Abstract
Although encystation (cyst formation) is important for the survival of Giardia lamblia outside its human host, the molecular events that prompt encystation have not been fully elucidated. Here, we demonstrate that sphingolipids (SLs), which are important for the growth and differentiation of many eukaryotes, play key roles in giardial encystation. Transcriptional analyses showed that only three genes in the SL biosynthesis pathways are expressed and transcribed differentially in nonencysting and encysting Giardia trophozoites. While the putative homologues of giardial serine palmitoyltransferase (gSPT) subunit genes (gspt-1 and -2) are differentially expressed in nonencysting and encysting trophozoites, the giardial ceramide glucosyltransferase 1 gene (gglct-1) is transcribed only in encysting cells. l-Cycloserine, an inhibitor of gSPT, inhibited the endocytosis and endoplasmic reticulum/perinuclear targeting of bodipy-ceramide in trophozoites, and this could be reversed by 3-ketosphinganine. On the other hand, d-threo-1-phenyl-2-palmitoylamino-3-morpholino-1-propanol (PPMP), an inhibitor of glucosylceramide synthesis, blocked karyokinesis and reduced cyst production in culture. PPMP also altered the expression of cyst wall protein transcripts in encysting cells. Phylogenetic analyses revealed that the gspt genes are paralogs derived from an ancestral spt sequence that underwent gene duplication early in eukaryotic history. This ancestral sequence, in turn, was probably derived from prokaryotic aminoacyl transferases. In contrast, gglct-1 is found in both prokaryotes and eukaryotes without any evidence of gene duplication. These studies indicate that SL synthesis genes are involved in key events in giardial biology and could serve as potential targets for developing new therapies against giardiasis.
Giardiasis, a clinical syndrome caused by the intestinal protozoan Giardia lamblia, is a reemerging waterborne infectious illness worldwide. Giardia exists in two morphological forms: (i) actively dividing trophozoites and (ii) relatively inactive cysts. The water-resistant dormant cysts are responsible for transmission of giardiasis via contaminated water and undergo excystation, a stage of the giardial life cycle in which a cyst differentiates into two trophozoites in the stomach. Newly emerged trophozoites then move down the small intestine and colonize below the bile duct (2). Components of the intestinal milieu, including dietary lipids, bile salts, intestinal pH (pH ∼7.8), and lactic acid, among others, trigger the process of encystation to complete the life cycle of Giardia in the small intestine (10, 16). During encystation, various molecular and cellular changes take place that allow this protozoan to transport cyst wall proteins (CWPs) through regulatory secretory pathways (35). In encysting cells, three encystation-specific CWPs (CWP-1, -2, and -3 encoded by cwp-1, -2, and -3, respectively) are synthesized and concentrated within encystation-specific vesicles (ESVs) before targeting into the cyst wall (17, 36, 44). Recent studies suggest that all three CWPs are essential for forming ESVs and that CWP-2 functions as an aggregation factor to regulate ESV formation by interacting with CWP-1 and CWP-3 via conserved regions (17). It has been proposed that transient Golgi body-like membranes synthesized during encystation are involved in modifying the CWPs and other membrane proteins (27-29).
As an obligate parasite, Giardia has lost lipid synthesis machinery and therefore has evolved a well-regulated lipid transport mechanism allowing it to acquire the majority of its membrane lipids from the small intestine environment (8, 23). Many of these lipids also play regulatory roles in inducing encystation (14). We have reported previously that Giardia also has the ability to carry out deacylation-reacylation and headgroup exchange reactions to generate new parasite-specific phospholipids, bypassing the synthesis of entirely new phospholipid molecules via de novo pathways (7, 43). More recently, we demonstrated that ceramide, which is not synthesized by Giardia de novo, is taken up by clathrin-mediated pathways and targeted intracellularly via the microtubule network (21), which suggests that ceramide and other sphingolipids (SLs) may play an important role in the giardial life cycle.
In the current study, we showed that for all known SL metabolic pathways only three synthesis-related genes and two metabolic genes are present in Giardia and expressed differentially in nonencysting and encysting parasites. We also showed that the inhibitors of SL synthesis affect endocytic functions and the production of cysts in vitro, which indicates the importance of SLs in giardial growth and differentiation.
MATERIALS AND METHODS
Materials.
Unless otherwise specified, all chemicals were purchased from Sigma Chemical Co. (St. Louis, MO) and were the highest purity available. Tetramethylrhodamine (TMR)-conjugated goat anti-mouse antibodies and bodipy-ceramide [N-(4,4-difluoro-5,7-dymethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)-sphingosine, bodipy FLC5-ceramide] were purchased from Invitrogen (Carlsbad, CA). Mouse anti-glucosylceramide (anti-GlcCer) antibody raised against fungal cerebrosides as described previously (34) was provided by Eliana Barreto-Bergter (Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil). GlcCer and other lipid standards were purchased from Matreya, Inc. (Pleasant Gap, PA).
Trophozoites, encysting cells, and in vitro cysts.
G. lamblia trophozoites (strain WB [= ATCC 30957]) were cultivated using the method of Diamond et al. (9) and TYI-S-33 medium supplemented with 10% adult bovine serum and 1% adult bovine bile (25). The antibiotic piperacillin (50 μg/ml) was added during routine culture of the parasite (15). Parasites were detached by ice chilling and were harvested by centrifugation at 1,500 × g for 10 min at 4°C, which was followed by washing and microscopic determination of the cell density using a hemocytometer. In vitro encystation was carried out by the method of Gillin et al. (15) by culturing trophozoites in TYI-S-33 medium (pH 7.8) supplemented with bovine serum (10%, vol/vol), lactic acid (5 mM), and porcine bile (250 mg/ml) for various times, as described below. In vitro-derived, water-resistant cysts were generated by cultivating trophozoites in TYI-S-33 medium (pH 7.8) supplemented with 10% bovine serum and bovine bile (high-bile medium) using the protocol of Kane et al. (24). Encystation was carried out for 24 h, and cells were isolated by centrifugation (1,500 × g for 5 min at 4°C), washed three times in cold distilled water, and kept in water for 3 days in a refrigerator (4 to 8°C). Water-resistant cysts were isolated by centrifugation and then counted and used for the microscopic experiments described below.
Identification of putative SL synthesis and metabolic genes.
Predicted open reading frames were obtained from the Giardia genome database (30; www.giardiaDB.org) and compared, using BLASTP (3), with Genprot and Swiss-Prot databases. A set of potential SL-metabolizing genes were identified, including the genes encoding giardial ceramide glucosyltransferase 1 (gGlcT-1) (gglct-1, ORF 11642), giardial serine palmitoyl transferase 1 (gSPT-1) (gspt-1, ORF 123015), giardial serine palmitoyl transferase 2 (gSPT-2) (gspt-2, ORF 14374), giardial acid sphingomyelinase B (gSmase B) (gsmase B, ORF 16737), and giardial acid sphingomyelinase 3b (gSmase 3b) (gsmase 3b, ORF 8360). The sequences of the encoded proteins (gSPT-1 and -2 encoded by gspt-1 and -2, respectively; gGlcT-1 encoded by gglct-1; and gSmases B and 3b encoded by gsmase B and 3b, respectively) were subjected to protein family (pfam) database analyses to predict functional homology to other families (4; http://pfam.wustl.edu). In order to predict the subcellular localization of gSPT-1, gSPT-2, gGlcT-1, gSmase B, and gSmase 3b, predicted sequences (proteins) were analyzed by using the ExPaSY/PSORT software as described previously (13, 31, 32).
Determination of mRNA levels of giardial SL metabolic genes by quantitative reverse transcription-PCR (qRT-PCR).
To determine whether gspt-1, gspt-2, gglct-1, gsmase B, and gsmase 3b are expressed in nonencysting and encysting trophozoites, cells were cultivated in growth medium and subjected to preencystation and then encystation for 6, 12, and 90 h using the two-step method described by Gillin et al. (15). RNA from trophozoites, preencysting cells, and 6-, 12-, and 90-h encysting cells were extracted and purified using TRIzol reagent from Invitrogen, Inc. (Carlsbad, CA). One to two microliters of total RNA was reverse transcribed using the standard protocol of a Reaction Ready first strand cDNA synthesis kit from Super Array Bioscience Corporation (Frederick, MD). Primers for PCR were designed using Primer 3 software (primer3_www.cgi v 0.2) (39) and were synthesized by Sigma Genosys (St. Louis, MO). The sequences of the primer pairs are as follows: for gspt-1, 5′-GAAACCAACCACGTGAGGAT-3′ and 3′-CATAGCCCATGTCACACCAG-5′; for gspt-1, 5′-CATGACAGCAGTGGCAAGTT-3′ and 3′-TCCATCGTCTTCCCTCAAAC-5′; for gglct-1, 5′-GCTGTCAACCGCATAAGTGA-3′ and 3′-TTGAGCTGTGAGTTCCATCG-5′; for gsmase B, 5′-TGAAAGCCTTGTTGATGCAG-3′ and 3′-TAGCTCGCTGGGTCATCTCT-5′; for gsmase 3b, 5′-TGTGGAGCAGTTGACAAAGC-3′ and 3′-ATTTAATCGCCTGGTCATGG-5′; for cwp-1, 5′-CCAATTGACGAACCTCCAGT-3′ and 3′-CATAAGGTAGGGGAGCGTCA-5′; for cwp-2, 5′-TCATCCTGTTTGCTGCTTTG-3′ and 5′-CATGCACCCCAGTTTCTTCT-3′; and for cwp-3, 5′-TTCGCTCATAGGGGATGTTC-3′ and 3′-GCGAGATCCAAGTGGCTAAA-5′.
cDNA samples were diluted 1:10, and 1 μl of each cDNA sample was used as a template in PCRs. The PCR products were run on a 2% agarose gel. For quantitative real-time PCR, cDNA samples were diluted 1:20, and 2 μl of each sample was used in a 20-μl PCR mixture with 10 μl of 2× SYBR green PCR master mixture (Superarray Biosciences, Frederick, MD). qRT-PCR was performed using an MYiQ version 1.0 thermal cycler (Bio-Rad, Hercules, CA). The relative standard curve method was used to quantify transcript levels.
Treatment of parasites with l-cycloserine.
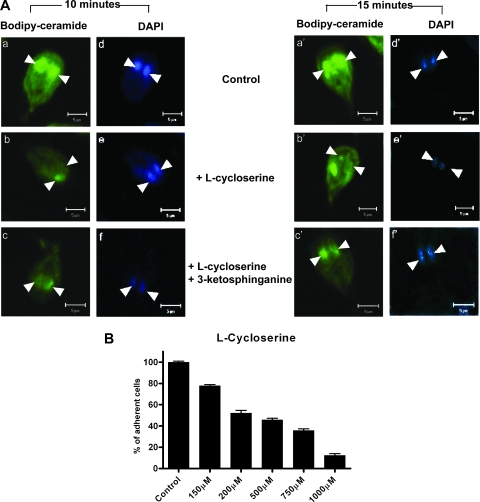
Giardia trophozoites were grown and harvested as described above. Approximately 1 × 107 cells were inoculated into 5-ml tubes. The cells were allowed to attach for 30 min at 37°C, and various concentrations (0, 150, 200, 500, 750, and 1,000 μM) of l-cycloserine were added to the culture tubes. The cells were incubated overnight at 37°C, and attached cells were counted using a hemocytometer and phase-contrast microscopy as described previously (21). To determine whether l-cycloserine interrupts ceramide uptake and targeting to nuclear/perinuclear membranes, trophozoites were treated with l-cycloserine (150 μM, ∼107 cells) for 30 min before labeling with bodipy-ceramide (100 nM) for 10 and 15 min, also as described previously (21). In some experiments, 50 μM 3-ketosphinganine (3-KS) (49) was added to reverse the effect of l-cycloserine, as described below (see Fig. 3). Cells were fixed using 4% paraformaldehyde (ethanol free) in phosphate-buffered saline (PBS) for 20 min, and trophozoites were then subjected to 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg/ml) staining for 5 min. Slides were washed and mounted using DAKO mounting medium (DAKO Corporation, Carpentaria, CA) and were observed using an LSM Pascal 5 Zeiss confocal microscope.
FIG. 3.
3-KS restores the endocytosis and ER/perinuclear targeting of bodipy-ceramide in nonencysting trophozoites. Attached trophozoites were treated with l-cycloserine (150 μM, ∼107 cells) and incubated for 30 min at 37°C. For rescue experiments, 3-KS was added and incubated with trophozoites for 5 min prior to l-cycloserine treatment. Trophozoites were labeled with bodipy-ceramide as described in Materials and Methods. (A) Confocal images showing ER/perinuclear labeling of bodipy-ceramide for 10 and 15 min in the presence and absence of l-cyloserine (150 μM, ∼107 cells) and l-cycloserine plus 3-KS (50 μM, ∼107 trophozoites), respectively. Bars = 5 μm. (B) Dose-response effects of l-cycloserine on adherence of Giardia trophozoites. The data are the means and standard deviations of three separate experiments, and each experiment was carried out in triplicate.
Effects of PPMP and PDMP on cyst production.
Approximately 1 × 107 trophozoites were inoculated into 5-ml tubes and incubated overnight at 37°C. The medium was decanted and then replaced with new medium (TYI-S-33 medium [pH 7.8] supplemented with 10% bovine serum and bile [high-bile medium]), as described by Kane et al. (24). The encystation medium was supplemented with d-threo-1-phenyl-2-decanoylamino-3-morpholinopropanol (PDMP) and d-threo-1-phenyl-2-palmitoylamino-3-morpholino-1-propanol (PPMP) at various concentrations (0, 30, 60, 90, and 120 μM) and incubated at 37°C for 24 h. The cysts were obtained by centrifugation at 1,500 × g for 5 min, resuspended in distilled water, and kept at 4°C for 72 h; this was followed by counting using a hemocytometer. For microscopic analyses, water-resistant cells were collected by centrifugation, fixed in 4% paraformaldehyde (ethanol free), and stained with DAPI (1 μg/ml) for observation with an LSM Pascal 5 Zeiss confocal microscope.
Effects of PPMP and PDMP on expression of the gglct-1 and CWP (cwp) genes.
Approximately 1 × 107 cells were first cultured in preencystation medium for 48 h before encystation was initiated, as described by Gillin et al. (15). PPMP or PDMP (final concentration, 10 μM) was added during encystation, and each preparation was incubated for 12 h at 37°C. The cells were harvested, and RNA was extracted using TRIzol reagent; this was followed by qRT-PCR as described above.
Labeling with anti-GlcCer antibody.
Nonencysting, encysting, and water-resistant cysts (∼1 × 107 cells) were suspended in PBS and incubated for 15 min at 37°C in chamber slides. Cells were then fixed with 4% paraformaldehyde (ethanol free) in PBS and blocked in 5% normal goat serum for 1 h. Slides were washed three times and incubated overnight with 10 μg/ml anti-GlcCer antibody diluted in 1% normal goat serum, which was followed by exposure to a rabbit anti-mouse antibody (1:500) conjugated with TMR for 1 h at room temperature (34). To confirm the specificity of serological reactions, primary antibodies were preabsorbed with 30 μg GlcCer or glucose (GlcCer-to-antibody or glucose-to-antibody ratio, 3:1) before they were incubated with Giardia as described above. Cells were subjected to DAPI (1 μg/ml) staining for 5 min, and then the slides were washed, mounted using DAKO mounting medium (DAKO Corp., Carpentaria, CA), and observed with an LSM Pascal 5 Zeiss confocal microscope.
Phylogenetic analysis of SPT genes.
For the phylogenetic analysis, the protein sequences of serine palmitoyl transferases (SPT) (encoded by spt genes) from Giardia were used as query sequences for BLAST searches in the NCBI GenBank database. The search was based on a BLOSUM62 amino acid substitution scoring matrix, and all available taxa in the GenBank database, both eukaryotic and prokaryotic, were searched for significant matches. As a heuristic cutoff rule, sequences with local alignment E values of 0.001 or less were used for the subsequent multiple-sequence alignment and phylogeny reconstruction.
Progressive, multiple-sequence alignment was performed using Clustal X, in which the scoring was also based on a BLOSUM62 matrix. The aligned sequences were analyzed with the PHYLIP package (11) using the PROML routine for maximum likelihood analysis of amino acid sequences (12). The maximum likelihood calculation assumed that the Jones-Taylor-Thornton model of equal substitution rates for all amino acids is a computationally convenient approximation, given the lack of any data for substitution rates in the sequences. Subsequent analysis of the robustness of the resulting likelihood-based trees was carried out with bootstrap analysis (100 samplings for the sequence data), using the bootseq and consense modules (for building consensus trees from the bootstrapped data) in PHYLIP. Finally, the phylogenies were edited and displayed using MrEnt (51).
Nucleotide sequence accession numbers.
The GenBank (http://www.ncbi.nlm.nih.gov) accession numbers (annotation date, May 2007) for the gene sequences described above are as follows: gspt-1, XP_779228; gspt-2, XP_767432; gglct-1, XP_767155; gsmase b, XP_768146.1; and gsmase 3b, XP_770759.1.
RESULTS
Genomic and molecular analyses reveal stage-specific expression of SL metabolic genes.
For identification and characterization of SL metabolic genes from G. lamblia, NCBI BLAST search and protein family (pfam) database analyses were performed. Five putative genes in the Giardia genome database (30; www.giardiaDB.org) were identified, including gspt-1 and gspt-2 (encoding the gSPT-1 and gSPT-2 subunits, respectively), gglcT-1 (encoding gGlcT-1), and the gsmase B and 3b genes (encoding gSmases B and 3b, respectively) (Table 1). The pfam analysis revealed that the gspt-1 and gspt-2 products belong to the aminoacyl transferase class I and II families, the gglct-1 product belongs to the eukaryotic glycosyl transferase 2 family, and the gsmase gene products resemble the calcineurinlike phosphoesterase family of enzymes.
TABLE 1.
Predicted open reading frames and pfam matches of giardial SL metabolic genesa
| Giardial open reading frame | GenBank accession no. | Match with pfam (motif location) | Species with best BLASTp match (E value) | pfam family match |
|---|---|---|---|---|
| SPT-1 | XP_779228 | 0.00012 (amino acids 137 to 523) | Danio rerio | Aminotransferase classes I and II |
| SPT-2 | XP_767472 | 6e−36 (amino acids 154 to 503) | Aspergillus terreus (6e−76) | Aminotransferase classes I and II |
| GlcT-1 | XP_767155 | 5.2e−05 (amino acids 148 to 256) | Arabidopsis thaliana (9e−30) | Glycosyltransferase family 2 |
| Smase B | XP_768146.1 | 2.4e−05 (amino acids 18 to 273) | Xenopus tropicalis (1e−25) | Calcineurinlike phosphoesterase |
| Smase 3b | XP_770759.1 | No match | Danio rerio (6e−19) | No match |
Five putative genes encoding SL metabolic enzymes were identified in Giardia using the NCBI and pfam databases.
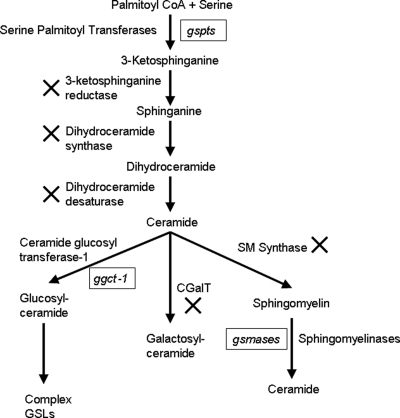
Figure 1 shows the SL metabolic pathways and genes that are present in Giardia. Two subunits of the gSPT enzyme (encoded by the gspt-1 and gspt-2 genes) catalyze a condensation reaction between serine and palmitoyl coenzyme CoA to synthesize 3-KS. gGlcT-1 (encoded by the glct-1 gene) transfers glucose from UDP-glucose to a ceramide acceptor molecule, and finally two acid sphingomyelinaselike phosphodiesterase (gSmase) enzymes (encoded by gsmase B and gsmase 3b) hydrolyze sphingomyelin to ceramide. To determine the predicted subcellular localization of SL metabolic enzymes, DNA sequences of gspt-1, gspt-2, gglct-1, gsmase B, and gsmase 3b were translated into proteins using the ExPaSY software (13), and the protein sequences were analyzed to determine the predicted subcellular localization using the same software. The subcellular localization prediction analysis suggested that gSPT-1 and -2 are transmembrane proteins in the endoplasmic reticulum (ER). gGlcT-1 is located in the luminal membrane of the ER, and gSmase B is predicted to be located in the parasite's cytoplasm, whereas gSmase 3b might be a secreted enzyme (not shown).
FIG. 1.
Identification of putative SL metabolic genes in Giardia. A search of the Giardia genome database (30; www.giardiaDB.org) revealed the presence of only five putative genes for SL metabolism, which encode gSPT, gGlct-1 (GlcT-1 is also known as glucosylceramide synthase), and acid sphingomyelinases. It is not known if Giardia has the ability to carry out the reactions indicated by a multiplication sign because the BLASTP search did not reveal any significant matches of these genes with genes of other organisms. CoA, coenzyme A; GSLs, glycosphingolipids; SM, sphingomyelin.
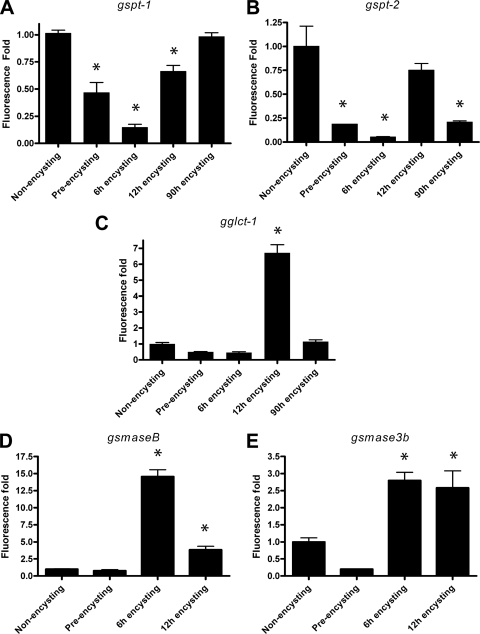
To evaluate the differential expression of SL genes, qRT-PCR were performed to measure the mRNA levels in encysting and nonencysting trophozoites. As shown in Fig. 2A and B, the gspt-1 and gspt-2 genes are transcribed mostly in trophozoites and then upregulated in encysting cells (12 h). Interestingly, after prolonged periods of cyst induction (90 h), the mRNA levels of gspt-2 are down-regulated, but the level of gspt-1 remains high. This result suggests that the two subunit genes are differentially expressed and encode enzymes that are required to perform specific functions in the nonencysting and encysting stages of the giardial life cycle. On the other hand, gglct-1 transcripts are generated in early (12-h) encysting cells (Fig. 2C), when visible ESVs for transporting cyst wall antigens are synthesized and targeted to plasma membranes (16). Both gsmase B and gsmase 3b are expressed in 6- and 12-h encysting cells (Fig. 2D and 2E), which may suggest that during encystation gsmase B and gsmase 3b encode gSmases to scavenge ceramide from dietary components present in the small intestine.
FIG. 2.
Differential expression of SL metabolic genes in nonencysting, preencysting, and encysting trophozoites: expression of gSPT-1 (gspt-1), gSPT-2 (gspt-2), gGlct-1 (gglct-1), gSmase B (gsmase B), and gSmase 3b (gsmase 3b) transcripts in preencysting and encysting trophozoites of G. lamblia relative to the expression in nonencysting (vegetative) trophozoites. The data are the means and standard deviations of three separate experiments, and each experiment was carried out in triplicate. The qRT-PCR analysis of SL genes was carried out as described in Materials and Methods. An asterisk indicates that there were significant differences compared with vegetative trophozoites (P < 0.05).
l-Cycloserine blocks ER/perinuclear targeting of bodipy-ceramide in trophozoites and 3-KS restores ceramide endocytosis.
As mentioned above, SPT catalyzes the synthesis of 3-KS, which is the first step in SL biosynthesis (19, 20). The reaction catalyzed by SPT can be inhibited by various serine analogues, including sphingofungin B, myriocin, and l-cycloserine (22). Previously, we showed that Giardia has actin-regulated endocytic pathways to import ceramide molecules from outside sources (21). Since mammalian and yeast spt mutants are defective in endocytosis and require sphingoid bases to overcome the defect (19, 47), we considered the possibility that the expression of gspt genes (Fig. 2A and 2B), especially in trophozoites, is important for regulating endocytosis through giardial actin filaments. The analysis of parasite growth demonstrated that l-cycloserine inhibits the adherence of Giardia trophozoites in a dose-dependent manner (Fig. 3B), with a 50% reduction in adherence at 200 μM (50% inhibitory concentration [IC50], 200 μM). In contrast, myriocin, a potent inhibitor of SPT in higher eukaryotes, was not at all effective in inhibiting the growth of trophozoites (not shown).
To confirm that ceramide internalization occurs through SPT (presumably encoded by gspt genes) and its reaction product 3-KS, trophozoites were treated with 50 μM 3-KS for 5 min, followed by addition of l-cyloserine (150 μM) to the culture medium (Fig. 3A). The 3-KS restored ceramide endocytosis and the ER/perinuclear targeting affected by l-cycloserine (Fig. 3A, panels c and c′).
Inhibitors of GlcCer synthesis interfere with cyst formation.
As shown in Fig. 2C, the conditions that induce encystation (pH 7.8, lactic acid, and porcine bile) also upregulate gglct-1 transcription, suggesting that increased GlcCer synthesis (facilitated by the gGlcT-1 enzyme) could be linked to encystation. To test this possibility, we evaluated the effects of PDMP and PPMP on encystation and cyst production. PPMP and PDMP, two well-known inhibitors of GlcT-1, differ only in the length of their fatty acyl chains. These structural analogues of ceramide compete with natural ceramide and inhibit GlcCer synthesis (26).
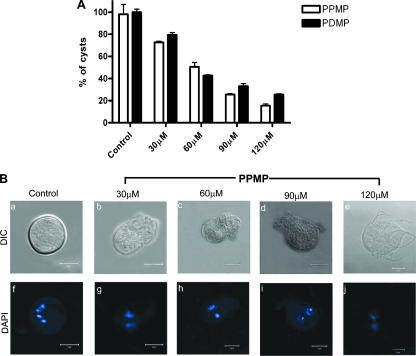
Giardia trophozoites were subjected to in vitro encystation in a medium containing a high level of bile, as described previously by Kane et al. (24), in the presence and absence of PPMP or PDMP for 24 h before collection of the water-resistant cysts. Both PDMP and PPMP inhibited cyst formation by Giardia trophozoites (Fig. 4A). The IC50 of the inhibitors were ∼60 μM. However, the inhibitors were active only when they were added within the first 2 h of encystation (not shown). Next, we asked if the reduction in cyst formation was specific for PDMP or PPMP, so encystation was also carried out in the presence of myriocin and l-cycloserine (SPT inhibitors). We found that these two inhibitors were not effective in reducing cyst formation (not shown). Figure 4B (panels a to e) shows the effect of PPMP (at various concentrations) on cyst morphology. At 30 μM PPMP, cysts were rectangular with two nuclei. At 60 and 90 μM PPMP, they were much smaller, with an indication of the presence of flagella, and at 120 μM PPMP, they resembled round trophozoites with apparent flagellar structure. It is also worth noting that all of the PPMP-treated cysts were binucleate (Fig. 4B, panels f to j). In a separate experiment, we found that PDMP and PPMP were more effective (IC50, ∼15 μM) in reducing cyst production (not shown) when encystation was carried out using a two-step method (15). However, in the current study cysts were generated by the high-bile method because it required less time (12 to 18 h versus 3 to 4 days) and yielded a large quantity of water-resistant cysts.
FIG. 4.
Inhibitors of GlcCer synthesis inhibit in vitro cyst production. Trophozoites were grown until the late log phase and subjected to encystation by culturing them in high-bile medium (24) for 24 h in the presence or absence of PDMP and PPMP, as described in Materials and Methods. (A) Dose-dependent inhibition of water-resistant cyst generation by inhibitors. The data are the means and standard deviations of four separate experiments, and each experiment was carried out in triplicate. (B) Alteration of the morphological shape of water-resistant cells by PPMP. DAPI staining shows that the cryptic cystlike structures contain two nuclei instead of the four nuclei present in the control. Bars = 5 μm. DIC, differential interference contrast microscopy.
Intracellular localization of GlcCer.
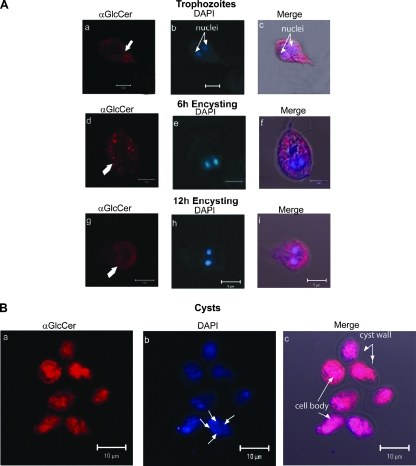
To test whether the morphological transition from nonencysting trophozoites to encysting trophozoites influences the cellular distribution of GlcCer, we used a monoclonal antibody raised against this lipid as a probe (34). As shown in Fig. 5, the antibody recognized different cellular sites in nonencysting and encysting trophozoites, as well as in cysts. In nonencysting trophozoites, GlcCer was localized in the plasma membranes (Fig. 5A, panels a and c), while in 6-h encysting cells GlcCer appeared to be localized in vesiclelike structures scattered throughout the cytoplasm (Fig. 5A, panels d and f). In late periods (12 h), there was characteristic cytoplasmic labeling (panels g and i) surrounding the ER/perinuclear regions, but in cysts GlcCer was localized mostly in the cell body. Since no labeling of the cyst wall was observed with anti-GlcCer antibody (Fig. 5B, panels a and c), it is reasonable to postulate that GlcCer is not present in cyst walls.
FIG. 5.
Stage-specific localization of GlcCer in Giardia. Nonencysting, encysting, and water-resistant cysts were labeled with monoclonal antibody against GlcCer, which was followed by reaction with TMR-conjugated rabbit anti-mouse antibody, as described in Materials and Methods. (A) Panels a and c show that in nonencysting trophozoites, GlcCer localizes to the plasma membranes. In 6- and 12-h encysting cells, the majority of the antibody labeling is localized in vesiclelike structures (panels d and f) and in the area surrounding the nuclei (panels g and i). (B) In water-resistant cysts (panels a to c), anti-GlcCer labeling is prominent in the cell body. Antigen-antibody reactions were considered to be specific because no labeling was observed with anti-GlcCer antibody pretreated with 30 μg/ml GlcCer (GlcCer-to-antibody ratio, 3:1). In contrast, glucose had no effect on antibody binding (not shown). “Merge” represents the colocalization of TMR, DAPI, and differential interference contrast microscopy images. αGlcCer, anti-GlcCer antibody. Bars = 5 μm (A) and 10 μm (B).
PPMP and PDMP affect the expression of cwp genes.
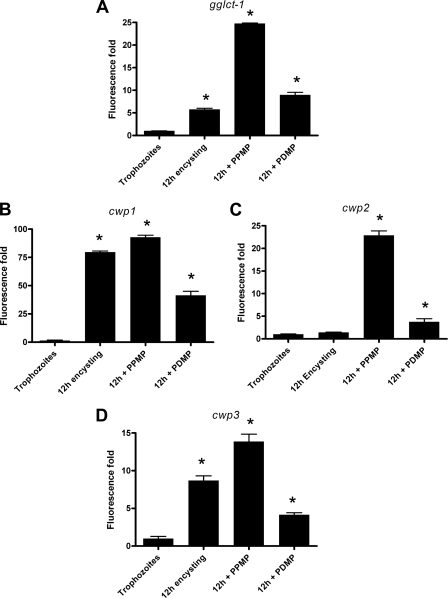
The hallmark of encystation in Giardia is the synthesis of ESVs that transport CWPs to the cell surface (16). During encystation, three encystation-specific CWPs (CWP-1, -2, and -3) are expressed and concentrated within ESVs before they are targeted to the cyst wall (17, 36, 44). Because the expression of the gglct-1 gene is upregulated (Fig. 2C) in encysting cells and, most importantly, because the inhibitors of GlcCer synthesis reduce the production of cysts (Fig. 4), we asked whether these two inhibitors also influence the transcription of CWP genes (cwp genes). Figure 6 shows that PDMP and PPMP upregulated the expression of the gglct-1 and cwp genes in 12-h encysting cells and that the stimulation by PPMP was greater than the stimulation by PDMP. These results suggest that GlcT-1 inhibitors influence the expression of not only gglct-1 but also the cwp genes. It is possible that the transcription of all of these genes is interlinked and regulated during encystation.
FIG. 6.
PPMP and PDMP alter gglct-1 and CWP (cwp) transcripts. Giardia trophozoites (∼1 × 107 cells) were cultured and subjected to preencystation for 48 h and then encystation for 12 h using the protocol of Gillin et al. (15) in the presence and absence of inhibitors (10 μM). RNA was isolated, reverse transcribed, and subjected to qRT-PCR analyses as described in Materials and Methods. The data are the means and standard deviations of three individual experiments, and each experiment was carried out in triplicate. An asterisk indicates that there were significant differences compared with vegetative trophozoites (P < 0.05).
gSPT is derived from bacterial aminoacyl transferase.
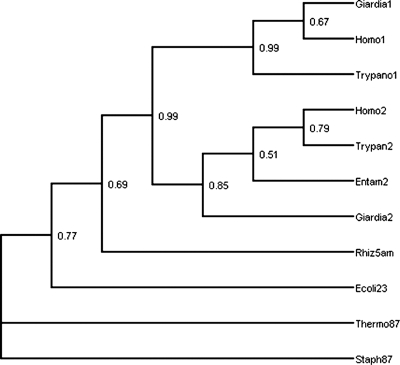
Since Giardia is phylogenetically basal (6, 33, 40, 41) and its growth is not affected by myriocin, a potent inhibitor of mammalian SPT (see above), we asked if homologs of the gspt genes are truly ancestral in the Eukaryota. Therefore, giardial protein sequences were used as the queries for BLAST searches in the NCBI GenBank database. Representative sequences were then analyzed to determine phylogenetic relationships using maximum likelihood methods. The initial BLAST output was used to determine the phylogenetic distribution of the spt-1 and spt-2 genes and their homologues. Both of these genes are present in virtually every eukaryote assayed, including metamonad taxa such as Giardia and Trichomonas, “excavate” taxa such as Trypanosoma and Leishmania, “alveolate” and “heterokont” taxa such as Paramecium and Plasmodium, the “amoebozoan” Dictyostelium, and the “higher” multicellular eukaryotes, such as plants, fungi, and metazoans (40). The one notable exception is the absence of spt-1 in Entamoeba (see Discussion). In contrast, the gspt genes were found to be completely absent in prokaryotes. However, a number of bacterial enzymes (specifically aminoacyl transferases) were found to be homologous to the spt-1 and -2 products. Examples of enzymes that gave highly significant E values with an SPT query (E < 0.001) include 5-aminolevulinic acid synthase (also present in certain eukaryotes), 2-amino-3-ketobutyrate coenzyme synthase (found in eubacteria), and 8-amino-7-oxononanoate synthase (found in both archaea and eubacteria). The other homologous enzyme, 5-aminolevulinate synthase, has a more restrictive phylogenetic distribution and is found in Rhizobiales and in some eukaryotes. Figure 7 shows the phylogeny of some representative eukaryotic spt gene products and bacterial aminoacyl transferases, which was obtained using maximum likelihood, including bootstrap values at each node for 100 replicates. We noted that the phylogeny derived using maximum parsimony (not shown) is largely consistent with the likelihood tree, except for the position of eukaryotic taxa within the spt paralog subclades. In all of the observed unrooted trees, 8-amino-7-oxononanoate synthase was found to be an outgroup with respect to the other enzymes, and therefore it was used to root the tree. Figure 7 shows that (i) the spt-1 and spt-2 lineages are monophyletic lineages with 99 and 85% support, respectively (i.e., all eukaryotic SPT-1 sequences form a subclade with other SPT-1 sequences, and the same is true for SPT-2 sequences), and (ii) although the SPT-1 and SPT-2 sequences are distinct, they are indeed paralogous sequences, together forming a monophyletic subclade (with 99% support) of SPTs with the various aminoacyl transferases as outgroups. Database searches and phylogenetic analysis also suggested that glct-1 is quite ancient, since it is present both in eukaryotes and in a number of bacteria. Thus, the data mirror what is known about the evolutionary relationships of eukaryotic organisms.
FIG. 7.
Phylogenetic analyses of gSPTs. The tree is a majority rule consensus tree constructed from 100 bootstrap replicates of maximum likelihood phylogenies for gspt-1 and gspt-2 sequences, along with homologous aminoacyl transferase sequences. The individual trees were constructed on the assumption of a Jones-Taylor symmetric amino acid substitution model. The taxon and sequence abbreviations are as follows: Giardia1 and Giardia2, gSPT-1 and gSPT-2, respectively; Entam2, SPT-2 from Entamoeba histolytica (lacking SPT-1); Trypano1 and Trypan2, Trypanosoma cruzi; Homo1 and Homo2, Homo sapiens. For aminoacyl transferases, the suffix “5am” indicates 5-amino leuvulinic acid synthase, “87” indicates 8-amino-7-oxononanoate synthase, and “23” indicates 2-amino-3-ketobutyrate coenzyme, and the archaeal and eubacterial taxa included are Thermophilus aquaticus (Therm), Staphylococcus aureus (Staph), Escherichia coli (Ecoli), and Rhizobium leguminosarum (Rhiz).
DISCUSSION
In this paper, we report that SL metabolic genes are involved in regulating encystation and cyst formation by Giardia. Over the past several years SLs have been recognized as major signaling molecules and inducers of apoptosis in mammalian cells. It has been proposed that SL-containing membrane microdomains function as clusters to receive and transduce extracellular signals downstream of the plasma membranes (18). Glycosphingolipids, on the other hand, have been shown to play critical roles in the development and differentiation of some embryonic tissues, as evidenced by the targeted disruption of the GlcCer gene, which blocks the normal differentiation through apoptosis (48). In Leishmania, SLs (inositol-phosphorylceramides) are essential for differentiation, infectivity, and vesicular trafficking but not for viability (50). Unlike the viability of Leishmania, however, the viability of Trypanosoma brucei has been shown to be dependent upon SL synthesis (45).
The transcriptional analyses of SL genes indicate that these genes are differentially expressed in the nonencysting and encysting stages of the giardial life cycle. The fact that spt expression is increased in trophozoites supports the notion that the product of the SPT reaction (3-KS) might modulate endocytic traffic, as shown in the yeast Saccharomyces cerevisiae (49). However, despite the expression of the five SL metabolic genes, Giardia exhibits limited lipid synthesis capacity de novo, and it takes up ceramide and SL from its host for survival, growth, and encystation (21). Therefore, it is conceivable that the products of these genes, especially the gspt genes and gglct-1, are required for cellular functions rather than for the synthesis of new SL bases per se. Our results show that l-cycloserine significantly affects the internalization and targeting of bodipy-ceramide into ER/perinuclear membranes. 3-KS, however, reverses the effect of l-cycloserine on bodipy-ceramide uptake and nuclear localization, which suggests that the synthesis and proper functioning of gSPT are important for ceramide uptake and targeting. A higher concentration of l-cycloserine is needed to inhibit gSPT, probably because of its ancestral nature (Fig. 7). This rationale could be supported by the fact that myriocin, a potent inhibitor of mammalian SPT (19), failed to inhibit the adherence of the parasite even at a concentration of 1 mM (not shown).
PPMP and PDMP are two well-recognized inhibitors of the GlcT-1 enzyme that have been extensively used to evaluate GlcCer functioning in other organisms (26). PDMP has been reported to induce the production of a variety of regulators in eukaryotic cells. For Plasmodium falciparum, for instance, PPMP has been described as a potent inhibitor of the intraerythrocytic maturation that leads to restriction of the development of the malaria parasite (1). We found that both PPMP and PDMP reduced the production of in vitro cysts by a mechanism yet to be delineated. As shown by differential interference contrast microscopy, PPMP interferes with the production of mature tetranucleated cysts (Fig. 4B). The Giardia cyst is a quadrinucleated (16N) structure that releases two binucleated (4N) trophozoites by excystation (5). The inhibition of cyst production by PPMP and PDMP and the localization of GlcCer in the cell body of the cyst, which consists of nuclei, chromosomes, and transcriptional machinery, support the idea that this glycosphingolipid is associated with nuclear division (karyokinesis) and linked to the expression of the cwp genes. This information is in full accordance with previous reports for fungal cells, in which GlcCer was directly linked with cell wall assembly (37) and cellular replication (38). More recently, Sonda et al. (42) reported that PPMP blocks the growth of Giardia trophozoites at a specific stage of late cytokinesis and also reduces the production of cysts. The increase in gglct-1 and cwp transcripts observed after treatment with GlcT-1 inhibitors (Fig. 6) could be due to the fact that functionally active GlcT-1 or the product of GlcT-1 may be associated with regulating transcriptional levels of these genes directly or indirectly, which means that inhibition of enzyme activity may relieve transcriptional repression. Nonetheless, more in-depth experiments should be carried out to obtain a better understanding of this phenomenon.
From the phylogenetic analyses (Fig. 7), several conclusions can be made about the origin and evolution of spt genes. The first conclusion concerns the monophyletic origin of all spt genes from a prokaryotic aminoacyl transferase precursor (the most closely related gene appears to be the 5-aminolevulinic acid synthase gene), a result strongly supported by the bootstrap value (46) at the node defining the spt subclade. In spite of the overall sequence similarity, the differences in function imply that there was divergence of the catalytic domains of bacterial aminoacyl transferases and eukaryotic SPTs. The second conclusion that can be made is that the early duplication event that gave rise to the spt-1 and spt-2 paralogs is more ancient than any of the modern protist lineages, as both paralogs are present in all of the eukaryotes assayed (only a representative subset of which were included in the phylogenetic analysis) except Entamoeba, which only has gspt-2. Taken together, these results imply that the divergent roles of SPT-1 and -2 in the metabolism of so-called “lower” protists were carried over into the SL metabolism of their more derived “higher” multicellular eukaryote relatives. Our analyses also shed light on the possible ancestry of the SPT enzyme. Data suggest that 5-aminolevulinic acid synthase falls out as the outgroup of the SPT-1/SPT-2 subclade, with 2-amino-3-ketobutyrate transferase as the next outgroup (note that the sequences identified in the BLAST search as glycine-C acetyltransferase and pyridoxal phosphate-dependent acyltransferase nest within the 2-amino-3-ketobutyrate synthase clade, which implies that they are in the same class of enzymes). 8-Amino-7-oxononanoate synthase is more distantly related to the SPT subclade than either 5-aminolevulinic acid synthase or 2-amino-3-ketobutyrate synthase. This result is not simply an artifact of the root choice (as previously noted, in unrooted trees, the 8-amino-7-oxononanoate synthase sequence is always an outgroup with respect to the other sequences). It would be of interest to determine whether the catalytic activity of the three prokaryotic aminoacyl transferases was coopted for its function in the SL pathway or if a novel catalytic function (in an entirely different region) evolved de novo.
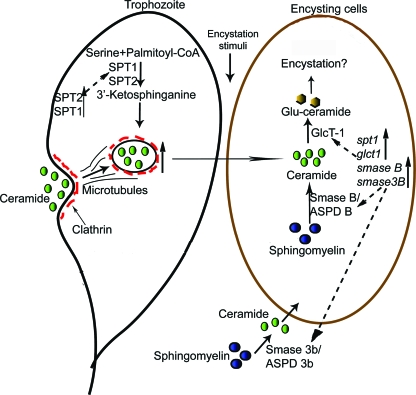
Based on the interpretation of findings presented in this study and the currently available data, we propose a comprehensive model (Fig. 8) that describes how SL metabolic genes regulate ceramide uptake and in vitro encystation by Giardia. In this model, a metabolically active trophozoite expresses the gspt genes encoding the gSPT enzymes for catalyzing the synthesis of 3-KS. 3-KS regulates ceramide uptake by interacting with actin and other endocytic machinery, as proposed previously for yeast (49). The inhibition of ceramide endocytosis by l-cycloserine and its reversal by 3-KS (Fig. 3A) support this notion. When a trophozoite undergoes encystation in the small intestine, the gglct-1 and gsmase genes are expressed as genes that encode the corresponding enzymes. We believe that stored ceramides (in the ER/perinuclear membranes [Fig. 3A]) are used as scaffolds for synthesizing GlcCer with the help of the gGlcT-1 enzyme located in the ER. The production of cystlike structures and the reduction in the total number of cysts by PPMP further support this idea (Fig. 4). To increase the pool of ceramide to synthesize GlcCer, both cytoplasmic and secreted gSmases degrade intestinal and cellular sphingomyelin to generate excess ceramide, as shown by the elevated transcription of the gsmase B and 3b genes during encystation (Fig. 2D and 2E).
FIG. 8.
Integrated model proposing the functions of SL genes in giardial encystation. The model proposes that ceramide uptake by Giardia is regulated by SPTs encoded by the gspt-1 and gspt-2 genes in nonencysting trophozoites. During encystation, the expression of gglct-1, which encodes GlcT-1, is upregulated. gGlcT-1 catalyzes the production of GlcCer by combining ceramide with glucose that is required for encystation. To ensure a steady supply of ceramide, Giardia synthesizes gSmases during encystation (encoded by gsmase B and gsmase 3b) to hydrolyze sphingomyelin to generate additional ceramide. gSmases were annotated as acid sphingomyelinase-like phosphodiesterases (ASPD) in the genome database. CoA, coenzyme A.
In summary, we demonstrated that Giardia has the ability to express selective SL genes that are involved in regulating ceramide endocytosis and encystation. Although the gene sequences for giardial enzymes are homologous to the gene sequences in other eukaryotes (including mammals), the catalytic domains could be sufficiently divergent that they could be used as targets to develop novel therapies against giardiasis.
Acknowledgments
We are thankful to everyone in the Das lab for valuable suggestions during this investigation. We also thank Gary Olsen for information on aminoacyl transferase nomenclature and evolution, Ifeanyi Nwokeabia for collaborative work on glucosyl transferase phylogenies, and Eliana Barreto-Bergter for anti-GlcCer antibody. Microscopy experiments were performed in the Analytical Cytology Facility at UTEP.
This work was supported by grant S06 GM 008012 to S.D. from the National Institutes of Health and by infrastructure development grant 5G112RR08124 to UTEP from NCRR/RCMI. T.T.D. and T.L.M. were supported by MBRS/RISE grant 2R25GM069621.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 21 April 2008.
REFERENCES
- 1.Abe, A., N. S. Radin, J. A. Shayman, L. L. Worting, R. E. Zipkin, R. Sivakumar, J. M. Ruggieri, K. G. Carson, and B. Ganem. 1995. Structural and stereochemical studies of potent inhibitors of glucosylceramide synthase and tumor cell growth. J. Lipid Res. 36611-621. [PubMed] [Google Scholar]
- 2.Adam, R. D. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14447-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 5403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshal, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 1138-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernander, R., J. E. Palm, and S. G. Svärd. 2001. Genome ploidy in different stages of the Giardia lamblia life cycle. Cell. Microbiol. 355-62. [DOI] [PubMed] [Google Scholar]
- 6.Cavalier-Smith, T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52297-354. [DOI] [PubMed] [Google Scholar]
- 7.Das, S., C. Castillo, and T. Stevens. 2001. Phospholipid remodeling/generation in Giardia: the role of the Lands cycle. Trends Parasitol. 17316-319. [DOI] [PubMed] [Google Scholar]
- 8.Das, S., T. Stevens, C. Castillo, A. Villasenõr, H. Arredondo, and K. Reddy. 2002. Lipid metabolism in mucus-dwelling amitochondriate protozoa. Int. J. Parasitol. 32655-675. [DOI] [PubMed] [Google Scholar]
- 9.Diamond, L. S., D. R. Harlow, and C. C. Cunnick. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 27487-488. [DOI] [PubMed] [Google Scholar]
- 10.Eckman, L., and F. D. Gillin. 2001. Microbes and microbial toxins: paradigms for microbial-mucosal interactions. I. Pathophysiological aspects of enteric infections with the lumen-dwelling protozoan pathogen Giardia lamblia. Am. J. Physiol. Gastrointest Liver Physiol. 280G 1-6. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5164-166. [Google Scholar]
- 12.Felsenstein, J. 2004. Inferring phylogenies. Sinauer Associates, Sunderland, MA.
- 13.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 13784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillin, F. D., D. S. Reiner, M. J. Gault, H. Douglas, S. Das, A. Wunderlich, and J. Sauch. 1987. Encystation and expression of cyst antigens by Giardia lamblia in vitro. Science 2351040-1045. [DOI] [PubMed] [Google Scholar]
- 15.Gillin, F. D., S. E. Boucher, S. S. Rossi, and D. S. Reiner. 1989. Giardia lamblia: the roles of bile, lactic acid and pH in the completion of the life-cycle in vitro. Exp. Parasitol. 69164-174. [DOI] [PubMed] [Google Scholar]
- 16.Gillin, F. D., D. S. Reiner, and J. M. McCaffery. 1996. Cell biology of the primitive eukaryote Giardia lamblia. Annu. Rev. Microbiol. 50679-705. [DOI] [PubMed] [Google Scholar]
- 17.Gottig, N., E. V. Elias, R. Quiroga, M. J. Nores, A. J. Solari, M. C. Touz, and H. D. Lujan. 2006. Active and passive mechanisms drive secretory granule biogenesis during differentiation of the intestinal parasite Giardia lamblia. J. Biol. Chem. 3018156-18166. [DOI] [PubMed] [Google Scholar]
- 18.Gulbins, E., and P. L. Li. 2006. Physiological and pathophysiological aspects of ceramide. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290R11-R26. [DOI] [PubMed] [Google Scholar]
- 19.Hanada, K. 2003. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta 1016-30. [DOI] [PubMed] [Google Scholar]
- 20.Hanada, K., T. Hara, M. Fukasawa, A. Yamaji, M. Umeda, and M. Nishijima. 1998. Mammalian cell mutants resistant to a sphingomyelin-directed cytolysin. Genetic and biochemical evidence for complex formation of the LCB1 protein with the LCB2 protein for serine palmitoyltransferase. J. Biol. Chem. 27333787-33794. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez, Y., C. Castillo, S. Roychowdhury, A. Hehl, S. B. Aley, and S. Das. 2007. Clathrin dependent pathways and the cytoskeleton network are involved in ceramide endocytosis by a parasitic protozoan, Giardia lamblia. Int. J. Parasitol. 3721-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikushiro, H., H. Hayashi, and H. Kagamiyama. 2004. Reactions of serine palmitoyltransferase with serine and molecular mechanisms of the actions of serine derivatives as inhibitors. Biochemistry 431082-1092. [DOI] [PubMed] [Google Scholar]
- 23.Jarroll, E. L., P. J. Muller, E. A. Meyer, and S. A. Morse. 1981. Lipid and carbohydrate metabolism of Giardia lamblia. Mol. Biochem. Parasitol. 2187-196. [DOI] [PubMed] [Google Scholar]
- 24.Kane, A. V., H. D. Ward, G. T. Keusch, and M. E. Pereira. 1991. In vitro encystation of Giardia lamblia: large-scale production of in vitro cysts and strain and clone differences in encystation efficiency. J. Parasitol. 77974-981. [PubMed] [Google Scholar]
- 25.Keister, D. B. 1983. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans. R. Soc. Trop. Med. Hyg. 77487-488. [DOI] [PubMed] [Google Scholar]
- 26.Kovacs, P., M. Pinter, and G. Csaba. 2000. Effect of glucosphingolipid synthesis inhibitor (PPMP and PDMP) treatment on Tetrahymena pyriformis: data on the evolution of the signaling system. Cell. Biochem. Funct. 18269-280. [DOI] [PubMed] [Google Scholar]
- 27.Lujan, H. D., M. R. Mowatt, J. T. Conrad, B. Bowers, and T. E. Nash. 1995. Identification of a novel Giardia lamblia cyst wall protein with leucine-rich repeats. Implications for secretory granule formation and protein assembly into the cyst wall. J. Biol. Chem. 829307-29313. [DOI] [PubMed] [Google Scholar]
- 28.Marti, M., A. Regos, Y. Li, E. M. Schraner, P. Wild, N. Muller, L. G. Knopf, and A. B. Hehl. 2003a. An ancestral secretory apparatus in the protozoan parasite Giardia intestinalis. J. Biol. Chem. 27824837-24848. [DOI] [PubMed] [Google Scholar]
- 29.Marti, M., Y. Li, E. M. Schraner, P. Wild, P. Kohler, and A. B. Hehl. 2003b. The secretory apparatus of an ancient eukaryote: protein sorting to separate export pathways occurs before formation of transient Golgi-like compartments. Mol. Biol. Cell 141433-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison, H. G., A. G. McArthur, F. D. Gillin, S. B. Aley, R. D. Adam, G. J. Olsen, A. A. Best, W. Z. Cande, F. Chen, M. J. Cipriano, B. J. Davids, S. C. Dawson, H. G. Elmendorf, A. B. Hehl, M. E. Holder, S. M. Huse, U. U. Kim, E. Lasek-Nesselquist, G. Manning, A. Nigam, J. E. Nixon, D. Palm, N. E. Passamaneck, A. Prabhu, C. I. Reich, D. S. Reiner, J. Samuelson, S. G. Svärd, and M. L. Sogin. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 3171921-1926. [DOI] [PubMed] [Google Scholar]
- 31.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 2434-36. [DOI] [PubMed] [Google Scholar]
- 32.Nakai, K., and P. Horton. 2007. Computational prediction of subcellular localization. Methods Mol. Biol. 390429-466. [DOI] [PubMed] [Google Scholar]
- 33.Nasmyth, K. 1996. An homage to Giardia. Curr. Biol. 61042. [DOI] [PubMed] [Google Scholar]
- 34.Nimrichter, L., E. Barreto-Bergter, R. R. Mendonça-Filho, L. F. Kneipp, M. T. Mazzi, P. Salve, S. E. Farias, R. Wait, C. S. Alviano, and M. L. Rodrigues. 2004. A monoclonal antibody to glucosylceramide inhibits the growth of Fonsecaea pedrosoi and enhances the antifungal action of mouse macrophages. Microbes Infect. 6657-665. [DOI] [PubMed] [Google Scholar]
- 35.Reiner, D. S., M. McCaffery, and F. D. Gillin. 1990. Sorting of cyst wall proteins to a regulated secretory pathway during differentiation of the primitive eukaryote, Giardia lamblia. Eur. J. Cell Biol. 53142-153. [PubMed] [Google Scholar]
- 36.Reiner, D. S., J. M. McCaffery, and F. D. Gillin. 2001. Reversible interruption of Giardia lamblia cyst wall protein transport in a novel regulated secretory pathway. Cell. Microbiol. 3459-472. [DOI] [PubMed] [Google Scholar]
- 37.Rodrigues, M. L., L. R. Travassos, K. R. Miranda, A. J. Franzen, S. Rozental W. de Souza, C. S. Alviano, and E. Barreto-Bergter. 2000. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect. Immun. 687049-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rittershaus, P. C., T. B. Kechichian, J. C. Allegood, A. H. Merrill, Jr., M. Hennig, M. C. Luberto, and M. Del Poeta. 2006. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J. Clin. Investig. 1161651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132365-386. [DOI] [PubMed] [Google Scholar]
- 40.Simpson, A. G., Y. Inagaki, and A. J. Roger. 2006. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol. Biol. Evol. 23615-625. [DOI] [PubMed] [Google Scholar]
- 41.Sogin, M. L. 1991. Early evolution and the origin of eukaryotes. Curr. Opin. Genet. Dev. 1457-463. [DOI] [PubMed] [Google Scholar]
- 42.Sonda, S., S. Štefanic′, and A. B. Hehl. 2008. A sphingolipid inhibitor induces a cytokinesis arrest and blocks stage differentiation in Giardia lamblia. Antimicrob. Agents Chemother. 52563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian, A., S. Navarro, R. Carrasco, M. Marti, and S. Das. 2000. Role of exogenous inositol and phosphatidylinositol in glycosylphosphatidylinositol synthesis of GP49 by Giardia lamblia. Biochim. Biophys. Acta 148369-80. [DOI] [PubMed] [Google Scholar]
- 44.Sun, C. H., J. M. McCaffery, D. S. Reiner, and F. D. Gillin. 2003. Mining the Giardia lamblia genome for new cyst wall proteins. J. Biol. Chem. 27821701-21708. [DOI] [PubMed] [Google Scholar]
- 45.Sutterwala, S. S., C. H. Creswell, S. Sanyal, A. K. Menon, and J. D. Bangs. 2007. De novo sphingolipid synthesis is essential for viability, but not for transport of glycosylphosphatidylinositol-anchored proteins, in African trypanosomes. Eukaryot. Cell 6454-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 244876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wells, G. B., and R. L. Lester. 1983. The isolation and characterization of a mutant strain of Saccharomyces cerevisiae that requires a long chain base for growth and for synthesis of phosphosphingolipids. J. Biol. Chem. 1010200-10203. [PubMed] [Google Scholar]
- 48.Yamashita, T., R. Wada, T. Sasaki, C. Deng, U. Bierfreund, K. Sandhoff, and R. L. Proia. 1999. A vital role for glycosphingolipid synthesis during development and differentiation. Proc. Natl. Acad. Sci. USA 969142-9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zanolari, B., S. Friant, K. Funato, C. Sutterlin, B. J. Stevenson, and H. Riezman. 2000. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 152824-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, K., M. Showalter, J. Revollo, F. F. Hsu, J. Turk, and S. M. Beverley. 2003. Sphingolipids are essential for differentiation but not growth in Leishmania. EMBO J. 176016-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuccon, A., and D. Zuccon. 2006. MrEnt v1.2. Department of Vertebrate Zoology and Molecular Systematics Laboratory, Swedish Museum of Natural History, Stockholm, Sweden.