Abstract
The rlrA pilus locus of Streptococcus pneumoniae is an example of a pathogenicity island acquired through genetic recombination. Many acquired genetic elements commandeer preexisting networks of the new organism for transcriptional regulation. We hypothesized that the rlrA locus has integrated into transcriptional regulatory networks controlling expression of virulence factors important in adhesion and invasion. To test this hypothesis, we determined the impact on pilus expression of known regulators controlling adherence, including the two-component systems CbpR/S and HK/RR03 and the transcriptional regulators of divalent cation transporters MerR and PsaR in vitro and in vivo. It was determined that the pilus locus is down-regulated by preexisting networks designed for adhesion and cation transport/response and that its regulation occurs through RlrA. The pilus locus was found to participate in invasion specifically restricted to lung epithelial cells in vitro. While expression of pili had only a small effect on virulence with an intranasal infection model, pili were critically important with an intratracheal infection model. Thus, expression of pili appears to have become integrated into the regulatory circuits for lung-specific invasion by pneumococci.
Streptococcus pneumoniae (the pneumococcus) is an important human pathogen manifesting itself in a number of diseases, including otitis media, pneumonia, sepsis, and meningitis. Worldwide the pneumococcus remains a significant cause of morbidity and mortality and is responsible for more than a million deaths annually. Normally a commensal, S. pneumoniae is an opportunistic pathogen and is able to spread from the nasopharynx to the lungs, blood, and central nervous system. During these processes, the environmental conditions encountered by the pneumococcus represent unique challenges in terms of coordinating nutrient acquisition and evading the host immune response.
Being naturally competent, the pneumococcus is able to take up foreign DNA for integration into its chromosome. While such genetic plasticity allows for great diversity, imported elements must be regulated appropriately to confer a fitness benefit (8, 31). The pilus locus is an acquired genetic element in many streptococci and other gram-positive pathogens (38). Recently a locus encoding a pilus was detected in pneumococci within a 14-kb pathogenicity island known as the rlrA islet (14, 23, 24), a locus present in about 30% of clinical strains (2). Flanked by inverted repeats indicative of a mobile genetic element (40), the IS1167 element harbors genes with homology to agents binding to extracellular matrix components (13). The pilus locus encodes the pilus subunits as well as putative sortases predicted to covalently link the pilus to the cell wall (see Fig. S1 in the supplemental material) (23). In two studies, the presence of the locus correlated with the ability of bacteria to associate with lung epithelial cells in vitro and replicate in the nasopharynx and lung but not the blood in vivo (1, 15). The associated sortases have also been shown to be required for proper assembly of the pilus and subsequent anchoring to the cell wall in group B streptococci (7).
The precise events that trigger pneumococcal pilus expression and assembly are currently unknown. The first gene in the locus, rlrA, encodes a transcriptional regulator required for pilus expression (13). Interestingly, rlrA shows a high degree of homology to the rofA/mga family of transcriptional regulators, which plays a major role in coordinating gene expression in other species of streptococci (3, 9). Signature-tagged mutagenesis and microarray analyses determined that another transcriptional regulator, MgrA, negatively regulates the rlrA islet (15, 34). Furthermore, there is evidence that divalent cation transporters and associated regulators play a role in pilus regulation, particularly in the case of manganese (20). Metalloregulators, including Fur, PerR, and MerR (29), often play vital roles in controlling metal homeostasis, a crucial factor in bacterial survival in metal-limited environments in the host. Finally, one of the most well-characterized mechanisms by which bacteria interact with the environment is through two-component systems (28). Such systems are particularly important in the streptococci, since these bacteria lack the typical alternative sigma factors for regulating gene expression. S. pneumoniae harbors 13 two-component systems that control diverse cellular processes, including various adhesins required for virulence (33). The regulatory networks themselves are complex, with some regulators showing strain-specific effects (16). It would be particularly important for adherence via pili to be coordinated with adherence by other adhesins. Bacterial regulatory networks often have a high degree of cross talk and coregulation to coordinate expression of various genes. We hypothesized that S. pneumoniae has adapted preexisting regulatory networks to control the expression of the acquired pilus locus.
MATERIALS AND METHODS
Media and growth conditions.
S. pneumoniae strains (see Table S1 in the supplemental material) were grown on tryptic soy agar (EMD Chemicals, NJ) supplemented with 3% sheep blood or in defined semisynthetic casein liquid medium (22) supplemented with 0.5% yeast extract (C+Y). Chloramphenicol (5 μg/ml), erythromycin (1 μg/ml), spectinomycin (500 μg/ml), and kanamycin (400 μg/ml) were added when appropriate. Cultures of S. pneumoniae were inoculated from frozen stock and incubated at 37°C in 5% CO2.
Primers/mutant construction.
Mutants of strains TIGR4 and TIGR4R were made by PCR-based overlap extension. Briefly, regions flanking upstream and downstream of the target gene were amplified by PCR and spliced to an antibiotic cassette. The final PCR product was transformed into the pneumococcus by conventional methods, replacing the targeted gene with the antibiotic cassette. To confirm transformation, primers outside of the transformed region were used for PCR and subsequent region sequencing. To construct Δhk03, ΔcbpS, ΔmerR, and pilus-negative mutants, each gene was replaced with the erythromycin cassette. The ΔpsaR and ΔrlrA mutants were constructed with replacement by spectinomycin. A list of the oligonucleotides used is presented in Table S2 in the supplemental material.
RNA isolation and analysis.
Bacterial RNA was harvested from mid-log-phase cultures using a Qiagen RNAeasy minikit. Microarray experiments were performed as described previously (32). Briefly, whole-genome S. pneumoniae cDNA microarrays obtained from the Pathogen Functional Genomics Resource Center (PFGRC) at The Institute for Genomic Research consisted of PCR products representing segments of the 2,131 open reading frames of TIGR4. Microarray experiments were performed by the Functional Genomics lab, Hartwell Center for Bioinformatics and Biotechnology, St. Jude Children's Research Hospital using standard protocols provided by PFGRC (http://pfgrc.tigr.org/protocols.shtml). Microarray experiments were performed in triplicate using independent RNA samples. Data represent means from triplicate experiments. Microarray data were confirmed using quantitative reverse transcription-PCR (qRT-PCR). Purified mRNA was quantitated via Nanodrop with appropriate standards. cDNA synthesis and quantitative PCR were done using the Superscript III Platinum SYBR Green two-step qRT-PCR kit (Invitrogen) on an ABI Prism 7300. The relative transcript abundance of rlrA and rrgB was normalized to that of gyrA, which served as an internal control. The qRT-PCR experiments were done in triplicate with independently isolated RNA samples.
Anti-RrgB antibody production.
Nucleotides encoding amino acids 40 to 600 of RrgB were amplified from TIGR4 DNA by PCR with oligonucleotides 463Eco and 463Xho. The PCR product was digested with EcoRI and XhoI and ligated into a prepared pET28a vector. The reaction product was transformed in Novablue competent cells (Novagen), and colonies were screened by sequencing. The construct was transformed into the BL21(DE3) expression cell system and induced overnight at 4°C with 0.07 mM isopropyl-β-d-thiogalactopyranoside. The cells were harvested, lysed for soluble protein with the Bugbuster HT reagent (Novagen), and purified over a His-Select Ni2+ column (Sigma). Five hundred micrograms of the RrgB protein was used for polyclonal antibody production in rabbits at Invitrogen.
Western blot analysis.
Logarithmically growing cells were pelleted by centrifugation and subjected to lysis in 0.1% Triton X-100. To ensure equal loading, the protein concentration was determined for each lysate via the A280 value and loaded accordingly. Duplicate gels stained with Coomassie were used to confirm measured protein concentrations to confirm equivalent loading. Lysates were run on 4 to 12% NuPAGE Bis-Tris gels (Invitrogen) for 5 to 6 h to resolve the higher-molecular-weight pilus polymers. Proteins were subsequently transferred to polyvinylidene difluoride membranes by Western blotting. Pilus proteins were detected using rabbit anti-RrgB (1:5,000) in phosphate-buffered saline (PBS)- 0.1% Tween 20-5% nonfat dry milk. Band intensities from three independent replicates were measured to obtain a quantitative measure of pilus expression.
Enzyme-linked immunosorbent assay (ELISA) analysis using equivalent numbers of bacteria, as determined by measuring CFU, was utilized to confirm the Western analysis. Strain TIGR4 and the CbpS-, HK03-, MerR-, and pilus-negative mutants were grown in C+Y to an optical density at 620 nm (OD620) of 0.5, diluted in 0.1 M carbonate buffer (pH 9.6), and transferred in serial dilutions to 96-well ELISA plates (Nunc). The plates were spun at 2,000 × g for 10 min, and the supernatant was removed. The plates were dried under a vent hood for 1 h before blocking in 10% fetal bovine serum for 2 h. Rabbit polyclonal antiserum against SPO463 (rrGB) was diluted 1:1,000 in 10% fetal bovine serum. The plates were washed three times with wash buffer (1% Tween 20, 1 mM Tris, 154 mM NaCl) and incubated with primary antibody for 1 h. The plates were washed five times and incubated with alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (Southern Biotech) (1:2,000) for 1 h. The plates were washed five times and incubated for 20 min in alkaline phosphatase yellow substrate (Sigma), and OD405 readings were taken in a Spectramax 340 plate reader (Molecular Devices).
Protein purification and promoter binding assays.
Full-length rr03, cbpR, and merR were amplified by PCR with primers GRRNde and GRREco, CbpRNde and CbpREco, and MerRNde and MerREco, respectively. The PCR products were digested with NdeI and EcoRI and cloned into expression vector pET28a (Novagen). All proteins carried the His tag at the C terminus. Clones were screened and selected by sequencing, transformed into Escherichia coli expression host BL21(DE3) competent cells, and grown on LB agar with kanamycin (25 μg/ml). An overnight liquid culture was diluted 1:25 into fresh LB media with antibiotics and grown to an OD600 of 0.5. Cultures were induced with 1 mM isopropyl-β-d-thiogalactopyranoside and shaken at 37°C for 2 h. The cells were lysed with Bugbuster HT (Novagen) and purified over a Ni2+ column (Sigma). Proteins were dialyzed overnight in PBS.
Electrophoretic mobility shift assay.
The promoter region of rlrA was amplified using primers RlrAF and RlrAR and used at a concentration of 20 fmol per reaction (see Table S2 in the supplemental material). The promoter region of L31 was used as a negative control and was amplified using oligonucleotides L31F and L31R. The DNA was biotin labeled using the Biotin 3′ End labeling kit (Pierce). Prior to labeling, the PCR products were boiled for 10 min to denature them and were incubated 1 h at room temperature after labeling for annealing. The purified proteins CbpR and RR03 were phosphorylated with 12.5 mM acetylphosphate for 30 min at 37°C (12). Binding reactions were carried out using the Lightshift electrophoretic mobility shift assay kit (Pierce) according to protocol. Briefly, increasing concentrations of protein (0 to 25 μg for RR03, 0 to 12.5 μg for CbpR, and 0 to 0.5 μg for MerR) were incubated for 20 min at room temperature in the binding reaction mixture and loaded on a prerun 6% Tris-borate-EDTA DNA retardation gel. The gel was transferred to a nylon membrane for 45 min at 100 V. The membrane was cross-linked, and the biotin-labeled DNA was detected using a chemiluminescent nucleic acid detection module (Pierce).
Promoter fusion assay.
The promoter region of the rlrA gene was cloned into the pneumococcal reporter plasmid pPP2 (11). Briefly, the promoter region of rlrA was amplified by PCR from TIGR4R chromosomal DNA using primers RlrApF1 (GCGGCATGCCACTTGTATACAATAGTATAG) and RlrApR (GCGGGATCCCGAATCTTAGTTGCATATAG). The PCR fragment, along with the pPP2 reporter plasmid, was digested with SphI and BamHI. The DNA was ligated and transformed into DH5α competent cells and plated on LB plates with tetracycline (10 μg/ml). The clones were screened by PCR and sequencing. The pPP2/RlrAp construct was transformed into the pneumococcal strains by conventional methods and grown on tryptic soy agar blood agar plates (tetracycline concentration, 3 μg/ml). Pneumococcal strains that contained the reporter construct pPP2/RlrAp were grown in C+Y to an OD620 of 0.5, and 1-ml samples were taken. The samples were pelleted and lysed for 5 min in 100 μl 0.1% Triton X-100. The lysates were used in triplicate wells with the High Sensitivity β-galactosidase assay kit (Stratagene) according to the manufacturer's protocol. Incubation time for the lysates with the substrate was 2 h.
Matrix protein adhesion assay.
Terasaki plates were coated with 0.5 μg collagen, fibronectin, or laminin (Sigma) overnight at 4°C and subsequently blocked for 3 h at 37°C with 5% bovine serum albumin. Pneumococcal cultures were grown to an OD620 of 0.5 and labeled with fluorescein isothiocyanate (1 mg/ml in 50 mM carbonate buffer, Sigma) for 30 min. The cells were washed three times with 1 ml PBS, and 1 × 105 CFU/ml were loaded per well in a volume of 10 μl. The plates were incubated for 30 min at 37°C, 5% CO2, washed four times with dPBS, and fixed with 2.5% glutaraldehyde (Sigma). Adherent fluorescent pneumococci were quantified under a fluorescence microscope (Nikon TE300).
A549 respiratory epithelial cell assays.
Adhesion and invasion properties of the mutants were assessed by several methods (27). Initially, adherence to A549 cells (ATCC) was quantitated by direct visualization of fluorescein isothiocyanate (FITC)-labeled bacteria. Pneumococcal strains were grown to an OD620 of 0.4 and resuspended for 30 min in FITC solution (1 mg/ml in carbonate buffer). The bacteria were washed three times and diluted to 1 × 107 CFU/ml in dPBS. A549 lung epithelial cells were seeded in 96-well Terasaki trays at 37°C in 5% CO2 at 80% confluence and activated for 2 h with tumor necrosis factor alpha (10 ng/ml) (27). Each well was infected for 30 min at 37°C with 1 × 105 CFU FITC-labeled bacteria. The cells were washed and fixed with 2.5% glutaraldehyde, and adherent FITC-labeled bacteria were visualized under a microscope and counted. For each experiment, four to six wells were counted per strain, and the experiment was performed three independent times.
Alternatively, adherence and invasion were independently assessed using unlabeled bacteria. A549 cells were grown in 24-well plates at 37°C in 5% CO2 to 80% confluence and activated for 2 h with tumor necrosis factor alpha (10 ng/ml). Pneumococcal cultures were grown to an OD620 of 0.5, washed, and then added to eukaryotic cells at 1 × 107 CFU/well. Three wells were used for each mutant, and the assays were repeated three to four times. For adherence assays, cells were incubated for 30 min with bacteria, a time chosen to minimize internalization of adherent bacteria. After three washes in dPBS, the cells were released from the plate with trypsin but not lysed before plating on blood agar plates. Colonies grown overnight were counted as bacteria adherent to cells. For invasion assays, cells were incubated with the bacteria for 2 h, washed three times in dPBS, and subjected to 1 h of penicillin (10 μg/ml) and gentamicin (200 μg/ml) to kill extracellular bacteria. The cells were again washed, trypsinized, and lysed with 0.025% Triton X-100. Colonies were incubated overnight on blood agar plates and counted to represent the intracellular (invasive) bacterial number.
Mouse challenge.
Virulence studies were performed as previously described (32). Exponential cultures were centrifuged, washed in sterile PBS, and resuspended at the appropriate concentration in PBS as confirmed by serial dilution and plating on blood agar plates. Female BALB/cJ mice (Jackson Laboratory, Bar Harbor, ME), age 6 weeks, were maintained in BSL2 facilities. All experiments were done under inhaled isoflurane (2.5%). Bacteria were introduced by intranasal administration of 107 CFU in 25 μl PBS. For intratracheal infections, 105 CFU in 100 μl PBS was used as the inoculum. Mice were monitored daily for signs of infection. For intratracheal infections, lungs were excised, washed in sterile PBS, and homogenized for serial dilution and enumeration of CFU. Nasal passages were lavaged and blood extracted from the tail vein at 24 and 72 h postinfection, diluted, and plated on blood agar to ascertain the extent of bacterial colonization and bacteremia.
RESULTS
Regulation of pilus expression by two-component systems.
The CbpR/S two-component regulator is encoded immediately upstream of the adhesin CbpA, and the hk/rr03 two-component system genes are upstream of the adherence-related protease CbpG (see Figure S1 in the supplemental material). To further understand the possible contributions of these two-component systems to pilus regulatory patterns, gene splicing by overlap extension was used to generate nonpolar deletion mutations in TIGR4. Clean knockouts in cbpS and hk03 were readily generated and confirmed by PCR and sequencing. To determine the effect of the deletion on global gene transcription, total RNA was subjected to microarray analysis to determine global regulatory patterns. A number of genes showed altered expression profiles in the absence of cbpS or hk03 as summarized in Tables S3 and S4 in the supplemental material.
In agreement with previous studies (25, 36, 37), cbpS deletion resulted in an overexpression of cbpA. However, in contrast to results in previous studies (25), other genes were also differentially regulated, including plcR, the hemolysin gene, and the entire locus encoding the pilus and associated sortases (see Table S3 in the supplemental material). Interestingly, the locus encoding the pilus was consistently among the most highly upregulated gene set when the ΔcbpS mutant was compared to the parental strain, TIGR4. The global transcriptional analysis of the HK/RR03 two-component system revealed that, as expected, cbpG and contiguous Sp0389 were under negative control of this regulator (see Table S4 in the supplemental material). The cation transporter encoded by Sp0729 was downregulated in the Δhk03 mutant, whereas this transporter was upregulated in the ΔcbpS strain. In addition, two putative iron transporters, encoded by Sp1062 and Sp1871, were also downregulated when hk03 was deleted. One locus that was consistently strongly upregulated was SP0461 to SP0468, a region which encodes the pilus subunits and the sortases required for their assembly.
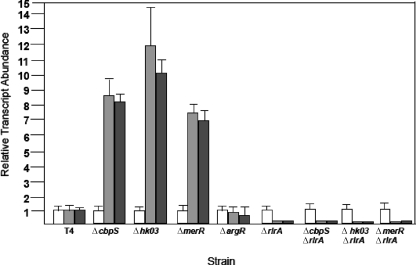
Increased expression of the pilus locus in the two-component system deletion mutants indicated negative regulation. To determine if this effect was mediated through the pilus locus regulator, RlrA, double mutants carrying mutations in the two-component systems and rlrA were constructed and analyzed for pilus locus expression. In the absence of RlrA, the overexpression of the pilus genes seen in the two-component system mutants was lost, as confirmed at the transcriptional level via qRT-PCR analysis of the rlrA and rrgB transcripts (Fig. 1). The ΔcbpS strain demonstrated an eightfold increase in rlrA and rrgB expression, respectively, while the Δhk03 strain showed a marked increase in rlrA and rggB expression of approximately 10-fold. Thus, the two-component systems appeared to negatively regulate pilus expression.
FIG. 1.
Relative transcript abundance of major pilus subunit. Quantitative PCR analysis of the transcript abundance of rlrA (gray), rrgB (black), and gyrA (white) in two-component system ΔcbpS and Δhk03 mutants and the ΔmerR metalloregulator mutant compared to results for the parental strain, TIGR4 (T4). Relative transcript abundance of rlrA and rrgB indicate more than sixfold upregulation of both genes in all mutants. RNA samples were isolated in two independent experiments and transcript abundance measured in triplicate using gyrA as a control transcript. Values represent means ± standard deviations.
Effect of cation transport on pilus expression.
We next sought to determine what other regulatory networks might converge on the pilus locus. The Psa operon comprises a manganese-specific transport system, PsaABC, along with a manganese-dependent repressor, PsaR (20). Disruption of this pathway results in a significant upregulation of the pilus locus (20). Interestingly, a number of cation transporters and response elements were identified as being differentially regulated in the ΔcbpS and Δhk03 mutants (see Tables S3 and S4 in the supplemental material). Hence, we decided to investigate whether regulation of the pilus by PsaABC/R was specific to this transporter or a characteristic of metal transporters in general. The MerR-type transcriptional regulators typically function as activators and regulate gene expression in response to a number of metals (5). Furthermore, recent studies have indicated a role for MerR (CzcD) in the regulation of divalent cation transport in S. pneumoniae (21). A deletion of merR (Sp1856) was generated by gene splicing by overlap extension and confirmed by PCR. Global gene expression of the merR knockout was then examined, with a summary of the results in Table S5 in the supplemental material. The pilus subunit genes were all upregulated in the absence of the MerR repressor. Quantitative RT-PCR confirmed an upregulation of 7.2- and 6.9-fold for rlrA and rrgB, respectively, in the ΔmerR strain compared to levels for parental TIGR4 (Fig. 1). Taken together, these data suggest that multiple regulatory pathways regulate pilus expression, and this appears to involve rlrA.
Convergence of regulatory networks on pilus locus.
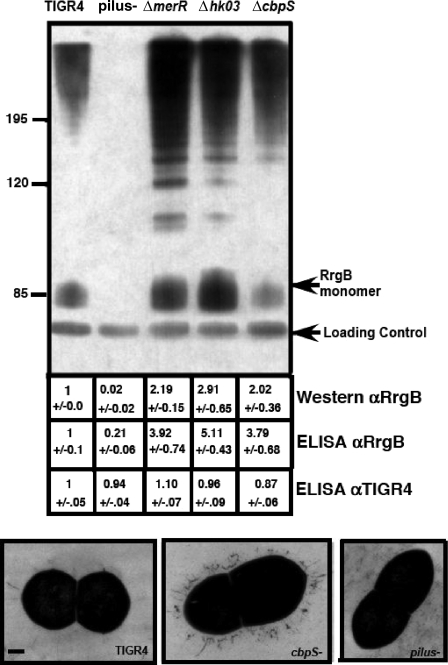
The array and quantitative RT-PCR data suggested the pilus locus might be repressed in part by two separate two-component systems involved in adhesin down-regulation and repression by metalloregulatory pathways. This was supported by Western blot analysis using antisera generated against the RrgB pilus subunit to probe whole-cell lysates of exponentially growing mutant cells. Pilus expression was greatly increased in both the ΔcbpS and Δhk03 mutants, indicating the increased transcript abundance corresponded to an increase in the total amount of pilus protein (Fig. 2). A similar pattern was observed with the ΔmerR mutant (Fig. 2). Note that the RrgB subunit was detected as both a monomer and higher-molecular-weight species that correspond to the assembled pilus (7). We also utilized all these mutants in ELISA assays probed with anti-RrgB antiserum to obtain a relative value for the increase in RrgB protein expression in whole cells (Fig. 2). All three mutants demonstrated at least twice the RrgB protein levels of the parental strain, TIGR4 (Fig. 2). To ensure all strains were equally adherent to the ELISA plate, samples were also probed with antiserum generated against TIGR4 (Fig. 2). Electron micrographs revealed a great increase in the number of pili on the surface of ΔcbpS cells compared to results for parental TIGR4 (Fig. 2). These data further support that the pilus locus is negatively controlled by a complex regulatory network encompassing both two-component systems and metalloregulatory proteins.
FIG. 2.
Multiple systems regulate pilus expression. Western blotting of whole-cell lysates using antisera generated against RrgB (αRrgB) confirmed pilus upregulation in the ΔmerR mutant and the two-component system ΔcbpS and Δhk03 mutants. A majority of the pilus was found as a high-molecular-weight ladder corresponding to the assembled pilus. A pilus-negative mutant was used as a negative control and showed no bands. A cross-reactive band running at a lower molecular weight served as a control to ensure equal loading. Numerical values below each lane indicate relative levels of expression as measured by band intensity and ELISA from at least three independent experiments. αTIGR4, anti-TIGR4. Bottom panels show representative transmission electron micrographs of TIGR4 and of the ΔcbpS and pilus-negative mutants (bar = 100 nm) showing an increase in the number of pilus structures observed on the surface of the ΔcbpS mutant compared to that for the parental strain, TIGR4. No pilus structures were observed for the pilus-negative mutant.
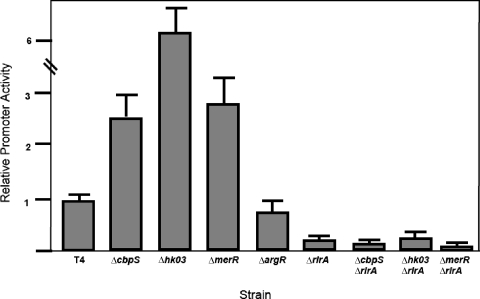
We further investigated the role of these regulators in the promoter activity controlling expression of the pilus locus. We utilized the recently described integrative plasmid pPP2 to measure the relative promoter activities in all the knockout strains generated (11). In this assay, the native beta-galactosidase gene is under control of the promoter of interest, allowing a measurement of promoter activities in various genetic backgrounds (12). Deletion of hk03, cbpS, or merR resulted in an increase in promoter activity corresponding to a minimum of double the parental levels (Fig. 3). This increase in activity was dependent upon rlrA, since in the double knockout mutants the increase in promoter activity was abrogated (Fig. 3). An irrelevant regulator knockout, ΔargR, was also included to show that the increase in promoter activity was not due to genetic manipulation of the strains. These data indicate that Rr03, CbpS, and MerR upregulate pilus expression through facilitating the promoter activity of rlrA.
FIG. 3.
Regulatory pathways converge on rlrA promoter activity. The rlrA promoter fusions were introduced into all genetic backgrounds studied. Deletion of cbpR, rr03, and merR resulted in an increase in rlrA promoter activity. The increased promoter activity was dependent upon RlrA, since the double mutants showed decrease promoter activity. T4, parental strain TIGR4.
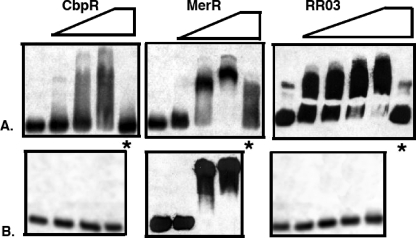
Although these regulators converged at the transcription of rlrA, there remained the alternative possibility of a downstream regulator. To differentiate between these possibilities, we investigated whether purified regulators directly bound the promoter region of rlrA. MerR, CbpR, and RR03 were expressed in E. coli and purified to homogeneity. The promoter sequence of rlrA was tested for direct binding to the regulators by electrophoretic mobility shift assay. Addition of any one of these purified regulators was able to cause a concentration-dependent shift in mobility of the rlrA promoter, indicating all these transcriptional regulators could function by direct binding to the rlrA promoter (Fig. 4). This interaction could be competitively inhibited using the unlabeled rlrA promoter (Fig. 4, lanes marked with asterisk). This interaction was found to be specific for only RR03 and CbpR, since both regulators failed to shift the L31 promoter at equivalent concentrations whereas the MerR protein resulted in an L31 shift identical to that for the rlrA promoter (Fig. 4, bottom panels). The DNA binding specificity of the MerR regulators is often governed by the binding of their cognate metal (17, 18); hence the specificity of this interaction may be improved by supplementing MerR with the cognate cation, which is currently unidentified. These data support a regulatory network in which the global regulators CbpR and RR03 negatively regulate rlrA via direct binding of the promoter region.
FIG. 4.
Regulators directly repress rlrA. Each of the regulators (CbpR, RR03, and MerR) was purified and assayed for direct binding to the rlrA promoter region via a gel shift assay. All three regulators showed a concentration-dependent shift of the migration of the rlrA promoter probe. An asterisk indicates the addition of unlabeled rlrA promoter DNA that specifically competes for the protein, causing the shift to collapse for CbpR and RR03 but not MerR.
Contribution of pilus regulators in adhesion and invasion in vitro.
The original description of the pilus locus indicated the predicted gene products showed homology to agents binding to extracellular matrix (13). The role of the pilus locus in adhering to matrix proteins was analyzed by using adhesion assays with immobilized collagen, fibronectin, and laminin. There was no significant loss of adherence to any of these components for the pilus locus deletion mutant; conversely, overexpression of pilus did not enhance adherence (see Fig. S2 in the supplemental material). Thus, homology of sequence did not appear to extend to function.
The pilus of S. pneumoniae has been suggested to play a role in adhesion to lung epithelial cells (1). Since the mutants carrying deletions in the cbpS, hk03, and merR regulatory systems exhibited pilus overexpression, we investigated the abilities of these overexpressing constructs to adhere to and invade lung epithelial cells in comparison to strains in which rlrA or all the pilus genes were deleted. To study adherence specifically, unlabeled or FITC-labeled bacteria were allowed to adhere to activated lung epithelial cells for 30 min, a time frame minimizing bacterial internalization. The attached bacteria were enumerated by visual counting with FITC-labeled bacteria or direct CFU enumeration. (see Fig. S3 in the supplemental material). No significant difference in adhesion between the mutants was observed, though the strains expressing no pili consistently showed slightly lower levels than parental TIGR4. Further, addition of N-acetylgalactosamine β1-4 galactose, a carbohydrate known to block pneumococcal adherence to lung cells, did not alter adhesion as a function of pilus expression (data not shown), indicating this moiety is not a receptor for pili.
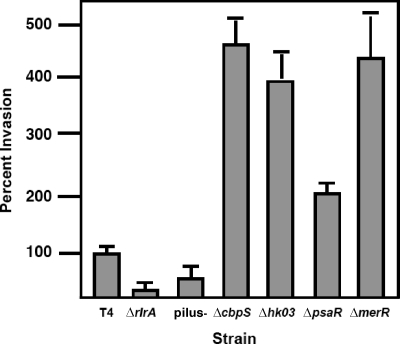
We next sought to determine if the pilus played a role in bacterial invasiveness. To assess bacterial invasion, unlabeled bacteria were incubated with activated cells for 2 h, followed by treatment with antibiotics to kill extracellular bacteria and enumeration of bacteria released from lysed A549 cells. All the strains in which the pilus was overexpressed, including the ΔcbpS, Δhk03, ΔmerR, and ΔpsaR strains, showed a dramatic increase in invasion (Fig. 5). Consistent with this data, the strains not expressing pili showed a reduced capability to invade cells (Fig. 5). These data suggested a role for the pilus in invasion into the pulmonary epithelium. This role appeared to be specific to the lung, since adherence and invasion studies using Detroit nasopharyngeal cells or microvascular endothelial cells showed no effect of the absence of the pilus (compared to results for TIGR4, ΔrlrA bacteria adhered at 116% ± 20% and invaded at 113% ± 18% with Detroit cells and adhered at 113% ± 31% and invaded at 189% ± 28% with endothelial cells.
FIG. 5.
Invasiveness of mutant constructs. Invasion of mutants affected in pilus expression was assessed in activated lung A549 cells. Data represent means and standard deviations from at least three independent experiments. Invasion levels were normalized to that of TIGR4 (T4) (100% = 150 CFU/well).
Contribution of pilus regulators to pathogenicity of S. pneumoniae.
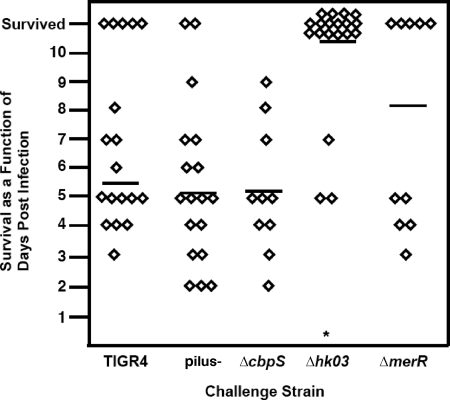
To ascertain the contribution of the effects of various regulators on pilus expression to the virulence of S. pneumoniae infection, BALB/c mice were challenged with 1 × 107 bacteria via intranasal inoculation and followed to assess survival. Titers in nasal lavage and blood samples were taken to determine the degree of colonization or bacteremia. Deletion of the pilus had no effect on survival with the intranasal model of infection; rather, there was a slight but consistent increase in virulence (Fig. 6 and 7). Of the pilus-overexpressing strains, only the Δhk03 mutant showed a decrease in mortality (Fig. 7).
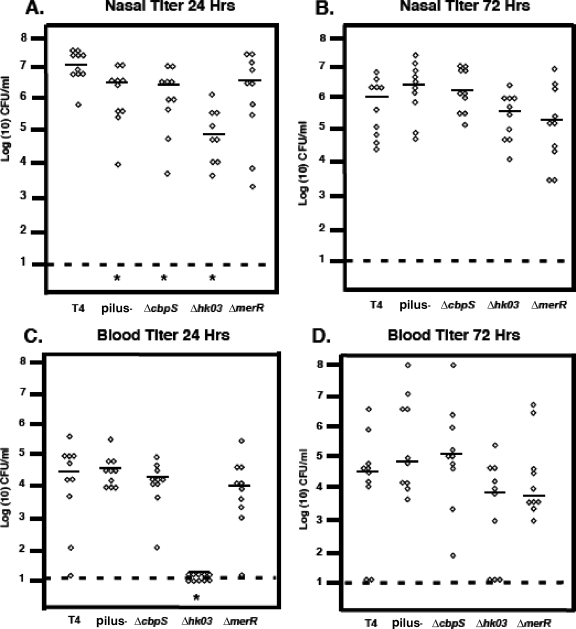
FIG. 6.
Colonization and bacteremia of mutants in mouse intranasal infection model. Bacterial titers in nasal lavage (A and B) or blood (C and D) were quantified at 24 and 72 h postchallenge. Each symbol represents titers from an individual animal. Bar, mean; dashed lines, limit of detection; *, statistically significant difference compared to results for TIGR4 (T4).
FIG. 7.
Virulence of mutants in mouse intranasal infection model. Mice were infected intranasally with 1 × 107 of the mutant construct, and survival was monitored daily for 10 days. Each symbol represents time to death for an individual mouse. The bar represents the mean.
At 24 h postinoculation, the pilus-negative, ΔcbpS, and ΔmerR constructs were all able to colonize the nasopharynx at levels comparable to that of parental TIGR4 (Fig. 6A). However, there was a pronounced defect in the ability of the hk03 mutant to colonize the nasopharynx, as evidenced by a decrease in recoverable bacteria by about 1.5 log at 24 h (Fig. 6A). Absence of the pilus locus did result in a slight, statistically significant decrease in nasal colonization. Similar results for nasal colonization were observed at a lower intranasal dose of 1 × 105, with the nonpiliated strain being slightly deficient in nasal colonization compared to TIGR4 (data not shown). These data indicate the ΔcbpS, pilus-negative, and merR mutants and to a lesser degree the Δhk03 mutant are able to successfully establish themselves in the nasal passages (Fig. 6A and B).
When the mutants were compared for progression from intranasal colonization to disease, most of the constructs, including the pilus-negative mutant and the ΔcbpS and ΔmerR mutants, showed no difference in average bacterial titers in the blood compared to the parental strain, TIGR4 (Fig. 6C). However, a dramatic difference was seen with the Δhk03 strain, which showed no detectable bacteria in the blood at 24 h (Fig. 6C). These relative values for the mutants remained constant at 48 h of infection (data not shown), while at 72 h the Δhk03 strain was eventually detected in the blood (Fig. 6D). These data indicate that of the strains examined, only the Δhk03 strain and to a lesser extent the ΔmerR strain exhibited attenuation with the intranasal model of pathogenesis (Fig. 7).
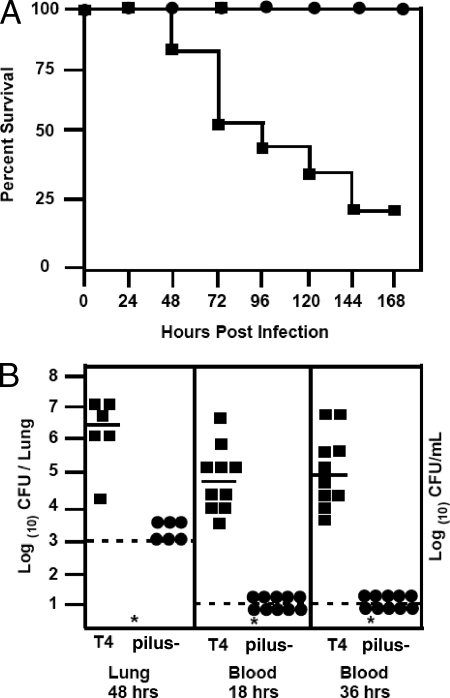
Since our intranasal data were in contrast to the findings of other studies that demonstrated a role for pili in pathogenesis (1), we further investigated the nonpiliated mutant with an intratracheal model of pathogenesis. The nonpiliated mutant showed a dramatic virulence defect when inoculated directly into the lung in terms of overall survival, with all mice inoculated with the pilus-negative mutant surviving challenge with 105 CFU, in marked contrast to results with the corresponding TIGR4 challenge (Fig. 8A). We also examined bacterial titers in infected lungs at 48 h and found the nonpiliated strain to have significantly lower bacterial loads than the parental strain, TIGR4 (Fig. 8B). This defect was also observed using a low (1 × 103) inoculum (data not shown). The observed attenuation was further confirmed by examining titers of bacteria in blood at both 18 and 36 h postinfection (Fig. 8B), which confirmed that the absence of the pilus resulted in a dramatic virulence defect when bacteria were introduced directly into the lung. This survival defect in the lung supports our earlier observation that the pilus is important for invasion of lung epithelial cells. These data indicate a niche-specific role for pili in Streptococcus pneumoniae.
FIG. 8.
Contribution of pili in a mouse intratracheal infection model. (A) Overall survival of mice challenged with 1 × 105 CFU of TIGR4 (T4) (squares) or the pilus-negative mutant (circles) via intratracheal inoculation (n = 10 mice per group). (B) Bacterial titers in the lung at 48 h and in blood at 18 and 36 h were quantified. Each symbol represents titers from an individual animal. Bar, mean; dashed lines, limit of detection; *, statistically significant difference.
DISCUSSION
Our understanding of how S. pneumoniae responds to the host environment will come from an understanding of the signals encountered by the bacterium during the course of infection and the response elicited by such stimuli. Crucial to this understanding is a clear picture of the regulatory cascades used by the pneumococcus for the control of virulence factor expression and the cross talk between such networks. Of considerable interest is how acquired genetic elements, such as the pilus locus, coopt the preexisting regulatory networks to control and integrate their own expression. To elucidate the role of some of these regulators, we utilized microarray analysis to compare transcript abundance for the pilus locus as a function of regulators known to be involved in adherence, including the two-component regulators that control the expression of the choline-binding proteins CbpA and CbpG (25, 27, 33, 37), the manganese-responsive regulator PsaR (20), and the MerR metalloregulator responsive to zinc and other metals (5, 21). Our findings clearly indicate that all of these systems repress not only known adherence factors but also pilus expression. They uniformly act on the pilus locus through the RlrA positive transcriptional regulator via direct binding of the promoter. This convergence of regulation suggests pilus functions in parallel with known mediators of host cell interactions rather than being specialized to a particular regulatory circuit.
It is of considerable interest that such diverse regulators converge on the pilus locus. Examination of the promoter region of rlrA revealed a putative PerR/Fur binding site with high homology to PerR binding sites in other streptococci, showing only a single mismatch (4). Interestingly, no sequences with even remote homology to PerR or Fur could be identified via BLAST searches of the sequenced S. pneumoniae genomes, though homologs in other streptococci were readily identified. It is not known whether the pilus locus is regulated by PerR in other streptococci that have this regulator. Streptococci also utilize two-component regulators to sense cation availability (10), and the orphan response regulator RitR of S. pneumoniae is involved in the regulation of iron transport (39). Given that a number of cation transporters were differentially regulated by the CbpR/S and HK/RR03 systems, there may be significant cross talk between these networks.
Careful attention was directed at distinguishing the role of the pilus in cellular adherence (extracellular bacteria) and invasion (intracellular bacteria). No significant defect in extracellular adherence was detected in bacteria without pili, and no increase in adherence was found with pilus overexpression. Although these regulators also regulate other adhesins which may obscure the relative contribution of the pili, we observed no adherence defect in the absence of pilus alone. In contrast, invasion directly paralleled the degree of pilus expression. Hemsley et al. reported that loss of pilus decreased adherence, and Barocchi et al. concluded that addition of pilus increased adherence (1, 15). However, both reports quantitated total cell-associated bacteria as adherent bacteria, enumerating both extracellular and intracellular bacteria. Our data suggest this increase may be specifically the result of an increased capability of piliated strains to invade host cells. It should also be noted that the contribution of the pilus to adherence to various cell lines may produce conflicting data, since the host receptor for the pilus has yet to be identified. In this regard, it appears that pilus is not targeted to carbohydrates known to influence bacterial adherence to lung cells and bacterial load in the lung (6, 19).
The relative contribution of pilus expression to host pathogenesis revealed little or no impact with the intranasal model of infection. The pilus-negative mutant showed no attenuation with our mouse model, similar to the modest change of only one log in virulence with models using competition of pilus-negative mutants and wild-type bacteria (1). However, a dramatic decrease in pathogenesis was observed upon direct inoculation of the nonpiliated strain into the lung. One reason for this attenuation may relate to the recent observation that pili in group B streptococci confer protection against killing mediated by macrophages and neutrophils, (26), phagocytes that are found in abundance in the lung. Loss of the RrgA subunit of the pilus has been shown to result in decreased virulence in pneumococcal pathogenesis with the murine sepsis model of infection (30). It should also be noted that different strains of mice were used in these two studies; hence the contribution of pilus to pathogenesis may be more apparent in certain genetic backgrounds. Another possible explanation is that in some previous studies the entire rlrA pathogenicity island, including rlrA, was deleted, whereas in our strain only the genes encoding the pilus subunits were deleted. It is possible that RlrA controls the expression of other loci besides the pilus that are involved in pathogenesis, since a number of differences in global transcription are observed in the absence of this regulator (15). Many studies investigating the role of pili in pathogenesis utilized mixed-infection models and measured competitive indexes (1); hence pili may play a more decisive role in the presence of nonpiliated streptococci or other species of bacteria.
The contribution of the pneumococcal pilus in human disease remains poorly understood. Genetic studies have indicated that expression of pili is a contributing factor to successful competition versus nonpiliated strains in the environment (35). Other studies found that the pneumococcal pilus is associated with certain capsule types but found no association with increased virulence (2). This would accord with the presence of the pilus locus in a region of diversity (RD4) that is not skewed in representation in clinical strains to either colonization or invasive disease (31). Our data indicate that the role of pili in pneumococcal pathogenesis is specific to the environmental niche of the lung. This work highlights the complex network of regulation controlling the acquired pilus locus of S. pneumoniae.
Supplementary Material
Acknowledgments
We thank TIGR for providing the S. pneumoniae microarrays and Geli Gao for excellent technical assistance with the mouse experiments. We thank Reinhold Bruckner for the generous gift of the pPP2 reporter plasmid.
We thank American Lebanese Syrian Associated Charities and NIH grant AI27913 to E.T. for funding support.
Editor: A. Camilli
Footnotes
Published ahead of print on 28 April 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 1032857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basset, A., K. Trzcinski, C. Hermos, K. L. O'Brien, R. Reid, M. Santosham, A. J. McAdam, M. Lipsitch, and R. Malley. 2007. Association of the pneumococcal pilus with certain capsular serotypes but not with increased virulence. J. Clin. Microbiol. 451684-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckert, S., B. Kreikemeyer, and A. Podbielski. 2001. Group A streptococcal rofA gene is involved in the control of several virulence genes and eukaryotic cell attachment and internalization. Infect. Immun. 69534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenot, A., K. Y. King, and M. G. Caparon. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55221-234. [DOI] [PubMed] [Google Scholar]
- 5.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27145-163. [DOI] [PubMed] [Google Scholar]
- 6.Cundell, D. R., and E. T. Tuomanen. 1994. Receptor specificity of adherence of Streptococcus pneumoniae to human type II pneumocytes and vascular endothelial cells in vitro. Microb. Pathog. 17361-374. [DOI] [PubMed] [Google Scholar]
- 7.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. C. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 601401-1413. [DOI] [PubMed] [Google Scholar]
- 8.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 9913855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granok, A. B., D. Parsonage, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site in Streptococcus pyogenes is utilized by multiple transcriptional pathways. J. Bacteriol. 1821529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gryllos, I., J. C. Levin, and M. R. Wessels. 2003. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+. Proc. Natl. Acad. Sci. USA 1004227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halfmann, A., R. Hakenbeck, and R. Brukner. 2007. A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268217-224. [DOI] [PubMed] [Google Scholar]
- 12.Halfmann, A., M. Kovacs, R. Hakenbeck, and R. Bruckner. 2007. Identification of gene directly controlled by the response regulator CiaR of Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 66110-126. [DOI] [PubMed] [Google Scholar]
- 13.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 451389-1406. [PMC free article] [PubMed] [Google Scholar]
- 14.Hava, D. L., C. J. Hemsley, and A. Camilli. 2003. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 185413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemsley, C., E. Joyce, D. L. Hava, A. Kawale, and A. Camilli. 2003. MgrA, an orthologue of Mga, acts as a transcriptional repressor of the genes within the rlrA pathogenicity islet in Streptococcus pneumoniae. J. Bacteriol. 1856640-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendriksen, W. T., N. Silva, H. J. Bootsma, C. E. Blue, G. K. Paterson, A. R. Kerr, A. de Jong, O. P. Kuipers, P. W. Hermans, and T. J. Mitchell. 2007. Regulation of gene expression in Streptococcus pneumoniae by response regulator 09 is strain dependent. J. Bacteriol. 1891383-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobman, J. L. 2007. MerR family transcriptional activators: similar designs, different specificities. Mol. Microbiol. 631275-1278. [DOI] [PubMed] [Google Scholar]
- 18.Hobman, J. L., J. Wilkie, and N. L. Brown. 2005. A design for life: prokaryotic metal-binding MerR family regulators. Biometals 18429-436. [DOI] [PubMed] [Google Scholar]
- 19.Idanpaan-Heikkila, I., P. M. Simon, D. Zopf, T. Vullo, P. Cahill, K. Sokol, and E. Tuomanen. 1997. Oligosaccharides interfere with the establishment and progression of experimental pneumococcal pneumonia. J. Infect. Dis. 176704-712. [DOI] [PubMed] [Google Scholar]
- 20.Johnston, J. W., D. E. Briles, L. E. Myers, and S. K. Hollingshead. 2006. Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect. Immun. 741171-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloosterman, T. G., M. M. van der Kooi-Pol, J. J. E. Bijlsma, and O. P. Kuipers. 2007. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+ resistance gene czcD in Streptococcus pneumoniae. Mol. Microbiol. 651049-1063. [DOI] [PubMed] [Google Scholar]
- 22.Lacks, S., and R. D. Hotchkiss. 1960. A study of the genetic material determining enzyme in the pneumococcus. Biochim. Biophys. Acta 39508-517. [DOI] [PubMed] [Google Scholar]
- 23.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in Group B Streptococcus. Science 309105. [DOI] [PubMed] [Google Scholar]
- 24.LeMieux, J., D. L. Hava, A. Basset, and A. Camilli. 2006. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect. Immun. 742453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma, Z., and J. R. Zhang. 2007. RR06 activates transcription of spr1996 and cbpA in Streptococcus pneumoniae. J. Bacteriol. 1892497-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maisey, H. C., D. Quach, M. E. Hensler, G. Y. Liu, R. L. Gallo, V. Nizet, and K. S. Doran. 15 January 2008, posting date. A group B streptococcal pilus protein promotes phagocyte resistance and systemic virulence. FASEB J. [Epub ahead of print.] doi: 10.1096/fj.07-093963. [DOI] [PMC free article] [PubMed]
- 27.Mann, B., C. Orihuela, J. Antikainen, G. Gao, J. Sublett, T. K. Korhonen, and E. Tuomanen. 2006. Multifunctional role of choline binding protein G in pneumococcal pathogenesis. Infect. Immun. 74821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCluskey, J., J. Hinds, S. Husain, A. Witney, and T. J. Mitchell. 2004. A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. Mol. Microbiol. 511661-1675. [DOI] [PubMed] [Google Scholar]
- 29.Moore, C. M., and J. D. Helmann. 2005. Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 8188-195. [DOI] [PubMed] [Google Scholar]
- 30.Nelson, A. L., J. Ries, F. Bagnoli, S. Dahlberg, S. Rounioja, J. Tschop, E. Morfeldt, I. Ferlenghi, M. Hilleringmann, D. Holden, R. Rappuoli, S. Normark, M. A. Barocchi, and B. Henriques-Normark. 2007. RrgA is a pilus-associated adhesin and virulence factor in S. pneumoniae. Mol. Microbiol. 66329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obert, C., J. Sublett, D. Kaushal, E. Hinojosa, T. Barton, E. I. Tuomanen, and C. J. Orihuela. 2006. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect. Immun. 744766-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orihuela, C. J., J. N. Radin, J. E. Sublett, G. Gao, D. Kaushal, and E. I. Tuomanen. 2004. Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 725582-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paterson, G. K., C. E. BLue, and T. J. Mitchell. 2006. Role of two-component systems in the virulence of Streptococcus pneumoniae. J. Med. Microbiol. 55355-363. [DOI] [PubMed] [Google Scholar]
- 34.Polussi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 665620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sjostrom, K., C. Blomberg, J. Fernebro, J. Dagerhamn, E. Morfeldt, M. A. Barocchi, S. Browall, M. Moschioni, M. Andersson, F. Henriques, B. Albiger, R. Rappuoli, S. Normark, and B. Henriques-Normark. 2007. Clonal success of piliated penicillin nonsusceptible pneumococci. Proc. Natl. Acad. Sci. USA 10412907-12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Standish, A. J., U. H. Streher, and J. C. Paton. 2005. The two-component signal transduction system RR06/HK06 regulates expression of cbpA in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 1027701-7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Standish, A. J., U. H. Stroher, and J. C. Paton. 2007. Pneumococcal two-component signal transduction system RR/HK06 regulates cbpA and pspA by two distinct mechanisms. J. Bacteriol. 1895591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Telford, J. L., M. A. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in Gram-positive pathogens. Nat. Rev. Microbiol. 4509-519. [DOI] [PubMed] [Google Scholar]
- 39.Ulijasz, A. T., D. R. Andes, J. D. Glasner, and B. Weisblum. 2004. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J. Bacteriol. 18681223-88136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, L., F. M. Hue, and D. A. Morrison. 1995. Characterization of IS1167, a new insertion sequence in Streptococcus pneumoniae. Plasmid 33127-138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.