column are not significantly different from each other at the P = 0.05 level (Tukey's t test).
FIG. 2.
Effect of AA on P. falciparum. (A) Inhibitory effect of sub-IC50s (5 and 10 μM) of AA on in vitro growth of parasite strains 3D7 and 7G8. Shown are average daily parasitemias determined from three replicates of synchronized culture with a 0.5% initial parasitemia. (B) Inhibition of intraerythrocytic growth of 3D7 P. falciparum. Shown is the gross morphology of Giemsa-stained parasites from control (DMSO) and AA-treated (at 20 or 50 μM) cultures. Note the growth retardation and vacuolated parasites, especially after 50 μM of AA treatment. (C) Proportions of each asexual erythrocytic stage at different times points in control (a) and AA-treated (b) cultures. Synchronized 12-h ring stage parasites at 0.5% parasitemia were treated with or without 20 μM of AA, and the proportion of each stage was obtained by counting 300 live parasites.
AA inhibits PfGCN5 in parasites.
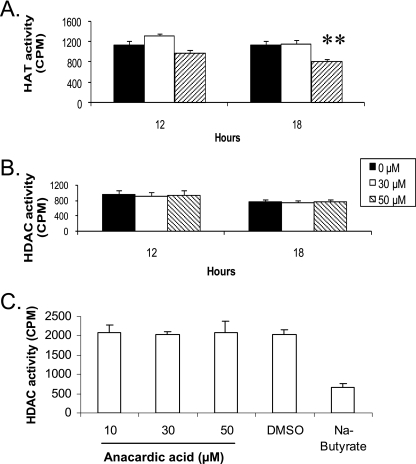
To establish whether AA could inhibit PfGCN5 activity in parasites, we treated parasites with 30 and 50 μM of AA (approximately IC50 and IC90) and evaluated HAT activity of the nuclear extracts using the filter assay. While 30 μM of AA did not result in significant changes in nuclear HAT activity for up to 24 h, 50 μM of AA led to only a 20% reduction in nuclear HAT activity (Fig. 3A). We further tested the effect of AA on parasite HDAC activity. Treatment of the 3D7 parasite with 30 and 50 μM of AA for 12 and 18 h had no visible effect on the HDAC activity of the nuclear extracts (Fig. 3B). When the HDAC assays were carried out with the addition of AA (10 to 50 μM) to a control nuclear extract, no changes in HDAC activity were observed compared to ∼70% reduction of HDAC activity with sodium butyrate (Fig. 3C).
FIG. 3.
Effects of AA on in vivo HAT and HDAC activity in P. falciparum. (A) HAT activity in equal amounts of nuclear extracts from control and AA-treated parasites for 12 and 18 h was measured using PfH3. The asterisks indicate that HAT activity in the nuclear extract of 50 μM-AA-treated parasites at 18 h was significantly different from that of the control (P < 0.01, ANOVA). (B) HDAC activity of the same nuclear extracts shown in panel A. There were no significant differences between the controls and AA-treated parasites at the two time points (P > 0.05, ANOVA). (C) HDAC assays were performed with a control nuclear extract incubated with DMSO, different concentrations of AA, or sodium butyrate. The means plus standard deviations (error bars) from three replicates are shown. AA (10 to 50 μM) had no significant effect on HDAC activity in the nuclear extract (P > 0.05, ANOVA).
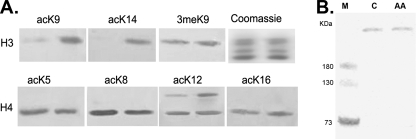
To establish the effect of PfGCN5 on histone acetylation, we determined the acetylation status of parasite histones after AA treatment using antibodies specific for lysine acetylation and methylation of H3 and H4. The results showed that treatment of parasites with 20 μM of AA for 12 h did not lead to obvious changes in acetylation levels at four lysine residues of histone H4, nor did it affect H3K9 trimethylation (Fig. 4A). However, AA treatment significantly reduced acetylation of H3K9 and H3K14, suggesting that AA might have affected PfGCN5 activity. In addition, AA treatment did not cause apparent changes in the amount of PfGCN5 protein in the parasite, indicating that alteration of histone acetylation was likely due to inhibition of PfGCN5 (Fig. 4B).
FIG. 4.
Effect of AA treatment (20 μM) for 12 h on histone acetylation and expression of PfGCN5. (A) Western blots of histone with a panel of antibodies specific for acetylated (ac) lysines (K) in histones H3 (K9 and K14) and H4 (K5, K8, K12, and K16) and antibodies for trimethylated H3K9 (3meK9). The Coomassie blue-stained gel indicates approximately equal loading. Histones were extracted from control (right lanes) and AA-treated (left lanes) parasites. (B) Western blot with antibodies against PfGCN5 using equal amounts of protein extracts (100 μg per lane) from control (C) and AA-treated parasites (AA). M indicates molecular markers.
Effects of AA on global gene expression.
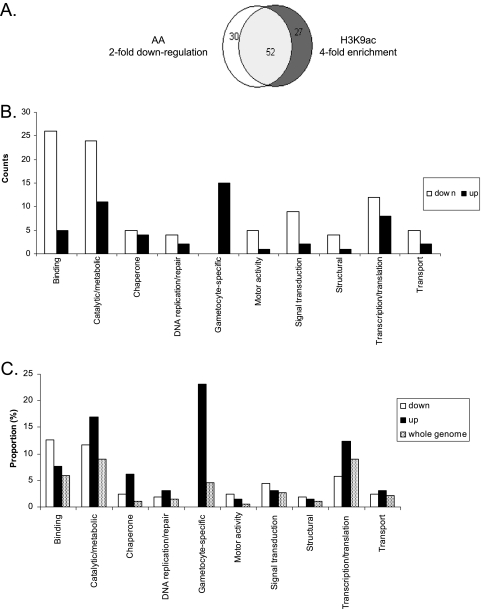
To study the effect of HAT inhibitors on parasite global gene expression, we performed a microarray analysis using AA. AA was favored over another HAT inhibitor, curcumin, in order to avoid additional complications resulting from the high levels of reactive oxygen species produced by curcumin treatment. Since gene expression during the asexual erythrocytic cycle of the parasite is stage specific (9, 39), we first tested conditions of AA treatment and selected 20 μM of AA for 12 h, as this treatment did not cause observable changes in parasite growth but resulted in considerable alteration of global gene expression (data not shown). Using this established condition, we measured global gene expression in P. falciparum 3D7 with spotted arrays and compared levels of gene expression in control and AA-treated parasites. The results were highly reproducible, and there were high correlations among the three independent experiments (Pearson correlation, R2 > 0.8, P < 0.001). To enhance the consistency of the study and evaluate the effect of different degrees of PfGCN5 inhibition, we tested three concentrations of AA for the microarray experiments. The results showed that treatment of 18-h late-ring stage parasites for 12 h with 5, 10, and 20 μM of AA led to a twofold or greater downregulation of 60, 109, and 207 genes and a twofold or greater upregulation of 66, 60, and 64 genes, respectively. Besides, among the genes downregulated at 5 and 10 μM of AA, 96.7% (58/60) and 86.9% (93/107) were also found in the downregulated gene pool at 20 μM of AA. These results also revealed that a greater number of genes were downregulated than upregulated, consistent with the effects of another HAT inhibitor, garcinol, and its derivatives on gene expression in HeLa cells (2, 42). The observed downregulation of a consistent set of genes agrees well with the expectation that reduced histone acetylation (possibly due to attenuation of PfGCN5 activity) would lead to downregulation of gene expression. To further support this argument, we compared the results from this study with those from our earlier H3K9ac ChIP-chip analysis (16). Based on the notion that H3K9ac is associated with the promoter regions, we selected only genes whose microarray oligonucleotides are located within 500 bp of the putative ATG start codons. These included 82 genes downregulated twofold or greater by 20 μM of AA and 79 genes with fourfold or greater H3K9ac enrichment over the input genomic DNA. This analysis showed that >63% of genes identified in one study were also found in the other (Fig. 5A), indicating that genes highly enriched with H3K9ac at the putative promoter regions in our earlier mapping were correlated with genes downregulated by AA treatment (16). In contrast, upregulation by AA treatment was not dosage dependent, with similar numbers of parasite genes being upregulated by different AA concentrations. While this was not expected, a plausible explanation could be that the upregulation was not sensitive to dosage variation of AA at the concentration range used. Moreover, the pools of upregulated genes had only 21 (32 to 35%) genes in common, suggesting that AA increased expression of a large number of genes in a nonspecific manner.
FIG. 5.
Microarray analysis to determine global expression response to 20 μM of AA treatment for 12 h. (A) Venn diagram showing the overlap of genes that were downregulated twofold or greater by AA treatment and those that had fourfold or greater enrichment with H3K9ac over input genomic DNA (16). Note that only genes whose microarray oligonucleotides are within 500 bp of the putative ATG codons were chosen for this analysis. (B) Numbers of different functional categories of genes with twofold or greater downregulation or upregulation by AA treatment. Note that genes encoding hypothetical proteins are not shown. (C) Proportion of genes within each functional category in AA-deregulated pools and the parasite genome.
Based on functional annotation of parasite genes in PlasmoDB, we classified the genes responsive to 20 μM AA treatment into potential functional categories. The largest category included 112 downregulated and 27 upregulated genes encoding hypothetical proteins with unknown functions. Genes encoding proteins with binding and catalytic/metabolic activity and genes involved in translation/transcription were among the most abundant groups shared between the down- and upregulated gene pools (Fig. 5B). The group of genes with binding activity that were downregulated by AA treatment includes quite a few members of the multigene families var, rif, and clag, whose expression is under epigenetic control (13, 21, 27, 56). Whereas most of the responsive genes are distributed on different chromosomes, a cluster of four genes encoding the SERA (serine repeat antigen) cysteine proteases were downregulated (see Table S2 in the supplemental material). Eight SERA genes are clustered on chromosome 2, and they have similar expression profiles, with peak expression at late erythrocytic stages, suggesting that they might be coordinately regulated (44). In addition, a significant number of genes (∼5%) encoding kinases and phosphatases that are potentially involved in signal transduction were repressed rather than activated by AA treatment. A comparison of the proportions of these groups of genes in the genome indicated that only genes involved in binding and signal transduction had high representation among the AA-downregulated genes, whereas genes whose products had catalytic/metabolic activity, genes encoding chaperones, and genes expressed in gametocytes were enriched among the AA-upregulated genes (Fig. 5C). Since microarray analysis showed that AA treatments did not affect the expression of the HAT and HDAC genes, the deregulation of gene expression in the parasites by AA may not be the direct result of altered expression of these chromatin regulators. It is noteworthy that AA treatment disturbed the expression of a number of chaperones, DNA repair enzymes, and peroxidoxin, which implies a general stress response in the parasite (Fig. 5B; see Table S2 in the supplemental material). Furthermore, such stress was reflected in the upregulation of two known early gametocyte genes (Pfg27/25 and Pfs16) and several gametocyte-specific genes, as gametocytogenesis is often indicative of a parasite's response to stress (11).
We have performed cluster analysis on the expression profiles of the genes with twofold or greater changes after 20 μM AA treatment (see Fig. S1 in the supplemental material). The results showed that the majority of the downregulated genes were active at the time of the microarray analysis according to the 48-h expression profile (9). As for the upregulated genes, only a small portion (13/64) were expressed; the majority of them were silent at the time of microarray analysis. Taken together, these results indicated that AA mostly downregulated active genes, whereas most of the upregulated genes were silent in late trophozoites.
Validation of differentially expressed genes after AA treatment.
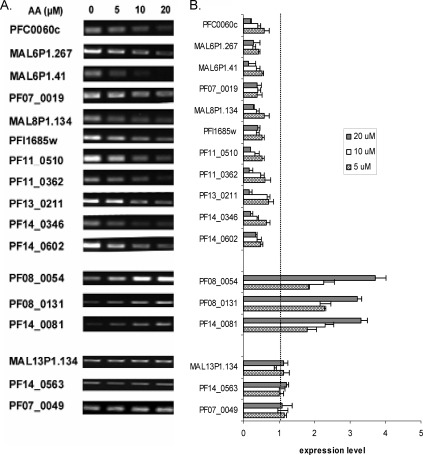
The relative expression levels of genes displaying alteration in response to AA treatment were further evaluated by real-time RT-PCR analysis (Fig. 6). We selected 11 downregulated, three upregulated, and three invariant genes for RT-PCR analysis. We were particularly interested in the category of genes potentially involved in signaling pathways. Among the 11 AA-downregulated genes selected for validation, 10 encode putative protein kinases or phosphatases. The semiquantitative and real-time RT-PCR results confirmed that AA treatment repressed the expression of the 11 genes, and their expression levels appeared inversely correlated with AA concentrations (Fig. 6A and B). In contrast, the expression levels of three AA-upregulated genes (heat shock protein 70, DNA repair helicase, and peroxidoxin genes) that are potentially involved in detoxification were all elevated in AA-treated parasites. Consistently, AA treatment did not affect the expression of the three unchanged genes from microarray analysis.
FIG. 6.
RT-PCR analysis of gene expression to validate results of the microarray analysis. (A) Semiquantitative RT-PCR was performed using cDNA synthesized from equal amounts of total RNA from parasites treated with 0, 5, 10, and 20 μM of AA for 12 h. Top, middle, and bottom panels show downregulated, upregulated, and invariant genes, respectively. For downregulated genes 22 cycles of PCR were performed, whereas 25 cycles were done for the rest of genes. (B) Real-time RT-PCR was performed, and the relative expression level was the 2−ΔΔCT value determined by comparing AA-treated parasites with untreated controls and using gene PF07_0047 as the internal control.
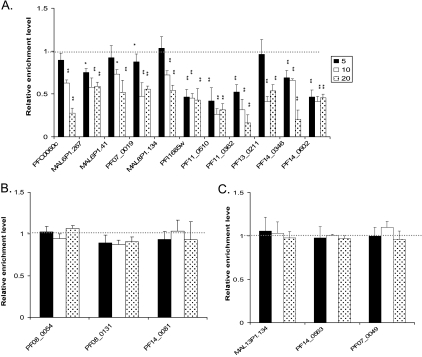
We have shown that PfGCN5-mediated H3K9ac is associated with active genes at the promoter region and that the reduced gene expression after treatment with another HAT inhibitor, curcumin, is partially due to reduced H3 acetylation at the promoters of these genes (15). ChIP and real-time PCR analysis of the same set of selected genes detected a dramatic reduction of H3K9ac in the upstream regions of downregulated genes compared to insignificant changes in the 5′ regions of most upregulated and invariant genes (Fig. 7). Compared with the control, all downregulated genes had significant reduction in H3K9ac at the 5′ regions at the higher AA concentrations (10 and 20 μM), whereas one-half of the selected genes also experienced a significant level of hypoacetylation at 5 μM of AA (Fig. 7A). These results indicated that AA treatment resulted in H3K9 hypoacetylation (possibly through inhibition of PfGCN5 activity) at the promoters of the downregulated genes.
FIG. 7.
Effect of AA treatment (0, 5, 10, and 20 μM) for 12 h on the levels of H3K9 acetylation at the putative promoter regions of selected downregulated (A), upregulated (B), and invariant (C) genes as shown in Fig. 6. ChIP was performed with anti-H3K9ac using equal numbers of parasites, and real-time PCR was done using a pair of primers around 500 bp upstream of the putative ATG codon of each gene. The relative enrichment level represents the 2−ΔΔCT value obtained by comparing AA-treated parasites with untreated controls and normalizing the data first with the control gene PF07_0047 and then with the relative nucleosome occupancy value determined by ChIP using anti-H4. * and **, significant difference at P < 0.05 and P < 0.01 levels, respectively (ANOVA), between the AA-treated parasites and controls. The break line at a relative enrichment level of 1 indicates the baseline value for the untreated control.
DISCUSSION
To date, a number of synthetic and natural compounds have recently been identified as HAT inhibitors (54). Of the naturally occurring compounds, AA and garcinol are potent inhibitors of both p300 and PCAF, while curcumin is only active against p300 (2-4). Target specificity is important for developing effective chemotherapeutics. However, the specificity of these HAT inhibitors for the broad spectrum of HAT families has not been fully tested. For example, AA has recently been found to also inhibit the MYST family HAT member Tip60, thus blocking the acetylation and activation of the ATM protein kinase in HeLa cells and sensitizing the cells to the cytotoxic effects of radiation (51). In addition, we have previously documented that curcumin, which is not active against human PCAF, is able to inhibit PfGCN5, a GNAT family member (15). This could result from the significant divergence of PfGCN5 from its mammalian counterparts (24). In this study, we have shown that AA possesses equally potent antiparasitic activity against both CQ-sensitive and -resistant parasite strains. Despite limited cell permeability in mammalian cells, AA still effectively blocked the development of intraerythrocytic parasites, suggesting that AA is accessible to the parasites. This enhanced accessibility possibly owes to the increased permeability of the red cell membrane due to extensive remodeling by the parasite (33). While our in vitro and in vivo data strongly suggest that the parasite-toxic effect of AA is due to its inhibition of PfGCN5 activity, we observed only insignificant reduction of HAT activity in nuclear extracts at concentrations of AA inhibitory for parasite growth. While the reason for this discrepancy is not clear, it is possible that the reversible binding of AA to PfGCN5 may have resulted in the partial loss of AA and restoration of PfGCN5 activity during the preparation of nuclear extracts. This argument is supported by the observation of H3K9 and H3K14 hypoacetylation at 20 μM of AA despite that fact that this concentration did not induce loss of PfGCN5 HAT activity in nuclear extracts. We have shown that H3K9 and H3K14 are the preferred sites for PfGCN5, whereas PfMYST preferentially acetylates H4K8 and H4K12 (15; J. Miao et al., unpublished data). The effect of AA and curcumin on PfGCN5 may also affect nonhistone targets in malaria parasites which remain to be identified. Moreover, the biological activity of AA may be in addition to its actions against other unidentified targets in the malaria parasites, since AA is known to inhibit a large spectrum of enzymes in other organisms (reference 32 and references therein). Therefore, at present, the parasiticidal effect of AA could be only partially attributed to its inhibition of PfGCN5.
Genome-wide expression profiling has been successfully used to identify the pathways that particular drugs act on and to elucidate the possible resistance mechanisms. In yeasts, this has led to the identification of a coordinated transcriptional response to specific antifungal drug categories as well as genes that belong to the general detoxification responses (see, e.g., references 1, 5, and 40). In some cases, the response to chemical perturbation is profound: chemical carcinogens could evoke significant changes in transcript level for more than one-third of the ∼6,200 genes in budding yeast (34). This is in sharp contrast to what is found for the malaria parasite, where attempts to relate global gene expression changes to perturbation with antimalarial drugs often detect either subtle changes in steady-state RNA levels or non-function-related changes as the result of altered growth rates of the treated parasites (18, 30, 31), which led to the speculation that the malaria parasite transcription regulatory network is probably “hardwired.” However, some studies did find consistent impact of drug treatments on specific metabolic pathways of the parasite, suggesting that the parasite has some capacity to respond at the transcript level to environmental perturbations (17, 18). In this study, we found that AA treatment at a sub-IC50s for a short period of time resulted in twofold or greater changes in ∼5% of the parasite genes in late trophozoites. Our finding also concurs with findings for other organisms such as baker's yeast, where the extent of transcriptional response was correlated with the concentrations of the chemicals (6, 34). Besides, most of the AA-responsive genes were downregulated, possibly due to induced hypoacetylation of H3K9 at the promoters of active genes. Interestingly, both CQ and AA affected a category of genes potentially participating in cell signaling and upregulated several genes involved in general stress response and detoxification (31). Regardless of the variability in the AA-upregulated genes among experiments, >32% of the AA-responsive loci are shared, suggesting that AA could potentially deregulate particular cellular pathways in the parasite. Moreover, the finding that AA treatment also downregulated members of multigene families (var, rif, and clag) encoding clonally variant surface antigens should be explored in future investigations. Taken together, these data demonstrate the potential utility of gene expression profiling in antimalarial drug development and elucidation of the underlying mechanisms.
Supplementary Material
Acknowledgments
We are grateful to Bill Sullivan at Indiana University for critically reading the manuscript.
We thank the NIH for grant support (AI064553) to L.C. and the Division of Intramural Research, NIAID, NIH, for support to X.S.
Footnotes
Published ahead of print on 16 May 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 27834998-35015. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanyam, K., M. Altaf, R. A. Varier, V. Swaminathan, A. Ravindran, P. P. Sadhale, and T. K. Kundu. 2004. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J. Biol. Chem. 27933716-33726. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanyam, K., V. Swaminathan, A. Ranganathan, and T. K. Kundu. 2003. Small molecule modulators of histone acetyltransferase p300. J. Biol. Chem. 27819134-19140. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramanyam, K., R. A. Varier, M. Altaf, V. Swaminathan, N. B. Siddappa, U. Ranga, and T. K. Kundu. 2004. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 27951163-51171. [DOI] [PubMed] [Google Scholar]
- 5.Barker, K. S., S. Crisp, N. Wiederhold, R. E. Lewis, B. Bareither, J. Eckstein, R. Barbuch, M. Bard, and P. D. Rogers. 2004. Genome-wide expression profiling reveals genes associated with amphotericin B and fluconazole resistance in experimentally induced antifungal resistant isolates of Candida albicans. J. Antimicrob. Chemother. 54376-385. [DOI] [PubMed] [Google Scholar]
- 6.Benton, M. G., S. Somasundaram, J. D. Glasner, and S. P. Palecek. 2006. Analyzing the dose-dependence of the Saccharomyces cerevisiae global transcriptional response to methyl methanesulfonate and ionizing radiation. BMC Genomics 7e305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-411. [DOI] [PubMed] [Google Scholar]
- 8.Biel, M., A. Kretsovali, E. Karatzali, J. Papamatheakis, and A. Giannis. 2004. Design, synthesis, and biological evaluation of a small-molecule inhibitor of the histone acetyltransferase Gcn5. Angew. Chem. Int. Ed. Engl. 433974-3976. [DOI] [PubMed] [Google Scholar]
- 9.Bozdech, Z., M. Llinas, B. L. Pulliam, E. D. Wong, J. Zhu, and J. L. DeRisi. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brower-Toland, B., D. A. Wacker, R. M. Fulbright, J. T. Lis, W. L. Kraus, and M. D. Wang. 2005. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J. Mol. Biol. 346135-146. [DOI] [PubMed] [Google Scholar]
- 11.Buckling, A., C. L. Ranford-Cartwright, A. Miles, and A. F. Read. 1999. Chloroquine increases Plasmodium falciparum gametocytogenesis in vivo. Parasitology 118339-346. [DOI] [PubMed] [Google Scholar]
- 12.Cary, C., D. Lamont, J. P. Dalton, and C. Doerig. 1994. Plasmodium falciparum chromatin: nucleosomal organisation and histone-like proteins. Parasitol. Res. 80255-258. [DOI] [PubMed] [Google Scholar]
- 13.Cortes, A., C. Carret, O. Kaneko, B. Y. S. Y. Lim, A. Ivens, and A. A. Holder. 2007. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PloS Pathog. 31023-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui, L., Q. Fan, L. Cui, and J. Miao. 2008. Histone lysine methyltransferases and demethylases in the malaria parasite Plasmodium falciparum. Int. J. Parasitol. 381083-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui, L., J. Miao, and L. Cui. 2007. Cytotoxic effect of curcumin on malaria parasite Plasmodium falciparum: inhibition of histone acetylation and generation of reactive oxygen species. Antimicrob. Agents Chemother. 51488-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui, L., J. Miao, T. Furuya, X. Li, X. Su, and L. Cui. 2007. PfGCN5 mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum. Eukaryot. Cell 61219-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahl, E. L., J. L. Shock, B. R. Shenai, J. Gut, J. L. DeRisi, and P. J. Rosenthal. 2006. Tetracyclines specifically target the apicoplast of the malaria parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 503124-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deitsch, K., M. Duraisingh, R. Dzikowski, A. Gunasekera, S. Khan, K. Le Roch, M. Llinás, G. Mair, V. McGovern, D. Roos, J. Shock, J. Sims, R. Wiegand, and E. Winzeler. 2007. Mechanisms of gene regulation in Plasmodium. Am. J. Trop. Med. Hyg. 77201-208. [PubMed] [Google Scholar]
- 19.de la Cruz, X., S. Lois, S. Sanchez-Molina, and M. A. Martinez-Balbas. 2005. Do protein motifs read the histone code? Bioessays 27164-175. [DOI] [PubMed] [Google Scholar]
- 20.Desjardins, R. E., C. J. Canfield, J. D. Haynes, and J. D. Chulay. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother. 16710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duraisingh, M. T., T. S. Voss, A. J. Marty, M. F. Duffy, R. T. Good, J. K. Thompson, L. H. Freitas-Junior, A. Scherf, B. S. Crabb, and A. F. Cowman. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 12113-24. [DOI] [PubMed] [Google Scholar]
- 22.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 9514863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eliseeva, E. D., V. Valkov, M. Jung, and M. O. Jung. 2007. Characterization of novel inhibitors of histone acetyltransferases. Mol. Cancer Ther. 62391-2398. [DOI] [PubMed] [Google Scholar]
- 24.Fan, Q., L. An, and L. Cui. 2004. Plasmodium falciparum histone acetyltransferase, a yeast GCN5 homologue involved in chromatin remodeling. Eukaryot. Cell 3264-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan, Q., L. An, and L. Cui. 2004. PfADA2, a Plasmodium falciparum homologue of the transcriptional coactivator ADA2 and its in vivo association with the histone acetyltransferase PfGCN5. Gene 336251-261. [DOI] [PubMed] [Google Scholar]
- 26.Felsenfeld, G., and M. Groudine. 2003. Controlling the double helix. Nature 421448-453. [DOI] [PubMed] [Google Scholar]
- 27.Freitas-Junior, L. H., R. Hernandez-Rivas, S. A. Ralph, D. Montiel-Condado, O. K. Ruvalcaba-Salazar, A. P. Rojas-Meza, L. Mancio-Silva, R. J. Leal-Silvestre, A. M. Gontijo, S. Shorte, and A. Scherf. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 12125-36. [DOI] [PubMed] [Google Scholar]
- 28.Furuya, T., J. Mu, K. Hayton, A. Liu, J. Duan, L. Nkrumah, D. A. Joy, D. A. Fidock, H. Fujioka, A. B. Vaidya, T. E. Wellems, and X. Z. Su. 2005. Disruption of a Plasmodium falciparum gene linked to male sexual development causes early arrest in gametocytogenesis. Proc. Natl. Acad. Sci. USA 10216813-16818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner, M. J., N. Hall, E. Fung, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunasekera, A. M., A. Myrick, K. Le Roch, E. Winzeler, and D. F. Wirth. 2007. Plasmodium falciparum: genome wide perturbations in transcript profiles among mixed stage cultures after chloroquine treatment. Exp. Parasitol. 11787-92. [DOI] [PubMed] [Google Scholar]
- 31.Gunasekera, A. M., S. Patankar, J. Schug, G. Eisen, and D. F. Wirth. 2003. Drug-induced alterations in gene expression of the asexual blood forms of Plasmodium falciparum. Mol. Microbiol. 501229-1239. [DOI] [PubMed] [Google Scholar]
- 32.Ha, T. J., and I. Kubo. 2005. Lipoxygenase inhibitory activity of anacardic acid. J. Agric. Food Chem. 534350-4354. [DOI] [PubMed] [Google Scholar]
- 33.Haldar, K., and N. Mohandas. 2007. Erythrocyte remodeling by malaria parasites. Curr. Opin. Hematol. 14203-209. [DOI] [PubMed] [Google Scholar]
- 34.Jelinsky, S. A., and L. D. Samson. 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA 961486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 36.Kang, S. K., S. H. Cha, and H. G. Jeon. 2006. Curcumin-induced histone hypoacetylation enhances caspase-3-dependent glioma cell death and neurogenesis of neural progenitor cells. Stem Cells Dev. 15165-174. [DOI] [PubMed] [Google Scholar]
- 37.Lambros, C., and J. P. Vanderberg. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65418-420. [PubMed] [Google Scholar]
- 38.Lau, O. D., T. K. Kundu, R. E. Soccio, S. Ait-Si-Ali, E. M. Khalil, A. Vassilev, A. P. Wolffe, Y. Nakatani, R. G. Roeder, and P. A. Cole. 2000. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell 5589-595. [DOI] [PubMed] [Google Scholar]
- 39.Le Roch, K. G., Y. Zhou, P. L. Blair, M. Grainger, J. K. Moch, J. D. Haynes, P. De la Vega, A. A. Holder, S. Batalov, D. J. Carucci, and E. A. Winzeler. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 3011503-1508. [DOI] [PubMed] [Google Scholar]
- 40.Liu, T. T., R. E. B. Barker, K. S. Lee, R. E. Lee, L. Wei, R. Homayouni, and P. D. Rogers. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 492226-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mai, A., D. Rotili, D. Tarantino, P. Ornaghi, F. Tosi, C. Vicidomini, G. Sbardella, A. Nebbioso, M. Miceli, L. Altucci, and P. Filetici. 2006. Small-molecule inhibitors of histone acetyltransferase activity: identification and biological properties. J. Med. Chem. 496897-6907. [DOI] [PubMed] [Google Scholar]
- 42.Mantelingu, K., B. A. Ashok Reddy, V. Swaminathan, A. H. Kishore, N. B. Siddappa, G. V. P. Kumar, G. Nagashankar, N. Natesh, S. Roy, P. P. Sadhale, U. Ranga, C. Narayana, and T. K. Kundu. 2007. Specific inhibition of p300-HAT alters global gene expression and represses HIV replication. Chem. Biol. 14645-657. [DOI] [PubMed] [Google Scholar]
- 43.Miao, J., Q. Fan, L. Cui, J. Li, J. Li, and L. Cui. 2006. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene 36953-65. [DOI] [PubMed] [Google Scholar]
- 44.Miller, S. K., R. T. Good, D. R. Drew, M. Delorenzi, P. R. Sanders, A. N. Hodder, T. P. Speed, A. F. Cowman, T. F. de Koning-Ward, and B. S. Crabb. 2002. A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J. Biol. Chem. 27747524-47532. [DOI] [PubMed] [Google Scholar]
- 45.Nandakumar, D. N., V. A. Nagaraj, P. G. Vathsala, P. Rangarajan, and G. Padmanaban. 2006. Curcumin-artemisinin combination therapy for malaria. Antimicrob. Agents Chemother. 501859-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ornaghi, P., D. Rotili, G. Sbardella, A. Mai, and P. Filetici. 2005. A novel Gcn5p inhibitor represses cell growth, gene transcription and histone acetylation in budding yeast. Biochem. Pharmacol. 70911-917. [DOI] [PubMed] [Google Scholar]
- 47.Reddy, R. C., P. G. Vatsala, V. G. Keshamouni, G. Padmanaban, and P. N. Rangarajan. 2005. Curcumin for malaria therapy. Biochem. Biophys. Res. Commun. 326472-474. [DOI] [PubMed] [Google Scholar]
- 48.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 7081-120. [DOI] [PubMed] [Google Scholar]
- 49.Stimson, L., M. G. Rowlands, Y. M. Newbatt, N. F. Smith, F. I. Raynaud, P. Rogers, V. Bavetsias, S. Gorsuch, M. Jarman, A. Bannister, T. Kouzarides, E. McDonald, P. Workman, and G. W. Aherne. 2005. Isothiazolones as inhibitors of PCAF and p300 histone acetyltransferase activity. Mol. Cancer Ther. 41521-1532. [DOI] [PubMed] [Google Scholar]
- 50.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 51.Sun, Y., X. Jiang, S. Chen, and B. D. Price. 2006. Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett. 5804353-4356. [DOI] [PubMed] [Google Scholar]
- 52.Tanner, K. G., M. R. Langer, and J. M. Denu. 2000. Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry 3911961-11969. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, P. R., H. Kurooka, Y. Nakatani, and P. A. Cole. 2001. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J. Biol. Chem. 27633721-33729. [DOI] [PubMed] [Google Scholar]
- 54.Varier, R. A., V. Swaminathan, K. Balasubramanyam, and T. K. Kundu. 2004. Implications of small molecule activators and inhibitors of histone acetyltransferases in chromatin therapy. Biochem. Pharmacol. 681215-1220. [DOI] [PubMed] [Google Scholar]
- 55.Voss, T. S., J. Healer, A. J. Marty, M. F. Duffy, J. K. Thompson, J. G. Beeson, J. C. Reeder, B. S. Crabb, and A. F. Cowman. 2006. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature 4391004-1008. [DOI] [PubMed] [Google Scholar]
- 56.Voss, T. S., C. J. Tonkin, A. J. Marty, J. K. Thompson, J. Healer, B. S. Crabb, and A. F. Cowman. 2007. Alterations in local chromatin environment are involved in silencing and activation of subtelomeric var genes in Plasmodium falciparum. Mol. Microbiol. 66139-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng, Y., K. Balasubramanyam, M. Cebrat, D. Buck, F. Guidez, A. Zelent, R. M. Alani, and P. A. Cole. 2005. Synthesis and evaluation of a potent and selective cell-permeable p300 histone acetyltransferase inhibitor. J. Am. Chem. Soc. 12717182-17183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.