Abstract
Iron is an essential nutrient that is severely limited in the mammalian host. Candida albicans encodes a family of 15 putative ferric reductases, which are required for iron acquisition and utilization. Despite the central role of ferric reductases in iron acquisition and mobilization, relatively little is known about the regulatory networks that govern ferric reductase gene expression in C. albicans. Here we have demonstrated the differential regulation of two ferric reductases, FRE2 and FRP1, in response to distinct iron-limited environments. FRE2 and FRP1 are both induced in alkaline-pH environments directly by the Rim101 transcription factor. However, FRP1 but not FRE2 is also induced by iron chelation. We have identified a CCAAT motif as the critical regulatory sequence for chelator-mediated induction and have found that the CCAAT binding factor (CBF) is essential for FRP1 expression in iron-limited environments. We found that a hap5Δ/hap5Δ mutant, which disrupts the core DNA binding activity of CBF, is unable to grow under iron-limited conditions. C. albicans encodes three CBF-dependent transcription factors, and we identified the Hap43 protein as the CBF-dependent transcription factor required for iron-limited responses. These studies provide key insights into the regulation of ferric reductase gene expression in the fungal pathogen C. albicans.
The mammalian host is an iron-poor environment due to the iron-withholding activities of the innate immune system. Thus, commensal and pathogenic microbes are in a chronic state of iron limitation, primarily through the action of lactoferrin and transferrin but also because of competition with other endogenous microbes (7, 51). To obtain iron in the host environment, Candida albicans has evolved several distinct systems, including high-affinity transporters, an Fe-siderophore uptake system, and a heme uptake system (23, 25, 48, 49). Both the high-affinity uptake and Fe-siderophore systems are required for pathogenesis, and the heme uptake system is predicted to be required for survival in the mammalian host (23, 48, 49). Thus, the ability of C. albicans to acquire iron in the mammalian host is critical for its ability to cause disease.
The importance of iron acquisition in C. albicans can also be inferred from analysis of the genomic sequence. The C. albicans genome encodes at least 2 iron permeases, 5 multicopper oxidases, and a family of 15 putative ferric reductases. In contrast, Saccharomyces cerevisiae encodes only one iron permease, two multicopper oxidases, and seven ferric reductases. This is particularly striking given that S. cerevisiae, but not C. albicans, underwent a whole-genome duplication ∼500 million years ago (22). Thus, the chronic state of iron starvation appears to have had a profound influence on C. albicans evolution.
The ferric reductases, which convert ferric iron (Fe3+) to the biologically available ferrous iron (Fe2+), are a central component of the three iron acquisition systems. Ferric reductases act in conjunction with a multicopper oxidase and iron permease to promote high-affinity iron transport (9, 32). Further, ferric reductase activity is required to remove iron from chelators, including siderophores (26, 61). Ferric reductases are also required to move internal stores of iron across the various organelle membranes (55).
Despite the fact that iron acquisition is essential for survival of C. albicans in the host, little is known about the regulatory control of genes involved in iron acquisition in this important opportunistic pathogen. Several genes encoding proteins involved in iron acquisition, including several members of the ferric reductase family, are repressed in iron-replete conditions by Sfu1 (34). Sfu1 is a transcriptional repressor conserved in the filamentous fungi and Schizosaccharomyces pombe but absent from S. cerevisiae (18, 46). Members of the ferric reductase gene family have also been shown to be negatively regulated by the Tup1 general repressor in C. albicans (6, 30). Finally, genes involved in iron acquisition are positively regulated by the pH-responsive transcription factor Rim101 in neutral-alkaline environments, although it is unclear if this is a direct or indirect effect (4, 10, 11). The induction of genes involved in iron acquisition in response to neutral-alkaline environments reflects the mass conversion of soluble ferrous iron to insoluble ferric iron, and this response appears to be conserved in other fungi (33, 53, 54). In S. cerevisiae, genes involved in iron acquisition are positively regulated by Aft1 (8). However, C. albicans lacks an apparent Aft1 homolog, suggesting that other transcription factors may be involved. Since ferric reductases play a central role in iron acquisition and mobilization, we have analyzed the expression and associated regulation of two ferric reductases, FRE2 (orf19.1264/CFL2) and FRP1 (orf19.5634).
We found that FRE2 and FRP1 are differentially regulated by distinct types of iron limitation, alkaline pH and the presence of chelators. We found that Rim101 directly induces expression of both FRE2 and FRP1 in response to alkaline pH. Further, FRP1 but not FRE2 was induced by chelators in a Rim101-independent fashion. Analysis of the FRP1 promoter revealed that the CCAAT-binding factor (CBF) is required for Rim101-independent chelator-dependent expression. CBF was also required for alkaline-pH induction of FRP1 but not FRE2. Further, we found that Hap43 harbors the transcriptional activation function of CBF in response to iron limitation and that CBF is essential for growth in iron-limited environments. Thus, we find that the ferric reductase gene family in C. albicans is regulated in a complex process that integrates different environmental signals through at least two positive regulatory pathways.
MATERIALS AND METHODS
Strains and plasmids.
All strains used in this study are listed in Table 1. The hap5Δ/hap5Δ mutant (DAY1062) was generated as follows. BWP17 was consecutively transformed with the hap5::ARG4 and hap5::URA3-dpl200 cassettes, which were amplified in a PCR using primers HAP5-5DR and HAP5-3DR (Table 2) to generate DAY1061 and DAY1062, respectively. Correct integration of each cassette was confirmed by the PCR using primers HAP5-5detect and HAP5-3detect, which bind to genomic sequences flanking the site of integration.
TABLE 1.
Strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| BWP17 | ura3Δ::λimm434his1::hisGarg4::hisG | 58 |
| ura3Δ::λimm434 his1::hisG arg4::hisG | ||
| DAY5 | ura3Δ::λimm434his1::hisGarg4::hisGrim101::URA3 | 58 |
| ura3Δ::λimm434 his1::hisG arg4::hisG rim101::ARG4 | ||
| DAY286 | ura3Δ::λimm434his1::hisGpARG4::URA3::arg4::hisG | 12 |
| ura3Δ::λimm434 his1::hisG arg4::hisG | ||
| DAY609 (CTA87.4) | ura3Δ::λimm434his1::hisGarg4::hisGftr1::ARG4 | 31 |
| ura3Δ::λimm434 his1::hisG arg4::hisG ftr1::URA3 | ||
| DAY1061 | ura3Δ::λimm434his1::hisGarg4::hisGhap5::ARG4 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisG HAP5 | ||
| DAY1062 | ura3Δ::λimm434his1::hisGarg4::hisGhap5::ARG4 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisG hap5::URA3-dpl200 | ||
| DAY1082 | ura3Δ::λimm434his1::hisGarg4::hisGhap41::ARG4 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisG HAP41 | ||
| DAY1083 | ura3Δ::λimm434his1::hisGarg4::hisGhap41::ARG4 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisG hap41::URA3 | ||
| DAY1084 | ura3Δ::λimm434his1::hisGarg4::hisGhap43::ARG4 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisG HAP43 | ||
| DAY1085 | ura3Δ::λimm434his1::hisGarg4::hisGhap43::ARG4 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisG hap43::URA3 |
TABLE 2.
Sequences of primers used in this study
| Primer | Sequencea |
|---|---|
| FRE2−22 5a | tgtacaattttcaacaCCAAGaagagtttcctaaa |
| FRE2−22 3a | tttaggaaactcttCTTGGtgttgaaaattgtaca |
| FRE2−22 mut5a | tgtacaattttcaacaGCCGGCagagtttcctaaa |
| FRE2−22 mut3a | tttaggaaactctGCCGGCtgttgaaaattgtaca |
| FRE2−443 5a | tttttataaattaatGCCAAGccatttttatttct |
| FRE2−443 3a | agaaataaaaatggCTTGGCattaatttataaaaa |
| FRE2−443 mut5a | tttttataaattaatGCCGGCccatttttatttct |
| FRE2−443 mut3a | agaaataaaaatggGCCGGCattaatttataaaaa |
| FRE2−876 5a | agaactaattgaaaGCCAAGaaaagttatatggaa |
| FRE2−876 3a | ttccatataacttttCTTGGCtttcaattagttct |
| FRE2−876 mut5a | agaactaattgaaaGCCGGCaaaagttatatggaa |
| FRE2−876 mut3a | ttccatataacttttGCCGGCtttcaattagttct |
| PFRP1−981 5a | ggtacatcatgaatCCAAGaattaccgatccgatt |
| PFRP1−981 3a | aatcggatcggtaattCTTGGattcatgatgtacc |
| PFRP1−981 mut 5a | ggtacatcatgaatGCCGGCattaccgatccgatt |
| PFRP1−981 mut 3a | aatcggatcggtaatGCCGGCttcatgatgtacc |
| PFRP1−636 5a | aacacaacgaagtaGCCAAGaatctggcactagca |
| PFRP1−636 3a | tgctagtgccagattCTTGGCtacttcgttgtgtt |
| PFRP1−636 mut 5a | aacacaacgaagtaGCCGGCaatctggcactagca |
| PFRP1−636 mut 3a | tgctagtgccagattGCCGGCtacttcgttgtgtt |
| PFRP1−164 5a | cgcagttttcggattCTTGGCatagaaacgataaa |
| PFRP1−164 3a | atttatcgtttctatGCCAAGaatccgaaaactgcg |
| PFRP1−164 mut 5a | cgcagttttcggattGCCGGCatagaaacgataaa |
| PFRP1−164 mut 3a | tttatcgtttctatGCCGGCaatccgaaaactgcg |
| PFRE2−876 m1 | ataatagaactaattgaaaGCCGGCaaaagttatatggaattgct |
| PFRE2−876 m2 | agcaattccatataacttttGCCGGCtttcaattagttctattat |
| PFRE2−443 m1 | tcctttttttataaattaatGCCGGCccatttttatttcttacag |
| PFRE2−443 m2 | ctgtaagaaataaaaatggGCCGGCattaatttataaaaaaagga |
| PFRP1−981 m1 | cgggaattcgaatGCCGGCattaccgatccgattcctcct |
| PFRP1−981 m2 | aggaggaatcggatcggtaatGCCGGCttcatgatgtacc |
| PFRP1−636 m1 | aactgaacacaacgaagtaGCCGGCaatctggcactagcaacatc |
| PFRP1−636 m2 | gatgttgctagtgccagattGCCGGCtacttcgttgtgttcagtt |
| PFRP1−164 m1 | aatttcgcagttttcggattGCCGGCatagaaacgataaatccac |
| PFRP1−164 m2 | gtggatttatcgtttctatGCCGGCaatccgaaaactgcgaaatt |
| FRP1-1 | ggcgaattgggcccgacgtcgcatgctcccggccgccatggcggccgcgggaattcgatttgatgagcctgtagatgacg |
| FRP1-2 | ggcgaattgggcccgacgtcgcatgctcccggccgccatggcggccgcgggaattcgattgcacttcctggagcagtaac |
| FRP1-3 | ggcgaattgggcccgacgtcgcatgctcccggccgccatggcggccgcgggaattcgattaagcaggcatcatccaagta |
| FRP1-4 | ggcgaattgggcccgacgtcgcatgctcccggccgccatggcggccgcgggaattcgattacattgaccaactaaaccag |
| FRP1-5 | ggcgaattgggcccgacgtcgcatgctcccggccgccatggcggccgcgggaattcgatttttgcacagtctggtaataa |
| FRP1-6 | ggcgaattgggcccgacgtcgcatgctcccggccgccatggcggccgcgggaattcgattcagcagatactaaaagaatc |
| FRP1-7 | ggcgaattgggcccgacgtcgcatgctcccggccgccatggcggccgcgggaattcgattgttaaacccgaaagaaaagt |
| FRP1-8 | ggcgaattgggcccgacgtcgcatgctcccggccgccatggcggccgcgggaattcgattgtaaaatttcgcagttttcg |
| FRP1-9 | ggcgaattgggcccgacgtcgcatgctcccggccgccatggcggccgcgggaattcgatttgcatgaatgaaaagtatta |
| LacZClaI+200c | tacggaaataatcagccatgtcagc |
| FRP1-8-1-5a′ | taaaaatgcttttgtgctgcgtaaaatttc |
| FRP1-8-1-3a′ | gaaattttacgcagcacaaaagcattttta |
| FRP1-8-2-5a′ | gtaaaatttcgcagttttcggattCTTGGC |
| FRP1-8-2-3a′ | GCCAAGaatccgaaaactgcgaaattttac |
| FRP1-8-3-5a′ | gattCTTGGCatagaaacgataaatccacc |
| FRP1-8-3-3a′ | ggtggatttatcgtttctatGCCAAGaatc |
| FRP1-8-4-5a′ | taaatccaccaattaaacgatggccgcatc |
| FRP1-8-4-3a′ | gatgcggccatcgtttaattggtggattta |
| FRP1-8-5-5a′ | tggccgcatcccttgtctgatgataataag |
| FRP1-8-5-3a′ | cttattatcatcagacaagggatgcggcca |
| FRP1-8-6-5a′ | tgataataaggtttattgaatgcatgaatg |
| FRP1-8-6-3a′ | cattcatgcattcaataaaccttattatca |
| FRP1-8-4-mut5a′ | taaatccaTGCGCtaaacgatggccgcatc |
| FRP1-8-4-mut3a′ | gatgcggccatcgtttaGCGCAtggattta |
| HAP5 5DR | aacaaacattcaattaagacgaagaaatacagaaagaccccccaaaacaatactcaaacatttcccagtcacgacgtt |
| HAP5 3DR | tattattacaaaatcaaacactatttttaaaatgaacgaaaaaaaaaaaaaaatcctctagtggaattgtgagcggata |
| HAP5 5detect | aaaatgcgacaacggaaagg |
| HAP5 3detect | caagtgaggaactgagaactg |
| 5′SnaBI CaHIS1 | gtagtggaagatattctttattgaaaaatagcttgtcaccggatcctggaggatgaggag |
| 3-detect | tgtggaattgtgagcggataacaatttcac |
| HAP41 5DR | tacagtatatttattttttcccccatacattaattattctaatcttatgaaattaccatttttcccagtcacgacgtt |
| HAP41 3DR | catctcatcacatcacatcacatcaaaaataaaatccgttaattttttactgttccttcagtggaattgtgagcggata |
| HAP43 5DR | caaaaaaaaacaacagagtccaaaaaaatacataataattagaatttcaatttgaacaactttcccagtcacgacgtt |
| HAP43 3DR | gaaaagaaaagaaaaaaaaaactgaagtgtcggaaatacttcatactgtaagtcaaactagtggaattgtgagcggata |
| HAP41 5detect new | ccactagttcactcaattac |
| HAP41 3detect new | ccaaatcgagcaaaatcaaacc |
| HAP43 5detect | tgtcttccccccaatagact |
| HAP43 3detect | tacctttttgaatacattgt |
| HAP41UPFWD2ND | tgcaaggcgattaagttgggtaacgccagggttttcccagtcacgacgttgtaaaacgactgaaatttctataattttagcagaa |
| HAP41UPREV2ND | ttttaaagaaagtagtaccagctcttttttttgtttccgtttataccatccaaatcccgcaatggtaatttcataagattagaat |
| HAP41DOWNFWD | aataattaaataagggtggtaattattactatttacaatcaaaggtggtccttctagatgtgaaggaacagtaaaaaattaacgg |
| HAP41DOWNREV | ttacactttatgcttccggctcctatgttgtgtggaattgtgagcggataacaatttcacgtgcgtagtgttcggaagtgttgag |
| HAP43UPFWD | tgcaaggcgattaagttgggtaacgccagggttttcccagtcacgacgttgtaaaacgacgctgacacaaacgggaacagaaata |
| HAP43UPREV2ND | ttttaaagaaagtagtaccagctcttttttttgtttccgtttataccatccaaatcccgcgttgttcaaattgaaattctaatta |
| HAP43DOWNFWD | aataattaaataagggtggtaattattactatttacaatcaaaggtggtccttctagatgtagtttgacttacagtatgaagtat |
| HAP43DOWNREV | ttacactttatgcttccggctcctatgttgtgtggaattgtgagcggataacaatttcactaaaggtattggtgctgccgcttat |
| URA3FWD2ND | gcgggatttggatggtataaacgg |
| URA3REV | catctagaaggaccacctttgattg |
Uppercase letters denote the endogenous or mutated putative Rim101 binding site.
To generate the hap41Δ/hap41Δ and hap43Δ/hap43Δ mutants, BWP17 was first transformed with the hap41:ARG4 and hap43:ARG4 cassettes, which were amplified in a PCR using primers HAP41-5DR and HAP41-3DR and primers HAP43-5DR and HAP43-3DR to generate DAY1082 and DAY1084, respectively. To delete the second wild-type copy of HAP41 and HAP43, cassettes were constructed that contained 1 kb of homology to the 5′ untranslated region (UTR) and 3′ UTR of HAP41 and HAP43 flanking the URA3 gene. For HAP41, the 5′ UTR was amplified in a PCR using primers HAP41UPFWD2ND and HAP41UPREV2ND; the 3′ UTR was amplified using primers HAP41DOWNFWD and HAP41DOWNREV; the URA3 cassette was amplified using primers URA3FWD2ND and URA3REV. For HAP43, the 5′ UTR was amplified in a PCR using primers HAP43UPFWD and HAP43UPREV2ND; the 3′ UTR was amplified using primers HAP43DOWNFWD and HAP43DOWNREV; the URA3 cassette was amplified using primers URA3FWD2ND and URA3REV. The PCR products for the HAP41 and HAP43 cassettes were then transformed into S. cerevisiae along with EcoRI/NotI-digested pDDB78 (40) to generate pDDB410 and pDDB411 by in vivo recombination. Plasmids pDDB410 and pDDB411 were recovered from Escherichia coli, digested with PvuII, and transformed into DAY1082 and DAY1084 to generate the homologs hap41Δ/hap41Δ and hap43Δ/hap43Δ mutants DAY1083 and DAY1085, respectively. Correct integration of each cassette was confirmed by the PCR.
The PFRP1-lacZ plasmid pDDB273 was generated as follows. Nine hundred eighty-three base pairs of the FRP1 promoter was amplified in a PCR using primers FRP1-Pdistal and FRP1-ATG, and the resulting product was ligated into pGEM-T Easy (Promega) to generate pDDB269. pDDB211 (2) was digested with AseI/MluI and ligated into NdeI/MluI-digested pDDB269 to generate pDDB271. The PFRP1-lacZ construct was cut from pDDB271 by SapI/NgoMIV digestion and transformed into a trp1 S. cerevisiae strain with NotI/EcoRI-digested pDDB78 to generate pDDB273 by in vivo recombination (40).
The −981, −636, and −164 single site-specific mutations within PFRP1 were generated as follows. PFRP1-lacZ was amplified in two fragments from pDDB273 using primers PFRP1−981m1, PFRP1−636m1, and PFRP1−164m1, respectively, with 3′-detect and PFRP1−981m2, PFRP1−636m2, and PFRP1−164m2, respectively, with 5′SnaBICaHis1. These two fragments were introduced into SacI/ClaI-digested pDDB273 by in vivo recombination, resulting in pDDB389, pDDB390, and pDDB391 for the −981, −636, and −164 site-specific mutations, respectively.
The −981 −636, −636 −164, and −981 −164 double mutations were generated by the same strategy used for the single site-specific mutations with the same primers using SacI/ClaI-digested pDDB389 for the −981 −636 and −981 −164 and pDDB390 for −636 −164 double mutations to yield pDDB392, pDDB393, and pDDB394, respectively, after in vivo recombination. The −981 −636 −164 triple mutation was generated using SacI/ClaI-digested pDDB392 to yield pDDB395.
A series of PFRP1-lacZ deletion constructs (PFRP1-6-lacZ to PFRP1-9-lacZ) was generated as follows. The PCR-amplified fragments of the FRP1 promoter (PFRP1-6-lacZ to PFRP1-9-lacZ) using primers FRP1-6 to FRP1-9 with LacZClaI+200c were introduced into NheI/ClaI-digested pDDB225 to generate pDDB396, pDDB397, pDDB398, and pDDB399, respectively. The −164 single site-specific mutation within PFRP1-8-lacZ was generated using SacI/ClaI-digested pDDB397 or pDDB398 to yield pDDB400 or pDDB401.
The site-specific CCAAT mutation within PFRP1-8-lacZ was generated from two PCR-amplified fragments using primers FRP1-8-4 mut 3a with 5′SnaBICaHis1 and FRP1-8-4 mut 5a with LacZClaI+200c to yield pDDB402.
The CCAAT-164 double mutation within PFRP1-8-lacZ was generated using SacI/ClaI-digested pDDB402 to yield pDDB403.
The PFRE2-lacZ construct pDDB274 was generated by the same strategy used for pDDB273. The pGEM-T-PFRE2 construct pDDB270 was generated from the 996-bp PCR product of the FRE2 promoter using primers FRE2-Pdistal and FRE2-ATG. pDDB272 was generated by ligation of AseI/MluI-digested pDDB211 and NdeI/MluI-digested pDDB270. The PFRE2-lacZ construct was cut from pDDB272 by SapI/NgoMIV digestion introduced into NotI/EcoRI-digested pDDB78 to generate pDDB274 by in vivo recombination.
The −876 and −443 single site-specific mutations within PFRE2 were generated as follows. PFRE2-lacZ was amplified in two fragments from pDDB274 using primers PFRE2−876m1 (or PFRE2−443m1) with 3′-detect and PFRE2−876m2 (or PFRE2−443m2) with 5′SnaBICaHis1. These two fragments were introduced into SacI/ClaI-digested pDDB274 by in vivo recombination, resulting in pDDB404 and pDDB405 for the −876 and −443 site-specific mutations, respectively. The −876 −443 double mutation was generated similarly using the PFRE2−443m1 and PFRE2−443m2 primer sets and SacI/ClaI-digested pDDB404 to yield pDDB406.
Plasmids pDDB389 through pDDB406 were digested with NruI and transformed into DAY286 (wild type) and/or appropriate mutants to generate strains for β-galactosidase assays. Correct integration was verified by PCR with primers 5-detect and LacZ+354c. The FRP1 and FRE2 promoter regions of all plasmid constructs were confirmed by DNA sequencing.
Media and growth conditions.
C. albicans was routinely grown in YPD plus uridine (2% Bacto peptone, 1% yeast extract, 2% dextrose, and 80 μg of uridine per ml). Selection for the Arg+ and Ura+ or His+ transformants was done on synthetic medium (0.67% yeast nitrogen base plus ammonium sulfate and without amino acids, 2% dextrose, with or without 80 μg of uridine per ml, and supplemented according to the auxotrophic needs of the cells). YPG (2% Bacto peptone, 1% yeast extract, 3% glycerol, and 80 μg of uridine per ml) plates with and without bathophenanthrolinedisulfonic acid (BPS) was used to monitor growth under iron-limited conditions.
β-Galactosidase assays.
β-Galactosidase assays were performed as previously described (2). Cell pellets grown for 7 h in 35 ml of M199 medium (Invitrogen) at pH 4 with or without 200 μM 2′2′dipyridyl (BIP) or at pH 8 with 150 mM HEPES were resuspended in 1 ml Z buffer (1) with 30 μl of cell suspension used to determine the optical density at 600 nm. Then, 150 μl of cell suspension was added to a mixture of 0.1% sodium dodecyl sulfate and chloroform for permeabilization. After a 10-min incubation at 37°C, 0.7 ml o-nitrophenyl-β-d-galactoside (1 mg/ml) was added to start the β-galactosidase reaction. Reactions were stopped by adding 0.5 ml 1 M Na2CO3 when the solution turned yellow, and the A420 was measured. Miller units were calculated as follows: (A420)/(OD600 × volume assayed × time) (1), where OD600 is the optical density at 600 nm. All experiments were conducted at least two times. Data were statistically analyzed by analysis of variance.
Protein preparation.
Cells grown overnight in pH 4 M199 medium with 150 mM HEPES were inoculated into M199 medium buffered with 150 mM HEPES to pH 4 (with or without 200 μM BIP) or pH 8 and grown for 7 h at 30°C. Cells were harvested and stored at −80°C prior to protein extraction. Cell pellets were resuspended in ice-cold radioimmunoprecipitation assay buffer containing 1 μg/ml leupeptin, 2 μg/ml aprotinin, 1 μg/ml pepstatin, 1 mM phenylmethylsulfonyl fluoride, and 10 mM dithiothreitol and transferred to glass test tubes containing acid-washed glass beads. Cells were lysed by vortexing four times for 2 min followed by 2 min on ice. Supernatants were taken after centrifugation and used for electrophoretic mobility shift assays (EMSAs) after protein quantitation.
EMSA.
EMSAs were performed as previously described (2). Briefly, 20 μg of cell extract was allowed to bind 40 fmol DNA probe in 20 μl of binding buffer [10 mM Tris-HCl, pH 7.5, 4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl, 2 μg of poly(dI-dC)] at room temperature for 30 min. DNA-protein complexes were then resolved through 4% nondenaturing polyacrylamide gels in 0.5× Tris-borate-EDTA buffer at room temperature overnight. After electrophoresis, the gels were dried and visualized with a phosphorimager. All DNA probes used were end labeled with [γ-32P]ATP using T4 polynucleotide kinase. EMSAs were analyzed using ImageJ (NIH).
RESULTS
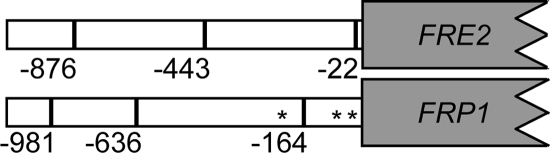
We previously identified two ferric reductase genes, FRE2 (orf19.1264) and FRP1 (orf19.5634), as being induced in alkaline environments and found that this induction was dependent on the Rim101 fungal pH-sensing pathway (4). However, our initial analysis did not establish whether Rim101 played a direct or indirect role in FRE2 and FRP1 induction at alkaline pH. The promoters of FRE2 and FRP1 each contain three putative Rim101 binding sites (Fig. 1) (2), suggesting that Rim101 may directly regulate FRE2 and FRP1.
FIG. 1.
Predicted FRP1 and FRE2 regulatory sites. Predicted Rim101 (lines) and Sfu1 (asterisk) regulatory sites within the FRP1 and FRE2 promoter regions are shown.
Rim101 directly regulates ferric reductase gene expression.
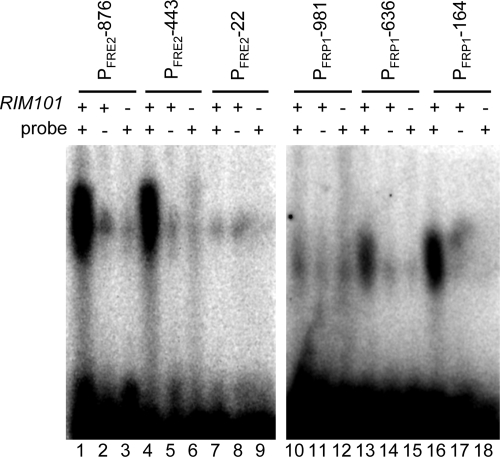
To test the hypothesis that Rim101 directly regulates FRE2 and FRP1, we first determined whether Rim101 could bind to the putative FRE2 and FRP1 Rim101 binding sites by using EMSAs. Radiolabeled 35-bp DNA probes that span the −876, −443, and −22 putative Rim101 binding sites of FRE2 and the −981, −636, and −164 sites of FRP1 were generated by annealing oligomers FRE2−876 5a and FRE2−876 3a, FRE2−443 5a and FRE2−443 3a, FRE2−22 5a and FRE2−22 3a, PFRP1−981 5a and PFRP1−981 3a, PFRP1−636 5a and PFRP1−636 3a, or PFRP1−164 5a and PFRP1−164 3a, respectively, and end labeling. The resulting probes were incubated with protein extracts from wild-type cells, separated by nondenaturing polyacrylamide gel electrophoresis, and analyzed on a phosphorimager (Fig. 2). EMSA of these probes incubated with protein extracts from wild-type cells revealed a distinct band of gel shift for the FRE2 −876 and −443 probes and the FRP1 −164 probe and to a lesser extent the −636 probe (Fig. 2, lanes 1, 4, 13, and 16). No significant gel shifts were observed with the FRE2 −22 and FRP1 −981 probes (Fig. 2, lanes 7 and 10).
FIG. 2.
EMSA of promoter regions containing putative Rim101 binding sites. Protein extracts from the wild-type (WT) (DAY286; lanes 1, 2, 4, 5, 7, 8, 10, 11, 13, 14, 16, and 17) or rim101Δ/rim101Δ (DAY5; lanes 3, 6, 9, 12, 15, and 18) strain grown at pH 8 were incubated with radiolabeled DNA probes for endogenous (+) (lanes 1, 3, 4, 6, 7, and 9) or mutated (−) (lanes 2, 5, and 8) FRE2 (−876, −443, −22) oligomers or endogenous (+) (lanes 10, 12, 13, 15, 16, and 18) or mutated (−) (lanes 11, 14, and 17) FRP1 (−981, −626, −164) oligomers and analyzed by EMSA.
To determine whether these gel shifts were dependent on Rim101, we repeated the EMSAs using probes in which the Rim101 binding site was mutated or using protein extracts lacking the Rim101 protein. Probes spanning the FRE2 −876 and −443 sites and the FRP1 −636 and −164 sites containing GCCGGC in place of the Rim101 binding site failed to yield any gel shift when incubated with protein extracts from wild-type cells (Fig. 2, lanes 2, 5, 14, and 17). Further, incubation of wild-type FRE2 and FRP1 probes with protein extracts obtained from rim101Δ/rim101Δ cells also failed to yield a gel shift (Fig. 2, lanes 3, 6, 15, and 18). These results suggest that Rim101 can bind to sites within the FRE2 and FRP1 promoters and thus serve as a direct regulator of gene expression.
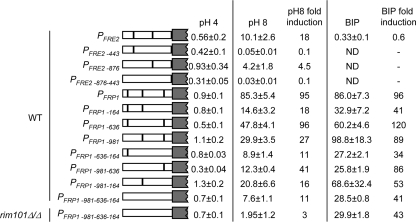
Although the Rim101 binding sites within the promoters of FRE2 and FRP1 could bind to Rim101, we wanted to determine if these sites were required for expression. Thus, we constructed PFRE2-lacZ and PFRP1-lacZ promoter fusions, integrated these constructs into wild-type cells, and analyzed LacZ expression at pH 4 and pH 8. In wild-type cells, LacZ expression was induced at pH 8 ∼20-fold and ∼100-fold from PFRE2-lacZ and PFRP1-lacZ, respectively, compared to expression levels at pH 4 (Fig. 3). When introduced into rim101Δ/rim101Δ cells, alkaline-pH induction of PFRE2-lacZ and PFRP1-lacZ fusions was drastically reduced (data not shown). These results demonstrate that the promoter fusions function appropriately (4).
FIG. 3.
FRE2 and FRP1 promoter lacZ fusions identify important Rim101 binding sites. β-Galactosidase assays were performed on PFRE2-lacZ and PFRP1-lacZ reporter strains grown at pH 4 or pH 8 or at pH 4 with 200 μM BIP. At least three independent transformants were used to determine average Miller units and standard deviation. n-fold induction was determined by comparison of the expression of a given construct in either pH-8- or BIP-grown cells to expression in pH-4-grown cells. rim101Δ/Δ, rim101Δ/rim101Δ.
To determine the role of the putative Rim101 binding sites in induction, site-specific mutations were made analogous to those used in the EMSA mutated probes (Fig. 2). In the PFRE2-lacZ construct, mutation of the −876 site resulted in only ∼5-fold induction at pH 8, compared to ∼20-fold induction with the nonmutated construct; mutation of the −443 site abolished induction at pH 8. These results suggest that the −443 Rim101 binding site within the FRE2 promoter is the critical site for alkaline pH induction.
In the PFRP1-lacZ construct, mutation of the −981 site resulted in ∼30-fold induction at pH 8, compared to ∼100-fold induction with the nonmutated construct. Mutation of the −636 site did not affect induction. Mutation of the −164 site resulted in ∼20-fold induction at pH 8 compared to that for the nonmutated construct. These results demonstrate that the Rim101 binding sites within the FRP1 promoter contribute to, but are not required for, wild-type levels of alkaline pH induction.
Since the single Rim101 binding-site mutations of PFRP1-lacZ all maintained pH 8 induction, we considered the possibility that multiple sites promote alkaline-pH induction of FRP1. To address this idea, we constructed double and the triple Rim101 binding site mutations and determined their effect on PFRP1-lacZ expression at alkaline pH (Fig. 3). The PFRP1−981,−636-lacZ construct was induced ∼40-fold at pH 8, which was similar to results for the PFRP1−981-lacZ single-mutant construct; the PFRP1−981,−164-lacZ construct was induced ∼20-fold at pH 8, which was similar to results for the PFRP1−164-lacZ single-mutant construct. The PFRP1−636,−164-lacZ and the PFRP1−981,−636,−164-lacZ constructs showed the least induction at pH 8, ∼10-fold. These results demonstrate that multiple Rim101 binding sites within the FRP1 promoter are required for expression at alkaline pH. Further, these results demonstrate that FRP1 but not FRE2 is induced at alkaline pH, albeit at reduced levels, in the absence of all identifiable Rim101 binding sites.
FRP1 is induced by iron chelation independently of Rim101.
While the environmental pH has profound effects on iron bioavailability, other conditions can dramatically affect iron availability, including the presence of chelators. Thus, we tested whether FRE2 and FRP1 expression is induced by the presence of an iron chelator in the medium (Fig. 3). Wild-type cells carrying the PFRE2-lacZ or PFRP1-lacZ construct were incubated in pH 4 medium, to avoid pH-dependent induction, with or without 200 μM BIP, an iron chelator. The PFRE2-lacZ construct was not induced by the presence of BIP; the PFRP1-lacZ construct was induced ∼100-fold in pH 4 medium containing BIP compared to results in pH 4 medium without a chelator (Fig. 3). Similar results were obtained using alternate iron chelators (data not shown), demonstrating that FRP1 but not FRE2 expression is induced in iron starvation conditions at acidic pH.
Analysis of effects of the Rim101 binding site mutations on BIP-induced expression of PFRP1-lacZ revealed that mutation of the −164 site but not the −636 or −981 site gave rise to ∼40-fold induction in the presence of a chelator, compared to 100-fold induction in the nonmutated construct. Analysis of the double- and triple-mutant constructs demonstrated that only the −164 site contributed to BIP-induced expression, although this effect was less than that observed for pH 8-induced expression.
Since the PFRP1−981,−636,−164-lacZ construct is still induced in alkaline pH and BIP-containing medium, we wanted to determine if this residual induction was Rim101 dependent. To address this question, the PFRP1−981,−636,−164-lacZ triple mutant construct was introduced into a rim101Δ/rim101Δ mutant background and assayed for expression. In pH-8 medium, the PFRP1−981,−636,−164-lacZ construct showed no significant induction in the rim101Δ/rim101Δ background, suggesting that Rim101 binds to sites distinct from the consensus or that Rim101 can also indirectly promote FRP1 induction. Regardless, alkaline-pH-dependent induction is completely dependent on Rim101. Conversely, in BIP-containing medium, the PFRP1−981,−636,−164-lacZ construct in the rim101Δ/rim101Δ background still retained ∼40-fold induction, which was indistinguishable from PFRP1−981,−636,−164-lacZ expression in the wild-type background. These results clearly demonstrate that FRE2 and FRP1 respond differently to distinct aspects of iron starvation: FRE2 is induced by alkaline pH; FRP1 is induced by alkaline pH and by chelation. These results also demonstrate that these distinct transcriptional responses are governed by discrete mechanisms: the response to alkaline pH is Rim101 dependent; the response to iron chelation is primarily Rim101 independent.
Identification of chelator-dependent regulatory sites.
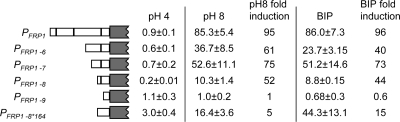
To define novel regions of the FRP1 promoter important for chelator-dependent regulation, a series of PFRP1-lacZ deletion constructs (PFRP1-6-lacZ to PFRP1-9-lacZ) was generated and assayed for expression (Fig. 4). The PFRP1-6-lacZ, PFRP1-7-lacZ, and PFRP1-8-lacZ constructs showed ∼40- to 70-fold induction in alkaline-pH and BIP-containing medium, suggesting that distal sites are not critical for induction. However, the PFRP1-9-lacZ construct was not induced at pH 8 or in BIP-containing medium. To explain this lack of induction, we considered the possibility that the PFRP1-9-lacZ construct lacks a binding site for core transcriptional machinery. However, potential TATA binding sites exist at −71 TATTAAA and −58 TATATT but not in the region between the PFRP1-8 and PFRP1-9 constructs. Thus, the critical region for FRP1 induction appears to lie between −100 and −200 in relation to the FRP1 START codon.
FIG. 4.
Deletion analysis of the FRP1 promoter. β-Galactosidase assays were performed with the PFRP1-lacZ and derivative reporter strains grown at pH 4 or pH 8 or at pH 4 with 200 μM BIP. At least three independent transformants were used to determine average Miller units and standard deviation. n-fold induction was determined by comparison of the expression a given construct in either pH-8- or BIP-grown cells to expression in pH-4-grown cells.
The 100-bp region of the FRP1 promoter essential for induction contains the −164 Rim101 binding site. To determine if this Rim101 binding site affected the induction observed in pH-8 or BIP-containing medium in the PFRP1-8 construct, a site-directed mutation of PFRP1-8, PFRP1-8*164, was generated and introduced into wild-type cells (Fig. 4). Wild-type cells expressing PFRP1-8*164 showed ∼5-fold and ∼15-fold induction in pH 8 and BIP-containing medium, respectively. While this represents a partial diminishment in induction compared to the that with the PFRP1-8-lacZ construct, this appears to be due to increased expression in noninducing pH 4 medium and not to reduced expression under inducing conditions. Thus, our results suggest that a BIP-dependent regulatory element exists within the −200 to −100 region of the FRP1 promoter that is distinct from the Rim101 binding site.
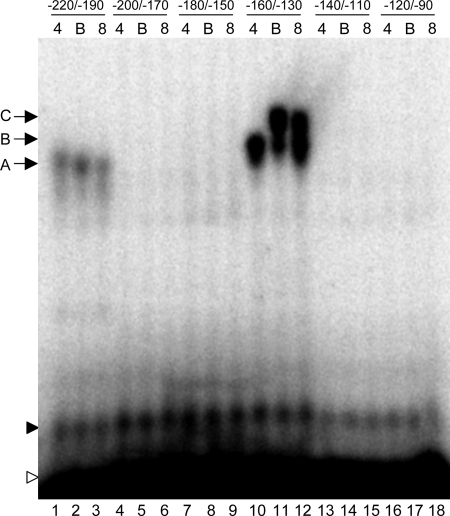
To identify potential regulatory sites within the −200 to −100 region of PFRP1, we constructed a set of six overlapping radiolabeled 30-bp oligomers that span this region (Fig. 5). These probes were then incubated with protein extracts from wild-type cells grown at pH 4, pH 4 with BIP, or pH 8 and separated by nondenaturing polyacrylamide gel electrophoresis. Using extracts from wild-type cells, distinct gel shifts were observed for two of the six probes (Fig. 5, lanes 1 to 3 and lanes 10 to 12). We also observed a nonspecific gel shift running just above the free probe in all lanes (Fig. 5). For extracts from wild-type cells grown at pH 4, the FRP1−220/−190 probe and the FRP1−160/−130 probe each revealed a distinct band (band A, lane 1; band B, lane 10). Somewhat surprisingly, the FRP1−180/−150 probe, which contains the −164 Rim101 binding site, did not give a gel shift. However, the Rim101 binding site is located close to one end of the probe and may lack sufficient flanking sequence to allow Rim101 binding. The gel shift observed with the FRP1−220/−190 probe did not appear to be affected by growth in either iron-limited medium (Fig. 5, compare lane 1 with lanes 2 and 3). However, the gel shift observed with the FRP1−160/−130 probe was altered in iron-limited media; we observed a diminished band B and the presence of a further-retarded band C in BIP-containing medium and, to a lesser extent, in pH 8 medium (Fig. 5, compare lane 10 with lanes 11 and 12).
FIG. 5.
EMSAs of the region predicted to contain the BIP-dependent site. Thirty-base-pair oligomers spanning −220/−190, −200/−170, −180/−150, −160/−130, −140/−110, and −120/−90 in relation to the FRP1 START codon were incubated with protein extracts from wild-type C. albicans (DAY286) grown at pH 4 (lanes 1, 4, 7, 10, 13, and 16), pH 4 with 200 μM BIP (lanes 2, 5, 8, 11, 14, and 17), or pH 8 (lanes 3, 6, 9, 12, 15, and 18). Free probe (open arrowhead), a nonspecific gel shift (closed arrowhead), and specific gel shifts (arrows A to C) are indicated.
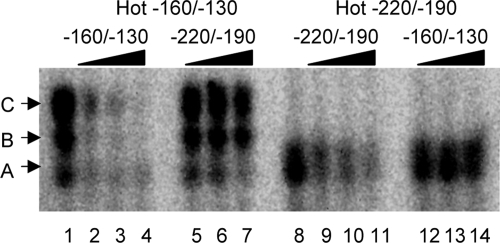
The gel shifts observed with the FRP1−220/−190 and FRP1−160/−130 probes ran in different positions, suggesting that distinct proteins are binding to these two probes. To address this possibility, we conducted a competition assay (Fig. 6). The gel shifts observed with a hot FRP1−160/−130 probe were efficiently competed using cold FRP1−160/−130 oligomers (Fig. 6, compare lane 1 with lanes 2 to 4). However, the gel shifts were not competed using cold FRP1−220/−190 oligomers (Fig. 6, compare lane 1 with lanes 5 to 7). Conversely, the gel shift observed with a hot FRP1−220/−190 probe were efficiently competed using cold FRP1−220/−190 oligomers (Fig. 6, compare lane 8 with lanes 9 to 11) but not when using cold FRP1−160/−130 oligomers (Fig. 6, compare lane 8 with lanes 12 to 14). These results demonstrate that the band A and band B/C gel shifts are due to the DNA probes interacting with distinct protein(s). Thus, we have identified two elements within the −200 to −100 region of the FRP1 promoter that can interact with at least two distinct cellular proteins or protein complexes.
FIG. 6.
Competition assays between the −220/−190 and −160/−130 regions. Radiolabeled probes for the −160/−130 region (lanes 1 to 7) and −220/−190 region (lanes 8 to 14) were incubated with protein extracts from wild-type (DAY286) cells and −160/−130 region (lanes 2 to 4 and 12 to 14) or −220/−190 region (lanes 5 to 7 and 9 to 11) cold competitor. Competitor was added at 50-fold (lanes 2, 5, 9, and 12), 100-fold (lanes 3, 6, 10, and 13), or 500-fold (lanes 4, 7, 11, and 14) excess. Control reactions lacking competitor are included (lanes 1 and 8).
CBF regulates FRP1 expression.
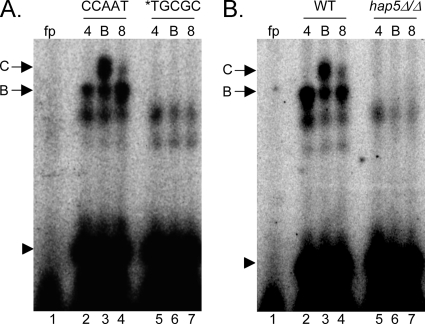
Since the FRP−160/−130 probe revealed an iron starvation-specific response, we focused on this region. Sequence analysis of this 30-bp region revealed a CCAAT sequence (−138 to −134), which is the consensus site for the transcriptional regulator CBF (3, 21). Interestingly, CBF has been implicated in iron starvation responses in Aspergillus nidulans and S. pombe (24, 38). To determine if the CCAAT site is required for the band B and/or C gel shifts, we conducted EMSAs using the *TGCGC probe, in which the CCAAT site of FRP1−160/−130 is changed to TGCGC (Fig. 7A). When the *TGCGC probe was incubated with protein extracts from wild-type cells grown in pH 4, pH 4 with BIP, or pH 8 medium, the band B and band C gel shifts were absent (Fig. 7, compare lanes 2 to 4 with 5 to 7). This result demonstrates that the CCAAT sequence is required for the gel shifts.
FIG. 7.
The FRP1 promoter is bound by Hap5 via the CCAAT site. (A) EMSAs of the −160/−130 region containing the endogenous CCAAT site (lanes 1 to 4) or mutated TGCGC site (lanes 5 to 7). Protein extracts were obtained from wild-type (WT) (DAY286) cells grown in M199 (pH 4) (lanes 2 and 5), M199 (pH 4) plus 200 μM BIP (lanes 3 and 6), or M199 (pH 8) medium (lanes 4 and 7). (B) EMSAs of the −160/−130 region using protein extracts from wild-type (DAY286; lanes 2 to 4) or hap5Δ/hap5Δ (DAY1062; lanes 5 to 7) cells grown as for panel A. fp, free probe lane with no protein extract.
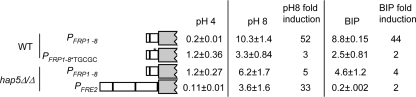
To determine if these gel shifts and the CCAAT site were biologically relevant, we introduced the *TGCGC mutation into PFRP1-8-lacZ to generate PFRP1-8*TGCGC-lacZ. While the PFRP1-8-lacZ construct was induced ∼40- to 50-fold in pH 8 and BIP-containing medium, the PFRP1-8*TGCGC-lacZ construct showed ∼2- to 3-fold induction in pH 8 and BIP-containing medium (Fig. 8). Thus, the CCAAT site, and presumably CBF, is required for induction of FRP1 under iron starvation conditions.
FIG. 8.
Hap5 via the CCAAT site governs FRP1 promoter activity. β-Galactosidase assays were performed on wild-type (WT) cells containing the PFRP1-8-lacZ (data also shown in Fig. 4) and PFRP1-8*TGCGC-lacZ reporters and on hap5Δ/hap5Δ cells containing the PFRE2-lacZ and PFRP1-8-lacZ reporters grown at pH 4 or pH 8 or at pH 4 with 200 μM BIP. At least three independent transformants were used to determine average Miller units and standard deviation. Induction (n-fold) was determined by comparison of the expression of a given construct in either pH 8- or BIP-grown cells to expression in pH 4-grown cells.
To test if CBF governs FRP1 expression via the CCAAT site, we constructed an insertion-deletion mutation of HAP5, a core component of CBF. In S. cerevisiae, Hap5, Hap2, and Hap3 form a heterotrimer with DNA binding activity (36, 37), which recruits the transcriptional activator Hap4 (36). Disruption of any member of the complex, including Hap5, abolishes CBF activity. When the FRP1−160/−130 probe was incubated with protein extracts from hap5Δ/hap5Δ cells grown in pH 4 medium, pH 4 medium with BIP, or pH 8 medium, no gel shifts were observed (Fig. 7B). Thus, Hap5 is required for the band B and band C gel shifts observed for the FRP1−160/−130 probe.
Although Hap5 is required for FRP1 promoter gel shifts, we wanted to determine if this interaction affected FRP1 regulation. To address this possibility, we introduced the PFRP1-8-lacZ construct into the hap5Δ/hap5Δ mutant and analyzed its expression. While wild-type cells induced PFRP1-8-lacZ expression ∼40- to 50-fold in pH 8 or BIP-containing medium, the hap5Δ/hap5Δ mutant only induced PFRP1-8-lacZ expression ∼3- to 5-fold (Fig. 8). Since Hap5 was required for FRP1 induction at alkaline pH, we tested whether FRE2 expression at alkaline pH was Hap5 dependent. We introduced the PFRE2-lacZ construct into the hap5Δ/hap5Δ mutant and found that LacZ was induced ∼30-fold in pH 8 medium and not induced in BIP-containing medium (Fig. 3), similar to results observed for PFRE2-lacZ expression in wild-type cells (Fig. 3). Thus, Hap5 is a critical regulator of some (FRP1) but not all (FRE2) ferric reductases in iron-limited environments. Further, this is the first demonstration that CBF governs iron starvation responses in the fungal pathogen C. albicans.
Hap43 is the iron starvation transcriptional activator.
The FRP1−160/−130 probe resulted in two distinct gel shifts: a constitutive gel shift (band B) and an iron starvation-dependent gel shift (band C). Based on the fact that CBF is essential for these two gel shifts, we considered the model where the DNA-binding Hap2/3/5 heterotrimer binds DNA constitutively, resulting in the band B gel shift. Upon iron starvation, the transcriptional activator, Hap4, is recruited to the Hap2/3/5 DNA binding complex at the promoter, resulting in the band C gel shift. The C. albicans genome contains three genes predicted to encode CBF transcriptional activators: HAP41, HAP42, and HAP43. Each of the predicted proteins contains a 20-amino-acid Hap2/Hap3/Hap5 interaction domain found in S. cerevisiae Hap4 and A. nidulans HapX. In S. cerevisiae, Hap4 governs expression of genes involved in respiration and is required for growth on nonfermentable carbon sources (15). In A. nidulans, HapX governs gene expression during iron starvation and is required for growth in iron-depleted conditions (24).
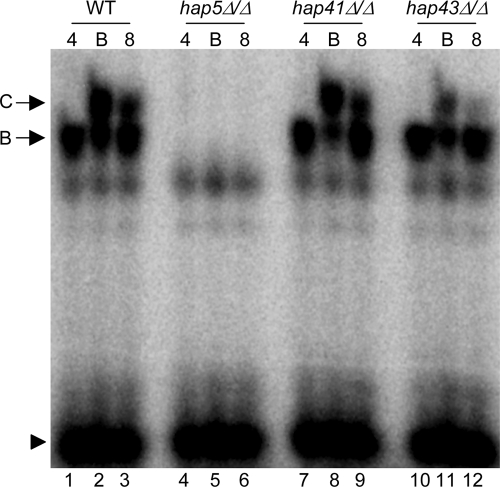
To determine if HAP41, HAP42, or HAP43 was involved in the gel shifts observed for the FRP1−160/−130 probe, we constructed insertion-deletion hap41Δ/hap41Δ and hap43Δ/hap43Δ mutants. Although we have not recovered a hap42Δ/hap42Δ mutant, based on the data presented below we do not expect that HAP42 is the transcription factor required for adaptation to iron-limited environments. Protein extracts from wild-type, hap5Δ/hap5Δ, hap41Δ/hap41Δ, and hap43Δ/hap43Δ cells grown in pH 4 medium, pH 4 plus BIP medium, or pH 8 medium were incubated with the radiolabeled FRP1−160/−130 probe and analyzed by EMSA (Fig. 9). Wild-type cell extracts gave the two gel shifts, which were Hap5 dependent, as observed previously. Protein extracts from the hap41Δ/hap41Δ mutant shifted the FRP1−160/−130 probe to band B and band C, as observed for the wild-type protein extracts (Fig. 9, lanes 7 to 9). However, protein extracts from the hap43Δ/hap43Δ mutant shifted the FRP1−160/−130 probe to the band B position similarly to wild-type extracts but showed an obvious reduction in the shift to the band C position in iron-starved cells (Fig. 9, lanes 10 to 12). These results suggest that Hap43, and not Hap41, is the transcription factor responsible for ferric reductase gene regulation.
FIG. 9.
Hap43 is required for iron limitation-specific promoter binding. EMSAs of the −160/−130 probe incubated with protein extracts derived from wild-type (WT) (DAY286; lanes 1 to 3), hap5Δ/hap5Δ (DAY1062; lanes 4 to 6), hap41Δ/hap41Δ (DAY1083; lanes 7 to 9), or hap43Δ/hap43Δ (DAY1085; lanes 10 to 12) cells are shown. Protein extracts were obtained from cells grown in M199 (pH 4) (lanes 1, 4, 7, and 10), M199 (pH 4) plus 200 μM BIP (lanes 2, 5, 8, and 11), or M199 (pH 8) medium (lanes 3, 6, 9, and 12).
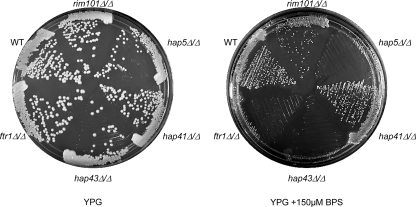
We have demonstrated that FRP1 but not FRE2 is regulated by CBF in C. albicans. We wanted to know if the role CBF plays in adaptation to iron starvation is more general. To address this idea, we grew wild-type, rim101Δ/rim101Δ, ftr1Δ/ftr1Δ, hap5Δ/hap5Δ, hap41Δ/hap41Δ, and hap43Δ/hap43Δ strains on rich medium with or without BPS and analyzed growth. On rich-medium plates, all strains were able to grow comparably to the wild-type strain (Fig. 10). On rich medium containing 150 μM BPS, the wild-type, rim101Δ/rim101Δ, and hap41Δ/hap41Δ strains showed reduced growth compared to that with rich medium without BPS but were able to form colonies. However, the hap5Δ/hap5Δ and hap43Δ/hap43Δ strains showed no growth. The growth defect in the hap5Δ/hap5Δ and hap43Δ/hap43Δ strains was more severe than that observed for the ftr1Δ/ftr1Δ high-affinity iron permease mutant, which was able to form visible microcolonies. These results demonstrate that Hap43 is the transcription factor responsible for CBF-dependent responses to iron starvation and that CBF plays a profound role in adaptation to iron limitation.
FIG. 10.
Growth of the wild-type (WT) (DAY286), rim101Δ/rim101Δ (DAY5), hap5Δ/hap5Δ (DAY1062), ftr1Δ/ftr1Δ (DAY609), hap41Δ/hap41Δ (DAY1083), and hap43Δ/hap43Δ (DAY1085) strains on YPG plates with or without 150 μM BPS after 3 days of incubation at 37°C.
DISCUSSION
The ability to acquire iron from the iron-limited host environment is essential for microbial survival (14, 44, 45, 60). C. albicans encodes at least three independent high-affinity uptake systems to obtain iron from the host environment: a high-affinity permease system, an iron-siderophore uptake system, and a heme uptake system, which are required for survival and pathogenesis in the host (23, 48, 49). Ferric reductases play a central role in each of these systems and function to mobilize internal stores of iron (20, 29, 57, 61). In this study, we have analyzed the regulation of two ferric reductases, FRE2 and FRP1. We demonstrated that FRE2 and FRP1 are differentially regulated by distinct environmental conditions of iron limitation; we identified transcriptional regulators that directly control this differential regulation, one of which is CBF; and we established that the CBF transcriptional activator for iron-dependent responses is governed by the A. nidulans HapX homolog, HAP43.
The C. albicans genome encodes 15 putative ferric reductases (http://www.candidagenome.org/). Of these, only FRE1/CFL1 and FRE10 have been shown to have ferric reductase activity, either by complementation of the S. cerevisiae fre1Δ mutant or by direct mutant analysis with C. albicans (20, 30, 31). However, microarray analyses have demonstrated that nine of the putative ferric reductases, including FRE1/CFL1 and FRE10, are regulated in response to environmental iron status, suggesting that these genes may encode bona fide ferric reductases (4, 34). Further, Bensen et al. found that while ferric reductases, like FRE2 and FRP1, are induced by pH-dependent iron starvation, FRE5 and FRE10 are preferentially expressed under iron-replete conditions and repressed by pH-dependent iron starvation (4). This represented the first demonstration that cells can differentially regulate ferric reductase expression, in this case in response to iron-replete versus iron starvation conditions. Here, however, we demonstrate that C. albicans cells also regulate ferric reductase gene expression in response to the environmental condition promoting iron starvation. FRE2 and FRP1 are both induced by alkaline pH, but only FRP1 is induced by the presence of a chelator in the environment. However, we do not yet know whether the cells are differentially responding to iron chelates, iron oxides, or other signals indirectly affected by iron availability. While the conditions used in our studies are by definition artificial, they reflect important conditions found within the host. Neutral-alkaline pH, which promotes FRE2 and FRP1 expression, is the pH range of most sites within the human body, and iron chelators, which promote expression of FRP1, are found in the tissues (transferrin) and on mucosal surfaces (lactoferrin and xenosiderophores). Thus, C. albicans cells respond not only to the presence or absence of iron but also to the manner in which iron is limited.
Previously ferric reductases were shown to be transcriptionally regulated by repression. Lan et al. showed that members of the ferric reductase gene family were induced by iron limitation by alleviation of Sfu1-dependent repression (34). Sfu1 is the C. albicans GATA-type transcriptional regulator homologous to A. nidulans SreA and S. pombe Fep1 (18, 34, 46, 47). Members of the ferric reductase gene family were also shown to be negatively regulated by the Tup1 general repressor and associated DNA binding proteins (6, 16, 30, 41). This situation is distinct from that described for S. cerevisiae, which regulates iron acquisition primarily through positive regulation via Aft1 and, to a lesser degree, Aft2 (5, 8, 50). Thus, the paradigm with C. albicans has been that iron acquisition genes, including the ferric reductases, are governed by negative regulation.
Our work demonstrates that regulation of the ferric reductases in C. albicans is more complex and also requires positive regulation. We found that pH-dependent induction of FRE2 and FRP1 is governed by the Rim101 transcription factor. A role for Rim101 in pH-dependent expression of iron acquisition genes has been suggested for C. albicans and other fungi; however, our studies represent the first demonstration that Rim101 directly regulates iron acquisition genes (4, 10, 13, 33). Direct regulation occurs through specific Rim101 binding sites within each promoter, the −443 site of the FRE2 promoter and primarily the −164 site of the FRP1 promoter. These two sites specify the classical Rim101 binding site, GCCAAG, found in C. albicans and other fungi (2, 56). While Rim101 was necessary and sufficient for FRE2 regulation, it was not necessary for FRP1 regulation in the presence of a chelator, nor was it sufficient for FRP1 regulation at an alkaline pH. We mapped a Rim101-independent regulatory element to a 100-bp region of the FRP1 promoter and identified the CCAAT motif as being essential for both chelation- and pH-dependent induction. The CCAAT motif is bound by CBF, a ubiquitous transcription factor in the eukaryotes (35); we found that the DNA binding activity of CBF is essential for FRP1 induction. Thus, we found that as with S. cerevisiae, expression of iron acquisition genes in C. albicans is also positively regulated.
In fungi, CBF activity appears to function in two steps. First, Hap2/3/5 form a heterotrimer with DNA binding activity but without transcription activation function. Second, Hap4 binds to the Hap2/3/5-DNA complex and promotes transcriptional activation (36). In higher eukaryotes, CBF appears to lack Hap4 and acts by stimulating or recruiting other transcription factors (17, 19, 42). Hap5 was previously identified in C. albicans as having a role in respiration. In S. cerevisiae, CBF is an activator of respiratory genes, but in C. albicans, CBF acts as a repressor (28, 36). We found that in C. albicans, Hap5 is additionally required for expression of FRP1 and growth in low-iron environments. A role for CBF in iron starvation has not been described for S. cerevisiae but has been suggested for other fungi (24, 38, 39). Based on these studies, we propose that under low-iron conditions, CBF induces expression of genes required for iron acquisition and represses expression of respiratory genes, which require iron as a cofactor.
For C. albicans, three genes have been identified that may encode CBF transcription factor activity, HAP41, HAP42, and HAP43 (28). Compared to S. cerevisiae Hap4, the only sequence with significant homology to the products of HAP41, HAP42, and HAP43 is a 20-amino-acid evolutionarily conserved domain specifying the Hap2/3/5 interaction domain (LVVRTSKHWVLPPRPRPGRR). Compared to all available protein sequences, there are no obvious homologs to Hap41, and homologues to Hap42 are found only in Lodderomyces elongisporus, Debaryomyces hansenii, and Pichia species. However, numerous fungal genomes encode homologs to Hap43, including HapX from A. nidulans. HapX is a transcriptional regulator of CBF-dependent genes required for growth in iron-limited environments (24). We found that Hap43 but not Hap41 is required for induction of FRP1 expression and growth in iron-limited environments. While we have not investigated a potential role of Hap42 in iron-dependent processes, our analyses suggest that all observed iron-dependent phenotypes can be attributed to Hap43. We did note that Hap43 and Hap5 were dispensable for FRE2 induction, demonstrating that CBF is not essential for all iron-dependent processes. However, growth defects observed for the hap43Δ/hap43Δ and hap5Δ/hap5Δ mutants in iron-limited medium demonstrate that CBF is a critical factor in adaptation to iron-limited environments in C. albicans.
The activity of CBF itself also appears to be regulated by iron availability. Previously HAP43 was found to be repressed under iron-replete conditions in an Sfu1-dependent manner (34). Further, C. albicans encodes two putative Hap3 homologs, Hap31 and Hap32 (28). HAP31 is expressed preferentially under iron-replete conditions. However, HAP32, like HAP43, is repressed in iron-replete medium in an Sfu1-dependent manner. HAP32 is also induced at an alkaline pH in a Rim101-dependent manner (4, 34). We initially considered that the gel shift bands B and C observed with the −160/−130 probe were due to the presence of either Hap41 or Hap43 in the protein complex. However, loss of Hap41 had no effect on the gel shifts, and loss of Hap43 drastically reduced but did not eliminate the band C gel shift. The difference in HAP31 and HAP32 expression provides an additional framework for explaining the band B and C gel shifts observed in our EMSAs. Under iron-replete conditions, Hap31 is expressed and the DNA binding component of CBF is composed of the Hap2/Hap31/Hap5 heterotrimer. This heterotrimer can bind to the promoters of iron-responsive genes, producing the gel shift band B observed in Fig. 5, 7, and 9, although this interaction may not actually occur in vivo. Under iron-limited conditions, Hap32 is expressed and the DNA binding component of CBF is composed of the Hap2/Hap32/Hap5 heterotrimer, which can bind to the promoters of CBF-dependent iron-responsive genes, producing the gel shift band C observed in Fig. 5, 7, and 9. Hap32 is ∼200 amino acids larger than Hap31, providing an explanation for the difference in mobility between bands B and C. In this model, the reduction in the band C gel shift in protein extracts obtained from hap43Δ/hap43Δ cells suggests that either Hap43 is a member of the band C complex or Hap43 is required for efficient formation of a Hap2/Hap32/Hap5 complex. Finally, we have noted that Hap43 contains several cysteine-rich domains, which can in principle bind iron, suggesting the possibility that Hap43 activity may be regulated posttranscriptionally by cellular iron availability.
Iron acquisition systems are generally considered virulence properties in pathogenic organisms (14, 25, 26, 44, 60). As discussed above, the C. albicans genome encodes a family of 15 putative ferric reductases, significantly more than related nonpathogenic fungi, such as S. cerevisiae, which encodes 7 ferric reductases. This difference is particularly striking given that the S. cerevisiae ancestor underwent a whole-genome duplication after splitting from the C. albicans ancestor (59). Gene families are tightly associated with virulence traits in C. albicans, including the ALS adhesion proteins (9 members), the secreted aspartyl proteases (10 members), and the lipases (10 members) (27, 43, 52). Thus, we propose that the ferric reductase gene family (15 members) also encodes a virulence trait, although further studies are needed to establish this. However, our studies demonstrate that Rim101 and CBF play critical roles in adaptation to iron starvation, a chronic state in the mammalian host, at least in part through the regulation of ferric reductase gene expression in the most prevalent pathogenic fungus for humans.
Acknowledgments
We thank Andrew Dancis for generously providing the ftr1Δ/ftr1Δ (CTA87.4) mutant. We thank Sandra Armstrong and Timothy Brickman for expert advice and critical reading of the manuscript and Lucia Zacchi, Julie Wolf, and the members of the Davis laboratory for additional comments and advice.
This work was supported by NIH National Institute of Allergy and Infectious Diseases award 1R01-AI064054-01.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Adams, A., D. E. Gotschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics, 1997 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Baek, Y. U., S. J. Martin, and D. A. Davis. 2006. Evidence for novel pH-dependent regulation of Candida albicans Rim101, a direct transcriptional repressor of the cell wall beta-glycosidase Phr2. Eukaryot. Cell 51550-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barberis, A., G. Superti-Furga, and M. Busslinger. 1987. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell 50347-359. [DOI] [PubMed] [Google Scholar]
- 4.Bensen, E. S., S. J. Martin, M. Li, J. Berman, and D. A. Davis. 2004. Transcriptional profiling in C. albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 541335-1351. [DOI] [PubMed] [Google Scholar]
- 5.Blaiseau, P. L., E. Lesuisse, and J. M. Camadro. 2001. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J. Biol. Chem. 27634221-34226. [DOI] [PubMed] [Google Scholar]
- 6.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 15631-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullen, J. J., H. J. Rogers, P. B. Spalding, and C. G. Ward. 2006. Natural resistance, iron and infection: a challenge for clinical medicine. J. Med. Microbiol. 55251-258. [DOI] [PubMed] [Google Scholar]
- 8.Casas, C., M. Aldea, C. Espinet, C. Gallego, R. Gil, and E. Herrero. 1997. The AFT1 transcriptional factor is differentially required for expression of high-affinity iron uptake genes in Saccharomyces cerevisiae. Yeast 13621-637. [DOI] [PubMed] [Google Scholar]
- 9.Dancis, A. 1998. Genetic analysis of iron uptake in the yeast Saccharomyces cerevisiae. J. Pediatr. 132S24-S29. [DOI] [PubMed] [Google Scholar]
- 10.Davis, D. 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 441-7. [DOI] [PubMed] [Google Scholar]
- 11.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, D. A., V. M. Bruno, L. Loza, S. G. Filler, and A. P. Mitchell. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 1621573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisendle, M., H. Oberegger, R. Buttinger, P. Illmer, and H. Haas. 2004. Biosynthesis and uptake of siderophores is controlled by the PacC-mediated ambient-pH regulatory system in Aspergillus nidulans. Eukaryot. Cell 3561-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkelstein, R. A., C. V. Sciortino, and M. A. McIntosh. 1983. Role of iron in microbe-host interactions. Rev. Infect. Dis. 5(Suppl. 4)S759-S777. [DOI] [PubMed] [Google Scholar]
- 15.Forsburg, S. L., and L. Guarente. 1989. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 31166-1178. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Sanchez, S., A. L. Mavor, C. L. Russell, S. Argimon, P. Dennison, B. Enjalbert, and A. J. Brown. 2005. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol. Biol. Cell 162913-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge, Y., M. A. Konrad, L. H. Matherly, and J. W. Taub. 2001. Transcriptional regulation of the human cystathionine beta-synthase-1b basal promoter: synergistic transactivation by transcription factors NF-Y and Sp1/Sp3. Biochem. J. 35797-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas, H., I. Zadra, G. Stoffler, and K. Angermayr. 1999. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J. Biol. Chem. 2744613-4619. [DOI] [PubMed] [Google Scholar]
- 19.Hackzell, A., H. Uramoto, H. Izumi, K. Kohno, and K. Funa. 2002. p73 independent of c-Myc represses transcription of platelet-derived growth factor beta-receptor through interaction with NF-Y. J. Biol. Chem. 27739769-39776. [DOI] [PubMed] [Google Scholar]
- 20.Hammacott, J. E., P. H. Williams, and A. M. Cashmore. 2000. Candida albicans CFL1 encodes a functional ferric reductase activity that can rescue a Saccharomyces cerevisiae fre1 mutant. Microbiology 146869-876. [DOI] [PubMed] [Google Scholar]
- 21.Hatamochi, A., B. Paterson, and B. de Crombrugghe. 1986. Differential binding of a CCAAT DNA binding factor to the promoters of the mouse alpha 2(I) and alpha 1(III) collagen genes. J. Biol. Chem. 26111310-11314. [PubMed] [Google Scholar]
- 22.Heckman, J. D. 2001. Expanding roles of the orthopaedic surgeon. Clin. Orthop. Relat. Res. 200146-51. [DOI] [PubMed] [Google Scholar]
- 23.Heymann, P., M. Gerads, M. Schaller, F. Dromer, G. Winkelmann, and J. F. Ernst. 2002. The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect. Immun. 705246-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hortschansky, P., M. Eisendle, Q. Al-Abdallah, A. D. Schmidt, S. Bergmann, M. Thon, O. Kniemeyer, B. Abt, B. Seeber, E. R. Werner, M. Kato, A. A. Brakhage, and H. Haas. 2007. Interaction of HapX with the CCAAT-binding complex—a novel mechanism of gene regulation by iron. EMBO J. 263157-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howard, D. H. 1999. Acquisition, transport, and storage of iron by pathogenic fungi. Clin. Microbiol. Rev. 12394-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howard, D. H. 2004. Iron gathering by zoopathogenic fungi. FEMS Immunol. Med. Microbiol. 4095-100. [DOI] [PubMed] [Google Scholar]
- 27.Hoyer, L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9176-180. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, D. C., K. E. Cano, E. C. Kroger, and D. S. McNabb. 2005. Novel regulatory function for the CCAAT-binding factor in Candida albicans. Eukaryot. Cell 41662-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight, S. A., and A. Dancis. 2006. Reduction of 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide inner salt (XTT) is dependent on CaFRE10 ferric reductase for Candida albicans grown in unbuffered media. Microbiology 1522301-2308. [DOI] [PubMed] [Google Scholar]
- 30.Knight, S. A., E. Lesuisse, R. Stearman, R. D. Klausner, and A. Dancis. 2002. Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology 14829-40. [DOI] [PubMed] [Google Scholar]
- 31.Knight, S. A., G. Vilaire, E. Lesuisse, and A. Dancis. 2005. Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect. Immun. 735482-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosman, D. J. 2003. Molecular mechanisms of iron uptake in fungi. Mol. Microbiol. 471185-1197. [DOI] [PubMed] [Google Scholar]
- 33.Lamb, T. M., W. Xu, A. Diamond, and A. P. Mitchell. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 2761850-1856. [DOI] [PubMed] [Google Scholar]
- 34.Lan, C. Y., G. Rodarte, L. A. Murillo, T. Jones, R. W. Davis, J. Dungan, G. Newport, and N. Agabian. 2004. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 531451-1469. [DOI] [PubMed] [Google Scholar]
- 35.Mantovani, R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 23915-27. [DOI] [PubMed] [Google Scholar]
- 36.McNabb, D. S., and I. Pinto. 2005. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryot. Cell 41829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNabb, D. S., Y. Xing, and L. Guarente. 1995. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 947-58. [DOI] [PubMed] [Google Scholar]
- 38.Mercier, A., B. Pelletier, and S. Labbe. 2006. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 51866-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mercier, A., S. Watt, J. Bahler, and S. Labbe. 2008. Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot. Cell 7493-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhlrad, D., R. Hunter, and R. Parker. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 879-82. [DOI] [PubMed] [Google Scholar]
- 41.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42981-993. [DOI] [PubMed] [Google Scholar]
- 42.Nagai, M., J. Sakakibara, Y. Nakamura, F. Gejyo, and T. Ono. 2002. SREBP-2 and NF-Y are involved in the transcriptional regulation of squalene epoxidase. Biochem. Biophys. Res. Commun. 29574-80. [DOI] [PubMed] [Google Scholar]
- 43.Naglik, J. R., S. J. Challacombe, and B. Hube. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67400-428, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nyilasi, I., T. Papp, M. Tako, E. Nagy, and C. Vagvolgyi. 2005. Iron gathering of opportunistic pathogenic fungi. A mini review. Acta Microbiol. Immunol. Hung. 52185-197. [DOI] [PubMed] [Google Scholar]
- 45.Payne, S. M. 1993. Iron acquisition in microbial pathogenesis. Trends Microbiol. 166-69. [DOI] [PubMed] [Google Scholar]
- 46.Pelletier, B., J. Beaudoin, Y. Mukai, and S. Labbe. 2002. Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J. Biol. Chem. 27722950-22958. [DOI] [PubMed] [Google Scholar]
- 47.Pelletier, B., J. Beaudoin, C. C. Philpott, and S. Labbe. 2003. Fep1 represses expression of the fission yeast Schizosaccharomyces pombe siderophore-iron transport system. Nucleic Acids Res. 314332-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pendrak, M. L., M. P. Chao, S. S. Yan, and D. D. Roberts. 2004. Heme oxygenase in Candida albicans is regulated by hemoglobin and is necessary for metabolism of exogenous heme and hemoglobin to alpha-biliverdin. J. Biol. Chem. 2793426-3433. [DOI] [PubMed] [Google Scholar]
- 49.Ramanan, N., and Y. Wang. 2000. A high-affinity iron permease essential for Candida albicans virulence. Science 2881062-1064. [DOI] [PubMed] [Google Scholar]
- 50.Rutherford, J. C., S. Jaron, E. Ray, P. O. Brown, and D. R. Winge. 2001. A second iron-regulatory system in yeast independent of Aft1p. Proc. Natl. Acad. Sci. USA 9814322-14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaible, U. E., and S. H. Kaufmann. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2946-953. [DOI] [PubMed] [Google Scholar]
- 52.Schaller, M., C. Borelli, H. C. Korting, and B. Hube. 2005. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 48365-377. [DOI] [PubMed] [Google Scholar]
- 53.Serrano, R., D. Bernal, E. Simon, and J. Arino. 2004. Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 27919698-19704. [DOI] [PubMed] [Google Scholar]
- 54.Serrano, R., A. Ruiz, D. Bernal, J. R. Chambers, and J. Arino. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 461319-1333. [DOI] [PubMed] [Google Scholar]
- 55.Singh, A., N. Kaur, and D. J. Kosman. 2007. The metalloreductase Fre6p in Fe-efflux from the yeast vacuole. J. Biol. Chem. 28228619-28626. [DOI] [PubMed] [Google Scholar]
- 56.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Penalva, and H. N. Arst, Jr. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timmerman, M. M., and J. P. Woods. 2001. Potential role for extracellular glutathione-dependent ferric reductase in utilization of environmental and host ferric compounds by Histoplasma capsulatum. Infect. Immun. 697671-7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing in Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1811868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387708-713. [DOI] [PubMed] [Google Scholar]
- 60.Wooldridge, K. G., and P. H. Williams. 1993. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 12325-348. [DOI] [PubMed] [Google Scholar]
- 61.Yun, C. W., M. Bauler, R. E. Moore, P. E. Klebba, and C. C. Philpott. 2001. The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J. Biol. Chem. 27610218-10223. [DOI] [PubMed] [Google Scholar]