Abstract
The Paf1 complex (Paf1, Ctr9, Cdc73, Rtf1, and Leo1) is normally associated with RNA polymerase II (Pol II) throughout the transcription cycle. However, the loss of either Rtf1 or Cdc73 results in the detachment of the Paf1 complex from Pol II and the chromatin form of actively transcribed genes. Using functionally tagged forms of the Paf1 complex factors, we have determined that, except for the more loosely associated Rtf1, the remaining components stay stably associated with one another in an RNase-resistant complex after dissociation from Pol II and chromatin. The loss of Paf1, Ctr9, or to a lesser extent Cdc73 or Rtf1 results in reduced levels of serine 2 phosphorylation of the Pol II C-terminal domain and in increased read through of the MAK21 polyadenylation site. We found that the cleavage and polyadenylation factor Cft1 requires the Pol II-associated form of the Paf1 complex for full levels of interaction with the serine 5-phosphorylated form of Pol II. When the Paf1 complex is dissociated from Pol II, a direct interaction between Cft1 and the Paf1 complex can be detected. These results are consistent with the Paf1 complex providing a point of contact for recruitment of 3′-end processing factors at an early point in the transcription cycle. The lack of this connection helps to explain the defects in 3′-end formation observed in the absence of Paf1.
In Saccharomyces cerevisiae, the Paf1 complex (Paf1C), composed of Paf1, Ctr9, Cdc73, Rtf1, and Leo1, is found associated with RNA polymerase II (Pol II) from the promoters to the poly(A) sites of actively transcribed genes (23, 31, 32). The homologous complex in humans is composed of human Paf1 (hPaf1), hCtr9, hCdc73, and hLeo1 (45, 56, 60). Although there is an hRtf1 homolog, it does not appear to be part of the hPaf1C. The yeast Paf1C is recruited to Pol II at an as-yet-uncharacterized early step in the transcription cycle, subsequent to initiation complex formation (16), which is dependent on the Bur1 kinase (27). The presence of Paf1C is required for the histone H2B monoubiquitylation activity of Rad6; the modified histone H2B subsequently serves as the necessary substrate for H3 lysine 4 (K4) methylation by Set1 and H3 K79 methylation by Dot1 (27, 33, 54, 55). Therefore, the loss of the Paf1C ultimately results in changes in histone modifications.
Transcription by Pol II includes a cycle of phosphorylation and dephosphorylation of the C-terminal domain (CTD) of the largest Pol II subunit (reviewed in references 4, 7, 36, 39, 42, and 61). The initiating form of the enzyme is unphosphorylated; initiation and promoter escape are associated with, although not dependent on (21), phosphorylation of serine 5 (Ser5) of the YSPTSPS repeat of the CTD. During elongation, there is additional phosphorylation of serine 2 (Ser2) of the repeat. At, or just after, termination, the CTD is dephosphorylated, resetting Pol II for another transcription cycle. These modifications create unique interaction sites for a large and growing collection of transcriptional and posttranscriptional factors, including the mRNA-capping enzymes, histone modification enzymes, and the factors required for mRNA 3′-end formation and transcription termination (4, 6, 58).
With these CTD modifications in mind, it is interesting to consider the Paf1C, which in both its yeast and human forms has been shown previously to associate with all of the various forms of Pol II (32, 45) and whose association with Pol II and chromatin in yeast is unaffected by the loss of Ser2 phosphorylation (Ser2-P) caused by the loss of the major CTD Ser2 kinase Ctk1 (1, 10, 37). In contrast, loss of the Paf1C results in a reduction of CTD Ser2-P on transcribed genes and a concomitant decrease in the association of a Ser2-P-dependent factor (Pcf11) (32) and, by extension, all factors sensitive to CTD Ser2-P. Of particular interest in this category are the polyadenylation factors, several of which have been shown previously to interact directly with the Ser2-P form of the CTD (reviewed in references 6, 8, and 43).
Yeast and humans have similar complexes involved in mRNA 3′-end formation (reviewed in references 8, 12, 43, 48, and 59). Yeast CPF is a subcomplex of cleavage and polyadenylation factors, including Cft1 (Yhh1) and Ydh1 (Cft2), which is homologous to the human cleavage and polyadenylation factor subcomplex CPSF. CPSF is known to make direct contacts with RNA sequences at the poly(A) site. The yeast CF1A complex, which includes Pcf11, is similar to human CstF. Recently, evidence implicating human CPSF-73, the homolog of Ydh1, as the endonuclease responsible for cleavage has accumulated (15, 46), and Pcf11 was found previously to be able to directly trigger termination by purified Pol II (58). Cft1 and Pcf11 are among the factors reported previously to associate directly with the Ser2-P form of the Pol II CTD (3, 14, 26, 28), establishing physical links between both the CPF and CF1A complexes and Pol II. Therefore, posttranscriptional 3′-end formation is linked to transcription both by direct interactions with the Pol II CTD and by affinity for sequences in the nascent mRNA.
The fact that the Paf1C is necessary for full levels of CTD Ser2-P on actively transcribed genes (32) provides an excellent explanation for why mutations in the Paf1C factors result in defects in posttranscriptional processing. The loss of Paf1 results in shortened poly(A) tails (32) and, for a subset of genes, changes in poly(A) site utilization and the formation of unstable transcripts (38). In addition, the Paf1C is critical for the proper formation of 3′ ends of nonpolyadenylated RNAs (49). However, there is evidence that the connection between the Paf1C and polyadenylation factors may be more than the indirect consequence of reduced CTD Ser2-P. For example, the loss of Rtf1 results in only partial reduction of CTD Ser2-P levels (32) but causes defects in the formation of 3′ ends of nonpolyadenylated RNAs (49). In addition, the loss of either Paf1 or Rtf1 is synthetically lethal in combination with the loss of Ctk1 (13), which implies that the Paf1C has functions in addition to its effects on Ser2-P. In this work, we demonstrate that the Paf1C interacts in a Pol II-independent fashion with the polyadenylation factor Cft1. We also found that the presence of the Paf1C on Pol II was critical for the association of Cft1 with the Ser5-P form of Pol II. We conclude that the Paf1C helps to recruit the polyadenylation factor Cft1 and that it forms an additional early point of contact for the assembly of an active polyadenylation complex on the elongating Pol II.
MATERIALS AND METHODS
Yeast strains.
The YJJ S. cerevisiae strains used in this study were all derived from YJJ662 (MATa leu2Δ1 his3Δ200 ura3-52) (50) and its isogenic D273-11b derivatives as listed in Table 1. Some strains from the S288c (YSB, obtained from O. Rozenblatt-Rosen) and W303 (YN, obtained from C. Logie) backgrounds were also utilized as noted in the text and Table 1. The CTK1 gene was deleted in diploid strain YJJ1035 through replacement with the kanamycin resistance marker by using the kanmx4 cassette (52) as described previously (5). Haploid strain YJJ1821 was derived from the heterozygous diploid by sporulation and tetrad dissection (17). The construction of strains containing single protein A (tandem affinity purification [TAP]) or hemagglutinin (HA) tags has been described previously (31, 32). Note that all of the tags used in these studies were encoded by completely functional gene replacements, expressed at normal levels from their natural promoters. Additional singly tagged and all of the doubly (TAP- and HA-) tagged strains were constructed either by the direct transformation of wild-type (WT), paf1Δ, cdc73Δ, or rtf1Δ strains or by the mating of appropriate parents, the sporulation of the diploids, and the dissection of the haploid spores (17). The identification of the correct combinations of genes and tagged constructs used assays of selectable markers and mutant phenotypes and, subsequently, PCR and Western blot confirmation of the presence of tagged proteins.
TABLE 1.
Yeast strains
| Strain | Genotypea |
|---|---|
| YJJ577 | MATα his3Δ200 ura3-52 leu2Δ1 paf1Δ::HIS3 |
| YJJ662 | MATa his3Δ200 ura3-52 leu2Δ1 |
| YJJ664 | MATa his3Δ200 ura3-52 leu2Δ1 paf1Δ::HIS3 |
| YJJ665 | MATa his3Δ200 ura3-52 leu2Δ1 cdc73Δ::HIS3 |
| YJJ1035 | MATa/MATα his3Δ200/his3Δ200 ura3-52/ura3-52 leu2Δ1/leu2Δ1 PAF1/paf1Δ::HIS3 |
| YJJ1197 | MATa his3Δ200 ura3-52 leu2Δ1 ctr9Δ::Kanr |
| YJJ1303 | MATa his3Δ200 ura3-52 leu2Δ1 rtf1Δ::Kanr |
| YJJ1329 | MATa his3Δ200 ura3-52 leu2Δ1 CTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) |
| YJJ1330 | MATa his3Δ200 ura3-52 leu2Δ1 trp1Δ::KanrLEO1::(LEO1-6HA) (KlTRP1) |
| YJJ1336 | MATa his3Δ200 ura3-52 leu2Δ1 leo1Δ::Kanr |
| YJJ1583 | MATa his3Δ200 ura3-52 leu2Δ1 trp1Δ::hisG CDC73::(CDC73-6HA) (KlTRP1) |
| YJJ1589 | MATa his3Δ200 ura3-52 leu2Δ1 trp1Δ::hisG RTF1::(RTF1-6HA) (KlTRP1) |
| YJJ1599 | MATa his3Δ200 ura3-52 leu2Δ1 trp1Δ::hisG CTR9::(CTR9-6HA) (KlTRP1) |
| YJJ1782 | MATa his3Δ200 ura3-52 leu2Δ1 PAF1::(PAF1-CBP-TEV-ProtA) (KlURA3) CTR9::(CTR9-6HA) (KlTRP1) |
| YJJ1784 | MATa his3Δ200 ura3-52 leu2Δ1 rtf1Δ::KanrPAF1::(PAF1-CBP-TEV-ProtA) (KlURA3) CTR9::(CTR9-6HA) (KlTRP1) |
| YJJ1786 | MATa his3Δ200 ura3-52 leu2Δ1 PAF1::(PAF1-CBP-TEV-ProtA) (KlURA3) LEO1::(LEO1-6HA) (KlTRP1) |
| YJJ1787 | MATa his3Δ200 ura3-52 leu2Δ1 cdc73Δ::HIS3 PAF1::(PAF1-CBP-TEV-ProtA) (KlURA3) LEO1::(LEO1-6HA) (KlTRP1) |
| YJJ1789 | MATa his3Δ200 ura3-52 leu2Δ1 PAF1::(PAF1-CBP-TEV-ProtA) (KlURA3) RTF1::(RTF1-6HA) (KlTRP1) |
| YJJ1791 | MATa his3Δ200 ura3-52 leu2Δ1 cdc73Δ::HIS3 PAF1::(PAF1-CBP-TEV-ProtA) (KlURA3) RTF1::(RTF1-6HA) (KlTRP1) |
| YJJ1803 | MATa his3Δ200 ura3-52 leu2Δ1 cdc73Δ::HIS3 PAF1::(PAF1-CBP-TEV-ProtA) (KlURA3) CTR9::(CTR9-6HA) (KlTRP1) |
| YJJ1809 | MATa his3Δ200 ura3-52 leu2Δ1 rtf1Δ::Kanrcdc73Δ::HIS3 PAF1::(PAF1-CBP-TEV-ProtA) (KlURA3) LEO1::(LEO1-6HA) (KlTRP1) |
| YJJ1821 | MATα his3Δ200 ura3-52 leu2Δ1 ctk1Δ::Kanr |
| YJJ1824 | MATα his3Δ200 ura3-52 leu2Δ1 trp1Δ::hisG CTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) |
| YJJ1832 | MATa his3Δ200 ura3-52 leu2Δ1 PAF1::(PAF1-CBP-TEV-ProtA) (KlURA3) CDC73::(CDC73-6HA) (KlTRP1) |
| YJJ1834 | MATa his3Δ200 ura3-52 leu2Δ1 rtf1Δ::KanrPAF1::(PAF1-CBP-TEV-ProtA) (KlURA3) CDC73::(CDC73-6HA) (KlTRP1) |
| YJJ1841 | MATa his3Δ200 ura3-52 leu2Δ1 rtf1Δ::KanrCTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) CDC73::(CDC73-6HA) (KlTRP1) |
| YJJ1849 | MATα his3Δ200 ura3-52 leu2Δ1 trp1Δ::hisG rtf1Δ::KanrCTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) CFT1::(CFT1-6HA) (KlTRP1) |
| YJJ1850 | MATa his3Δ200 ura3-52 leu2Δ1 trp1Δ::hisG cdc73Δ::HIS3 CTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) CFT1::(CFT1-6HA) (KlTRP1) |
| YJJ1857 | MATa his3Δ200 ura3-52 leu2Δ1 CTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) RPB3::(RPB3-6HA) (KlTRP1) |
| YJJ1859 | MATa his3Δ200 ura3-52 leu2Δ1 rtf1Δ::KanrCTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) RPB3::(RPB3-6HA) (KlTRP1) |
| YJJ1861 | MATa his3Δ200 ura3-52 leu2Δ1 cdc73Δ::HIS3 CTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) RPB3::(RPB3-6HA) (KlTRP1) |
| YJJ1864 | MATa his3Δ200 ura3-52 leu2Δ1 CTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) CFT1::(CFT1-6HA) (KlTRP1) |
| YJJ1871 | MATα his3Δ200 ura3-52 leu2Δ11 ctk1Δ::KanrCTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) CFT1::(CFT1-6HA) (KlTRP1) |
| YJJ1873 | MATα his3Δ200 ura3-52 leu2Δ1 CTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) CTK1::(CTK1-6HA) (KlTRP1) |
| YJJ1875 | MATα his3Δ200 ura3-52 leu2Δ1 rtf1Δ::KanrCTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) CTK1::(CTK1-6HA) (KlTRP1) |
| YJJ1882 | MATα his3Δ200 ura3-52 leu2Δ1 paf1Δ::HIS3 CTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) CTK1::(CTK1-6HA) (KlTRP1) |
| YJJ1897 | MATα his3Δ200 ura3-52 leu2Δ1 cdc73Δ::HIS3 CTR9::(CTR9-CBP-TEV-ProtA) (KlURA3) CTK1::(CTK1-6HA) (KlTRP1) |
| YN96 | MAT his3-11-15 ura3-1 leu2-3,112 trp1-1 ADE2 LEO1::(LEO1-CBP-TEV-ProtA) |
| YSB2116 | MATa his3Δ1 ura3Δ0 leu2Δ0 met15Δ0 trp1Δ::LEU2 (CFT1-CBP-TEV-ProtA)::URA3 |
| YSB2118 | MATa his3Δ1 ura3Δ0 leu2Δ0 met15Δ0 trp1Δ::LEU2 (CFT1-CBP-TEV-ProtA)::URA3 paf1Δ::Kanr |
KlURA3 and KlTRP1, Kluyveromyces lactis URA3 and TRP1 genes; CBP-TEV-ProtA, TAP tag comprising CBP, tobacco etch virus protease (TEV), and Staphylococcus aureus protein A (ProtA); 6HA, six-HA tag.
Yeast co-IP and Western blot analyses.
Yeast strains bearing both TAP and HA tags in WT and mutant backgrounds were grown to mid-log phase (∼1.5 × 107 cells/ml) in yeast extract-peptone-dextrose (YEPD) with 2% glucose (17). Cells were harvested and processed for protein analysis as described previously (31). Protein abundance was determined by the Bradford assay (Bio-Rad), and 1.5 mg of protein was used for immunoprecipitation (IP). In some cases, 50 μg of RNase A (Sigma) was added prior to the IP. RNA digestion was confirmed by the total loss of rRNAs from the treated samples. After preincubation with Sepharose CL-2B beads (Sigma-Aldrich), HA-tagged proteins were precipitated by incubation with 10 μg of anti-HA (α-HA [12CA5; Roche]), followed by the addition of protein A-coupled Sepharose CL-4B (Amersham); TAP-tagged proteins were precipitated by incubation with rabbit immunoglobulin G agarose (Sigma-Aldrich) essentially as described previously (32). After the separation of the cell extracts or immunoprecipitates on 4 to 12% bis-Tris NOVEX gels (Invitrogen), the proteins were transferred onto Immobilon P (Millipore) or nitrocellulose (Protran, Schleicher and Schuell) membranes. Immobilized proteins were detected with α-HA (12CA5 [1:1,000; Roche]), α-TAP tag (α-protein A [1:1,000; U.S. Biological] or α-protein A-1-Step horseradish peroxidase [1:10,000; Abcam]), α-Pol II (8WG16 [1:500], H5 [α-Ser2-P; 1:500], or H14 [α-Ser5-P; 1:500], all from Covance, or BL2894 [α-Ser2-P; 1:10,000] or BL2896 [α-Ser5-P; 1:10,000], both from Bethyl Labs), or α-G6PD (1:20,000; Sigma) primary antibodies. Appropriate horseradish peroxidase-conjugated secondary antibodies were used prior to detection by chemiluminescence with a Western Lightning chemiluminescence kit (Perkin-Elmer). The linear range of exposure for the chemiluminescent reaction was determined by dilution series, and exposures were quantitated using Quantity One software (Bio-Rad).
TAP tag purifications and mass spectrometry.
Two-liter cultures of yeast strains were grown to an A600 of 1 at 30°C in YEPD, harvested, and processed for TAP tag purification essentially as described previously (44). Cells were extracted in buffer containing 33 mM Tris-HCl (pH 7.5), 8% glycerol, 330 mM NaCl, 0.07% Triton X-100, 0.3 mM dithiothreitol, 3.3 mM EDTA, and protease inhibitor cocktail (44). Aliquots of fractions eluted from calmodulin beads were analyzed by silver staining after electrophoresis on sodium dodecyl sulfate-10% polyacrylamide gels. Peak fractions were pooled, loaded onto sodium dodecyl sulfate-polyacrylamide gels, and run briefly to remove detergent. The gel lane was fixed, cut in pieces, reduced, and alkylated. Proteins were digested overnight with trypsin, eluted from the gel with trifluoroacetic acid, and concentrated. Peptide identification experiments were performed using a nano-high-performance liquid chromatography Agilent 1100 nanoflow system connected online to a 7-tesla linear quadrupole ion trap-Fourier transform mass spectrometer (Thermo Electron, Bremen, Germany) essentially as described previously (35).
RT-PCR.
RNA was isolated from cells grown in YEPD to mid-log phase (∼1.5 × 107cells/ml) by the hot-phenol method (47). Reverse transcription (RT) reactions using 5 μg of DNase I-treated total RNA were performed using a SuperScript III kit (Invitrogen) and random hexamers. PCRs used 1/10 to 1/20 of the RT reaction mixture under linear-range conditions. MAK21 primer sequences were as follows: forward, 5′-GACGATGAACCTAAGCTGGAGGCAA-3′, and reverse, 5′-ATAGTTGTAGCTAAAATCCGTTTCTATTTCCG-3′. PMA1 primer sequences were as follows: forward, 5′-CTATTATTGATGCTTTGAAGACCTCCAG-3′, and reverse, 5′-TGCCCAAAATAATAGACATACCCCATAA-3′.
RESULTS
The loss of Rtf1 or Cdc73, which reduces the association of the Paf1C with Pol II, does not disrupt interactions between the remaining Paf1C components.
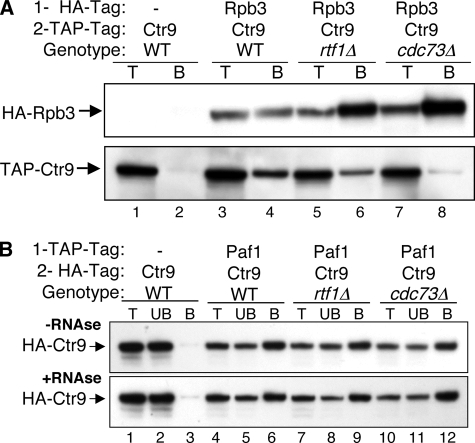
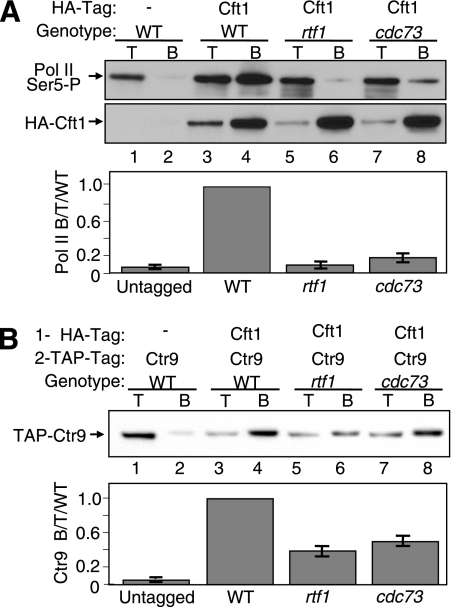
Cdc73 and Rtf1 are critical for connecting the Paf1C to transcribing Pol II, which was determined using chromatin IP (ChIP) and the co-IP of Paf1 with a tagged Pol II Rpb3 subunit (32). To analyze the fate of the Paf1C factors in the absence of Rtf1 or Cdc73 when the complex is dissociated from Pol II, we constructed a series of isogenic, doubly tagged strains using HA and TAP tags on Paf1C factors in WT and rtf1Δ and cdc73Δ mutant backgrounds (Table 1). To confirm that the tagged forms did not disrupt Paf1C function, we observed phenotypes, including the growth rates and sensitivities to temperature and drugs, of all the tagged strains, comparing them to those of untagged strains (5). None of the tagged constructs described in this work demonstrated any defect in function in these assays (data not shown). In addition, we used the tagged constructs to recapitulate and extend the previous co-IP analysis of the Paf1C and Pol II, as shown in Fig. 1A. Using strains doubly tagged on Pol II subunit Rpb3 (HA tag) and Ctr9 (TAP tag), we confirmed that the full association of Pol II and the Paf1C is dependent on the presence of Rtf1 and Cdc73 (compare the amount of TAP-tagged Ctr9 [Fig. 1A, lower panel] coimmunoprecipitated with Rpb3 from the WT [lane 4] to that from the rtf1Δ [lane 6] or cdc73Δ [lane 8] strain). As described previously (32), we found that the effect of loss of Cdc73 is greater than that of loss of Rtf1. Note that the background of nonspecifically coimmunoprecipitated TAP-tagged Ctr9 was very low for the control strain lacking an HA tag on Rpb3 (Fig. 1A, lower panel, lane 2). Similar appropriate untagged controls were included for all of the experiments in this study.
FIG. 1.
The mutation of RTF1 or CDC73 results in the dissociation of Ctr9 from Pol II but not from Paf1. (A) The loss of Rtf1 or Cdc73 reduces the co-IP of TAP-tagged Ctr9 with HA-tagged Rpb3. Protein extracts from strains bearing the indicated combinations of tags and deletion mutations were subjected to IP using an α-HA antibody as described in Materials and Methods. Ten-microgram samples of total protein (T) and the bound pellets (B) were probed for the immunoprecipitated HA-tagged Rpb3 (upper panel) and the coimmunoprecipitated TAP-tagged Ctr9 (lower panel). The strains used were YJJ1329 (lanes 1 and 2), YJJ1857 (lanes 3 and 4), YJJ1859 (lanes 5 and 6), and YJJ1861 (lanes 7 and 8). − represents the absence of protein with the indicated tag. (B) The association of Paf1 with Ctr9 is not affected by dissociation from Pol II or by RNase treatment. TAP-tagged Paf1 was immunoprecipitated, and the presence of coimmunoprecipitated HA-tagged Ctr9 was detected in samples incubated without (−RNAse; upper panel) or with (+RNAse; lower panel) RNase A treatment prior to the IP. Ten-microgram samples of total protein (T), an equal volume of the unbound IP supernatant (UB), and the bound pellets (B) were probed as indicated. The strains used were YJJ1599 (lanes 1 to 3), YJJ1782 (lanes 4 to 6), YJJ1784 (lanes 7 to 9), and YJJ1803 (lanes 10 to 12).
When the Paf1C is detached from Pol II by the loss of Cdc73 or Rtf1, all of the remaining factors redistribute and are found both in the nucleoplasm and in the nucleolus (41). To determine if the detached and redistributed factors are still in a complex with one another, we used co-IP analyses of the strains bearing distinguishable tags on two Paf1C components. As shown in Fig. 1B, we found that the loss of Rtf1 or Cdc73 did not affect the interaction between TAP-tagged Paf1 and HA-tagged Ctr9, consistent with Paf1C retaining its integrity after dissociation from Pol II and chromatin (Fig. 1B, compare lanes 9 and 12 to lane 6). In addition, the Paf1-Ctr9 interaction was not affected by the treatment of the extract with RNase prior to the IP (Fig. 1B, compare upper and lower panels). We also found that the co-IP interactions between the Paf1C components and Pol II were not affected by treatment with RNase (data not shown). The resistance of the Paf1C interactions to RNase argues that the complex does not depend on RNA for its association with Pol II or for association of the factors within the complex.
In similar experiments using appropriate doubly tagged strains, we found that HA-tagged Leo1 was still associated with TAP-tagged Paf1 in rtf1Δ and cdc73Δ strains and in an rtf1Δ cdc73Δ double mutant strain (Fig. 2A and data not shown). In addition, the association of HA-tagged Cdc73 with TAP-tagged Paf1 was not dependent on Rtf1 (Fig. 2B). The interaction between HA-tagged Cdc73 and TAP-tagged Ctr9 was somewhat reduced by the loss of Rtf1 in the experiment presented in Fig. 2B (lane 12), but in other experiments (data not shown), little effect on the Cdc73-Ctr9 interaction or the interaction between Cdc73 and Leo1 was seen. These interactions were further confirmed by changing the order of the coimmunoprecipitates and probing antibodies and by using strains bearing different combinations of tags (data not shown). The interactions among Cdc73, Leo1, Paf1, and Ctr9 were also not sensitive to RNase treatment (data not shown).
FIG. 2.
There is still an association of Leo1 and Cdc73 with Paf1 and Ctr9 after the Paf1C is dissociated from Pol II. Protein extracts from strains bearing the indicated combinations of tags and mutations were subjected to IP using an α-TAP antibody, and coimmunoprecipitates were detected with an α-HA antibody as described in Materials and Methods. Lane labeling is as described in the legend to Fig. 1B. − represents the absence of protein with the indicated tag. (A) HA-tagged Leo1 coimmunoprecipitates with TAP-tagged Paf1 in the absence of Cdc73 or Rtf1. The strains used were YJJ1330 (lanes 1 to 3), YJJ1786 (lanes 4 to 6), YJJ1787 (lanes 7 to 9), and YJJ1809 (lanes 10 to 12). (B) HA-tagged Cdc73 coimmunoprecipitates with TAP-tagged Paf1 and Ctr9 in the absence of Rtf1. The strains used were YJJ1583 (lanes 1 to 3), YJJ1832 (lanes 4 to 6), YJJ1834 (lanes 7 to 9), and YJJ1841 (lanes 10 to 12). The asterisk denotes an HA-cross-reacting band.
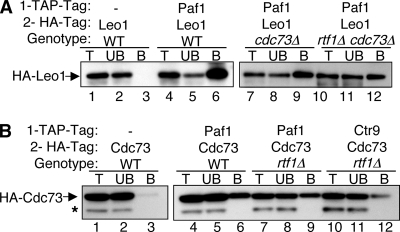
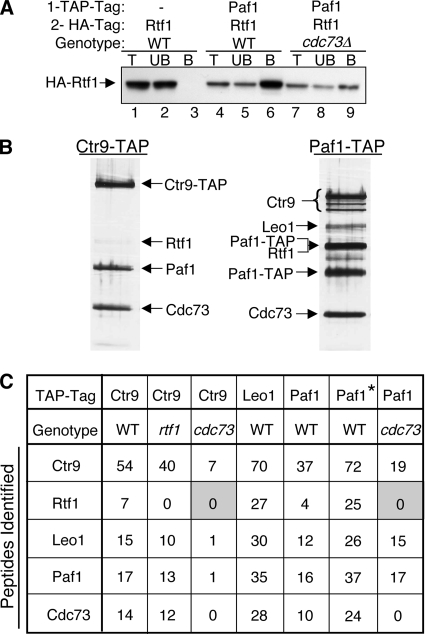
Rtf1 association with the Paf1C is partially disrupted by the loss of Cdc73.
In contrast to the stable association of Paf1, Ctr9, and Leo1 with one another in rtf1 and cdc73 mutant backgrounds and with Cdc73 in an rtf1 mutant background (Fig. 1B and 2 and data not shown), the association of Rtf1 with Paf1 was reduced, although still apparent, in the absence of Cdc73 (Fig. 3A, compare lane 6 to lane 9). This reduced association was not further diminished by treatment with RNase (data not shown). This result is consistent with our previous observation that loss of Cdc73 reduces, but does not abolish, the association of Rtf1 with chromatin (32). Rtf1 therefore appears to form weak attachments to both the Paf1C and some part of the Pol II transcription complex that together create a stable Rtf1-chromatin complex.
FIG. 3.
The association of Rtf1 with the Paf1C is reduced in the absence of Cdc73. (A) TAP-tagged Paf1 was immunoprecipitated, and the presence of coimmunoprecipitated HA-tagged Rtf1 was detected. The strains used were YJJ1589 (lanes 1 to 3), YJJ1789 (lanes 4 to 6), and YJJ1791 (lanes 7 to 9). Lane labeling is as described in the legend to Fig. 1B. − indicates the absence of TAP-tagged Paf1. (B) Large-scale purification of TAP-tagged Ctr9 (YJJ1857) and Paf1 (YJJ1782) from WT cells was performed as described in Materials and Methods. Copurifying proteins were separated by gel electrophoresis, silver stained, and identified by mass spectrometry as indicated. (C) Proteins associated with TAP-tagged Ctr9, Leo1, and Paf1 in strains with the indicated genetic backgrounds were analyzed by mass spectrometry as described in Materials and Methods. The numbers of identified peptides of the indicated Paf1C components are listed. The asterisk marking the second column corresponding to Paf1-TAP and the WT genotype indicates a preparation done using a buffer with a lower salt concentration (150 mM NaCl) than that in the other samples. The lower-molecular-weight Paf1-TAP band was the result of the proteolytic degradation of the amino terminus. Shading highlights the lack of detection of Rtf1 peptides. The strains used were Ctr9-TAP-expressing WT YJJ1873, Ctr9-TAP-expressing rtf1 mutant YJJ1875, Ctr9-TAP-expressing cdc73 mutant YJJ1861, Leo1-TAP-expressing WT YN96, Paf1-TAP-expressing WT YJJ1782, and Paf1-TAP-expressing cdc73 mutant YJJ1787.
To further test the idea that Rtf1 may be more weakly associated with the Paf1C than the other components of the complex, we used strains containing TAP-tagged forms of the Paf1C components to isolate the complexes from WT, rtf1Δ, and cdc73Δ strains and analyzed the complexes by mass spectrometry. As shown in Fig. 3B, Rtf1 could be detected on silver-stained gels of the purified TAP-tagged complexes from WT cells. In addition, peptides from Rtf1 were detected in the complexes isolated from WT cells using TAP-tagged Paf1, Ctr9, and Leo1 (Fig. 3C). In contrast, when either TAP-tagged Ctr9 or Paf1 was used to isolate the Paf1C from cdc73 mutant cells, no Rtf1 peptides were detected (Fig. 3C). Also consistent with Rtf1 being more weakly associated with the Paf1C was our observation that reducing the salt in the buffer used for isolating proteins associated with TAP-tagged Paf1 from WT cells from 330 to 150 mM KCl increased the recovery of Rtf1 peptides (compare the two columns for TAP-tagged Paf1 from WT cells in Fig. 3C). We conclude that although Rtf1 clearly is part of the Paf1C, its association with the complex is weaker than that of other complex components, and in the absence of Cdc73 this weak association is disrupted further. Our results establish that the Paf1C components Paf1, Ctr9, Cdc73, Rtf1, and Leo1 are still associated in a stable, RNase-resistant complex when dissociated from Pol II.
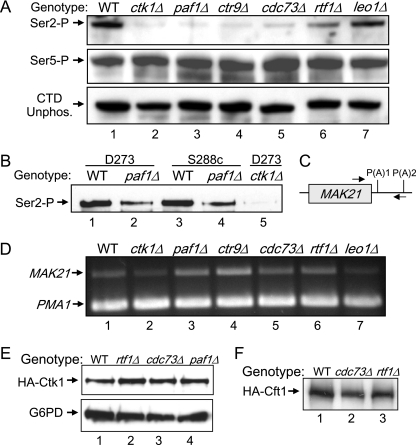
The effects of Paf1C mutation on Ser2-P levels correlate with phenotypes.
Using ChIP experiments, we have previously demonstrated that the loss of Paf1 and, to a lesser extent, Rtf1 results in lower levels of chromatin-associated Pol II CTD Ser2-P, which correlates with the reduced association of the Ser2-P-dependent cleavage and polyadenylation factor Pcf11 with chromatin (32). To investigate whether the total amount of Ser2-P in yeast cells is reduced when Paf1C components are missing, we compared extracts from isogenic strains bearing deletions of each Paf1C component to extracts from WT cells and from a strain lacking the major CTD Ser2 kinase Ctk1 (37). As shown in Fig. 4A (upper panel), by using the H5 monoclonal antibody, which detects primarily Ser2-P and the double Ser2-P, Ser5-P CTD (19), the total levels of Ser2-P in paf1Δ and ctr9Δ strains were reduced to levels similar to that in the ctk1Δ strain. The level of Ser2-P in the cdc73Δ strain was slightly higher than that in the ctk1Δ strain, and that in the rtf1Δ strain was higher yet, about one-third to one-half that in the WT. The level of Ser2-P in the leo1Δ mutant was indistinguishable from that in the WT. None of the mutations had any obvious effect on the abundance of the Ser5-P form of Pol II or the unphosphorylated CTD, detected with the H14 and 8WG16 monoclonal antibodies (Fig. 4A, lower panels). We also used another commercially available antibody specific for the CTD Ser2-P modification, which has greater sensitivity in immunoblotting (see Materials and Methods). As shown in Fig. 4B, with this antibody, the Ser2-P signal from the ctk1Δ strain was reduced nearly 100-fold and that from an isogenic paf1Δ strain was reduced about 10-fold compared to that from the WT based on quantitation as described in Materials and Methods (compare lane 1 to lanes 2 and 5). We also confirmed that the reduction of Ser2-P was not a strain-dependent phenomenon by comparing paf1Δ strains to WT strains of both the D273-11b (Fig. 4B, lanes 1 and 2) and S288c (Fig. 4B, lanes 3 and 4) backgrounds. Similar results were obtained by comparing a paf1Δ strain to a WT strain with a W303 background (data not shown). It is striking that the relative changes in Ser2-P levels seen in Fig. 4A correlate perfectly with the severity of each Paf1C component mutant phenotype, with paf1Δ and ctr9Δ mutants having identical and the most deleterious phenotypes, cdc73Δ and rtf1Δ mutants having intermediate phenotypes, and the leo1Δ mutant being almost indistinguishable from the WT (5). It is therefore likely that the reduction of Ser2-P and the concomitant loss of Ser2-P-dependent factors are the explanation for at least part of the deleterious phenotype associated with Paf1C mutations.
FIG. 4.
(A) The loss of some Paf1C components reduces Pol II CTD Ser2-P. Protein extracts prepared from isogenic strains bearing the indicated mutations were separated by electrophoresis, blotted, and probed for the indicated antigens as described in Materials and Methods. The strains used were YJJ662 (WT), YJJ1821 (ctk1Δ), YJJ577 (paf1Δ), YJJ1197 (ctr9Δ), YJJ665 (cdc73Δ), YJJ1303 (rtf1Δ), and YJJ1336 (leo1Δ). Antibodies used for Pol II were H5 (α-Ser2-P), H14 (α-Ser5-P), and 8WG16 (α-unphosphorylated CTD [CTD unphos.]). (B) The reduction of Pol II CTD Ser2-P by the loss of Paf1 is not yeast strain dependent. Protein extracts from pairs of isogenic strains with the indicated genetic backgrounds were analyzed for the Pol II Ser2-P CTD as described in the legend to panel A by using the BL2894 antibody. The strains used were YJJ662 (WT; lane 1), YJJ664 (paf1Δ; lane 2), YSB2116 (WT; lane 3), YSB2118 (paf1Δ; lane 4), and YJJ1871 (ctk1Δ; lane 5). (C) Schematic of the MAK21 gene showing the locations of primers spanning the first poly(A) site [P(A)1] used for RT-PCRs. (D) The loss of some Paf1C factors, but not Leo1 or Ctk1, increases the read through of the MAK21 poly(A) site. RT-PCR was used to measure the amount of the MAK21 read-through transcript, which was normalized to the signal from the PMA1 gene. Strains used for RNA isolation were the same as those listed in the legend to panel A. (E) The loss of Rtf1 or Cdc73 does not affect the abundance of HA-tagged Ctk1. Protein extracts from isogenic HA-tagged Ctk1 strains were analyzed as described in the legend to panel A. The strains used were YJJ1873 (WT), YJJ1875 (rtf1Δ), YJJ1897 (cdc73Δ), and YJJ1882 (paf1Δ). Extracts in the upper panel were probed with α-HA. Those in the lower panel were probed with α-G6PD. (F) The loss of Rtf1 or Cdc73 does not affect the abundance of HA-tagged Cft1. Protein extracts from isogenic HA-tagged Cft1 strains were analyzed as described in the legend to panel A and probed with α-HA. The strains used were YJJ1864 (WT), YJJ1850 (cdc73Δ), and YJJ1849 (rtf1Δ).
We also asked if the changes in CTD Ser2-P correlated with functions known to be associated with the loss of Paf1. We have shown previously that in the absence of Paf1, Pol II reads through the poly(A) site of the MAK21 gene, resulting in the production of a longer, unstable transcript (38). As shown in Fig. 4C, we used appropriate primers to detect the read-through transcript in RNA isolated from the same yeast strains used to analyze Ser2-P levels in Fig. 4A. As reported by Penheiter et al. (38), we observed an increase of about twofold in the levels of the read-through transcript from the MAK21 gene in the absence of Paf1 (Fig. 4D, compare lanes 1 and 3) when the levels were normalized to the signal from the control from an internal region of the PMA1 gene. The loss of Ctr9 resulted in a two- to threefold increase in read through. The loss of Cdc73 or Rtf1 resulted in slight elevations in read through, from 20 to 40% in multiple repetitions of the RT-PCR (Fig. 4D, lanes 5 and 6), but we did not observe an increase of poly(A) site read though due to the loss of either Ctk1 or Leo1 (Fig. 4D, lanes 2 and 7). These preliminary results indicate that the loss of the Paf1C may have a greater effect on mRNA 3′-end formation than does the loss of Ctk1-dependent CTD Ser2-P.
The reduction in Ser2-P levels may be due to a reduction in Ctk1 abundance or activity, the association of Ctk1 with chromatin, or an increase in the dephosphorylation of the CTD. To begin to address these possibilities, we constructed strains bearing an HA-tagged form of Ctk1 and TAP-tagged Ctr9 in isogenic WT, rtf1Δ, cdc73Δ, and paf1Δ backgrounds (Table 1; Fig. 4E). The double tags did not impair function, as determined by phenotypic testing (data not shown). As shown in Fig. 4E, the loss of Rtf1, Cdc73, or Paf1 did not reduce the abundance of HA-tagged Ctk1. Using both co-IP and ChIP techniques, we did not detect any significant reduction in the interaction of HA-tagged Ctk1 with Pol II or with chromatin in the absence of Cdc73 or Rtf1 (data not shown). Although we could detect greater interaction between HA-tagged Ctk1 and TAP-tagged Ctr9 than between untagged controls in co-IPs, this interaction also was not reduced by the loss of Rtf1 or Cdc73 (data not shown). It remains possible that the loss of Paf1 or Ctr9 and, to a lesser extent, the loss of Cdc73 or Rtf1 may reduce the activity of Ctk1 or the stability of the CTD Ser2-P modification.
The Paf1C interacts with polyadenylation factor Cft1 and is important for the association of Cft1 with Pol II.
The reduction of CTD Ser2-P levels in Paf1C mutants is at best a partial explanation for the reduced association of cleavage and polyadenylation factors, especially because the loss of Rtf1 results in only a partial reduction of Ser2-P (Fig. 4A, lane 6) but causes a much larger decrease in Pcf11 association with chromatin (32) and both paf1 and rtf1 mutations are lethal in combination with the loss of Ctk1 (13; M. G. Hoffman, unpublished results). In addition, the loss of the Paf1C results in changes in mRNA 3′-end formation that appear to be independent of effects on Ser2-P (Fig. 4D). To further determine the possible role of the Paf1C in recruiting cleavage and polyadenylation factors, we created doubly tagged strains containing TAP-tagged Ctr9 and an HA-tagged form of Cft1, a component of the yeast CPF complex and the homolog of human poly(A) binding factor CPSF-160 (51), in WT, rtf1Δ, and cdc73Δ backgrounds (Table 1; Fig. 5). The tagged strains demonstrated no deleterious phenotypes, and the abundance of HA-tagged Cft1 was not affected by the loss of Cdc73 or Rtf1 (Fig. 4F). Cft1 is associated with the chromatin of genes actively transcribed from the 5′ to the 3′ end, with a peak of abundance near the poly(A) site (23). In addition, Cft1 has been shown to interact with the Ser2-P form of Pol II (14). Because the loss of Cdc73 or Rtf1 causes only a partial loss of Ser2-P, we were not surprised to find little change in the association between HA-tagged Cft1 and the Ser2-P form of Pol II in the mutant strains relative to that in the WT (data not shown). We also did not observe any Rtf1- or Cdc73-dependent changes in the association between Cft1 and the unphosphorylated form of Pol II. However, as shown in the upper panel of Fig. 5A, we observed that the strong co-IP signal for the association of the Ser5-P form of Pol II with HA-tagged Cft1 in WT cells (lane 4) was reduced nearly 10-fold in the absence of Rtf1 (lane 6) or Cdc73 (lane 8), to levels just over the background found in untagged cells (lane 2). The results of multiple repetitions of these co-IPs are quantitated in the lower panel of Fig. 5A. None of the co-IP signals were affected by RNase treatment (data not shown). Therefore, the detachment of the Paf1C from Pol II appears to have a direct effect on the interaction of Cft1 with the Ser5-P form of Pol II. Because Cft1 is part of the CPF complex, we expect that this dependence on the Paf1C pertains to the entire complex.
FIG. 5.
Dissociation of the Paf1C from Pol II reduces the association of polyadenylation factor Cft1 with Pol II and reveals a direct association between the Paf1C and Cft1. Extracts from strains bearing the indicated tagged proteins and mutations were immunoprecipitated with α-HA antibody and then probed with appropriate antibodies. Signals in the lower panels were quantitated as described in Materials and Methods as the ratio of Pol II bound to total protein-, and the results are expressed relative to the WT ratio, which was set at 1. T, 10 μg of total protein; B, bound IP pellet; −, absence of HA-tagged Cft1. (A) The loss of Rtf1 or Cdc73 reduces the association of Cft1 with Pol II. HA-tagged Cft1 was immunoprecipitated, and the presence of the Ser5-P CTD form of Pol II was detected with the α-Ser5-P CTD H14 antibody (upper panel). The protein blot was stripped and reprobed with α-HA for the presence of HA-tagged Cft1 (middle panel). The error bars in the graph in the lower panel represent the standard deviations of data from four repetitions of the experiment. The strains used were YJJ1824 (lanes 1 and 2), YJJ1864 (lanes 3 and 4), YJJ1849 (lanes 5 and 6), and YJJ1850 (lanes 7 and 8). (B) The Paf1C and Cft1 interact in the absence of Rtf1 or Cdc73. HA-tagged Cft1 was immunoprecipitated, and the presence of coimmunoprecipitated TAP-tagged Ctr9 was detected with α-TAP (upper panel). The solid bars representing quantitated data in the lower panel indicate the averages and the error bars show the ranges of results obtained from replicate experiments. The strains used were YJJ1824 (lanes 1 and 2), YJJ1864 (lanes 3 and 4), YJJ1849 (lanes 5 and 6), and YJJ1850 (lanes 7 and 8).
We next asked if there is a direct association between the Paf1C and Cft1 independent of their associations with Pol II by assaying this interaction in the rtf1 and cdc73 mutants, which detach the Paf1C from Pol II. The answer to this question is shown in the upper and lower panels of Fig. 5B, where we analyzed the association between HA-tagged Cft1 and TAP-tagged Ctr9 in WT, rtf1Δ, and cdc73Δ cells. In WT cells, a strong Cft1-Ctr9 co-IP signal was observed, more than 10-fold higher than that in the untagged control (Fig. 5B, compare lane 4 to lane 2). Although removing the Paf1C from Pol II reduced the level of association between Cft1 and Ctr9 two- to threefold, the level of association was still significantly above the background in the untagged strain (quantitated in the lower panel of Fig. 5B) and much higher than the association seen between Cft1 and the Ser5-P form of Pol II in the same mutant strains (compare Fig. 5B to A). These intercomplex interactions were also not sensitive to RNase (data not shown). Therefore, in yeast there is an interaction between the Paf1C and cleavage and polyadenylation factor Cft1 and, presumably, the CPF complex that is independent of their associations with Pol II or RNA. This interaction appears to be critical for full levels of association of Cft1 with the Ser5-P form of Pol II.
DISCUSSION
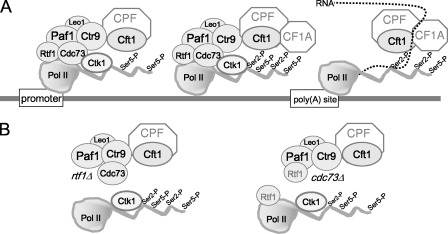
In this work, we have provided an additional molecular explanation for the link between the Paf1C and mRNA 3′-end formation and polyadenylation. Our discovery that there is a direct interaction between the Paf1C and a cleavage and polyadenylation factor provides one more piece of the complicated puzzle that links transcriptional and posttranscriptional processes. Our results can be summarized by the model shown in Fig. 6. When Pol II is at the promoter (Fig. 6A), it is joined by the Paf1C at an early step after the dissociation from the mediator and the phosphorylation of the CTD Ser5 (16, 23, 31). Cleavage and polyadenylation factors also begin to associate with Pol II at this early point in the transcription cycle, based on their localization to promoter regions of chromatin (23), their interactions with some of the general transcription factors at the promoter (reviewed in references 8 and 43), and our observations of robust interactions between Cft1 and the Ser5-P, initiating form of Pol II. Our data are consistent with Paf1C helping to recruit Cft1 and, by extension, the CPF complex to Pol II. Because we can detect direct contacts between the Paf1C and Cft1 in the absence of Pol II, under conditions that disrupt the interaction between Cft1 and Ser5-P Pol II (schematized in Fig. 6B), we speculate that the presence of Cft1 is more dependent on the Paf1C at this early stage of transcription than on Pol II or its modifications.
FIG. 6.
The Paf1C provides one of several contacts to recruit polyadenylation factors to elongating Pol II. Shown is a simplified model describing the interactions elucidated in this work within the Paf1C and between the Paf1C and Ctk1 and Cft1 in WT, rtf1Δ, and cdc73Δ strains as described in Discussion.
Our model shows Cft1/CPF but not CF1A at the promoter region for two reasons. First, we have not directly measured the association between a CF1A factor, like Pcf11, and the Paf1C as we have for Cft1, although we have shown previously that the lack of Rtf1 reduces the association of Pcf11 with chromatin at both the 5′ and 3′ ends of a gene (32). Second, based on the data of Kim et al. (23), there is significantly more Cft1 than Pcf11 at the promoter. Although the abundance of both factors peaks near the poly(A) site, the level of Cft1 increases only 3- to 4-fold from promoter to poly(A) site of different genes, but that of Pcf11 increases from 5- to 20-fold over this interval. It is possible that it is the connection between Cft1/CPF and the Paf1C that result in the different distribution of the two components of the cleavage and polyadenylation apparatus. It is also possible that the Paf1C does help to recruit CF1A factors early in the transcription cycle.
As Pol II moves into its elongation mode, the CTD is modified by the phosphorylation of Ser2, primarily by Ctk1 (37). This modification is linked to the dissociation of the mediator (29) and serves as an important point of contact for many Pol II-associated factors, including Pcf11 (28, 30) and Cft1 (14). It is interesting that although these factors have been shown to form complexes with the Ser2-P CTD in selective assays and structural studies, the direct interactions are relatively weak and these factors, unlike the components of the Paf1C, have not been identified in large-scale biochemical screens for CTD-interacting factors (40). Therefore, it is possible that during elongation, direct contacts with the Paf1C make contributions to the association of CPFs with Pol II and chromatin even greater than those of the Ser2-P-dependent interactions. This scenario would help to explain the dramatic diminution of Pcf11 association with chromatin in the absence of Rtf1 under conditions in which CTD Ser2-P levels are only modestly reduced (32; this work) and the apparently greater effects of the loss of Paf1 than of the loss of Ctk1 on transcriptional read through.
As schematized in Fig. 6, unlike our results for Cft1, we have not detected a role for the Paf1C in recruiting Ctk1 to Pol II. However, the distribution of Ctk1 on chromatin mirrors that of the Paf1C (23), and we have shown in this work that Paf1, Ctr9, and to a lesser extent, Cdc73 and Rtf1 are necessary for full levels of CTD Ser2-P. It is therefore possible that the presence of the Paf1C is required for the full activity of Ctk1, as it is for the histone H2B monoubiquitylation activity of Rad6 (54, 55). This possible reduction in activity is unlikely to be due to the loss of the preferred Ctk1 substrate, the Ser5-P form of the CTD (39), because this form of Pol II was not affected by the loss of Paf1 (this work). Another possibility is that the loss of the Paf1C somehow destabilizes the Ser2-P modification, perhaps by increasing accessibility to phosphatases.
It is interesting that the reductions in CTD Ser2-P levels that we observed in the different Paf1C mutants correlated perfectly with, and provide a molecular mechanism for, the reductions in histone H3 K36 trimethylation recently reported by Chu et al. (11). There were discrepancies in earlier reports of a link between the Paf1C and H3 K36 trimethylation, catalyzed by the Set2 methyltransferase. Krogan et al. reported that both Rtf1 and Cdc73 are required for Set2 recruitment and H3 K36 trimethylation (25), whereas Ng et al. found that the loss of Rtf1 has no effect on this modification (33). Chu et al. recently reported testing mutations in all of the Paf1C components and found that, just as demonstrated by our measurements of Ser2-P levels, the loss of Paf1 or Ctr9 has the greatest effect on reducing H3 K36 trimethylation, with the effect of the loss of Cdc73 being intermediate and the loss of Rtf1 or Leo1 having little effect (11). Because Set2 association with Pol II is known to be dependent on the CTD Ser2-P modification (24), it is very likely that the effects observed by Chu et al. on H3 K36 trimethylation are the indirect consequence of the loss of Ser2-P in the absence of Paf1 or Ctr9. Therefore, most of the currently known effects of the Paf1C on histone H3 modifications are indirect: the Set1 and Dot1 methylation of H3 K4 and K79, respectively, require the Paf1C-dependent Rad6 monoubiquitylation of histone H2B (55), and the Set2 methylation of H3 K36 requires the Paf1C-dependent phosphorylation of the Pol II CTD Ser2 (11; this work). Rtf1 appears to have some effects on histone modifications independent of its function in the Paf1C via interactions with the chromatin-remodeling factor Chd1 (53).
As Pol II transits the poly(A) site, some dramatic and still not fully characterized transitions occur (Fig. 6A). The appearance of the poly(A) site in the RNA creates a binding site for Cft1/CPF and leads to the highest levels of association of the cleavage and polyadenylation factors with chromatin (23). It may be that a three-way combination of contacts with the RNA, the CTD, and the Paf1C results in this peak of association. Yet another contact point may be provided by the chromatin protein Sin1/Spt2 (18). After passage through the poly(A) site, the abundance of the Bur1 kinase decreases (22), probably linked to the observed three- to fivefold decrease in the abundance of the Paf1C and Ctk1 relative to those found in the coding region (23). Clearly, beyond this point the Paf1C is no longer a major contact point for cleavage and polyadenylation factors, but the defects we and others have observed in 3′-end formation in the absence of the Paf1C indicate that its presence contributes to accurate 3′-end processing (38, 49). Our data are consistent with both the requirement for direct contacts between the Paf1C and cleavage and polyadenylation factors and the more indirect effects on CTD Ser2-P levels being important for accurate 3′-end formation. The fact that combining mutations in the Paf1C with the loss of Ctk1 (13) leads to lethality is a strong argument that these two contact points are independently necessary for the essential function of cleavage and polyadenylation. In addition, our observation that the loss of the Paf1C results in greater defects in 3′-end formation of the MAK21 gene than the loss of Ctk1 is preliminary support for a significant role of the Paf1C-provided contacts.
Detaching the Paf1C from Pol II destabilizes the association among the cleavage and polyadenylation factors, Pol II, and chromatin but does not affect the integrity of the Paf1C (Fig. 6B). We observed essentially unchanged interactions between Paf1, Ctr9, and Leo1 in the absence of Cdc73 or Rtf1 and normal interactions between Cdc73 and the other Paf1C factors in the absence of Rtf1. The integrity of the complex is consistent with the relocalization of all of the Paf1C factors to the nucleolus when the Paf1C is detached from Pol II, as we observed previously (41). The interactions among Paf1C components and those with Pol II and Cft1 are also not dependent on RNA, which indicates that it is not the nascent transcript that holds all of these components of the elongation complex together.
Although Rtf1 is clearly part of the Paf1C in yeast, it also has independent functions and an independent association with chromatin, at least in part through its direct interactions with Chd1, which are distinct from its interactions with Paf1 and Ctr9 (53). Rtf1 also differs in that its human homolog may not be part of the hPaf1C (45, 60). The Paf1C in multicellular organisms is also different in that the genes that encode it are essential (20), as opposed to the nonessential nature of the yeast genes demonstrated by the phenotypes of yeast Paf1C mutants (5). We would argue, however, that the Paf1C does perform an essential function in yeast in providing an important contact for recruiting the cleavage and polyadenylation factors in concert with Pol II modifications and the appearance of the RNA substrate. Mutations in Paf1C components have been linked to changes in gene expression in several multicellular organisms (2, 34, 57), which in humans can result in cancer (9, 45). It will be interesting to learn whether the Paf1C-dependent changes in gene expression in more complex eukaryotes are the result of alterations, not of transcription initiation, but rather in the later stages of mRNA production as in yeast.
Acknowledgments
We thank C. Campsteijn for performing the TAP-Leo1 purification, C. Logie and H. Stunnenberg for advice and materials for the mass spectrometry, T. Blumenthal and D. Bentley for many useful suggestions and comments on the manuscript, P. Megee for his more than generous help with ChIP and PCR and his comments on the manuscript, K. Penheiter and R. Leon for their help and suggestions, and R. Sclafani and the members of his lab for providing a productive work environment. We also thank O. Rozenblatt-Rosen and M. Meyerson for suggestions that were critical for the initiation of these studies, for sharing unpublished observations, and for supplying reagents and strains.
This work was supported by grants from the NIH: RO1-GM038101 to J.A.J. and R15-GM076099 to J.L.B.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 1367-76. [DOI] [PubMed] [Google Scholar]
- 2.Akanuma, T., S. Koshida, A. Kawamura, Y. Kishimoto, and S. Takada. 2007. Paf1 complex homologues are required for Notch-regulated transcription during somite segmentation. EMBO Rep. 8858-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barilla, D., B. A. Lee, and N. J. Proudfoot. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley, D. L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17251-256. [DOI] [PubMed] [Google Scholar]
- 5.Betz, J. L., M. Chang, T. M. Washburn, S. E. Porter, C. L. Mueller, and J. A. Jaehning. 2002. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268272-285. [DOI] [PubMed] [Google Scholar]
- 6.Buratowski, S. 2005. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 17257-261. [DOI] [PubMed] [Google Scholar]
- 7.Buratowski, S. 2003. The CTD code. Nat. Struct. Biol. 10679-680. [DOI] [PubMed] [Google Scholar]
- 8.Calvo, O., and J. L. Manley. 2003. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 171321-1327. [DOI] [PubMed] [Google Scholar]
- 9.Carpten, J. D., C. M. Robbins, A. Villablanca, L. Forsberg, S. Presciuttini, J. Bailey-Wilson, W. F. Simonds, E. M. Gillanders, A. M. Kennedy, J. D. Chen, S. K. Agarwal, R. Sood, M. P. Jones, T. Y. Moses, C. Haven, D. Petillo, P. D. Leotlela, B. Harding, D. Cameron, A. A. Pannett, A. Hoog, H. Heath III, L. A. James-Newton, B. Robinson, R. J. Zarbo, B. M. Cavaco, W. Wassif, N. D. Perrier, I. B. Rosen, U. Kristoffersson, P. D. Turnpenny, L. O. Farnebo, G. M. Besser, C. E. Jackson, H. Morreau, J. M. Trent, R. V. Thakker, S. J. Marx, B. T. Teh, C. Larsson, and M. R. Hobbs. 2002. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat. Genet. 32676-680. [DOI] [PubMed] [Google Scholar]
- 10.Cho, J. H., Y. K. Lee, and C. B. Chae. 2001. The modulation of the biological activities of mitochondrial histone Abf2p by yeast PKA and its possible role in the regulation of mitochondrial DNA content during glucose repression. Biochim. Biophys. Acta 1522175-186. [DOI] [PubMed] [Google Scholar]
- 11.Chu, Y., R. Simic, M. H. Warner, K. M. Arndt, and G. Prelich. 2007. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 264646-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 112755-2766. [DOI] [PubMed] [Google Scholar]
- 13.Costa, P. J., and K. M. Arndt. 2000. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dichtl, B., D. Blank, M. Ohnacker, A. Friedlein, D. Roeder, H. Langen, and W. Keller. 2002. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 101139-1150. [DOI] [PubMed] [Google Scholar]
- 15.Dominski, Z., X. C. Yang, and W. F. Marzluff. 2005. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 12337-48. [DOI] [PubMed] [Google Scholar]
- 16.Gao, L., and D. S. Gross. 2008. Sir2 silences gene transcription by targeting the transition between RNA polymerase II initiation and elongation. Mol. Cell. Biol. 283979-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 1941-931. [PubMed] [Google Scholar]
- 18.Hershkovits, G., H. Bangio, R. Cohen, and D. J. Katcoff. 2006. Recruitment of mRNA cleavage/polyadenylation machinery by the yeast chromatin protein Sin1p/Spt2p. Proc. Natl. Acad. Sci. USA 1039808-9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, J. C., H. P. Phatnani, T. A. Haystead, J. A. MacDonald, S. M. Alam, and A. L. Greenleaf. 2004. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 27924957-24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421231-237. [DOI] [PubMed] [Google Scholar]
- 21.Kanin, E. I., R. T. Kipp, C. Kung, M. Slattery, A. Viale, S. Hahn, K. M. Shokat, and A. Z. Ansari. 2007. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc. Natl. Acad. Sci. USA 1045812-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keogh, M. C., V. Podolny, and S. Buratowski. 2003. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 237005-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kizer, K. O., H. P. Phatnani, Y. Shibata, H. Hall, A. L. Greenleaf, and B. D. Strahl. 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 253305-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11721-729. [DOI] [PubMed] [Google Scholar]
- 26.Kyburz, A., M. Sadowski, B. Dichtl, and W. Keller. 2003. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Res. 313936-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laribee, R. N., N. J. Krogan, T. Xiao, Y. Shibata, T. R. Hughes, J. F. Greenblatt, and B. D. Strahl. 2005. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol. 151487-1493. [DOI] [PubMed] [Google Scholar]
- 28.Licatalosi, D. D., G. Geiger, M. Minet, S. Schroeder, K. Cilli, J. B. McNeil, and D. L. Bentley. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 91101-1111. [DOI] [PubMed] [Google Scholar]
- 29.Max, T., M. Sogaard, and J. Q. Svejstrup. 2007. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J. Biol. Chem. 28214113-14120. [DOI] [PubMed] [Google Scholar]
- 30.Meinhart, A., and P. Cramer. 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430223-226. [DOI] [PubMed] [Google Scholar]
- 31.Mueller, C. L., and J. A. Jaehning. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 221971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mueller, C. L., S. E. Porter, M. G. Hoffman, and J. A. Jaehning. 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14447-456. [DOI] [PubMed] [Google Scholar]
- 33.Ng, H. H., S. Dole, and K. Struhl. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 27833625-33628. [DOI] [PubMed] [Google Scholar]
- 34.Oh, S., H. Zhang, P. Ludwig, and S. van Nocker. 2004. A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 162940-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsen, J. V., S. E. Ong, and M. Mann. 2004. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics 3608-614. [DOI] [PubMed] [Google Scholar]
- 36.Palancade, B., and O. Bensaude. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 2703859-3870. [DOI] [PubMed] [Google Scholar]
- 37.Patturajan, M., N. K. Conrad, D. B. Bregman, and J. L. Corden. 1999. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J. Biol. Chem. 27427823-27828. [DOI] [PubMed] [Google Scholar]
- 38.Penheiter, K. L., T. M. Washburn, S. E. Porter, M. G. Hoffman, and J. A. Jaehning. 2005. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell 20213-223. [DOI] [PubMed] [Google Scholar]
- 39.Phatnani, H. P., and A. L. Greenleaf. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 202922-2936. [DOI] [PubMed] [Google Scholar]
- 40.Phatnani, H. P., J. C. Jones, and A. L. Greenleaf. 2004. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 4315702-15719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porter, S. E., K. L. Penheiter, and J. A. Jaehning. 2005. Separation of the Saccharomyces cerevisiae Paf1 complex from RNA polymerase II results in changes in its subnuclear localization. Eukaryot. Cell 4209-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prelich, G. 2002. RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot. Cell 1153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proudfoot, N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16272-278. [DOI] [PubMed] [Google Scholar]
- 44.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 171030-1032. [DOI] [PubMed] [Google Scholar]
- 45.Rozenblatt-Rosen, O., C. M. Hughes, S. J. Nannepaga, K. S. Shanmugam, T. D. Copeland, T. Guszczynski, J. H. Resau, and M. Meyerson. 2005. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol. Cell. Biol. 25612-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan, K., O. Calvo, and J. L. Manley. 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10565-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 183091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shatkin, A. J., and J. L. Manley. 2000. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 7838-842. [DOI] [PubMed] [Google Scholar]
- 49.Sheldon, K. E., D. M. Mauger, and K. M. Arndt. 2005. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell 20225-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi, X., M. Chang, A. J. Wolf, C. H. Chang, A. A. Frazer-Abel, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 171160-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stumpf, G., and H. Domdey. 1996. Dependence of yeast pre-mRNA 3′-end processing on CFT1: a sequence homolog of the mammalian AAUAAA binding factor. Science 2741517-1520. [DOI] [PubMed] [Google Scholar]
- 52.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 101793-1808. [DOI] [PubMed] [Google Scholar]
- 53.Warner, M. H., K. L. Roinick, and K. M. Arndt. 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 276103-6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 27834739-34742. [DOI] [PubMed] [Google Scholar]
- 55.Xiao, T., C. F. Kao, N. J. Krogan, Z. W. Sun, J. F. Greenblatt, M. A. Osley, and B. D. Strahl. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25637-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yart, A., M. Gstaiger, C. Wirbelauer, M. Pecnik, D. Anastasiou, D. Hess, and W. Krek. 2005. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol. Cell. Biol. 255052-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Youn, M. Y., H. S. Yoo, M. J. Kim, S. Y. Hwang, Y. W. Choi, S. V. Desiderio, and J. Y. Yoo. 2007. hCTR9, a component of Paf1 complex, participates in the transcription of interleukin 6-responsive genes through regulation of STAT3-DNA interactions. J. Biol. Chem. 28234727-34734. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Z., J. Fu, and D. S. Gilmour. 2005. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 191572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu, B., S. S. Mandal, A. D. Pham, Y. Zheng, H. Erdjument-Bromage, S. K. Batra, P. Tempst, and D. Reinberg. 2005. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 191668-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zorio, D. A., and D. L. Bentley. 2004. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 29691-97. [DOI] [PubMed] [Google Scholar]