Abstract
Loss of the protein kinase Sch9p increases both the chronological life span (CLS) and the replicative life span (RLS) of Saccharomyces cerevisiae by mimicking calorie restriction, but the physiological consequences of SCH9 deletion are poorly understood. By transcriptional profiling of an sch9Δ mutant, we show that mitochondrial electron transport chain genes are upregulated. Accordingly, protein levels of electron transport chain subunits are increased and the oxygen consumption rate is enhanced in the sch9Δ mutant. Deletion of HAP4 and CYT1, both of which are essential for respiration, revert the sch9Δ mutant respiratory rate back to a lower-than-wild-type level. These alterations of the electron transport chain almost completely blocked CLS extension by the sch9Δ mutation but had a minor impact on the RLS. SCH9 thus negatively regulates the CLS and RLS through inhibition of respiratory genes, but a large part of its action on life span seems to be respiration independent and might involve increased resistance to stress. Considering that TOR1 deletion also increases respiration and that Sch9p is a direct target of TOR signaling, we propose that SCH9 is one of the major effectors of TOR repression of respiratory activity in glucose grown cells.
The SCH9 gene of the yeast Saccharomyces cerevisiae encodes a serine-threonine protein kinase with a catalytic domain very similar to that of human AKT1 (12). This kinase participates in a conserved signaling network involving upstream acting kinases: Pkh1/2p, which phosphorylate its activation loop, and Tor1/2p, which target its hydrophobic motif (34, 36, 38).
There are several lines of evidence that Sch9p plays an important role in glucose signaling in the budding yeast. Early work showed parallelism and complementarity of SCH9 signaling with the cyclic AMP-dependent protein kinase (PKA) pathway, which signals hexose abundance (25, 29, 37, 40). Overexpression of SCH9 suppresses the growth defect of many PKA pathway component mutations (37), and the sch9Δ null mutant is synthetically lethal with each of gpr1Δ, gpa2Δ, and ras2Δ mutations, which act upstream of PKA (25, 29, 37). It was also recently shown that Sch9p integrates nutrient signals with cell size regulation. In fact, the sch9Δ mutation was one of the most potent modifiers of cell size identified in a genome-wide screen for pathways coupling cell growth and division in yeast (18). A subsequent study showed that Sch9p is an activator of ribosomal protein and ribosomal biogenesis regulons and is required for carbon source modulation of cell size (19).
SCH9 is also a negative regulator of both the chronological life span (CLS) and the replicative life span (RLS), both of which are influenced by glucose availability (12, 21, 22, 24). The CLS is the relative survival over time of yeast cells grown to stationary phase in liquid culture, while the RLS is defined as the number of daughter cells produced by a given yeast mother cell before senescence (11, 16, 21). In addition to the sch9Δ deletion, targeting of either the TOR or cyclic AMP/PKA pathway also extends the CLS and RLS (12, 27, 33). As well as these genetic manipulations, calorie restriction (CR) lengthens the S. cerevisiae CLS and RLS (9, 17, 24, 27). Although this is currently controversial, respiration is suggested to be critical for the physiology of life span because (i) the effect of CR on the RLS is mimicked by overexpressing HAP4, which activates expression of electron transport chain genes and is blocked by deletion of CYT1 (28), and (ii) affecting respiration by blocking mitochondrial transcription, ATP synthesis, or the electron transport chain (atp2Δ, coq3Δ, or ndi1Δ deletions or antimycin A treatment) causes a reduction of the yeast CLS (5, 10). HAP4 overexpression also causes an increased CLS, suggesting that increasing respiration may promote RLS and CLS extension (32). Recently, it was convincingly shown that both the sch9Δ and the tor1Δ mutations mimic CR and that reduction in TOR signaling increases yeast respiratory activity (4, 24). However, the physiological effects of perturbing Sch9p signaling remain largely uncharacterized. Here we show that the sch9Δ mutation is required for a derepression of respiration in glucose-grown cells and that this causes part of the extended CLS and RLS phenotype of the sch9Δ mutant.
MATERIALS AND METHODS
Yeast strains and growth conditions.
All Saccharomyces cerevisiae strains used in this study are BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and its isogenic derivatives (39). The sch9Δ heterozygous mutant was provided by Mike Tyers (18). The hap4Δ and cyt1Δ mutants were obtained from the Research Genetics yeast knockout library (Research Genetics). Double mutants were generated by standard yeast genetic manipulations. All strains were cultivated in SD-complete containing 2% dextrose, following standard protocols (1). The plasmid pMT3569, encoding hemagglutinin-Sch9p was a gift of Mike Tyers (19). This construct was subjected to PCR site-directed mutagenesis to introduce a kinase-disabling mutation (K441A). Transformations were performed with the lithium acetate method (13).
RNA extraction and labeling.
All cultures used for RNA extractions were grown to an optical density at 600 nm OD600 of 1, and the glucose concentration in the medium was quantitated with a glucose assay kit (Sigma) to ensure that culture conditions were the same for all analyzed strains. Total RNA was extracted by the hot-phenol extraction method (6) with the difference that glass beads (Sigma) were added to samples. Poly(A)+ RNAs were purified using the Invitrogen MicroFast Track kit (Invitrogen). For labeling, 5 μg of poly(A)+ RNA was reverse transcribed using oligo(dT)21 in the presence of Cy3 or Cy5-dCTP (Perkin-Elmer-Cetus/NEN) and Superscript II reverse transcriptase (Invitrogen). Reverse transcription reaction mixtures were treated with a mix of RNase H (Sigma) and RNase A (1 mg/ml) for 15 min at 37°C. The labeled cDNAs were purified with the QIAquick PCR purification kit (Qiagen).
Microarray analysis.
Saccharomyces cerevisiae Y6.4k cDNA arrays containing 6,218 genes printed in duplicate were obtained from University Health Networks (Toronto, Canada) and used for all hybridizations. The microarray slides were prehybridized for 2 h at 42°C with DIGeasy hybridization buffer containing yeast tRNA and salmon sperm DNA and subsequently washed with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and air dried. The slides were then hybridized with labeled cDNAs overnight at 42°C in DIGeasy hybridization buffer with yeast tRNA and salmon sperm DNA. The slides were washed twice with SSC-0.2% sodium dodecyl sulfate (SDS), once with 0.1× SSC-0.2% SDS, and three times with 0.1× SSC. A final wash with isopropanol was performed, and slides were air dried and scanned with a ScanArray 5000 scanner (Perkin-Elmer-Cetus) at 10-μm resolution. Signal intensity was quantified with QuantArray software (Perkin-Elmer-Cetus), and final normalization and inspection of the data were done with the GeneSpring package.
Southern and Northern blotting.
DNA extracts used for Southern blotting were obtained by the rapid glass beads method (15). Genomic DNA was digested with HaeIII and run on a 1% agarose gel. DNA was then transferred by capillarity action to a Zeta-Probe nylon membrane (Bio-Rad). To obtain Northern blots, total RNA (80 μg) was electrophoresed on a 7.5% formaldehyde-1% agarose gel and transferred by blotting to a Zeta-Probe nylon membrane (Bio-Rad).
Southern probes were a XbaI-PstI restriction fragment of the ACT1 gene from plasmid pGEM-ACT1 (41) and a COX2 probe generated by PCR. Northern probes were all generated by PCR. All probes were labeled by random priming with the RediPrime kit (Amersham Biosciences). Southern and Northern blot hybridizations were carried out as described previously (30).
Protein methods.
Whole-cell extracts were obtained by bead beating in a buffer containing 0.1% NP-40, 250 mM NaCl, 50 mM NaF, 5 mM EDTA, and 50 mM Tris-HCl, pH 7.5. Complete protease inhibitor cocktail (Roche) was added just before use. Protein concentrations were determined by the standard Bio-Rad protein assay (Bio-Rad). Proteins were separated on a 10% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Millipore). Antibodies were prepared in phosphate-buffered saline-0.05% Tween 20-5% skim milk powder. Rabbit polyclonal antibodies directed against Atp1p, Atp2p, and Atp7p were kind gifts of Jean Velours. They were used at dilutions of 1:100,000, 1:100,000 and 1:10,000, respectively. Antibodies directed against Cox4p, Cox5p, and Cyt1p were obtained from Alexander Tzagoloff and were all used at a dilution of 1:1000 (2). Horseradish peroxidase (HRP)-conjugated anti-rabbit and anti-mouse secondary antibodies (Santa Cruz) were used at 1:10,000. The HRP signal was revealed with Immobilon HRP substrate (Millipore).
Oxygen consumption measurements.
Cells were grown to an OD600 of 1, spun down, and kept on ice. One hour before the measurements, cells were resuspended in SD-2% glucose and put on a shaker at 30°C for 1 hour. OD600 measurements and cell counts were performed immediately before putting cells in the oxygen sensing setup. The oxygen depletion rate was monitored by using a lab-built respirometer comprising a polarographic dissolved oxygen probe in a glass syringe with constant agitation (26). Oxygen consumption rates were expressed as percent O2 per minute per 1 × 106 cells.
Microscopy.
Wild-type or sch9Δ mutant cells expressing Cox4p-red fluorescent protein (RFP) (3) were stained with DAPI (4′,6′-diamidino-2-phenylindole) (10 μg/ml in phosphate-buffered saline plus 2% glucose) for 10 min at room temperature, washed twice, and visualized with a Leica DM-IRE2 inverted microscope with a 63× objective and a 10× projection lens. Pictures were acquired with a Sensys charge-coupled-device camera. Images were manipulated with the Openlab software (Improvision).
Longevity analysis.
CLS and RLS analyses were performed in YPD as described previously (10, 22). RLS differences from the wild-type control were tested using the Wilcoxon rank sum test (21).
RESULTS
The sch9Δ mutation upregulates electron transport chain gene expression.
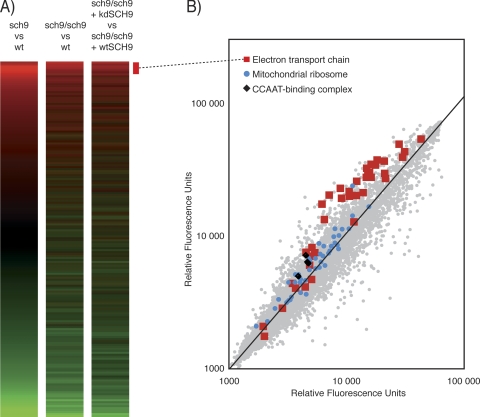
We profiled the sch9Δ mutant by microarray analysis in the haploid and diploid states (Fig. 1A). We also compared sch9Δ/sch9Δ diploid mutants transformed with a wild-type SCH9 or a kinase-dead SCH9 version (kd-SCH9; K441A). The profiles obtained are highly reproducible (Fig. 1A) and reveal that numerous genes are upregulated in the sch9Δ mutant (643 genes; P < 0.05). The list of upregulated genes was systematically tested for enrichment in gene ontology (GO) terms from the SGD database (http://www.yeastgenome.org/). A randomized set of GO annotations was used to set the threshold P value (P < 1 × 10−4). Most of the nonredundant GO terms significantly associated with the upregulated gene list are related to energy derivation and ATP synthesis (Table 1). The other GO terms are associated with chromatin organization and RNA polymerase II-dependent transcription. Genes involved in mitochondrial functions that are upregulated in the sch9Δ mutant are listed in Table 2 (P < 0.05). Almost all components of the five respiratory complexes are upregulated with fold changes that range from 1.2 to 2.5. In addition to canonical members of the respiratory complexes, cytochrome c oxidase chaperones (PET100, PET191, COX14, and COX17), heme biosynthesis enzymes (HEM4, HEM12, and HEM15), holocytochrome c synthases (CYC3 and CYT2), and mitochondrial ribosomal subunit genes are upregulated (Table 2; Fig. 1B). These observations suggest that the sch9Δ mutation affects respiratory physiology. We therefore decided to study the mitochondrial compartment of the sch9Δ mutant in more detail.
FIG. 1.
Gene expression is massively affected by deletion of SCH9. (A) The transcription profile of the sch9Δ cells is reproducible in haploid, diploid, or transformed strains. wt, wild type. (B) Electron transport chain components, the mitochondrial ribosome, and the CCAAT box complex proteins are systematically upregulated in the sch9Δ mutant.
TABLE 1.
GO term annotations that are significantly upregulated in the sch9Δ mutant (P < 1 × 10−4)
| GO category | GO no. | P value | GO description |
|---|---|---|---|
| Biological process | GO:0006119 | 2.78E−15 | Oxidative phosphorylation |
| GO:0006118 | 3.12E−11 | Electron transport | |
| GO:0042775 | 4.98E−11 | ATP synthesis-coupled electron transport (sensu Eukarya) | |
| GO:0006122 | 6.49E−08 | Mitochondrial electron transport, ubiquinol to cytochrome c | |
| GO:0006091 | 1.32E−07 | Energy pathways | |
| GO:0045333 | 1.11E−06 | Cellular respiration | |
| GO:0006754 | 5.44E−06 | ATP biosynthesis | |
| GO:0016310 | 5.45E−06 | Phosphorylation | |
| GO:0009206 | 1.59E−05 | Purine ribonucleoside triphosphate biosynthesis | |
| GO:0009201 | 2.57E−05 | Ribonucleoside triphosphate biosynthesis | |
| GO:0006796 | 8.63E−05 | Phosphate metabolism | |
| GO:0006123 | 1.26E−04 | Mitochondrial electron transport, cytochrome c to oxygen | |
| Cellular component | GO:0005746 | 3.79E−13 | Mitochondrial electron transport chain |
| GO:0005750 | 6.95E−09 | Respiratory chain complex III (sensu Eukarya) | |
| GO:0005743 | 9.42E−07 | Mitochondrial inner membrane | |
| GO:0005740 | 5.05E−06 | Mitochondrial membrane | |
| GO:0005753 | 5.44E−06 | Proton-transporting ATP synthase complex (sensu Eukarya) | |
| GO:0005751 | 4.16E−05 | Respiratory chain complex IV (sensu Eukarya) | |
| GO:0016602 | 8.98E−05 | cCAAT-binding factor complex | |
| Molecular function | GO:0008121 | 6.49E−08 | Ubiquinol-cytochrome c reductase activity |
| GO:0046933 | 2.72E−05 | Hydrogen-transporting ATP synthase activity, rotational mechanism | |
| GO:0004129 | 4.16E−05 | Cytochrome c oxidase activity | |
| GO:0015075 | 6.26E−05 | ion transporter activity |
TABLE 2.
Mitochondrial genes that are significantly upregulated in the sch9Δ mutant (P < 0.05)
| Function | Gene
|
Increase in expression (fold) | P value | |
|---|---|---|---|---|
| Systematic name | Common name | |||
| Complex I | YML120C | NDI1 | 1.27 | 2.99E-02 |
| Electron transport chain complex | YKL148C | SDH1 | 1.27 | 3.71E-02 |
| II, succinate dehydrogenase | YKL141W | SDH3 | 1.65 | 1.33E-04 |
| YDR178W | SDH4 | 1.83 | 8.61E-07 | |
| Electron transport chain complex | YBL045C | COR1 | 1.66 | 8.72E-05 |
| III (GO:0005750), cytochrome | YHR001W-A | QCR10 | 2.59 | 1.71E-05 |
| c reductase | YPR191W | QCR2 | 1.60 | 2.63E-04 |
| YFR033C | QCR6 | 2.04 | 6.20E-07 | |
| YDR529C | QCR7 | 2.03 | 1.62E-05 | |
| YJL166W | QCR8 | 2.01 | 1.14E-06 | |
| YGR183C | QCR9 | 1.48 | 2.43E-03 | |
| YEL024W | RIP1 | 1.36 | 9.92E-03 | |
| YJR048W | CYC1 | 1.89 | 1.11E-04 | |
| YAL039C | CYC3 | 1.52 | 1.31E-03 | |
| YML054C | CYB2 | 1.21 | 8.59E-03 | |
| YOR065W | CYT1 | 1.92 | 7.83E-04 | |
| YKL087C | CYT2 | 1.24 | 6.64E-03 | |
| Electron transport chain complex | YGL191W | COX13 | 1.88 | 5.54E-05 |
| IV (GO:0005751), cytochrome | YML129C | COX14 | 1.16 | 2.49E-02 |
| c oxidase | YLL009C | COX17 | 1.85 | 5.40E-04 |
| YDR231C | COX20 | 1.25 | 2.24E-03 | |
| YGL187C | COX4 | 1.69 | 3.07E-05 | |
| YNL052W | COX5A | 1.79 | 1.27E-05 | |
| YHR051W | COX6 | 1.92 | 1.45E-07 | |
| YMR256C | COX7 | 2.42 | 3.69E-06 | |
| YLR395C | COX8 | 1.89 | 7.15E-05 | |
| YDL067C | COX9 | 1.58 | 2.85E-08 | |
| YDR079W | PET100 | 1.54 | 8.61E-04 | |
| YJR034W | PET191 | 1.29 | 2.77E-02 | |
| Electron transport chain complex | YBL099W | ATP1 | 1.43 | 5.27E-03 |
| V (GO:0005753), F1F0 ATP | YLR295C | ATP14 | 1.96 | 1.11E-05 |
| synthase | YPL271W | ATP15 | 1.56 | 1.07E-02 |
| YDL004W | ATP16 | 1.74 | 4.45E-04 | |
| YDR377W | ATP17 | 1.47 | 1.12E-03 | |
| YPR020W | ATP20 | 2.61 | 1.10E-07 | |
| YBR039W | ATP3 | 1.87 | 2.94E-04 | |
| YPL078C | ATP4 | 1.59 | 1.95E-03 | |
| YDR298C | ATP5 | 1.28 | 7.69E-03 | |
| YKL016C | ATP7 | 2.01 | 6.28E-04 | |
| YDL181W | INH1 | 1.66 | 3.32E-04 | |
| YJR077C | MIR1 | 1.58 | 9.57E-06 | |
| Heme biosynthesis | YDR047W | HEM12 | 1.29 | 3.58E-02 |
| YOR176W | HEM15 | 1.28 | 1.64E-03 | |
| YOR278W | HEM4 | 1.31 | 3.34E-03 | |
| Mitochondrial ribosome | YPR166C | MRP2 | 1.42 | 2.11E-04 |
| YDR405W | MRP20 | 1.30 | 1.44E-02 | |
| YKL167C | MRP49 | 1.21 | 3.06E-02 | |
| YKL142W | MRP8 | 1.82 | 2.03E-02 | |
| YDL202W | MRPL11 | 1.31 | 6.45E-08 | |
| YBL038W | MRPL16 | 1.22 | 3.25E-02 | |
| YNL252C | MRPL17 | 1.17 | 2.11E-02 | |
| YOR150W | MRPL23 | 1.16 | 1.14E-02 | |
| YMR193W | MRPL24 | 1.17 | 1.03E-02 | |
| YGR076C | MRPL25 | 1.19 | 3.17E-03 | |
| YBR282W | MRPL27 | 1.45 | 4.72E-05 | |
| YKL138C | MRPL31 | 1.30 | 6.53E-03 | |
| YCR003W | MRPL32 | 1.20 | 2.79E-02 | |
| YBR268W | MRPL37 | 1.12 | 9.07E-03 | |
| YKL170W | MRPL38 | 1.24 | 1.96E-02 | |
| YLR439W | MRPL4 | 1.25 | 1.53E-02 | |
| YDR337W | MRPS28 | 1.34 | 1.08E-04 | |
| YOR158W | PET123 | 1.13 | 2.55E-02 | |
| mtDNA packaging | YMR072W | ABF2 | 1.79 | 1.04E-04 |
| YJR144W | MGM101 | 1.69 | 3.12E-05 | |
| Tricarboxylic acid cycle | YDR148C | KGD2 | 1.44 | 4.89E-03 |
| YLR304C | ACO1 | 1.52 | 3.80E-03 | |
| Retrograde signaling | YNL076W | MKS1 | 1.36 | 2.24E-05 |
| CCAAT-binding complex | YGL237C | HAP2 | 1.25 | 1.12E-04 |
| (GO:0016602) | YBL021C | HAP3 | 1.45 | 1.15E-06 |
| YKL109W | HAP4 | 1.44 | 5.08E-03 | |
| YOR358W | HAP5 | 1.16 | 1.02E-02 | |
| YLR256W | HAP1 | 0.99 | 9.17E-01 | |
Increased respiration in the sch9Δ strain.
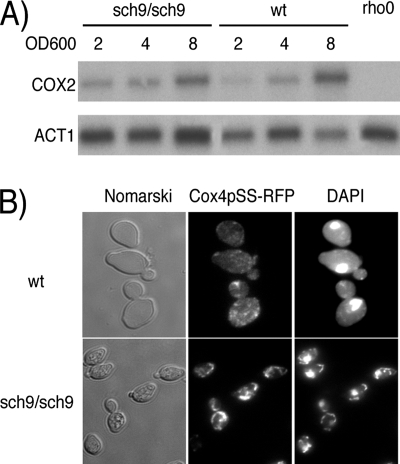
The sch9Δ mutant has a slow-growth phenotype that is a hallmark of petite mutants. However, our sch9Δ strain could grow on ethanol, glycerol, pyruvate, and succinate as sole carbon sources, suggesting that it is not a petite (data not shown). Since ABF2, MGM101, KGD2, and ACO1 are upregulated in the sch9Δ mutant (Table 2) and these genes are involved in mitochondrial DNA (mtDNA) maintenance, we also tested whether the sch9Δ mutant displays modified mtDNA abundance (7, 31, 35, 41). For this, Southern blot analysis with probes for a mitochondrial gene (COX2) and a single-copy nuclear gene (ACT1) was performed. As expected, the mtDNA/genomic DNA ratio increased as the culture density increased, but we observed no differences in mtDNA amounts between the sch9Δ mutant and the wild-type strain (Fig. 2A). We also microscopically examined the mitochondrial compartment in sch9Δ and observed that it is more densely stained and speckled than in wild-type cells, where DAPI staining and Cox4p-RFP distribution are diffuse and reticulated (Fig. 2B).
FIG. 2.
mtDNA segregation is not affected in the sch9Δ mutant. (A) Southern blotting of mtDNA and a one-copy nuclear gene (ACT1). wt, wild type. (B) Mitochondrial morphology in the sch9Δ mutant observed by Cox4p-RFP localization and DAPI staining.
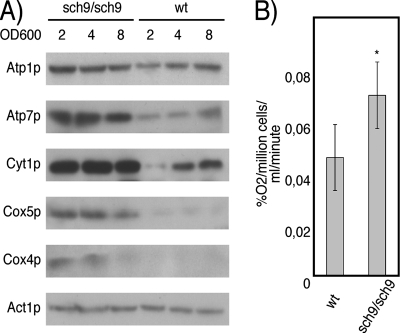
We validated that the increased expression of the respiratory regulon in sch9Δ cells corresponds to enhanced respiratory activity. First, Western blotting of cultures at different cell densities established that protein levels of Atp1p, Atp2p, Atp7p, Cox4p, Cox5p, and Cyt1p are higher in sch9Δ cells throughout culture growth (Fig. 3A). At all time points evaluated, glucose availability was the same in the wild-type and sch9Δ cultures (data not shown). Since the expression of electron transport chain mRNAs and proteins is increased in the sch9Δ mutant, we measured oxygen consumption rates in the sch9Δ/sch9Δ diploid and found that it consumes oxygen faster than the isogenic wild-type BY4743 cells, as predicted (Fig. 3B).
FIG. 3.
SCH9 deletion increases respiratory activity. (A) Electron transport chain proteins are more abundant throughout the course of culture growth in the sch9Δ strain. wt, wild type. (B) The oxygen consumption rate of the sch9Δ/sch9Δ mutant is significantly increased compared to that of the wild type (*, P < 0.05). Error bars indicate standard errors of the means.
The sch9Δ mutant respiratory phenotype is dependent on Hap4p.
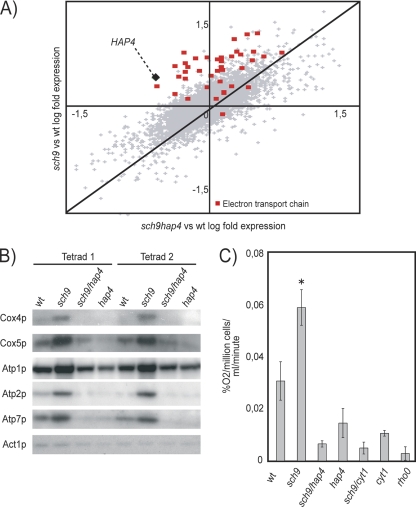
We intersected the list of genes of the sch9Δ mutant that were highly significantly upregulated (P < 0.001) with ChIP/CHIP and with conserved cis-regulatory element data for all transcription factors available (http://fraenkel.mit.edu/yeast_map_2004/) (8, 14). Hap4p-bound intergenic regions and conserved CCAAT box elements were highly enriched upstream of genes whose expression was perturbed in sch9Δ cells (Table 3). Accordingly, we observed a modest but significant increase in expression of all CCAAT-binding factor subunits in the sch9Δ mutant (Tables 1 and 2; Fig. 1B). It thus seems likely that the CCAAT box-binding complex is influenced by SCH9. Since HAP4 overexpression has been associated with an extended RLS, we investigated the role of HAP4 in the sch9Δ mutant. We first confirmed that the hap4Δ and sch9Δ/hap4Δ strains are unable to grow on nonfermentable carbon sources (data not shown). We then tested the effect of the hap4Δ mutation on the sch9Δ upregulation of respiratory genes and observed that it is reverted (Fig. 4A). Protein levels of many components of the electron transport chain and of the ATP synthase were decreased in the sch9Δ/hap4Δ mutant compared to the wild type (Fig. 4B). Increased oxygen consumption by sch9Δ cells was reverted by deletion of HAP4 or of the cytochrome c1 gene (CYT1) to levels close to that of a rho0 strain (Fig. 4C).
TABLE 3.
Transcription factors potentially regulating sch9Δ-dependent upregulated genes (P < 1 × 10−4)
| Transcription factor | Motif |
P value
|
|
|---|---|---|---|
| ChIP-CHIP bound | Motif conservation | ||
| HAP4 | CCAAT | 1.35E−14 | 2.79E−05 |
| HAP3 | CCAAT | 3.20E−04 | 2.79E−05 |
| HAP2 | CCAAT | 2.84E−02 | 2.79E−05 |
| HAP5 | CCAAT | NAa | 2.79E−05 |
| HAP1 | CGGNNNNNNCGG | 5.91E−06 | 8.43E−06 |
| SKN7 | ATTTGGCYGGSCC | 3.83E−01 | 6.78E−05 |
| RGT1 | NA | NA | 8.54E−05 |
NA, not applicable.
FIG. 4.
The increased respiration of the sch9Δ mutant is HAP4 and CYT1 dependent. (A) Scatter plot comparing expression profiles obtained in microarray experiments with the sch9Δ mutant versus the wild type (wt) and with the sch9Δ/hap4Δ mutant versus the wild type. (B) The sch9Δ upregulation of mitochondrial electron transport chain proteins is reverted by deletion of HAP4. (C) Respiratory activity of the sch9Δ mutant is reverted by the hap4Δ and cyt1Δ mutations (*, P < 0.001). Error bars indicate standard errors of the means.
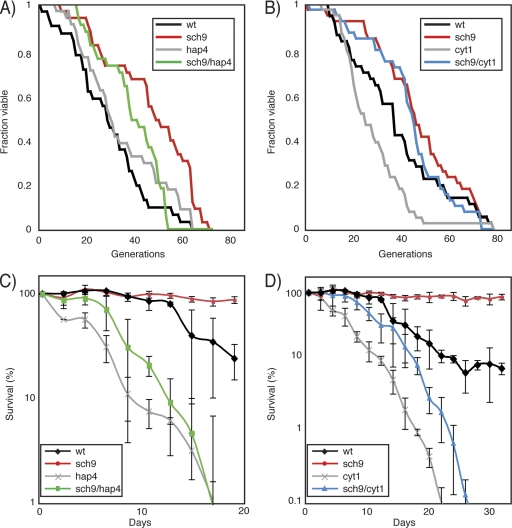
Increased life span of the sch9Δ mutant is partially dependent on increased respiration.
As previously observed for CR cells, the sch9Δ mutation is associated with increased respiratory activity. We thus tested whether the CLS and RLS of the sch9Δ mutant are dependent on intact electron transport chain regulation or function. For this, we assessed the RLS of the sch9Δ mutant with or without HAP4 and CYT1. A hap4Δ single mutant has essentially the same replicative longevity as the wild-type congenic strain, while the sch9Δ mutant and the sch9Δ/hap4Δ double mutant have significantly increased RLSs (P = 1.8 × 10−4 and P = 8.2 × 10−3, respectively) (Table 4; Fig. 5A). Similarly, the sch9Δ and sch9Δ/cyt1Δ mutants both show RLS extension compared to wild-type cells (P = 4.4 × 10−4 and P = 8 × 10−3, respectively) (Table 4), while we confirmed the previous finding that the cyt1Δ mutation causes a significant decrease in the RLS (Fig. 5B) (28). Since the effect of shutting down respiration in sch9Δ cells was small in terms of RLS, we tested whether the hap4Δ or cyt1Δ mutation could perturb the inability of the sch9Δ mutation to extend the CLS. As previously reported, we observed a dramatic extension of the CLS in the sch9Δ mutant (Fig. 5C and D). In contrast, the CLS of hap4Δ and cyt1Δ mutants was dramatically reduced compared to that of wild-type cultures, while the sch9Δ/hap4Δ and sch9Δ/cyt1Δ double mutants had slightly elongated CLSs compared to the hap4Δ and cyt1Δ single mutants (Fig. 5C and D). Interestingly, the sch9Δ/hap4Δ and sch9Δ/cyt1Δ double mutants fail to phenocopy hap4Δ and cyt1Δ mutants, respectively, in both CLS and RLS assays. Our data show that sch9Δ has respiration-independent effects on both the CLS and RLS. However, it seems that respiration or its proper regulation is required for full CLS extension in the sch9Δ mutant, while RLS extension by this mutation seems mostly respiration independent.
TABLE 4.
Effect of HAP4 and CYT1 deletion on sch9 mutant life span
| Strain | Expt 1
|
Expt 2
|
Expt 3
|
P value | |||
|---|---|---|---|---|---|---|---|
| Median RLS | n | Median RLS | n | Median RLS | n | ||
| Wild type | 30 | 33 | 33 | 35 | 31 | 40 | 1 |
| sch9 mutant | 45 | 42 | 47 | 35 | 48 | 40 | 1.8E−04 |
| hap4 mutant | 34 | 34 | 29 | 35 | 29.5 | 40 | 7.5E−01 |
| sch9 hap4 mutant | 37 | 46 | 38 | 33 | 38 | 40 | 8.2E−03 |
| Wild type | 36 | 22 | NDa | ND | 37 | 37 | 1 |
| sch9 mutant | 44 | 25 | ND | ND | 46 | 40 | 4.4E−04 |
| cyt1 mutant | 25 | 25 | ND | ND | 25 | 40 | 1.3E−05 |
| sch9 cyt1 mutant | 46 | 23 | ND | ND | 45 | 40 | 8.0E−03 |
ND, not determined.
FIG. 5.
The sch9Δ mutation increases CLS and RLS in a respiration-dependent manner. (A and B) The replicative longevity of the sch9Δ mutant is affected by the hap4Δ (A) and the cyt1Δ (B) deletions. wt, wild type. (C and D) The increased CLS of the sch9Δ mutant is blocked by deletion of HAP4 (C) or CYT1 (D). Error bars indicate standard errors of the means of three independent biological replicates.
DISCUSSION
The sch9Δ mutation causes significant changes in expression of 643 genes, including a systematic increase in mitochondrial respiratory chain gene expression. Here, we have shown that mitochondrial activity of the sch9Δ mutant is increased. This finding was previously reported for CR yeast and in the tor1Δ strain (4).
Increasing respiration has drawbacks for cellular physiology by producing detrimental by-products such as reactive oxygen species (ROS) that limit the life span of cells in stationary-phase cultures, likely by oxidizing proteins, lipids, and nucleic acids (16). Besides ROS, ethanol also limits the CLS of yeast cells (9). Previous work has shown that the sch9Δ deletion causes an increase in heat and oxidative stress resistance, and SOD2 was reported to be a downstream effector of the sch9Δ deletion increase in oxidative stress resistance and CLS (10, 12). Our expression profiling experiments show that many stress response genes are upregulated in sch9Δ cells (Table 5): we confirm increased expression of the mitochondrial superoxide dismutase gene SOD2 and of several heat shock protein genes, and we show increased transcription of ethanol-degrading enzyme genes ADH1, -2, and -3. This provides additional molecular evidence for the sch9Δ mutant's resistance to stress and accelerated ethanol depletion (9).
TABLE 5.
Stress response genes and other metabolic genes that are significantly modulated in the sch9Δ mutant (P < 0.05)
| Function | Systematic name | Common name | Increase in expression (fold) | P value |
|---|---|---|---|---|
| Stress response | YHR008C | SOD2 | 1.62 | 8.41E−03 |
| YGL073W | HSF1 | 1.13 | 3.25E−02 | |
| YFL014W | HSP12 | 3.06 | 5.35E−03 | |
| YDR171W | HSP42 | 1.71 | 3.96E−02 | |
| YFL016C | MDJ1 | 1.46 | 8.56E−03 | |
| YNL064C | YDJ1 | 1.44 | 3.16E−03 | |
| Alcohol metabolism | YOL086C | ADH1 | 1.13 | 2.62E−02 |
| YMR303C | ADH2 | 1.91 | 2.77E−03 | |
| YMR083W | ADH3 | 1.54 | 4.72E−03 |
Our study shows that not only is the sch9Δ mutant a genetic mimic of CR, but it also recapitulates many molecular hallmarks of CR in yeast. In fact, inactivation of SCH9 provokes shunting of the fermentative metabolism of yeast to a more respirative mode and promotes the heat shock response and the turnover of ROS and alcohol. We find that a large part of the SCH9 effect on the CLS is mediated by respiration, since deletion of HAP4 or CYT1 extensively abrogated sch9Δ-dependent extension of the CLS while RLS extension of sch9Δ was only slightly reduced by blocking respiration. Therefore, our data suggest that the sch9Δ mutation influences the CLS and RLS by both respiration-dependent and -independent mechanisms. Our data also indicate that the conditions required for extending the CLS and RLS are probably encountered by different means in the sch9Δ mutant, since disrupting respiration has a more dramatic effect on the CLS than on the RLS.
Our data support a recent finding that has challenged the relationship between CR-induced RLS extension and respiration (20). Those authors demonstrated that CR causes RLS extension in respiration-deficient yeast strains. Since sch9Δ is thought to be a CR genetic mimic and since it does not fully rely on respiration to promote CLS and RLS, our work further weakens the evidence linking CR-induced RLS and increased respiration.
The respiration-independent effect of the sch9Δ mutation might be explained by it being resistant to oxidative and heat stress through increased ROS and ethanol turnover, which we detect in expression analysis of exponentially growing cultures. Thus, reduced production of detrimental by-products during the growth phase caused by a respirative metabolism and faster turnover of ethanol and ROS might lead to increased survival of the sch9Δ mutant during CLS analysis.
It is believed that Sch9p, Tor1p, and PKA kinases belong to highly integrated signaling pathways that are all involved in nutrient sensing (19, 24). SCH9 and TOR inhibition increase CLS and RLS and are thought to mimic CR (23, 24). Recent findings show that Sch9p is the direct target of the TORC1 complex and that the tor1Δ strains display increased CLS and respiration (4, 38). This, in addition to our findings, supports the view that the TOR-SCH9 pathway feeds into the regulation of respiration and that Sch9p might be one of the major effectors of TOR repression of respiratory activity, as it is the major effector of translational activation by the TOR pathway. The CCAAT box-binding complex (Hap2/3/4/5p) is a likely downstream effector of this nutrient-dependent signal transduction pathway. Hap4p was originally proposed to be the regulatory moiety of the CCAAT box-binding complex, and the TOR-SCH9 pathway could thus reduce respiratory chain expression in high-glucose conditions by repressing Hap4p by a direct or indirect mechanism; in contrast, when glucose is depleted, decreased Sch9 signaling would induce the respiratory regulon as observed in our sch9Δ mutant (19).
It has been proposed that one of the major mitochondrial targets of TOR is translation (4). Here, we suggest that the TOR-SCH9 pathway not only impinges on mitochondrial translation but also affects transcriptional activity of the nuclear respiratory regulon through Sch9p.
Acknowledgments
We thank Mario Jolicoeur and Steve Hisiger for technical help with the oxygen consumption experiments. We thank Hervé Hogues for help with setting up a database for statistical analysis of gene ontology and transcription factor binding enrichment. We are grateful to Mike Tyers for providing the sch9Δ heterozygous mutant and the hemagglutinin-tagged Sch9 plasmid. Thanks go to Alexander Tzagolof for providing anti-Cyt1p, anti-Cox4p, and anti-Cox5p antibodies and to Jean Velours for anti-Atp1p, anti-Atp2p, and anti-Atp7p. The Cox4-RFP-expressing plasmid was provided by Benjamin Glick.
This work was supported by grants from the Canadian Institute for Health Research (CIHR). H.L. was supported by a CIHR fellowship. H.L also acknowledges partial support from an NCIC and CNRC fellowship. This is NRCC publication 49523.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 2.Barrientos, A., D. Pierre, J. Lee, and A. Tzagoloff. 2003. Cytochrome oxidase assembly does not require catalytically active cytochrome C. J. Biol. Chem. 2788881-8887. [DOI] [PubMed] [Google Scholar]
- 3.Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 2083-87. [DOI] [PubMed] [Google Scholar]
- 4.Bonawitz, N. D., M. Chatenay-Lapointe, Y. Pan, and G. S. Shadel. 2007. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 5265-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonawitz, N. D., M. S. Rodeheffer, and G. S. Shadel. 2006. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol. Cell. Biol. 264818-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson, M., and D. Botstein. 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28145-154. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X. J., X. Wang, B. A. Kaufman, and R. A. Butow. 2005. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307714-717. [DOI] [PubMed] [Google Scholar]
- 8.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 30171-76. [DOI] [PubMed] [Google Scholar]
- 9.Fabrizio, P., C. Gattazzo, L. Battistella, M. Wei, C. Cheng, K. McGrew, and V. D. Longo. 2005. Sir2 blocks extreme life-span extension. Cell 123655-667. [DOI] [PubMed] [Google Scholar]
- 10.Fabrizio, P., L. L. Liou, V. N. Moy, A. Diaspro, J. Selverstone Valentine, E. B. Gralla, and V. D. Longo. 2003. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 16335-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabrizio, P., and V. D. Longo. 2003. The chronological life span of Saccharomyces cerevisiae. Aging Cell 273-81. [DOI] [PubMed] [Google Scholar]
- 12.Fabrizio, P., F. Pozza, S. D. Pletcher, C. M. Gendron, and V. D. Longo. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292288-290. [DOI] [PubMed] [Google Scholar]
- 13.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 35087-96. [DOI] [PubMed] [Google Scholar]
- 14.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 43199-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57267-272. [DOI] [PubMed] [Google Scholar]
- 16.Jazwinski, S. M. 2005. Yeast longevity and aging—the mitochondrial connection. Mech. Ageing Dev. 126243-248. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, J. C., E. Jaruga, M. V. Repnevskaya, and S. M. Jazwinski. 2000. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 142135-2137. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen, P., J. L. Nishikawa, B. J. Breitkreutz, and M. Tyers. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297395-400. [DOI] [PubMed] [Google Scholar]
- 19.Jorgensen, P., I. Rupes, J. R. Sharom, L. Schneper, J. R. Broach, and M. Tyers. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 182491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaeberlein, M., D. Hu, E. O. Kerr, M. Tsuchiya, E. A. Westman, N. Dang, S. Fields, and B. K. Kennedy. 2005. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 1e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaeberlein, M., and B. K. Kennedy. 2005. Large-scale identification in yeast of conserved ageing genes. Mech. Ageing Dev. 12617-21. [DOI] [PubMed] [Google Scholar]
- 22.Kaeberlein, M., K. T. Kirkland, S. Fields, and B. K. Kennedy. 2004. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2E296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaeberlein, M., K. T. Kirkland, S. Fields, and B. K. Kennedy. 2005. Genes determining yeast replicative life span in a long-lived genetic background. Mech. Ageing Dev. 126491-504. [DOI] [PubMed] [Google Scholar]
- 24.Kaeberlein, M., R. W. Powers, 3rd, K. K. Steffen, E. A. Westman, D. Hu, N. Dang, E. O. Kerr, K. T. Kirkland, S. Fields, and B. K. Kennedy. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 3101193-1196. [DOI] [PubMed] [Google Scholar]
- 25.Kraakman, L., K. Lemaire, P. Ma, A. W. Teunissen, M. C. Donaton, P. Van Dijck, J. Winderickx, J. H. de Winde, and J. M. Thevelein. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 321002-1012. [DOI] [PubMed] [Google Scholar]
- 26.Lamboursain, L., F. St.-Onge, and M. Jolicoeur. 2002. A lab-built respirometer for plant and animal cell culture. Biotechnol. Prog. 181377-1386. [DOI] [PubMed] [Google Scholar]
- 27.Lin, S. J., P. A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 2892126-2128. [DOI] [PubMed] [Google Scholar]
- 28.Lin, S. J., M. Kaeberlein, A. A. Andalis, L. A. Sturtz, P. A. Defossez, V. C. Culotta, G. R. Fink, and L. Guarente. 2002. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418344-348. [DOI] [PubMed] [Google Scholar]
- 29.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martchenko, M., A. M. Alarco, D. Harcus, and M. Whiteway. 2004. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol Cell 15456-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meeusen, S., Q. Tieu, E. Wong, E. Weiss, D. Schieltz, J. R. Yates, and J. Nunnari. 1999. Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J. Cell Biol. 145291-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piper, P. W., N. L. Harris, and M. MacLean. 2006. Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mech. Ageing Dev. 127733-740. [DOI] [PubMed] [Google Scholar]
- 33.Powers, R. W., III, M. Kaeberlein, S. D. Caldwell, B. K. Kennedy, and S. Fields. 2006. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20174-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roelants, F. M., P. D. Torrance, and J. Thorner. 2004. Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology 1503289-3304. [DOI] [PubMed] [Google Scholar]
- 35.Sato, H., A. Tachifuji, M. Tamura, and I. Miyakawa. 2002. Identification of the YMN-1 antigen protein and biochemical analyses of protein components in the mitochondrial nucleoid fraction of the yeast Saccharomyces cerevisiae. Protoplasma 21951-58. [DOI] [PubMed] [Google Scholar]
- 36.Sobko, A. 2006. Systems biology of AGC kinases in fungi. Sci. STKE 2006re9. [DOI] [PubMed] [Google Scholar]
- 37.Toda, T., S. Cameron, P. Sass, and M. Wigler. 1988. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev. 2517-527. [DOI] [PubMed] [Google Scholar]
- 38.Urban, J., A. Soulard, A. Huber, S. Lippman, D. Mukhopadhyay, O. Deloche, V. Wanke, D. Anrather, G. Ammerer, H. Riezman, J. R. Broach, C. De Virgilio, M. N. Hall, and R. Loewith. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26663-674. [DOI] [PubMed] [Google Scholar]
- 39.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 40.Xue, Y., M. Batlle, and J. P. Hirsch. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 171996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zelenaya-Troitskaya, O., S. M. Newman, K. Okamoto, P. S. Perlman, and R. A. Butow. 1998. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics 1481763-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]