Abstract
Perturbations in secretory function activate stress response pathways critical for yeast survival. Here we report the identification of the Hos2p/Set3p deacetylase complex (SET3C) as an essential component of the secretory stress response. Strains lacking core components of the Hos2p/Set3p complex exhibit hypersensitivity to secretory stress. Although not required for the unfolded protein response (UPR) and ribosomal gene repression, the Hos2p complex is required for proper activation of the Mpk1p/Slt2p cell integrity kinase cascade. Disruption of the Hos2p complex results in abrogated Mpk1p phosphorylation, whereas constitutive activation of the Mpk1p pathway rescues the hos2Δ mutant growth defect in response to secretory stress. Furthermore, Hos2p activity is required for the Mpk1p-mediated activation of stress-responsive transcription factor Rlm1p, but not for the stress-induced degradation of the C-type cyclin Ssn8p. Our results identify the Hos2p complex as a critical component of the secretory stress response and support the existence a coordinated stress response consisting of the UPR, ribosomal gene repression, and mitogen-activated protein kinase signaling in response to defects in secretory function.
The budding yeast Hos2p protein is a member of the histone deacetylase (HDAC) family related to the founding class I HDAC Rpd3p (30, 37, 38, 40, 44). Biochemical purification has revealed that Hos2p is a component of the multisubunit Set3p complex (SET3C), which includes the SET domain-containing protein Set3p and several uncharacterized components, including Sif2p, Snt1p, and Yil112w (37). Further biochemical analysis indicated that Hos2p, Set3p, Sif2p, and Snt1p form the core of this complex, while three additional proteins, Hst1p, Sum1p, and Cpr1p, appear to be more peripherally associated (37). A functional role for SET3C was shown in the transcriptional repression of the early/middle class of sporulation-specific genes including the key meiotic regulators IME2 and NDT80 (37). In addition, genome-wide chromatin immunoprecipitation (ChIP) analysis has revealed that Hos2p is associated with genes that are actively transcribed, including those that encode small and large ribosomal subunits (44). It is interesting that ribosome biogenesis and meiotic induction are regulated by nutritional signals (18, 28). These studies suggest that the Hos2p complex might play a role in the responses to specific environmental stimuli. However, any role for the Hos2p complex in a signaling capacity has not been elucidated.
Maintaining a functional secretory pathway is critical for cell survival. Eukaryotic cells have evolved multiple means to respond to perturbations in the secretory pathway. One such signaling pathway feeds into the endoplasmic reticulum (ER), which ensures that secretory proteins are properly folded and modified prior to ER exit and entry into the secretory pathway (36, 39). Defects in this pathway lead to the toxic accumulation of misfolded proteins in the ER and the activation of a survival response known as the unfolded protein response (UPR) (7, 29, 36, 41). This occurs through the activation of an ER-based RNase/kinase called Ire1p, which catalyzes the nonconventional splicing of the HAC1 transcript, a bZIP transcription factor (7). Upon proper splicing, Hac1p is translated, enters the nucleus, and activates UPR genes to appropriately respond to the accumulating ER stress.
Interestingly, one major class of genes induced by the UPR are those involved in the secretory pathway (42). Furthermore, inhibition of secretory function by brefeldin A or secretory pathway-deficient mutants (sec− mutants), such as temperature-sensitive alleles of both COPI and COPII transport proteins, activates the UPR (2, 31, 32). These observations suggest that the UPR is intimately linked to the secretory pathway. In addition to UPR activation, secretory stress represses the machinery required for ribosomal biogenesis and protein synthesis (24, 25, 28, 33). By coupling the secretory status to UPR activation and protein synthesis pathways, cells may quickly and efficiently monitor the secretory network and establish the proper balance between cell growth and protein production.
It was proposed that perturbations in the secretory pathway may be sensed ultimately at the cell membrane through deficiencies in membrane lipids and/or essential membrane proteins, leading to Pkc1p activation (33). How Pkc1p relays the secretory stress signal to elicit a stress response remains unknown. However, previous studies indicated that the secretory stressor tunicamycin activates the yeast mitogen-activated protein (MAP) kinase Mpk1p (1, 4), a critical component of the yeast cell integrity pathway, which is a linear kinase cascade comprising Pkc1p → Bck1p (MEK kinase) → Mkk1/2p (MEK) → Mpk1p/Slt2p (MAP kinase) (22, 23, 35, 47). The Mpk1p cell integrity pathway is activated in response to a variety of environmental stimuli, including heat shock, cell wall stress, actin depolymerization, hypo-osmotic stress, and mating pheromone (10). Mpk1p phosphorylates and activates downstream transcriptional effectors including Rlm1p (15, 16, 45, 46) and triggers the destruction of the transcriptional repressor cyclin C (Ssn8p/Ume3p/Srb11p) (5, 6). Whether the Mpk1p cell integrity pathway regulates the response to secretory stress remains unknown.
Here, we provide evidence that the deacetylase Hos2p and its associated complex are required for an efficient response to secretory stress. This response requires a core set of proteins within SET3C including Hos2p, Set3p, Sif2p, and Snt1p. Unexpectedly, we demonstrate that two branches of secretory stress signaling, the UPR and the ribosomal repression response, remain intact in the absence of SET3C function. In addition, we have discovered that the Hos2p-dependent stress signal is transduced via the Mpk1p MAP kinase cascade leading to activation of Rlm1p-dependent transcription. Our results identify the Hos2p/Set3p complex (SET3C) as a novel component critical for cell survival in response to secretory stress.
MATERIALS AND METHODS
Media, strains, and plasmids.
Yeast strains were grown and cultured in rich medium containing 20 g glucose (Sigma) and 30 g YEP broth (QBiogene) per liter. Selective medium was made with 20 g glucose and 6.7 g yeast nitrogen base (with ammonium sulfate), as well as amino acids supplied in CSM mix (QBiogene). CSM lacking inositol was purchased from QBiogene. All drugs were purchased from Sigma and used at the following concentrations in yeast plates unless otherwise noted: 0.2 μg/ml tunicamycin (1 μg/ml in liquid culture), 5 mM 2-deoxyglucose, 20 mM dithiothreitol (DTT), 1 μg/ml bafilomycin A1, 100 μg/ml brefeldin A, 1 mM hydrogen peroxide, and 50 mM chlorpromazine. The yeast strains used in this study are shown in Table 1. Plasmids containing activated alleles of BCK1 (BCK1-20), the 2μm MPK1 expression vector, and the YIL117C-lacZ/YIL117C-rlm1Δ-lacZ plasmids were kindly provided by D. Levin (16).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| YDS2 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 44 |
| AWY1 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hos2::KAN | 44 |
| AWY1202 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hos2::KAN/pAW202 (TRP1) HOS2-13X-Myc | 44 |
| AWY1203 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hos2::KAN/pAW202 (TRP1) HOS2-13X-Myc-H195,196A | 44 |
| JN284 | MATaleu2 his7 ise1 | 34 |
| TCY1 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 set3::KAN | This study |
| TCY2 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 snt1::KAN | This study |
| TCY3 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sif2::KAN | This study |
| TCY4 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 cpr1::KAN | This study |
| TCY5 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 yil112w::KAN | This study |
| TCY6 | MATaleu2 his7 ise1 hos2::HIS | This study |
| TCY7 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hos2::KAN hda1::TRP | This study |
| TCY8 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hos2::KAN rpd3::TRP | This study |
| TCY9 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hac1::TRP | This study |
| TCY10 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hos2::KAN hac1::TRP | This study |
Chromosomal deletion/epitope tagging.
UME3/SRB11 deletion plasmid pVC329 was digested with BamHI/NotI, liberating the UME3::TRP1 cassette, which was transformed into strains YDS2 and AMY1 to generate UME3 deletion strains, and confirmed by genomic PCR. The generation of SET3C deletion strains was accomplished by PCR-mediated strategies by using plasmid pFA6a-KAN as a template (26). These modified alleles were verified by PCR analysis of genomic DNA (data not shown). The PCR primers used were as follows (5′ 224 3′): set3::KAN, CAGTTTTAGATCGTACTTCACAAAATACGAGAACTGAATCCGGATCCCCGGGTTAATTAA and TACTTAAGTTTATATAGGTGTAAGAAGGAAATGTCCATGTGAATTCGAGCTCGTTTAAAC; sif2::KAN, TAAAAATAACAGAAACAAAAAAAAGGTAGGGAAGGCCCATCACACGGAAACGGATCCCCGGGTTAATTAA and ATAACAATAAGAATGATAAAATTCATCTGTTTATGTACTGTACCTAGTTAGAATTCGAGCTCGTT TAAAC; yil112w::KAN, GATATTATTATATGTGACAGAGAAGAATTGCTGTAGAGATTCATGACAATCGGATCCCCGGGTTAATTAA and TCGAAAATGCAACTATGTATGAGCATATGCCAACGGACCGATGAATTGTTGAATTCGAGCTCGTTTAAAC; snt1::KAN, GCCTACTAACTTGTGCATAGAACAGCAAACAGAAACAAAGCGTAAGAAACCGGATCCCCGGGTTAATTAA and TTGGATGGAAAAGAAGTAGAGCATATGTATTGCCCGTCTCAGCCGTTTGTGAATTCGAGCTCGTTTAAAC; cpr1::KAN, TCTTGAATTTAATATCTCAACTCAATCCAAACTCAACCGCTAATACTACCCGGATCCCCGGGTTAATTAA and AGAGAGAATAGTTCGTTTCAATTTTTGCTGTATTGTTCCAGGCAGAGCGGGAATTCGAGCTCGTTTAAAC; rpd3::TRP, ACAATTGCGCCATACAAAACATTCGTGGCTACAACTCGATATCCGTGCAGCGGATCCCCGGGTTAATTAA and TTCTTTTGTTTCACATTATTTATATTCGTATATACTTCCAACTCTTTTTTGAATTCGAGCTCGTTTAAAC; hda1::TRP, ATATTGAGAAAGGGAAAGTTGAGCACTGTAATACGCCGAACAGATTAAGCCGGATCCCCGGGTTAATTAA and CATAAGGCATGAAGGTTGCCGAAAAAAAATTATTAATGGCCAGTTTTTCCGA ATTCGAGCTCGTTTAAAC.
Stress sensitivity and spotting assays.
The strains with the indicated genotypes were grown to mid-log phase (5 × 106 cells/ml) and serially diluted 1:10. Five microliters of each dilution was spotted onto plates containing the indicated inhibitors, incubated for 2 days at 30 or 37°C as indicated, and then photographed. Strains harboring BCK1-20 or MPK1 plasmids were spotted onto selective medium containing 0.2 μg/ml tunicamycin.
Cell lysis and Western blotting.
The indicated strains were grown to mid-log phase (5 × 106 cells/ml), at which point the t = 0 time point was taken and the remainder of the culture was treated with 1 μg/ml tunicamycin for 1 h. Cell pellets were gently centrifuged for 5 min and resuspended in NP-40 lysis buffer (50 mM Tris [pH 7.5], 5 mM EDTA [pH 8], 150 mM NaCl, and 1% NP-40 supplemented with leupeptin, aprotinin, phenylmethylsulfonyl fluoride, sodium fluoride, sodium vanadate, and sodium pyrophosphate). Acid-washed glass beads (G-8772; Sigma) were added, and the mixture was vortexed on ice intermittently for six rounds of 30 s on/30 s off to prevent excessive heating. Lysate was siphoned away from beads, transferred to an Eppendorf tube, and centrifuged for 15 min at 14,000 rpm at 4°C. Protein concentration was determined by the Bradford assay. Equal amounts of lysate (50 μg) were loaded onto a 10% polyacrylamide gel for Western blotting. Gel electrophoresis was performed, followed by transfer to polyvinylidene difluoride membrane with a semidry transfer apparatus for 2 h at room temperature. Membranes were blocked with 2% milk for 30 min and then treated with Mpk1p or P-Mpk1 polyclonal antibodies (1:500 dilution; Cell Signaling) overnight. For Ume3p Western blot assays, cells expressing pLR101 (contains the myc epitope-tagged wild-type SRB11 allele, Srb11p-myc) were monitored for Ume3p expression by Western blot analysis of immunoprecipitates from 250 μg of soluble protein. Blots were washed for 3 × 10 min each and treated for 30 min with secondary antibody conjugated to horseradish peroxidase (Promega). Blots were then washed for an additional 3 × 10 min and treated with ECL reagent (Amersham) for 1 min before exposure to film.
RNA analysis. (i) Northern blotting.
Cells were grown in the absence or presence of tunicamycin, aliquots were taken at specified time intervals, and RNA was isolated by the glass bead procedure. Briefly, cells were washed in water and mixed with 0.2 ml YRLB (0.5 M NaCl, 0.2 M Tris [pH 7.5], 10 mM EDTA, 1% sodium dodecyl sulfate), 0.2 ml phenol-chloroform-isoamyl alcohol (PCI), and 0.4 g glass beads (Sigma). Cells were vortexed for 2.5 min, and then an additional 0.3 ml YRLB and 0.2 ml PCI were added and the mixture was vortexed again for 2.5 min. Samples were centrifuged for 2 min, and lysates were removed and added to 0.4 ml PCI to remove additional protein contaminants. Samples were vortexed and centrifuged for 2 min at 14,000 rpm. The top aqueous solution was removed and added to 1 ml 100% ethanol, mixed, and centrifuged for 10 min at full speed at 4°C. The RNA pellet was washed with 70% ethanol and resuspended in 50 μl Tris-EDTA. Tris-EDTA and ethanol were diethyl pyrocarbonate treated (0.1%) to prevent RNase contamination, and the RNA concentration was determined by measuring the optical density at 260 and 280 nm. Five micrograms of isolated RNA was loaded onto RNA denaturing gel (6% formaldehyde, 10% morpholinepropanesulfonic acid [MOPS], 1% agarose) and underwent gel electrophoresis for 5 h. The gel was washed twice in water for 10 min each time and transferred onto nucleic acid membrane (Hybond N+) overnight at room temperature in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The membrane was then auto-cross-linked (Stratagene) and washed twice for 5 min each time in 2× SSC. Prehybridization buffer (Amersham) was added to the membrane, and it was incubated at 60°C for 2 h while gently rocking in a hybridization oven (model 400; Robbins Scientific). Radioactive probes were generated with the Prime-It II radioactive primer labeling kit (Stratagene) to label a 500- to 900-bp PCR product from genomic DNA with the following primers for KAR2, INO1, HAC1, RPL3, RPL30, and RPL28: HAC1, CGCAATCGAACTTGGCTATCCC and GGGTAGACTGTTTCCCGC; KAR2, CGCTGGCAAGCTGCTGGTAC and CAATACGGGTGGACATTTGGCTGG; INO1, CGAAGACAGCTAGTGGCCGC and CTGCATCCACTAAGAACTGG; RPL30, GGCCCCAGTTAAATCCCAAG and GCCAAGGTGGTCAAGATATC; RPL3, CGAAGCACCTCACGGTC and GCTTCATCATCACCCTTACC; RPL28, CCTTCCAGATTCACTAAGAC and GCGATCAATTCAACAACACC.
(ii) Quantitative PCR.
Total RNA was isolated from mid-log-phase wild-type and hos2Δ mutant cells treated for 4 h with 1 μg/ml tunicamycin. Total RNA was DNase treated with a DNA-free kit (Ambion), and 1 μg DNase-treated RNA was used for a cDNA synthesis reaction with an iScript reverse transcriptase kit (Bio-Rad). Samples were diluted 1:50, and 5 μl cDNA was used per reverse transcription (RT)-PCR. Real-time quantitative PCR was performed with iQ Syber green supermix on the iCycler iQ detection system (Bio-Rad). The sample volume was 20 μl per reaction. The efficiency and specificity of primers were confirmed by standard PCR and DNA electrophoresis. The PCR program on the iCycler was 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. All real-time PCR values were normalized to actin as indicated and represented as relative induction afterward. All reaction mixtures were analyzed in triplicate. The following primers were used: actin, GACTGATCTGTAATAACCACG (forward) and CAATCGATGTTAGTACATGAG (reverse); YIL117C, CCAAGTATTACTCCTCCCTCT (forward) and GTTGTTATTACCCACCATAGC (reverse); MPK1, GAATGTGATATGCACCAAATC (forward) and CAATTGACAATCTGCATTGAC (reverse).
RESULTS
Hos2p complex integrity is required for the cellular response to tunicamycin-induced stress.
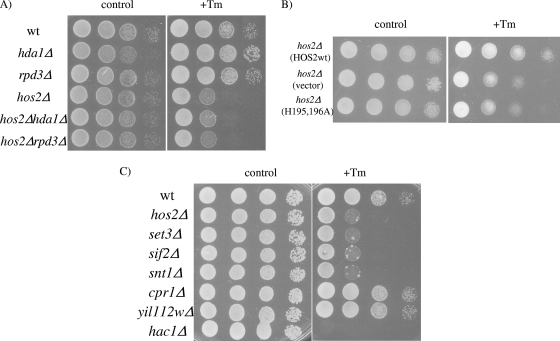
Previous work on HDACs suggested that mammalian HDACs are involved in a cellular response to misfolded protein stress (19). Since HDACs are evolutionarily conserved, a potential role for a yeast deacetylase in a similar stress response was evaluated. A collection of yeast strains with both the HDAC and SIR2 family proteins deleted were screened for hypersensitivity to the drug tunicamycin, an N-linked glycosylation inhibitor that causes an accumulation of improperly modified and misfolded proteins in the ER. Tunicamycin is commonly used to induce ER stress and activate the UPR. Among 12 deacetylase deletion mutants tested, the hos2Δ mutant was identified as the only strain exhibiting hypersensitivity to tunicamycin (Fig. 1A). Importantly, deletion of HDA1 or RPD3 in the hos2Δ mutant strain did not enhance sensitivity to tunicamycin, suggesting a dominant role for HOS2 in the response to tunicamycin (Fig. 1A). To determine whether Hos2p deacetylase activity is required for the cellular response to tunicamycin stress, a hos2Δ mutant strain harboring an episomal plasmid containing the vector or wild-type HOS2 or the catalytically inactive hos2 (H195,196A) mutant was tested for sensitivity to tunicamycin-induced stress. As shown in Fig. 1B, the catalytically inactive mutant shows the same degree of tunicamycin sensitivity as the hos2Δ mutant strain, indicating that enzymatic activity is required for Hos2p function in response to tunicamycin.
FIG. 1.
The Hos2p/Set2p complex is required for the cellular response to tunicamycin-induced stress. (A) Wild-type (wt) and hos2Δ, hda1Δ, rpd3Δ, hos2Δ hda1Δ, and hos2Δ rpd3Δ deletion mutant yeast strains were grown to mid-log phase, adjusted to an optical density of 1, serially diluted 1:10, spotted onto control and 0.2 μg/ml tunicamycin (Tm)-containing plates, and incubated for 48 h. (B) Cells of the hos2Δ mutant strain harboring episomal plasmids containing the vector alone or wild-type HOS2 or an enzymatically inactive hos2Δ (H195,196A) mutant were spotted described as above onto selective medium containing 0.2 μg/ml tunicamycin. (C) Hos2p/Set3p complex deletion strains were evaluated for tunicamycin sensitivity as described above.
Hos2p exists as part of multisubunit protein complex, which includes three additional core components, Set3p, Sif2p, and Snt1p, and three peripherally associated proteins, Cpr1p, Yil112wp, and Hst1p. The core, but not peripheral, subunits are required for the structural integrity of the SET3C complex (37). To test whether the Hos2p/Set3p complex is required for a proper tunicamycin response, single-deletion strains were evaluated for sensitivity to tunicamycin. The core subunit set3Δ, sif2Δ, and snt1Δ mutant strains all displayed hypersensitivity to tunicamycin-induced stress, similar to the hos2Δ mutant strain. In contrast, strains with deletions in the peripheral components of the complex, cpr1Δ, yil112wΔ, and hst1Δ, showed no phenotype (Fig. 1C; data not shown). We conclude that the Hos2p/Set3p deacetylase complex is required for the proper cellular response to tunicamycin.
The Hos2p/Set3p complex is essential for the secretory stress response.
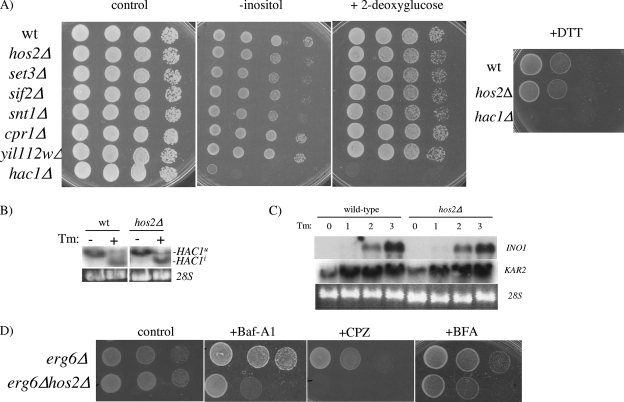
Tunicamycin is commonly used to induce ER stress and activate the UPR. Proper activation of the UPR is essential for conferring tunicamycin resistance. We therefore determined whether the Hos2p-Set3p complex is required for the activation of the UPR. To test this, Hos2p complex deletion strains were tested for sensitivity to multiple ER stress agents known to induce the UPR, including 2-deoxyglucose, DTT, and medium lacking inositol (Fig. 2A). Surprisingly, the hos2Δ deletion strain does not display hypersensitivity to 2-deoxyglucose or DTT but does display a partial sensitivity to medium lacking inositol. Similarly, all SET3C core complex deletion strains showed an ∼10-fold increase in sensitivity to inositol deficiency (Fig. 2A). Supporting a UPR-independent role for Hos2p, HAC1 splicing occurs normally and the UPR target genes INO1 and KAR2 are properly induced in the hos2Δ mutant strain in response to tunicamycin treatment (Fig. 2B and C). As expected, the hac1Δ deletion strain, which cannot mount a UPR, did not grow under any of the stress conditions tested. Thus, the tunicamycin and inositol hypersensitivity displayed by the hos2Δ mutant strain is likely caused by a UPR-independent mechanism.
FIG. 2.
The Hos2p/Set3p complex is required for a secretory stress response. (A) Hos2p complex deletion strains were spotted as described in the legend to Fig. 1 onto medium containing 2-deoxyglucose or DTT or medium lacking inositol. (B) UPR activation was monitored by Northern analysis from wild-type (wt) or hos2Δ mutant yeast strains challenged with 1 μg/ml tunicamycin (Tm) for 1 h. Northern analysis was performed with a HAC1-specific probe detecting both the full-length (HAC1u) and spliced (HAC1i) isoforms of HAC1. (C) Northern analysis was performed on wild-type and hos2Δ mutant strains treated with tunicamycin for the indicated times with INO1- and KAR2-specific probes. (D) erg6Δ or erg6Δ hos2Δ cells were serially diluted onto complete medium containing 100 μg/ml brefeldin A (BFA), 1 μg/ml bafilomycin A1 (Baf-A1), or 50 μM chlorpromazine (CPZ) for stress sensitivity analysis.
Tunicamycin can induce ER stress, as well as secretory failure, by inhibiting protein transport into the secretory pathway. We therefore investigated whether Hos2p is required for the proper response to secretory stress. The hos2Δ mutant strain was examined for its response to secretory stress agents including brefeldin A (ER-Golgi transport inhibitor), bafilomycin A1 (vacuolar ATPase inhibitor), and chlorpromazine (induces membrane stretching). Since many secretory inhibitors are relatively impermeable to yeast cells, an erg6Δ strain was used to increase the cellular permeability to these agents (34). As shown in Fig. 2D, the hos2Δ mutant strain exhibits hypersensitivity to inhibitors across the spectrum of the secretory network. These data indicate that Hos2p is selectively required for the proper response to secretory stress rather than an ER stress response.
Hos2p and Hac1p cooperate in response to membrane stress.
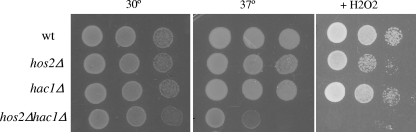
We propose that a Hos2p-dependent secretory pathway and the ER-sensing UPR operate coordinately in response to secretory stress, as both pathways are required for the proper response to secretory stressors (2, 31, 32; this study). UPR activation, in combination with the Hos2p-dependent secretory stress response, could confer additive protection against secretory defects. Ultimately, defects in secretory capacity are likely resulting in impaired membrane and/or cell wall integrity (33). If Hos2p and the UPR are independently required for transducing secretory stress signals, then cells lacking both pathways might display enhanced lethality in response to perturbations in cell membrane integrity. To test this hypothesis, wild-type cells and hos2Δ, hac1Δ, and hos2Δ hac1Δ deletion mutant cells were evaluated for hypersensitivity to elevated temperature and oxidative stress, conditions that alter membrane fluidity and result in loss of membrane integrity (9, 11, 17). As shown in Fig. 3, neither the hos2Δ nor the hac1Δ single mutant is sensitive to these stresses. In contrast, the hos2Δ hac1Δ double mutant has a severe growth defect under these conditions. These results suggest that Hos2p-dependent secretory function and the UPR work independently to promote cell survival in response to secretory stress. In addition, these findings suggest that these pathways are potentially monitoring secretory stress indirectly through cell membrane and/or cell wall integrity.
FIG. 3.
Genetic interactions between hos2Δ and UPR components. Wild-type (wt) and hos2Δ, hac1Δ, and hos2Δ hac1Δ mutant strains were serially diluted, as described in the legend to Fig. 2, onto complete medium at 30 or 37°C or complete medium containing 1 mM hydrogen peroxide (H2O2).
Hos2p is not required for ribosomal repression in response to secretory stress.
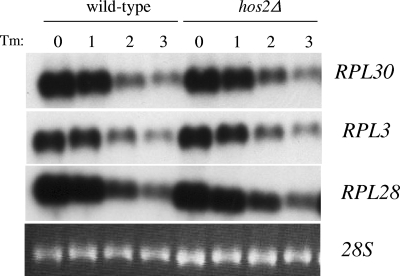
In yeast, one prominent cellular response to secretory failure is the down-regulation of ribosome biogenesis through transcriptional repression (28). Previous studies using global ChIP analysis indicated that Hos2p could potentially regulate ribosome biogenesis through direct binding to small and large ribosomal subunit gene promoters (38). Since ribosome biosynthesis is directly coupled to the secretory status of the cell, we tested whether Hos2p might control ribosomal gene expression in response to secretory stress by Northern analysis. As shown in Fig. 4, wild-type cells treated with tunicamycin exhibited dramatically reduced RPL3 and RPL30 mRNA expression levels after 2 h. In the hos2Δ mutant strain, the expression of all of the ribosomal mRNAs tested was similarly down-regulated within 2 h of tunicamycin treatment. These data show that although Hos2p appears to bind ribosomal genes under normal conditions (38), Hos2p is not required for the proper down-regulation of ribosomal gene expression during the cellular response to secretory stress.
FIG. 4.
Hos2p is not required for the proper repression of ribosomal gene expression during secretory failure. Northern analysis was performed with the wild-type or hos2Δ mutant strain challenged with 1 μg/ml tunicamycin (Tm) for 0, 1, 2, or 3 h. Northern analysis was performed with labeled probes detecting RPL3, RPL28, and RPL30 previously shown to respond to secretory defects. 28S rRNA served as a loading control.
The Hos2p-Set3p complex is required for proper activation of the Mpk1p/Slt2p MAP kinase signaling pathway.
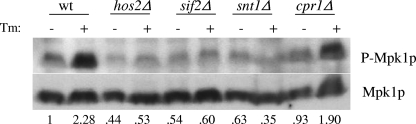
Since Hos2p is not required for either the UPR or the ribosomal repression response, we searched for an alternative stress response pathway requiring Hos2p activity. One possible target is the Pkc1p-directed MAP kinase pathway, as previous studies have shown that Mpk1p activation is affected in response to tunicamycin (1, 4). We thus investigated whether Hos2p is required for the activation and phosphorylation of MAP kinase signaling in response to secretory failure. Wild-type and SET3C deletion strains (both peripheral and core deletion mutants) were treated with tunicamycin, and Western blot analysis was performed with an antibody that detects the phosphorylated, active form of Mpk1p, which represents a readout of MAP kinase activation. As shown in Fig. 5, tunicamycin treatment induces Mpk1p phosphorylation in wild-type cells. In stark contrast, the hos2Δ mutant strain, as well as the core snt1Δ and sif2Δ mutant strains, displayed severely diminished Mpk1p phosphorylation. These findings demonstrate that the Hos2p complex is required for proper Mpk1p activation in response to secretory stress.
FIG. 5.
The Hos2p complex is required for Mpk1p activation following secretory stress. The wild-type (wt) and hos2Δ mutant strains were grown to mid-log phase and challenged with 1 μg/ml tunicamycin (Tm) for 1 h. Western analysis was carried out with antibodies against total Mpk1p and activated phospho-Mpk1p.
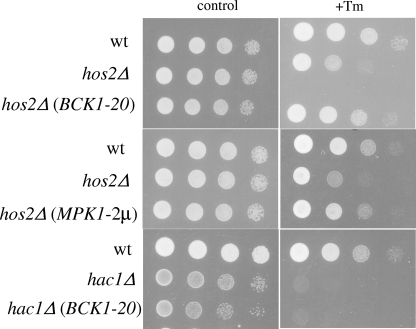
Ectopic activation of the Pkc1p MAP kinase cascade is sufficient to suppress the hos2Δ mutant's tunicamycin hypersensitivity phenotype.
Our working model predicts that SET3C-dependent stimulation of the Mpk1p MAP kinase is important for cell survival following secretory stress. If this is correct, then activation of the Mpk1p MAP kinase cascade would rescue the hos2Δ mutant's hypersensitivity phenotype. A constitutively active allele of the MEK kinase gene BCK1 (BCK1-20) was introduced into wild-type and hos2Δ mutant strains that were then serially spotted onto selective medium containing tunicamycin. Expression of BCK1-20 completely rescued the growth defect of the hos2Δ mutant strain in response to tunicamycin treatment (Fig. 6, top panels). Importantly, the effect of BCK1-20 is specific, as it does not rescue the growth defect of a hac1Δ mutant in the presence of tunicamycin (Fig. 6, bottom panels). Further supporting a role for Mpk1p in secretory stress signaling, increased MPK1 expression with a high-copy plasmid also suppresses the tunicamycin hypersensitivity in the hos2Δ mutant strain (Fig. 6, middle panels). Thus, activation of the Mpk1p MAP kinase pathway is sufficient to restore the proper secretory stress response in the absence of Hos2p activity. Together, these results strongly suggest that Hos2p regulates the secretory stress response through Mpk1p-dependent signaling.
FIG. 6.
Ectopic activation of Mpk1p signaling rescues the hos2Δ secretory growth defect. The wild-type (wt) and hos2Δ and hac1Δ mutant strains harboring the control vector, BCK1-20, or MPK1-2μm plasmid were grown to mid-log phase and serially spotted, as described in the legend to Fig. 5, onto selective medium containing 1 μg/ml tunicamycin (Tm). Plates were incubated at 30°C for 48 h.
Hos2p is required for full Mpk1p-dependent activation of Rlm1p.
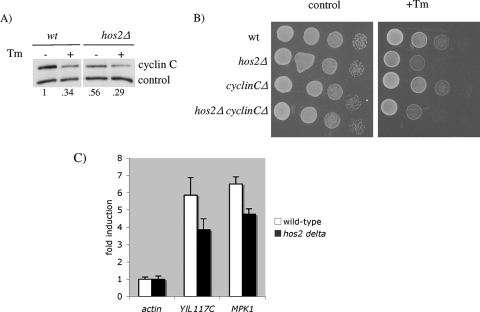
To identify potential downstream effectors that transduce the Hos2p-dependent stress signal, we considered two downstream effectors of the Mpk1p pathway. First, Mpk1p is required for the ROS-dependent destruction of the transcriptional repressor cyclin C (Ssn8p/Ume3p/Srb11p) (5, 6, 21), thus permitting stress response gene induction. We therefore determined if cyclin C degradation occurs in response to tunicamycin. Wild-type and hos2Δ mutant strains harboring a cyclin C expression plasmid were treated with tunicamycin, and cyclin C levels were monitored by Western blot analysis. As shown in Fig. 7A, cyclin C levels were indeed reduced in the hos2Δ mutant strain following tunicamycin treatment, indicating that secretory stress does trigger its destruction. In addition, epistasis analysis was performed to determine whether deletion of cyclin C rescues the hos2Δ mutant's growth defect during secretory stress. As shown in Fig. 7B, the hos2Δ cyclin CΔ double mutant and hos2Δ single mutant displayed similar sensitivities to tunicamycin. These results indicate that Hos2p-dependent activation of the Mpk1p pathway in response to secretory stress does not promote Ume3p/Srb11p degradation and increased cell survival.
FIG. 7.
Hos2p is not required for secretory stress-induced cyclin C degradation and cell survival but is required for activation of Rlm1p target genes. (A) The wild-type (wt) and hos2Δ mutant strains expressing myc-tagged UME3/SRB11 were grown to mid-log phase, treated with 1 μg/ml tunicamycin (Tm) for 1 h, and harvested for protein analysis by immunoprecipitation-Western blot assay. (B) The wild-type and hos2Δ, cyclinCΔ, and hos2Δ cyclinCΔ mutant strains were serially diluted and spotted onto control and tunicamycin-containing plates. (C) The wild-type and hos2Δ mutant strains were grown to mid-log phase and treated with 1 μg/ml tunicamycin for 4 h. Real-time RT-PCR analysis was performed with primers specific for the Rlm1p target genes YIL117C and MPK1. Values represent mRNA induction levels relative to control actin. Errors are reported as the standard error of the mean.
We next examined whether the activation of Rlm1p, a transcription factor required for cell wall integrity (16, 45), occurs normally in hos2Δ mutant cells. To monitor Rlm1p activity, we evaluated the expression of two known Rlm1p target genes, YIL117C and MPK1 (16), in response to tunicamycin-induced secretory stress. Real-time RT-PCR analysis was performed on RNA isolated from wild-type and hos2Δ mutant cells challenged with tunicamycin. As shown in Fig. 7C, YIL117C and MPK1 are robustly induced approximately sixfold in response to tunicamycin treatment. However, hos2Δ mutant cells displayed an ∼33% reduction in Rlm1p target gene activation, suggesting a partial impairment of Rlm1p-dependent gene activation. These results suggest that Mpk1p → Rlm1p signaling is a key effector pathway downstream of the Hos2p complex in response to secretory failure.
DISCUSSION
The ability to mount proper stress responses to secretory failure and membrane stress is critical for cell viability. We have identified the Hos2p-containing multiprotein deacetylase complex as a novel component of the response to secretory stress. We show that Hos2p deacetylase activity, as well as the integrity of the entire Hos2p/Set3p complex, is specifically required for the activation of the secretory stress-induced Mpk1p-cell integrity pathway. In contrast, the Hos2p complex is dispensable for the activation of the UPR and the ribosomal gene repression program, two additional stress response pathways commonly induced by secretory failure. Indeed, enhanced Mpk1p signaling was sufficient to rescue the hos2Δ mutant's growth defect in response to tunicamycin. Our study identifies the Hos2p/Set3p complex as a novel component essential for the proper response to secretory defects.
The use of tunicamycin as an ER stress reagent has been instrumental in uncovering the signaling components of the UPR (20). However, as protein glycosylation plays a critical role in protein transport in the secretory pathway beyond the ER, tunicamycin treatment induces secretory and membrane failure. Although the effect of tunicamycin has been analyzed almost exclusively in the context of ER stress and the UPR, several studies, including this one, have identified two additional pathways that are activated by tunicamycin, namely, the repression of ribosome-associated gene expression and the activation of the MAP kinase cell integrity pathway (1, 24, 25).
Our results clearly demonstrate a role for the Hos2p complex in tunicamycin-induced Mpk1p activation (Fig. 5 and 6). Since the Mpk1p/Slt2p MAP kinase pathway is required for cell survival in response to membrane stress (17, 22, 23, 35), the failure to activate Mpk1p → Rlm1p signaling is likely the key reason that SET3C mutants display growth defects in the presence of secretory stress. Indeed, strains lacking MAP kinase components including BCK1, MKK1, MPK1, and notably RLM1 are hypersensitive to tunicamycin-induced secretory stress (4). Activation of Mpk1p with the constitutively active upstream kinase BCK1-20 or overexpression of MPK1 itself can fully rescue the growth defect of the hos2Δ mutant in response to tunicamycin treatment. These results suggest a few possibilities. Hos2p may regulate Mpk1p activity by either inducing an activator of Mpk1p signaling, such as upstream components in the MAP kinase pathways, or repressing the expression of a protein that inactivates Mpk1p. Interestingly, Hos2p has been shown to function as both a transcriptional activator and a repressor in different contexts, suggesting that both possibilities may exist to fine-tune the levels of MAP kinase signaling (40, 44). Another interesting possibility is that Hos2p regulates Mpk1p activity independently of gene transcription. We have found that Hos2p is a nuclear protein before or after tunicamycin treatment (data not shown). Interestingly, Mpk1p has been localized to both the cytoplasm and the nucleus (27, 43), supporting the possibility that Hos2p directly regulates Mpk1p function and/or activity.
Hos2p is not required for the activation of the UPR or repression of ribosome-associated gene expression; however, it is important to note that these distinct pathways activated by secretory stress are, in fact, functionally connected. The enhanced sensitivity to membrane stresses in the absence of both Hac1p and Hos2p strongly supports this supposition (Fig. 3). Our data suggest that Hos2p-mediated regulation of MAP kinase signaling functions in parallel with the UPR and, in conjunction, promotes an efficient survival response to secretory failure. Supporting this view, recent reports have suggested that secretory failure is intimately associated with activation of the UPR (2, 3, 42). Indeed, we have observed low-level activation of the UPR in the absence of Hos2p function (data not shown). Although it remains unclear how secretory stress elicits a stress signal, Nierras and Warner (33) have proposed that secretory stress leads to protein and lipid deficiencies at the membrane, which ultimately activates a Pkc1p-mediated pathway. Our results support the existence of three distinct pathways that respond to secretory stress, i.e., the UPR, ribosomal gene repression, and the Mpk1p cell integrity pathway, with Hos2p being critical for Mpk1p activation.
Previous studies have shown that Hos2p exists as part of a multisubunit complex (37). Within this complex, Set3p, Snt1p, and Sif2p make up a core complex whereas Cpr1p, Yil112w, and Hst1p peripherally associate with the complex. It was further shown that mutations in core elements disrupt complex formation while mutations in peripheral components have no effect (37, 44). Indeed, we found that mutation of any of the core, but not the peripheral, components of this complex prevents the activation of Mpk1p and renders yeast hypersensitive to tunicamycin treatment (Fig. 1 and 5). Thus, similar to their roles in meiosis and galactose utilization, Hos2p, Set3p, Snt1p, and Sif2p likely work as a functional unit to regulate the secretory stress response (37, 44). Hos2p has HDAC activity and has the capacity to regulate gene expression (37, 38, 44). Importantly, a proper response to secretory stress also requires Hos2p deacetylase activity, which likely catalyzes histone deacetylation, resulting in changes in gene expression important for mounting a secretory stress response (Fig. 1B). Since Hst1p is also present within the complex, the two enzymes may provide histone specificity during the regulation of transcription. Genome-wide ChIP analysis revealed that Hos2p binds genes encoding ribosomal subunits, as well as the SEC31, INO1, and ERG11 genes (38, 44), whose products are responsible for protein synthesis, secretory function, and the synthesis of inositol and ergosterol (two major constituents of the plasma membrane and membranous organelles). However, we have found that the expression of rRNA, SEC31, INO1, and ERG11 is unaffected in the hos2Δ mutant strain challenged by secretory stress (Fig. 4; data not shown).
Diverse environmental stimuli activate MAP kinase signaling, eliciting distinct cellular responses. For example, heat stress, cytoskeletal perturbation, and hypo-osmotic stress enter the MAP kinase cascade at different lateral stages of the MAP kinase pathway (13). Additionally, distinct downstream outputs have been identified, including Mpk1p-mediated degradation of Srb11p/Ume3p in response to oxidative stress (21). Although tunicamycin-induced stress similarly promotes Ume3p/Srb11p degradation, it is not dependent on Mpk1p activation (Fig. 5 and 7), suggesting MAP kinase pathway specificity elicited by tunicamycin versus oxidative stress. These studies support the idea that diverse stimuli activate Mpk1p signaling, leading to the activation of distinct effector pathways, also referred to as a “multiple input/multiple output” model previously proposed for MAP kinase signaling (13).
The importance of a functional secretory stress response will likely be critical for higher eukaryotes as well. In particular, cells specialized in secretory function, such as insulin-secreting β islet cells, would require stress-monitoring mechanisms to ensure the proper assembly, transport, and secretion of insulin output in response to nutritional cues (12). Similarly, β-cell lymphocytes require a rapid and highly regulated system for antibody assembly and transport to the cell surface (8). Failure of this quality control pathway could lead to cell death and disease (14). In this regard, it is worth noting that homology searches suggest that the Hos2p/Set3p complex is highly related to the mammalian HDAC3/SMRT complex (37), suggesting that the secretory function of Hos2p may be conserved in higher eukaryotes. It will be of great interest to determine whether the HDAC3/SMRT complex in humans may regulate the complex secretory functions of high-output cells, including the pancreatic β cell and lymphocytic cells.
Acknowledgments
We thank D. Levin for kindly providing plasmids and M. Grunstein and J. Nitiss for kindly providing deletion strains.
This work was supported by the Leukemia and Lymphoma Society (T.P.Y.).
Footnotes
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Bonilla, M., and K. W. Cunningham. 2003. Mitogen-activated protein kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell 144296-4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, H. J., S. A. Jesch, M. L. Gaspar, and S. A. Henry. 2004. Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics 1681899-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, H. J., E. W. Jones, and S. A. Henry. 2002. Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics 16229-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y., D. E. Feldman, C. Deng, J. A. Brown, A. F. De Giacomo, A. F. Gaw, G. Shi, Q. T. Le, J. M. Brown, and A. C. Koong. 2005. Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Cancer Res. 3669-677. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, K. F., M. J. Mallory, J. B. Smith, and R. Strich. 1997. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p). EMBO J. 164665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper, K. F., M. J. Mallory, and R. Strich. 1999. Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol. Cell. Biol. 193338-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, J. S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87391-404. [DOI] [PubMed] [Google Scholar]
- 8.Gass, J. N., N. M. Gifford, and J. W. Brewer. 2002. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 27749047-49054. [DOI] [PubMed] [Google Scholar]
- 9.Gray, J. V., J. P. Ogas, Y. Kamada, M. Stone, D. E. Levin, and I. Herskowitz. 1997. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 164924-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 621264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutteridge, J. M., and B. Halliwell. 1990. The measurement and mechanism of lipid peroxidation in biological systems. Trends Biochem. Sci. 15129-135. [DOI] [PubMed] [Google Scholar]
- 12.Harding, H. P., and D. Ron. 2002. Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes 51(Suppl. 3)S455-S461. [DOI] [PubMed] [Google Scholar]
- 13.Harrison, J. C., T. R. Zyla, E. S. Bardes, and D. J. Lew. 2004. Stress-specific activation mechanisms for the “cell integrity” MAPK pathway. J. Biol. Chem. 2792616-2622. [DOI] [PubMed] [Google Scholar]
- 14.Huang, C. J., C. Y. Lin, L. Haataja, T. Gurlo, A. E. Butler, R. A. Rizza, and P. C. Butler. 2007. High expression rates of human islet amyloid polypeptide induce endoplasmic reticulum stress mediated beta-cell apoptosis, a characteristic of humans with type 2 but not type 1 diabetes. Diabetes 562016-2027. [DOI] [PubMed] [Google Scholar]
- 15.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 341049-1057. [DOI] [PubMed] [Google Scholar]
- 16.Jung, U. S., A. K. Sobering, M. J. Romeo, and D. E. Levin. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46781-789. [DOI] [PubMed] [Google Scholar]
- 17.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 91559-1571. [DOI] [PubMed] [Google Scholar]
- 18.Kassir, Y., N. Adir, E. Boger-Nadjar, N. G. Raviv, I. Rubin-Bejerano, S. Sagee, and G. Shenhar. 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224111-171. [DOI] [PubMed] [Google Scholar]
- 19.Kawaguchi, Y., J. J. Kovacs, A. McLaurin, J. M. Vance, A. Ito, and T. P. Yao. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115727-738. [DOI] [PubMed] [Google Scholar]
- 20.Kohno, K., K. Normington, J. Sambrook, M. J. Gething, and K. Mori. 1993. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 13877-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krasley, E., K. F. Cooper, M. J. Mallory, R. Dunbrack, and R. Strich. 2006. Regulation of the oxidative stress response through Slt2p-dependent destruction of cyclin C in Saccharomyces cerevisiae. Genetics 1721477-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin, D. E., and E. Bartlett-Heubusch. 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 1161221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin, D. E., B. Bowers, C. Y. Chen, Y. Kamada, and M. Watanabe. 1994. Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae. Cell. Mol. Biol. Res. 40229-239. [PubMed] [Google Scholar]
- 24.Li, B., C. R. Nierras, and J. R. Warner. 1999. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol. Cell. Biol. 195393-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Y., R. D. Moir, I. K. Sethy-Coraci, J. R. Warner, and I. M. Willis. 2000. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol. 203843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 27.Mattison, C. P., S. S. Spencer, K. A. Kresge, J. Lee, and I. M. Ota. 1999. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol. Cell. Biol. 197651-7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuta, K., and J. R. Warner. 1994. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol. Cell. Biol. 142493-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori, K., T. Kawahara, H. Yoshida, H. Yanagi, and T. Yura. 1996. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1803-817. [DOI] [PubMed] [Google Scholar]
- 30.Mou, Z., A. E. Kenny, and M. J. Curcio. 2006. Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics 1722157-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng, D. T. 2005. Screening for mutants defective in secretory protein maturation and ER quality control. Methods 35366-372. [DOI] [PubMed] [Google Scholar]
- 32.Ng, D. T., E. D. Spear, and P. Walter. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 15077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nierras, C. R., and J. R. Warner. 1999. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 27413235-13241. [DOI] [PubMed] [Google Scholar]
- 34.Nitiss, J., and J. C. Wang. 1988. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc. Natl. Acad. Sci. USA 857501-7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paravicini, G., M. Cooper, L. Friedli, D. J. Smith, J.-L. Carpentier, L. S. Klig, and M. A. Payton. 1992. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol. Cell. Biol. 124896-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13349-355. [DOI] [PubMed] [Google Scholar]
- 37.Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. F. Stewart. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 152991-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang, N. Suka, and M. Grunstein. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109437-446. [DOI] [PubMed] [Google Scholar]
- 39.Ron, D., and P. Walter. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8519-529. [DOI] [PubMed] [Google Scholar]
- 40.Sharma, V. M., R. S. Tomar, A. E. Dempsey, and J. C. Reese. 2007. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol. Cell. Biol. 273199-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidrauski, C., R. Chapman, and P. Walter. 1998. The unfolded protein response: an intracellular signalling pathway with many surprising features. Trends Cell Biol. 8245-249. [DOI] [PubMed] [Google Scholar]
- 42.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101249-258. [DOI] [PubMed] [Google Scholar]
- 43.van Drogen, F., and M. Peter. 2002. Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr. Biol. 121698-1703. [DOI] [PubMed] [Google Scholar]
- 44.Wang, A., S. K. Kurdistani, and M. Grunstein. 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 2981412-1414. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe, Y., K. Irie, and K. Matsumoto. 1995. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 155740-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe, Y., G. Takaesu, M. Hagiwara, K. Irie, and K. Matsumoto. 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 172615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarzov, P., C. Mazzoni, and C. Mann. 1996. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 1583-91. [PMC free article] [PubMed] [Google Scholar]