Abstract
Type 1 pili mediate binding, invasion, and biofilm formation of uropathogenic Escherichia coli (UPEC) in the host urothelium during urinary tract infection (UTI) via the adhesin FimH. In this study, we characterized the molecular basis of functional differences between FimH of the UPEC isolate UTI89 and the Klebsiella pneumoniae cystitis isolate TOP52. Type 1 pili characteristically mediate mannose-sensitive hemagglutination of guinea pig erythrocytes. Although the adhesin domain of K. pneumoniae TOP52 FimH (FimH52) is highly homologous to that of E. coli, with an identical mannose binding pocket and surrounding hydrophobic ridge, it lacks the ability to agglutinate guinea pig erythrocytes. In addition, FimH-dependent biofilm formation in K. pneumoniae is inhibited by heptyl mannose, but not methyl mannose, suggesting the need for contacts outside of the mannose binding pocket. The binding specificity differences observed for FimH52 resulted in significant functional differences seen in the pathogenesis of K. pneumoniae UTI compared to E. coli UTI. Infections in a murine model of UTI demonstrated that although the K. pneumoniae strain TOP52 required FimH52 for invasion and IBC formation in the bladder, FimH52 was not essential for early colonization. This work reveals that a limited amount of sequence variation between the FimH of E. coli and K. pneumoniae results in significant differences in function and ability to colonize the urinary tract.
Bacterial adherence to host mucosal surfaces is often an important first step in the infection process. This is especially true in the case of urinary tract infections (UTIs) (59). It is estimated that half of all women will experience at least one UTI in their lifetime (49), the vast majority of which are caused by uropathogenic Escherichia coli (UPEC) and other Enterobacteriaceae (48). An essential step in UPEC infection of the bladder is adherence to the host urothelial surface via type 1 pili (2, 27, 29). Type 1 pili are assembled via the chaperone/usher pathway (3, 30, 53). They are adhesive hair-like fibers consisting of cylindrical pilus rods composed of FimA pilin subunits and small-tip fibrillae composed of FimF, FimG, and the adhesin FimH (6, 31). The FimH adhesin recognizes mannosylated uroplakins and β-1 and α-3 integrin receptors on the luminal surface of bladder urothelial cells (17, 29, 63). Binding of UPEC to host cells induces a cascade of signaling events that ultimately leads to bacterial internalization and the formation of biofilm-like intracellular bacterial communities (IBCs) (1, 17, 22, 32, 39, 43, 51). IBC formation is also dependent on type 1 pili (62). Ultimately bacteria disperse from this intracellular niche and progress to infect other urothelial cells.
Type 1 piliated bacteria have historically been characterized by their ability to agglutinate guinea pig red blood cells (RBCs) in a mannose-sensitive manner (14, 15, 52). This mannose-sensitive hemagglutination (MSHA) phenotype of E. coli expressing type 1 pili requires the FimH adhesin. FimH consists of two domains: an amino-terminal adhesin domain (AD; receptor binding domain) and a carboxy-terminal pilin domain (PD) (8, 29, 31). FimH recognizes mannosylated glycoproteins, including those present on the host urothelium through its AD. FimH-mediated adhesion can be inhibited by d-mannose or oligosaccharides containing terminal mannose residues (5, 19-21). Additionally, it has been demonstrated that the FimH AD binds more tightly to α-d-mannosides with longer alkyl chains. Heptyl mannose was found to have the highest affinity for FimH (5). In animal models, neutralization of the adhesin by FimH-specific antibodies protects from UPEC cystitis (35, 36). X-ray crystal structures of FimH reveal a highly conserved mannose binding pocket at the tip of the FimH AD surrounded by a distal hydrophobic ridge (8, 29). Minor sequence differences in E. coli FimH, many of which are not located in close proximity to the mannose binding pocket, have been found to correlate with differential binding phenotypes (54-56).
Klebsiella pneumoniae is the second leading cause of gram-negative UTI but is a much less prevalent etiologic agent than UPEC. K. pneumoniae genes encode numerous chaperone/usher pili, including type 1 pili and type 3 pili (23). While type 1 pili have historically been defined by their MSHA phenotype, type 3 pili display a mannose-resistant hemagglutination (MRHA) with tannin-treated RBCs (47). Type 1 pili of K. pneumoniae are highly homologous to those of UPEC (23) and have been previously implicated in UTI pathogenesis (18, 40). The fim operon of K. pneumoniae, encoding type 1 pili, contains a terminal fimK gene, not present in UPEC, which plays a role in suppressing the expression of type 1 pili (50). K. pneumoniae binds, invades, and forms IBCs within host urothelial cells, albeit less efficiently than UPEC in the murine cystitis model. Similar to UPEC, K. pneumoniae also expresses type 1 pili within these IBCs (50). In this study, we discovered that type 1-piliated K. pneumoniae cells are unable to mediate hemagglutination of guinea pig erythrocytes despite the presence of wild-type FimH containing an identical mannose binding pocket to E. coli FimH. We analyzed functional and structural differences in FimH of the K. pneumoniae strain TOP52 and the effects of these differences on UTI pathogenesis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A complete list of bacterial strains and plasmids used in this study can be found in Table 1. The clinical strains used include UTI89, a UPEC cystitis isolate (43), and TOP52 1721 (abbreviated TOP52 in this article), a K. pneumoniae cystitis isolate (50). Bacteria were cultured at 37°C in Luria-Bertani (LB) broth containing, as appropriate, 20 μg/ml chloramphenicol and 0.4% arabinose as indicated.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| UTI89 | UPEC cystitis isolate | 43 |
| UTI89 ΔfimH | Knockout of fimH in UTI89 | This study |
| TOP52 1721 | K. pneumoniae cystitis isolate | 50 |
| TOP52 ΔfimH | Knockout of fimH in TOP52 1721 | This study |
| TOP52 ΔfimK | Knockout of fimK in TOP52 1721 | 50 |
| Plasmids | ||
| pBAD33 | Empty expression vector; Para Cmr | 24 |
| pfimX | fimXUTI89 expression vector; Para Cmr | 25 |
| pfimH89 (pfimH-AD89PD89) | fimHUTI89 expression vector; Para Cmr | This study |
| pfimH-AD89PD52 | fimH chimera expression vector; Para Cmr | This study |
| pfimH-AD52PD89 | fimH chimera expression vector; Para Cmr | This study |
| pfimH52 (pfimH-AD52PD52) | fimHTOP52 expression vector; Para Cmr | This study |
K. pneumoniae TOP52 and E. coli UTI89 mutant construction and complementation.
A targeted deletion of fimH in the K. pneumoniae isolate TOP52 was constructed with the pKOV vector as described previously (37). Flanking sequences of approximately 1,000 bp on each side of the targeted gene were amplified with the indicated primers (Table 2) and cloned into pKOV. Potential knockouts were screened by PCR, and the knockout region was sequenced. Growth curves were performed for mutant strains and showed no differences in growth compared to the wild type.
TABLE 2.
Primer sequences
| Name | Use | Sequence |
|---|---|---|
| NotIFimHF1 | Amplify 1-kb K. pneumoniae fimH 5′ flanking region | 5′-CGGTAACGCGGCCGCGGCGAATCGAAGGATTTTACT-3′ |
| BamHIFimHR2 | Amplify 1-kb K. pneumoniae fimH 5′ flanking region | 5′-AACGGATCCAAAGGACCAGGCGTTCATC-3′ |
| BamHIFimHF3 | Amplify 1-kb K. pneumoniae fimH 3′ flanking region | 5′-AACGGATCCACGCGATCTTCACCAATACC-3′ |
| SalIFimHR4 | Amplify 1-kb K. pneumoniae fimH 3′ flanking region | 5′-AACGTCGACGTCTGGGGGTGAAGTACCTG-3′ |
| FimHCheckF1 | Check TOP52 ΔfimH | 5′-GGACAGCACCGGCTATTACA-3′ |
| FimHCheckR2 | Check TOP52 ΔfimH | 5′-GGGATCGTCAGGGAGATACA-3′ |
| UTI89fimH KO F | Knock out fimH in E. coli UTI89 | 5′-ATGAAACGAGTTATTACCCTGTTTGCTGTACTGCTGATGGGCTGGTCGGTAGTGTAGGCTGGAGCTGCTTC-3′ |
| UTI89fimH KO R | Knock out fimH in E. coli UTI89 | 5′-TTATTGATAAACAAAAGTCACGCCAATAATCGATTGCACATTCCCTGCAGTCATATGAATATCCTCCTTAG-3′ |
| UTI89fimHcheckF | Check UTI89 ΔfimH | 5′-CAATCAGCGCACTTCCCGTTACAGG-3′ |
| UTI89fimHcheckR | Check UTI89 ΔfimH | 5′-CTCGAATTATAAACAACCCGCGGCG-3′ |
| 2KlebphaseF | Amplify fimS region for phase assay | 5′-GGGACAGATACGCGTTTGAT-3′ |
| 2KlebphaseR | Amplify fimS region for phase assay | 5′-GGGACAGATACGCGTTTGAT-3′ |
| ADecoliF | Amplify E. coli fimH adhesin domain | 5′-AATTCCATGGGATGAAACGAGTTATTACCCTGTTTGCTG-3′ |
| ADecoliR | Amplify E. coli fimH adhesin domain | 5′-TCGCAGCCGCCAGTGGGCACCACCAC-3′ |
| PDecoliF | Amplify E. coli fimH pilin domain | 5′-GTGGTGGTGCCCACTGGCGGCTGCGA-3′ |
| PDecoliR | Amplify E. coli fimH pilin domain | 5′-AATTGGTACCATTGATAAACAAAAGTCACGCCAATAATCG-3′ |
| ADklebF | Amplify K. pneumoniae fimH adhesin domain | 5′-AATTCCATGGGATGATGAAAAAAATAATCCCCCTGTTCACC-3′ |
| ADklebR | Amplify K. pneumoniae fimH adhesin domain | 5′-TCGCAGCCGCCGGTGGGGACCACCAC-3′ |
| PDklebF | Amplify K. pneumoniae fimH pilin domain | 5′-GTGGTGGTCCCCACCGGCGGCTGCGA-3′ |
| PDklebR | Amplify K. pneumoniae fimH pilin domain | 5′-AATTGGTACCCATTGATAGACAAAGGTGATGCCGATG-3′ |
| pBADF | Check pBAD33 clones | 5′-TATCGCAACTCTCTACTGTTTCTCCA-3′ |
| pBADR | Check pBAD33 clones | 5′-CTGTATCAGGCTGAAAATCTTCTCTCA-3′ |
UTI89 ΔfimH was constructed using the red recombinase method as previously described (10, 44), with pKD4 as a template and the primers indicated (Table 2) followed by expression of the FLP recombinase to eliminate the kanamycin cassette. PCR using flanking primers was used to confirm the deletion.
For complementation studies, the ADs and PDs of both UTI89 fimH and TOP52 fimH were amplified using the primers indicated (Table 2). Subscripts 89 and 52 were used to indicate a given domain was from UTI89 or TOP52, respectively. Single ADs and PDs were added together as templates in a PCR to create a full-length fimH gene that was subsequently cloned into the arabinose-inducible pBAD33 vector (abbreviated pBAD). The four permutations of the ADs and PDs yielded fimH complementation vectors pAD89PD89 (pfimH89), pAD89PD52, pAD52PD89, and pAD52PD52 (pfimH52). All constructs were verified and sequenced using pBAD plasmid primers.
HAs.
Hemagglutination assays (HAs) were performed with guinea pig RBCs (optical density at 640 nm [OD640] of 2.0; Colorado Serum Company) as previously described using serial dilutions in microtiter plates with and without the addition of 100 mM methyl α-d-mannopyranoside (28).
Biofilm assays.
Bacteria were grown in LB broth in wells of microtiter plates in the presence of 0.01% arabinose and either no mannose, 1 mM methyl mannose, 100 mM methyl mannose, or 1 mM heptyl mannose. After 48 h of growth at room temperature, wells were rinsed and then stained with crystal violet, and biofilms were quantified as previously described (46).
Modeling of K. pneumoniae FimH.
K. pneumoniae TOP52 FimH was modeled onto the X-ray crystal structure of E. coli FimH from the J96 isolate FimC-H complex structure (PDB identification 1KLF) (29) using the protein structure threading program Phyre (4). The resulting model was compared to the J96 structure and UTI89 amino acid sequence. (A structure for UTI89 FimH has not been solved to date, and J96 FimH differs by only 4 amino acids [aa] from UTI89 FimH.) Figures were rendered in the molecular modeling program Pymol (11).
Mouse infections.
Bacterial strains were used to inoculate 8-week-old female C3H/HeN mice (National Cancer Institute) by transurethral catheterization as previously described (42). Twenty-five-milliliter static cultures were inoculated from freezer stocks and grown at 37°C for 18 h and then subcultured 1:250 into 25 ml fresh medium. These cultures were then grown statically at 37°C for 18 h and centrifuged for 5 min at 5,800 rpm, and the resultant pellet was resuspended in phosphate-buffered saline (PBS) and diluted to approximately 2 × 108 CFU/ml. Fifty microliters of this suspension was used to infect each mouse with an inoculum of 1 × 107 to 2 × 107 CFU. All studies were approved by the Animal Studies Committee at Washington University School of Medicine.
Organ titers, gentamicin protection assays, and IBC enumeration.
To quantify bacteria present in mouse organs, bladders and kidneys were aseptically harvested at the indicated times postinfection, homogenized in PBS, serially diluted, and plated onto LB agar plates. Luminal and intracellular bacteria were quantified using an ex vivo gentamicin protection assay as previously described (33). For ex vivo enumeration of IBCs, infected bladders were harvested at 1 h postinfection, bisected, splayed, washed with PBS, fixed with 3% paraformaldehyde for 1 h at room temperature and lacZ stained as previously described (33). IBCs were visualized and counted using an Olympus SZX12 dissecting microscope (Olympus America).
fim operon phase assay.
To determine the orientation of the type 1 pilus phase-variable promoter switch (fimS) in UTI89 ΔfimH, a phase assay was performed as previously described (58). Briefly, PCR primers were used to amplify a 589-bp DNA region including fimS. The PCR product was then digested with the restriction endonuclease HinfI (New England Biolabs) and was separated on a 2.5% agarose gel. A phase-on switch results in products of 489 and 70 bp and a phase-off switch results in products of 359 and 200 bp.
Immunoelectron microscopy.
Bacteria were prepared as described above for mouse infection, fixed with 1% paraformaldehyde for 10 min, and absorbed onto Formvar/carbon-coated copper grids for 2 min. Grids were washed two times with PBS, blocked with 1% fetal bovine serum for 5 min, and incubated with rabbit anti-FimH antibody (1:100) for 30 min at room temperature. The rabbit anti-FimH antibody was raised against the FimH adhesin domain (positions 1 to 159, T2) of E. coli J96 (26) (SigmaGenesis). Grids were subsequently washed two times with PBS, blocked with 1% fetal bovine serum for 5 min, and incubated with 18-nm colloidal gold particle-conjugated anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories) for 30 min at room temperature. Following two PBS washes, grids were rinsed in distilled water and stained with 1% aqueous uranyl acetate (Ted Pella, Inc.) for 1 min. Excess liquid was gently wicked off, and grids were air dried. Samples were viewed on a JEOL 1200EX transmission electron microscope (JEOL USA) at 80 kV accelerating voltage.
Statistical analysis.
Continuous variables were compared using the Mann-Whitney U test since these variables were not normally distributed. All tests were two-tailed, and a P value of <0.05 was considered significant. Analyses were performed using GraphPad Prism (version 4.03) and SAS (version 9.0).
Nucleotide sequence accession number.
The TOP52 fimH nucleotide sequence has been deposited in the GenBank database under accession no. EU327536.
RESULTS
Type 1-piliated K. pneumoniae TOP52 is hemagglutination negative.
In contrast to UPEC, statically passaged K. pneumoniae TOP52 produced no detectable hemagglutination of guinea pig RBCs despite expression of type 1 pili (Table 3). The MSHA titer of the UPEC strain UTI89 was 1:512. Deletion of fimH abolished the ability of UTI89 ΔfimH to produce MSHA. UTI89 ΔfimH produced a low MRHA titer of 1:4, unlike UTI89. Wild-type K. pneumoniae TOP52 did not agglutinate guinea pig erythrocytes. Deletion of fimH to create TOP52 ΔfimH was also negative for hemagglutination. Recently, we discovered that deletion of fimK, a gene unique to Klebsiella fim gene clusters, resulted in a hyper-type 1-piliated phenotype (50). The hyperpiliated TOP52 ΔfimK was also hemagglutination negative. The fimX recombinase has been shown to have fimB-like properties (7, 25), and its overexpression results in increased expression of type 1 pili in both E. coli (25) and K. pneumoniae (50). The hyperpiliated TOP52/pfimX was also hemagglutination negative. Thus, type 1-piliated K. pneumoniae TOP52 is unable to mediate MSHA.
TABLE 3.
FimH52-specific inability of K. pneumoniae TOP52 to agglutinate guinea pig RBCs
| Strain | HA titer (1:2x) in guinea pig RBCsa:
|
|
|---|---|---|
| Without mannose | With mannose | |
| UTI89 | 9 | 2 |
| UTI89 ΔfimH | 2 | 2 |
| TOP52 | 0 | 0 |
| TOP52 ΔfimH | 0 | 0 |
| TOP52 ΔfimK | 0 | 0 |
| TOP52/pBAD | 0 | 0 |
| TOP52/pfimX | 0 | 0 |
| TOP52 ΔfimH/pBAD | 0 | 0 |
| TOP52 ΔfimH/pfimH52 | 0 | 0 |
| TOP52 ΔfimH/pfimH89 | 4 | 0 |
HA titer data are representative of three independent experiments.
FimH52 and FimH89 are highly similar in amino acid sequence and predicted structure.
To further investigate the functional differences of K. pneumoniae TOP52 type 1 pili, we sequenced fimH52 (GenBank accession no. EU327536) and compared it to other known K. pneumoniae FimH sequences and the sequence of E. coli UTI89 FimH (FimH89). The FimH52 amino acid sequence shares 100% identity with the FimH adhesin domain of K. pneumoniae strain IA565 (23), 99.6% amino acid identity with the FimH of K. pneumoniae ATCC 700721 strain (41), and 85.3% amino acid identity with K. pneumoniae strain IA551 (16). FimH52 has 86.4% amino acid identity to FimH of E. coli UTI89 (FimH89) (Fig. 1A) and maintains the general bidomain composition of E. coli FimH with an amino-terminal adhesin domain (amino acids [aa] 1 to 157) and a carboxy-terminal pilin domain (aa 161 to 279) separated by a short linker region.
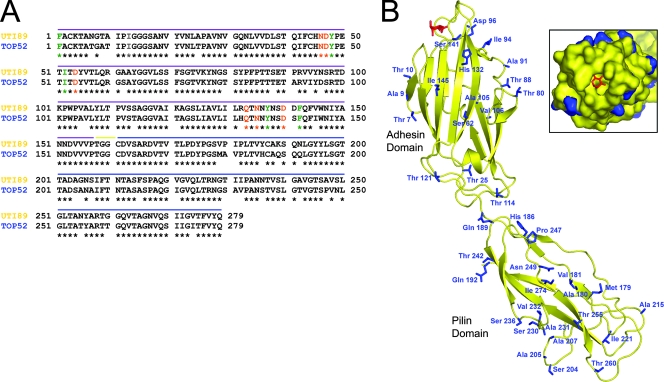
FIG. 1.
FimH52 and FimH89 are highly conserved in sequence and structure. (A) The FimH amino acid sequences of E. coli UTI89 and K. pneumoniae TOP52 are shown. Residues known to interact with mannose (orange) and form the hydrophobic ridge (green) are fully conserved. The purple line denotes sequence of the AD, the blue line denotes sequence of the PD, and the yellow line denotes the short linker region. Amino acid numbers refer to the mature protein without signal sequence. (B) Structure of E. coli FimH (yellow) (from the J96 strain FimC-H complex; PDB identification no. 1KLF) bound to mannose (red) overlaid with a threaded model of K. pneumoniae TOP52 FimH. Side chains of TOP52 amino acids that vary from the UTI89 sequence are shown as blue sticks. (Inset) Space-filling model of a view into the mannose binding pocket (same colors described above). This shows that all residues in direct contact with the mannose moiety and those that form the hydrophobic ridge are fully conserved between E. coli UTI89 and K. pneumoniae TOP52.
We threaded K. pneumoniae FimH onto the X-ray crystal structure of FimH from the complex structure of FimC-H from the J96 E. coli isolate (29). We then overlaid J96 FimH and TOP52 FimH and compared the positions and identities of amino acid differences in FimH89 and FimH52 (Fig. 1B). This comparison assumes that residues conserved between J96 FimH and UTI89 FimH have the same conformation as shown in the three-dimensional J96 FimH structure. There are only four amino acid differences between FimH of these two strains. Seventeen AD amino acid differences and 21 PD amino acid differences exist between FimH89 and FimH52. Interestingly, FimH52 displays full conservation of the residues known to interact with mannose in the mannose binding pocket (orange) and those that form the surrounding hydrophobic ridge (green in Fig. 1). Residue differences exist in areas adjacent to the receptor binding site and in other more distal parts of the molecule, which may together alter the molecular details of the interaction with mannose. Two differences in FimH52 primary sequence exist in residues adjacent to known mannose-binding residues (His132 and Ser141, changed from Arg and Asp, respectively, in FimH89). The threaded model FimH52 suggests that these residues would lie ∼8.5 Å away from the bound mannose moiety. Arg132 and Asp141 form a salt bridge in E. coli FimH that helps stabilize the structure of the FG loop that contains mannose binding residues Gln133, Asn135, and Asp140 and forms part of the hydrophobic ridge. Arg132 NH1 also makes two hydrogen bonds to Gln59 OE1 and Glu89 OE1. In FimH52, His132 is only able to make a single hydrogen bond with Glu89 OE2. Differences in these and other residues may help explain the inability of K. pneumoniae TOP52 to agglutinate guinea pig RBCs.
The inability of K. pneumoniae TOP52 to agglutinate guinea pig RBCs is specific to the adhesin domain of FimH52.
Although all residues involved in direct interactions with the mannose moiety and all those in the surrounding hydrophobic ridge are identical between FimH52 and FimH89, nearly 14% of amino acids differ between the two proteins. We hypothesized that if this variation in FimH sequence accounts for the inability to agglutinate guinea pig erythrocytes, then complementation of TOP52 ΔfimH with fimH cloned from E. coli UTI89 (fimH89), should restore the MSHA phenotype. Thus, TOP52 ΔfimH was complemented with the fimH gene of K. pneumoniae TOP52 (fimH52) or fimH89 on inducible plasmids. While the TOP52 ΔfimH/pBAD vector control and TOP52 ΔfimH/pfimH52 were hemagglutination negative, TOP52 ΔfimH/pfimH89 had an MSHA titer of 1:16 (Table 3). Thus, FimH89 is able to participate in type 1 pilus biogenesis with the Fim proteins of K. pneumoniae TOP52 and confers a gain of MSHA function.
To test the expression of exogenous fimH in the UTI89 ΔfimH background, phase assays were conducted analyzing the phase-variable promoter switch of type 1 pili (Fig. 2). The fim operon of wild-type E. coli UTI89 was primarily phase on after static growth; however, loss of fimH in UTI89 ΔfimH and the UTI89 ΔfimH/pBAD vector control resulted in bacterial populations that were primarily in the phase-off orientation. Complementation with either pfimH89 or pfimH52 did not result in a robust off-to-on switch as the populations remained primarily phase off with similarly low levels of piliated bacteria. However, enough phase-on bacteria were present to detect an MSHA titer with pfimH89 complementation (Table 3).
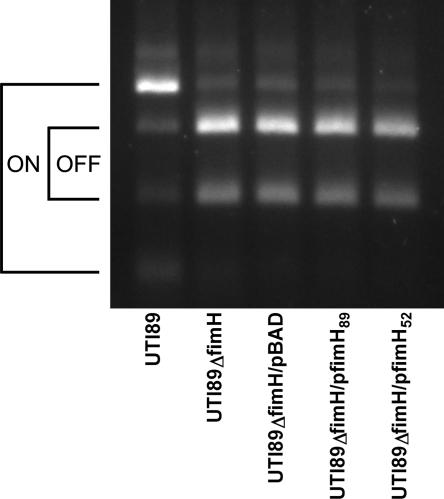
FIG. 2.
The fim operon of UTI89 ΔfimH is primarily in the phase-off orientation. Phase assays of the fimS invertible promoter region of the fim operon were done for E. coli UTI89, UTI89 ΔfimH, UTI89 ΔfimH/pBAD, UTI89 ΔfimH/pfimH89, and UTI89 ΔfimH/pfimH52. UTI89 was largely phase on, while the UTI89 ΔfimH strains were all primarily phase off despite complementation.
FimH52 and FimH89 function was investigated further by constructing FimH chimeras. We used the chimeras to complement fimH-knockout strains and then examined the final pilus assembly on each strain by immunoelectron microscopy. The ADs and PDs of each strain were amplified and expressed in different combinations on the arabinose-inducible pBAD33 vector. This resulted in four different fimH construct-expressing plasmids: pAD89PD89 (pfimH89), pAD89PD52, pAD52PD89, and pAD52PD52 (pfimH52).
The incorporation of pfimH89, pAD89PD52, pAD52PD89, and pfimH52 into pili was confirmed by immunoelectron microscopy using anti-FimH antibodies. Consistent with the phase switch being primarily off in these complementations, the majority of bacteria were bald without noticeable pili. However, similar subpopulations of bacteria existed in each sample that were moderately piliated and immunolabeling at the tips of pili was observed for UTI89 ΔfimH complemented with each construct (Fig. 3). The low level of type 1 pilus expression explains the inability to fully complement UTI89ΔfimH to wild-type E. coli UTI89 MSHA titers.
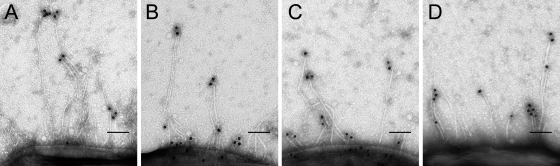
FIG. 3.
fimH constructs in UTI89 ΔfimH are expressed in some bacteria and localized at the tips of pili. Immunoelectron microscopy using an anti-FimH antibody was performed against UTI89 ΔfimH/pfimH89 (A), UTI89 ΔfimH/pAD89PD52 (B), UTI89 ΔfimH/pAD52PD89 (C), and UTI89 ΔfimH/pfimH52 (D). While the majority of bacteria did not appear to be expressing type 1 pili, piliated bacteria could be found in all four samples. Piliated bacteria displayed FimH immmunostaining at the distal tips of pili.
TOP52 ΔfimH was also complemented with each FimH chimera and the MSHA titers for all strains were analyzed (Table 4). Wild-type UTI89 produced an MSHA titer of 1:512, while wild-type TOP52 was hemagglutination negative. UTI89 ΔfimH, UTI89 ΔfimH/pBAD, TOP52 ΔfimH, and TOP52 ΔfimH/pBAD all lacked the ability to agglutinate guinea pig RBCs. UTI89 ΔfimH/pfimH89 and UTI89 ΔfimH/pAD89PD52 both produced an MSHA titer of 1:32, while UTI89 ΔfimH/pAD52PD89 and UTI89 ΔfimH/pfimH52 did not produce a hemagglutination titer. This trend was recapitulated in the TOP52 ΔfimH background. While TOP52 ΔfimH/pfimH52 and TOP52 ΔfimH/pAD52PD89 both lacked hemagglutination titers, TOP52 ΔfimH/pfimH89 and TOP52 ΔfimH/pAD89PD52 both produced MSHA titers of 1:16.
TABLE 4.
Adhesin domain-specific hemagglutination deficiency of K. pneumoniae TOP52 FimH52
| Strain | HA titer (1:2x) in guinea pig RBCsa:
|
|
|---|---|---|
| Without mannose | With mannose | |
| UTI89 | 9 | 3 |
| UTI89 ΔfimH | 2 | 2 |
| UTI89 ΔfimH/pBAD | 0 | 0 |
| UTI89 ΔfimH/pfimH89 | 5 | 0 |
| UTI89 ΔfimH/pAD89PD52 | 5 | 0 |
| UTI89 ΔfimH/pAD52PD89 | 0 | 0 |
| UTI89 ΔfimH/pfimH52 | 0 | 0 |
| TOP52 | 0 | 0 |
| TOP52 ΔfimH | 0 | 0 |
| TOP52 ΔfimH/pBAD | 0 | 0 |
| TOP52 ΔfimH/pfimH89 | 4 | 0 |
| TOP52 ΔfimH/pAD89PD52 | 4 | 0 |
| TOP52 ΔfimH/pAD52PD89 | 0 | 0 |
| TOP52 ΔfimH/pfimH52 | 0 | 0 |
HA titer data are representative of three independent experiments.
These results demonstrate that the K. pneumoniae TOP52 FimH inability to agglutinate guinea pig RBCs is specific to its AD. The AD of E. coli UTI89 FimH is capable of agglutinating guinea pig RBCs with the native UTI89 PD or with the PD of K. pneumoniae TOP52. Thus, variations between the FimH ADs of E. coli UTI89 and K. pneumoniae TOP52 are likely responsible for their differences in function.
K. pneumoniae TOP52 FimH-dependent biofilms are inhibited by heptyl mannose but not methyl mannose.
K. pneumoniae TOP52 is able to form biofilm at room temperature when the production of type 1 pili is induced via the E. coli recombinase, coded for by fimX (50). Thus, we investigated whether this biofilm was FimH dependent. Biofilms were stained with crystal violet and quantified after 48 h of incubation. Wild-type TOP52 and TOP52/pBAD vector control did not form biofilm, while TOP52/pfimX formed biofilm. TOP52 ΔfimH, TOP52 ΔfimH/pBAD, and TOP52 ΔfimH/pfimX did not form biofilm, suggesting that the TOP52/pfimX biofilm is FimH dependent. Thus, although FimH52 is unable to mediate hemagglutination, it is capable of mediating biofilm formation. E. coli UTI89 formed a robust biofilm, while UTI89 ΔfimH did not. The formation of E. coli UTI89 biofilm was fully inhibited by 100 mM methyl mannose or 1 mM heptyl mannose (Fig. 4). In contrast, TOP52/pfimX biofilm formation was not affected by the presence of 100 mM methyl mannose, but was fully inhibited by 1 mM heptyl mannose.
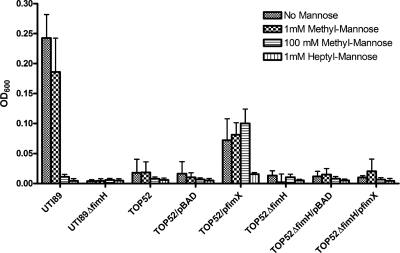
FIG. 4.
Heptyl mannose, but not methyl mannose, inhibits FimH-dependent biofilm formation of K. pneumoniae TOP52/pfimX. A 48-h biofilm assay was used to quantify biofilms produced by E. coli UTI89 and K. pneumoniae TOP52 strains in the presence of no mannose, 1 mM methyl mannose, 100 mM methyl mannose, or 1 mM heptyl mannose. TOP52 forms a FimH-dependent biofilm with induced expression of type 1 pili via the E. coli recombinase, fimX. This biofilm formation is inhibited by heptyl mannose but not high concentrations of methyl mannose. UTI89 forms robust biofilm without mannose but is inhibited by heptyl mannose or high concentrations of methyl mannose. Error bars represent standard deviations.
Therefore, TOP52/pfimX forms a FimH-dependent biofilm that is inhibited by heptyl mannose but not methyl mannose. This phenotype is distinct from those of E. coli UTI89 FimH-dependent biofilms, which are fully inhibited by the presence of 100 mM methyl mannose.
E. coli UTI89 and K. pneumoniae TOP52 both require fimH for effective persistence in the urinary tract.
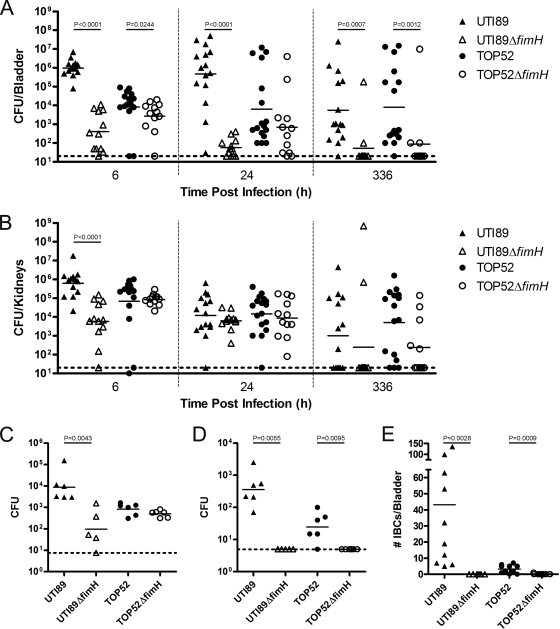
To analyze the respective roles of FimH52 and FimH89 in urinary tract infection, 107 CFU of E. coli UTI89, UTI89 ΔfimH, K. pneumoniae TOP52 or TOP52 ΔfimH were inoculated into the bladders of C3H/HeN mice by transurethral catheterization. Bladders and kidneys were harvested at various time points postinoculation, and bacterial titers were determined. In the bladder (Fig. 5A), E. coli UTI89 had significantly higher titers than UTI89 ΔfimH at 6 h (P < 0.0001), 24 h (P < 0.0001), and 336 h (P = 0.0007) postinfection. UTI89 ΔfimH was cleared from the bladder as time progressed. K. pneumoniae TOP52 had slightly but significantly higher titers (P = 0.0244) than TOP52 ΔfimH in the bladders of mice at 6 h postinoculation. By 24 h, there was no significant difference between TOP52 and TOP52 ΔfimH bladder titers. However, by 336 h postinfection, TOP52 ΔfimH had significantly lower titers than wild-type TOP52 (P = 0.0012). In the kidneys (Fig. 5B), UTI89 had significantly higher titers than UTI89 ΔfimH at 6 h postinfection (P < 0.0001), however the two strains had similar titers at both 24 and 336 h postinfection. TOP52 and TOP52 ΔfimH had similar levels of bacterial burden in the kidneys at all time points tested. Thus, FimH in K. pneumoniae TOP52 does not play a critical role early in bladder infection as is the case with E. coli UTI89; however, FimH is required for effective persistence in the bladder in both strains.
FIG. 5.
FimH of K. pneumoniae TOP52 is required for invasion, IBC formation, and persistence but not colonization in the murine model of UTI. Female C3H/HeN mice were inoculated with 107 E. coli UTI89 (▴), UTI89 ΔfimH (▵), K. pneumoniae TOP52 (•), or TOP52 ΔfimH (○) by transurethral inoculation. For organ titers, bladders (A) and kidneys (B) were harvested at various time points postinfection and CFU were calculated. Titer data are combined from three independent experiments. For ex vivo gentamicin protection assays, bladders were harvested at 1 h postinfection and luminal (C) and intracellular (D) populations of bacteria were quantified. IBCs were quantified (E) after visualization by LacZ staining at 6 h postinoculation. Short bars represent geometric means of each group, and horizontal dotted lines represent limits of detection. Significant P values, as calculated using the Mann-Whitney U test, are displayed.
FimH52 is required for K. pneumoniae TOP52 bladder invasion and IBC formation.
In order to further assess the role of FimH52 in acute K. pneumoniae TOP52 cystitis, bladder invasion assays were performed at 1 h postinfection with UTI89, UTI89 ΔfimH, TOP52, or TOP52 ΔfimH. In these assays, luminal bacteria were collected by successive bladder washes (Fig. 5C), prior to gentamicin treatment of the bladder to kill extracellular bacteria, as previously described (33). After 1.5 h of incubation in gentamicin, bladders were washed and homogenized and cell titers were determined to reveal the intracellular bacterial burden (Fig. 5D). UTI89 had 100-fold-higher luminal bacterial counts compared to UTI89 ΔfimH at 1 h postinfection (P = 0.0043). However, TOP52 and TOP52 ΔfimH had similar levels of luminal colonization. At this 1-h time point, UTI89 had significantly higher levels of intracellular bacteria than UTI89 ΔfimH (P = 0.0055), which did not have any intracellular titers above the limit of detection (5 CFU). TOP52 also invaded into the bladder tissue and had intracellular bacterial titers that were significantly higher than TOP52 ΔfimH (P = 0.0095), which did not have titers above the limit of detection.
To determine if the presence of FimH52 affects the ability of TOP52 to form IBCs, we visualized and quantified IBCs by lacZ staining of whole, mounted, fixed bladders as described previously (33) at 6 h postinoculation of UTI89, UTI89 ΔfimH, TOP52, or TOP52 ΔfimH (Fig. 5E). UTI89 formed a wide range of IBCs with a median of 25.5 per bladder, while UTI89 ΔfimH formed no detectable IBCs. TOP52 had a median of 2.0 IBCs per bladder, while TOP52 ΔfimH was unable to produce detectable IBCs (P = 0.0009).
These data suggest that K. pneumoniae TOP52 FimH52, in contrast to E. coli UTI89 FimH89, does not play a significant role in early bladder colonization. However, FimH52 is required for TOP52 invasion and IBC formation in the murine bladder, as is the case for UTI89.
fimH52 does not restore the ability of UTI89 ΔfimH to effectively infect the bladder.
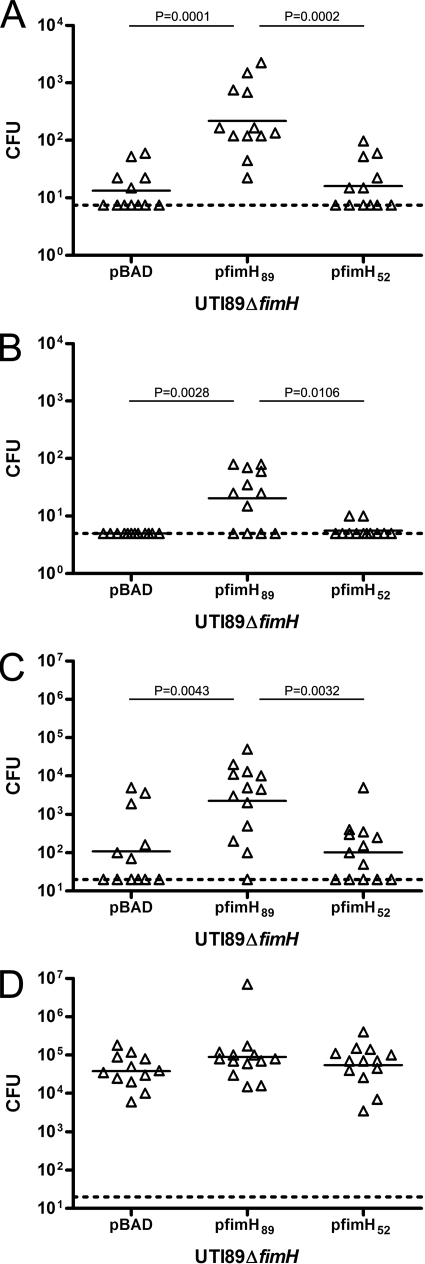
E. coli UTI89 relies on FimH to successfully cause UTI in the murine model. The ability of fimH52 to restore the ability of UTI89 ΔfimH to bind, invade, and infect murine bladders was investigated by using UTI89 ΔfimH complemented with pBAD vector control, pfimH89, and pfimH52. In 1-h gentamicin protection assays (Fig. 6A and B), UTI89 ΔfimH complemented with pfimH89 had significantly higher luminal titers than the same strain complemented with vector control (Fig. 6A, P = 0.0001) despite the known expression deficiencies observed above. Additionally, UTI89 ΔfimH/pfimH89 had significantly higher 1-h luminal titers than UTI89 ΔfimH/pfimH52, which had colonization levels similar to those of the vector control. Examination of the intracellular population at 1 h (Fig. 6B) revealed that the UTI89 ΔfimH/pBAD vector control did not have titers above the limit of detection, whereas UTI89 ΔfimH/pfimH89 did produce significantly higher burdens of intracellular bacteria (P = 0.0028). UTI89 ΔfimH/pfimH52 was able to invade the bladder tissue, but at significantly lower levels compared to UTI89 ΔfimH/pfimH89 (P = 0.0106). At 6 h postinoculation, UTI89 ΔfimH/pfimH89 had significantly higher burdens of bacteria in the bladder than both UTI89 ΔfimH/pBAD (P = 0.0043) and UTI89 ΔfimH/pfimH52 (P = 0.0032). Complementation of UTI89 ΔfimH with fimH from either UTI89 or TOP52 did not significantly affect 6-h kidney titers compared to those of the vector control.
FIG. 6.
Complementation of UTI89 ΔfimH with pfimH89 but not pfimH52 leads to increased bacterial burden in the murine model of UTI. Female C3H/HeN mice were inoculated with 107 cells of the UTI89 ΔfimH/pBAD vector control, UTI89 ΔfimH/pfimH89, or UTI89 ΔfimH/pfimH52 by transurethral inoculation. Ex vivo gentamicin protection assays were performed in which bladders were harvested at 1 h postinfection and luminal (A) and intracellular (B) populations of bacteria were quantified. For organ titers, bladders (C) and kidneys (D) were harvested at 6 h postinfection and CFU were enumerated. Short bars represent geometric means of each group, and horizontal dotted lines represent limits of detection. Significant P values, as calculated using the Mann-Whitney U test, are displayed.
The fimH52 gene was not able to restore UTI89 ΔfimH to levels above that of the vector control, while complementation with fimH89 yielded higher bacterial burdens at 1 and 6 h. This suggests a potential defect in the function of FimH52 in the bladder compared to FimH89.
DISCUSSION
FimH of the K. pneumoniae strain TOP52 (FimH52) has an amino acid sequence highly homologous to the sequence encoded by dozens of fimH genes that have been sequenced from E. coli (34, 54, 56). The residues that form the mannose binding pocket (Asn46, Asp47, Asp54, Gln133, Asn135, and Asp140) and hydrophobic ridge (Phe1, Ile13, Try48, Ile52, Tyr137, and Phe142) are completely identical between FimH52 and all known FimH adhesins of E. coli. Despite this identity, FimH52 has a receptor specificity unique from that of UPEC FimH. FimH52 is unable to mediate agglutination of guinea pig erythrocytes, whereas all known UPEC FimH adhesins are defined by their ability to mediate MSHA. Different E. coli FimH variants have been classified as high-affinity monomannose binders or lower-affinity trimannose binders (45, 54). However, both trimannose and monomannose variants display MSHA of guinea pig erythrocytes.
E. coli FimH recognizes mannose and has been shown to be able to interact with Manα1, 3Manβ1, 4GlcNAcβ1, 4GlcNAc in an extended binding site (61). These additional interactions between FimH and extended oligomannose moieties are mimicked by butyl α-d-mannose (61). Extended alkyl-α-mannosides have higher affinities for E. coli FimH compared to methyl-α-d-mannopyranoside (methyl mannose), with heptyl α-d-mannopyranoside (heptyl mannose) having the lowest dissociation constant (Kd) of 5 nM (5). K. pneumoniae FimH-dependent biofilms could only be inhibited by heptyl mannose and not methyl mannose, arguing that K. pneumoniae FimH requires additional contacts of the alkyl chain outside of the mannose binding pocket.
FimH52 differs at 17 positions from E. coli FimH and was threaded onto the three-dimensional structure of E. coli FimH. In the DE loop, adjacent to the hydrophobic ridge, Val94 and Asn96 of E. coli UTI89 FimH (FimH89) are changed to Ile and Asp, respectively, in FimH52. In the G strand, immediately C terminal to key residues in the hydrophobic ridge, Val145 in FimH89 is changed to Ile in FimH52. Combined, these differences may alter the structural stability of the hydrophobic ridge of FimH52 through changes in hydrophobic and hydrogen bond contacts. Thus, although FimH52 is unable to bind methyl mannose, these amino acid changes may facilitate interactions with longer oligomannose substrates (61).
The inability of FimH52 to mediate hemagglutination may be due to amino acid changes in proximity to the mannose binding pocket. Gln133 and Asp140 E. coli FimH residues are required for HA titers and mannose binding (29). Two differences in K. pneumoniae TOP52 primary sequence exist in residues adjacent to these mannose-binding residues at positions 132 and 141. Data from the threaded model suggest that at least two hydrogen bonds are lost in FimH52 with the combined differences in residues 132 and 141, which may have a destabilizing effect on interactions at the mannose site around the Asp140 and Gln133 mannose-binding residues. Sequence variation in regions of FimH not in close proximity to the mannose binding pocket may also significantly affect FimH function (56, 57).
Studies have suggested that fimbrial shafts can influence binding specificities of type 1 pili (16, 38). These effects do not account for the binding specificity differences observed for FimH52. FimH52 assembled into E. coli UTI89 type 1 pili was also hemagglutination negative, and FimH89 assembled into K. pneumoniae TOP52 type 1 pili produced an MSHA titer. Thus, the major functional disparities between E. coli and K. pneumoniae type 1 pili were specific to the AD of FimH, not the strain background or fimbrial shaft. However, fimbrial shafts may influence FimH binding in more subtle ways that could have been missed in this study due to the lower expression of type 1 pili in fimH-knockout backgrounds.
The binding specificity differences observed for FimH52 result in dramatic functional differences seen in K. pneumoniae UTI pathogenesis compared to E. coli UTI pathogenesis. Although K. pneumoniae TOP52 requires FimH for invasion and IBC formation in the murine bladder, FimH is not essential for early colonization. TOP52 and TOP52 ΔfimH have similar 1-h luminal bladder titers, 24-h whole-bladder titers, and only modest titer differences at 6 h postinfection. The small but significant differences at 6 h likely represent the intracellular population of bacteria in IBCs within TOP52-infected bladders that are absent in TOP52 ΔfimH-infected bladders. K. pneumoniae may use a different, non-type 1 pilus adhesin for initial binding to the bladder surface that E. coli lacks. This would explain why TOP52 ΔfimH had higher 1-h luminal titers and 6-h whole-bladder titers compared to UTI89 ΔfimH. K. pneumoniae contains a gene that encodes type 3 pili; however, these pili have not been implicated in binding to the bladder surface and are thought to mediate attachment to the basolateral surface of tracheal epithelial cells and basement membrane components (60). In addition to type 1 and type 3 pili, K. pneumoniae genes encode at least two other non-pilus adhesins. The CF29K and KPF-28 adhesins may play important roles in mediating attachment within the mammalian intestine, but their role in UTI has not been investigated (9, 12).
For many years, the glycoprotein uroplakin Ia has been considered the main receptor mediating FimH-dependent adhesion in the bladder (42, 63). Recently, it has been shown that host cell integrins also can mediate type 1 pilus-dependent invasion of urothelial cells (17). We currently do not know if K. pneumoniae FimH52 is capable of binding these receptors. It is possible that FimH52 may only be capable of binding integrin receptors (and not uroplakin Ia) for invasion of urothelial cells but not necessarily mediating significant adhesion to the uroplakin-coated bladder surface. Alternatively, K. pneumoniae FimH may have evolved for binding to a receptor in a different environment from the bladder.
This work focused on a single uropathogenic isolate of K. pneumoniae, and it is important to extend this work to other strains. The sequence of TOP52 FimH was almost identical to those of other sequenced K. pneumoniae FimH proteins and thus may be representative. The inability of K. pneumoniae TOP52 to agglutinate guinea pig RBCs is not an isolated finding. The ATCC 700721 strain also lacks an MSHA titer. The first studies of fimbriae and adhesive properties of 154 K. pneumoniae isolates found that 57.6% of strains produced little or no MSHA titer (13). Many researchers considered this to be due to poor type 1 expression in K. pneumoniae. However, K. pneumoniae TOP52 remained hemagglutination negative when expression of type 1 pili was increased by deletion of fimK or overexpression of fimX. Additionally, expression of E. coli type 1 pili at similar levels to TOP52 type 1 pili resulted in a positive MSHA.
This study suggests that limited sequence variation between the FimH of E. coli and K. pneumoniae results in differences in function and ability to colonize the urinary tract. Despite its poor adhesive properties in the urinary tract, FimH of K. pneumoniae remains an important virulence factor. It enables K. pneumoniae to progress through an IBC pathway during UTI and ultimately persist in the host. K. pneumoniae FimH likely requires ligand-receptor contacts outside of the mannose binding pocket for efficient binding. Further insight into these structural determinants will aid in our understanding of the altered host-pathogen interactions of K. pneumoniae UTI.
Acknowledgments
We thank Stefan Oscarson for the heptyl mannose. We also thank Wandy Beatty for assistance with electron microscopy experiments, Chia Hung for help with mouse experiments, and Melissa Kraus for statistical support.
This work was supported by the National Institutes of Health, Office of Research on Women's Health: Specialized Center of Research on Sex and Gender Factors Affecting Women's Health, grant DK64540; National Institute of Diabetes and Digestive and Kidney Diseases, grant R01 DK051406; and National Institute of Allergy and Infectious Diseases, grants R01 AI29549 and R01 AI48689.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 12 May 2008.
REFERENCES
- 1.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301105-107. [DOI] [PubMed] [Google Scholar]
- 2.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 451079-1093. [DOI] [PubMed] [Google Scholar]
- 3.Barnhart, M. M., F. G. Sauer, J. S. Pinkner, and S. J. Hultgren. 2003. Chaperone-subunit-usher interactions required for donor strand exchange during bacterial pilus assembly. J. Bacteriol. 1852723-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett-Lovsey, R. M., A. D. Herbert, M. J. Sternberg, and L. A. Kelley. 2008. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70611-625. [DOI] [PubMed] [Google Scholar]
- 5.Bouckaert, J., J. Berglund, M. Schembri, E. De Genst, L. Cools, M. Wuhrer, C. S. Hung, J. Pinkner, R. Slattegard, A. Zavialov, D. Choudhury, S. Langermann, S. J. Hultgren, L. Wyns, P. Klemm, S. Oscarson, S. D. Knight, and H. De Greve. 2005. Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin. Mol. Microbiol. 55441-455. [DOI] [PubMed] [Google Scholar]
- 6.Brinton, C. C., Jr. 1965. The structure, function, synthesis, and genetic control of bacterial pili and a model for DNA and RNA transport in Gram negative bacteria. Trans. N. Y. Acad. Sci. 271003-1165. [DOI] [PubMed] [Google Scholar]
- 7.Bryan, A., P. Roesch, L. Davis, R. Moritz, S. Pellett, and R. A. Welch. 2006. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect. Immun. 741072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 2851061-1066. [DOI] [PubMed] [Google Scholar]
- 9.Darfeuille-Michaud, A., C. Jallat, D. Aubel, D. Sirot, C. Rich, J. Sirot, and B. Joly. 1992. R-plasmid-encoded adhesive factor in Klebsiella pneumoniae strains responsible for human nosocomial infections. Infect. Immun. 6044-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, Palo Alto, CA. http://www.pymol.org.
- 12.Di Martino, P., V. Livrelli, D. Sirot, B. Joly, and A. Darfeuille-Michaud. 1996. A new fimbrial antigen harbored by CAZ-5/SHV-4-producing Klebsiella pneumoniae strains involved in nosocomial infections. Infect. Immun. 642266-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duguid, J. P. 1959. Fimbriae and adhesive properties in Klebsiella strains. J. Gen. Microbiol. 21271-286. [DOI] [PubMed] [Google Scholar]
- 14.Duguid, J. P., and D. C. Old. 1980. Adhesive properties of Enterobacteriacae, p. 186-217. In E. H. Beachy (ed.), Bacterial adherence receptors and recognition. Chapman & Hall, London, United Kingdom.
- 15.Duguid, J. P., I. W. Smith, G. Dempster, and P. N. Edmunds. 1955. Non-flagellar filamentous appendages (“fimbriae”) and hemagglutinating activity in bacterium coli. J. Pathol. Bacteriol. 70335-348. [DOI] [PubMed] [Google Scholar]
- 16.Duncan, M. J., E. L. Mann, M. S. Cohen, I. Ofek, N. Sharon, and S. N. Abraham. 2005. The distinct binding specificities exhibited by enterobacterial type 1 fimbriae are determined by their fimbrial shafts. J. Biol. Chem. 28037707-37716. [DOI] [PubMed] [Google Scholar]
- 17.Eto, D. S., T. A. Jones, J. L. Sundsbak, and M. A. Mulvey. 2007. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog. 3e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fader, R. C., and C. P. Davis. 1980. Effect of piliation on Klebsiella pneumoniae infection in rat bladders. Infect. Immun. 30554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Firon, N., S. Ashkenazi, D. Mirelman, I. Ofek, and N. Sharon. 1987. Aromatic alpha-glycosides of mannose are powerful inhibitors of the adherence of type 1 fimbriated Escherichia coli to yeast and intestinal epithelial cells. Infect. Immun. 55472-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firon, N., I. Ofek, and N. Sharon. 1984. Carbohydrate-binding sites of the mannose-specific fimbrial lectins of enterobacteria. Infect. Immun. 431088-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Firon, N., I. Ofek, and N. Sharon. 1983. Carbohydrate specificity of the surface lectins of Escherichia coli, Klebsiella pneumoniae and Salmonella typhimurium. Carbohydr. Res. 120235-249. [DOI] [PubMed] [Google Scholar]
- 22.Garofalo, C. K., T. M. Hooton, S. M. Martin, W. E. Stamm, J. J. Palermo, J. I. Gordon, and S. J. Hultgren. 2007. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect. Immun. 7552-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerlach, G.-F., S. Clegg, and B. L. Allen. 1989. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J. Bacteriol. 1711262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannan, T. J., I. U. Mysorekar, S. L. Chen, J. N. Walker, J. M. Jones, J. S. Pinkner, S. J. Hultgren, and P. C. Seed. 2007. LeuX tRNA-dependent and -independent mechanisms of Escherichia coli pathogenesis in acute cystitis. Mol. Microbiol. 67116-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hull, R. A., R. E. Gill, P. Hsu, B. H. Minshew, and S. Falkow. 1981. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect. Immun. 33933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hultgren, S. J., T. N. Porter, A. J. Schaeffer, and J. L. Duncan. 1985. Role of type 1 pili and effects of phase variation on lower urinary tract infections produced by Escherichia coli. Infect. Immun. 50370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hultgren, S. J., W. R. Schwan, A. J. Schaeffer, and J. L. Duncan. 1986. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect. Immun. 54613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung, C.-S., J. Bouckaert, D. Hung, J. Pinkner, C. Widberg, A. De Fusco, C. G. Auguste, B. Strouse, S. Langerman, G. Waksman, and S. J. Hultgren. 2002. Structure basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol. Microbiol. 44903-915. [DOI] [PubMed] [Google Scholar]
- 30.Hung, D. L., and S. J. Hultgren. 1998. Pilus biogenesis via the chaperone/usher pathway: an integration of structure and function. J. Struct. Biol. 124201-220. [DOI] [PubMed] [Google Scholar]
- 31.Jones, C. H., J. S. Pinkner, R. Roth, J. Heuser, A. V. Nicholes, S. N. Abraham, and S. J. Hultgren. 1995. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 922081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Justice, S. S., C. Hung, J. A. Theriot, D. A. Fletcher, G. G. Anderson, M. J. Footer, and S. J. Hultgren. 2004. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA 1011333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Justice, S. S., S. R. Lauer, S. J. Hultgren, and D. A. Hunstad. 2006. Maturation of intracellular Escherichia coli communities requires SurA. Infect. Immun. 744793-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klemm, P., and G. Christiansen. 1987. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol. Gen. Genet. 208439-445. [DOI] [PubMed] [Google Scholar]
- 35.Langermann, S., R. Mollby, J. E. Burlein, S. R. Palaszynski, C. G. Auguste, A. DeFusco, R. Strouse, M. A. Schenerman, S. J. Hultgren, J. S. Pinkner, J. Winberg, L. Guldevall, M. Soderhall, K. Ishikawa, S. Normark, and S. Koenig. 2000. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J. Infect. Dis. 181774-778. [DOI] [PubMed] [Google Scholar]
- 36.Langermann, S., S. Palaszynski, M. Barnhart, G. Auguste, J. S. Pinkner, J. Burlein, P. Barren, S. Koenig, S. Leath, C. H. Jones, and S. J. Hultgren. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276607-611. [DOI] [PubMed] [Google Scholar]
- 37.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 1796228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madison, B., I. Ofek, S. Clegg, and S. N. Abraham. 1994. Type 1 fimbrial shafts of Escherichia coli and Klebsiella pneumoniae influence sugar-binding specificities of their FimH adhesins. Infect. Immun. 62843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez, J. J., M. A. Mulvey, J. D. Schilling, J. S. Pinkner, and S. J. Hultgren. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 192803-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matatov, R., J. Goldhar, E. Skutelsky, I. Sechter, R. Perry, R. Podschun, H. Sahly, K. Thankavel, S. N. Abraham, and I. Ofek. 1999. Inability of encapsulated Klebsiella pneumoniae to assemble functional type 1 fimbriae on their surface. FEMS Microbiol. Lett. 179123-130. [DOI] [PubMed] [Google Scholar]
- 41.McClelland, M., L. Florea, K. Sanderson, S. W. Clifton, J. Parkhill, C. Churcher, G. Dougan, R. K. Wilson, and W. Miller. 2000. Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi, and Paratyphi. Nucleic Acids Res. 284974-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 2821494-1497. [DOI] [PubMed] [Google Scholar]
- 43.Mulvey, M. A., J. D. Schilling, and S. J. Hultgren. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 694572-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nilsson, L. M., W. E. Thomas, E. Trintchina, V. Vogel, and E. V. Sokurenko. 2006. Catch bond-mediated adhesion without a shear threshold: trimannose versus monomannose interactions with the FimH adhesin of Escherichia coli. J. Biol. Chem. 28116656-16663. [DOI] [PubMed] [Google Scholar]
- 46.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 47.Ottow, J. C. G. 1975. Ecology, physiology and genetics of fimbriae and pili. Annu. Rev. Microbiol. 2979. [DOI] [PubMed] [Google Scholar]
- 48.Ronald, A. R., L. E. Nicolle, E. Stamm, J. Krieger, J. Warren, A. Schaeffer, K. G. Naber, T. M. Hooton, J. Johnson, S. Chambers, and V. Andriole. 2001. Urinary tract infection in adults: research priorities and strategies. Int. J. Antimicrob. Agents 17343-348. [DOI] [PubMed] [Google Scholar]
- 49.Ronald, A. R., and A. L. Pattullo. 1991. The natural history of urinary infection in adults. Med. Clin. N. Am. 75299-312. [DOI] [PubMed] [Google Scholar]
- 50.Rosen, D. A., J. S. Pinkner, J. M. Jones, J. N. Walker, S. Clegg, and S. J. Hultgren. 2008. Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infect. Immun. 763337-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosen, D. A., T. M. Hooton, W. E. Stamm, P. A. Humphrey, and S. J. Hultgren. 2007. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 4e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salit, I. E., and E. C. Gotschlich. 1977. Hemagglutination by purified type 1 Escherichia coli pili. J. Exp. Med. 1461169-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saulino, E. T., D. G. Thanassi, J. S. Pinkner, and S. J. Hultgren. 1998. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 172177-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokurenko, E. V., V. Chesnokova, R. J. Doyle, and D. L. Hasty. 1997. Diversity of the Escherichia coli type 1 fimbrial lectin. Differential binding to mannosides and uroepithelial cells. J. Biol. Chem. 27217880-17886. [DOI] [PubMed] [Google Scholar]
- 55.Sokurenko, E. V., H. S. Courtney, S. N. Abraham, P. Klemm, and D. L. Hasty. 1992. Functional heterogeneity of type 1 fimbriae of Escherichia coli. Infect. Immun. 604709-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sokurenko, E. V., H. S. Courtney, J. Maslow, A. Siitonen, and D. L. Hasty. 1995. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J. Bacteriol. 1773680-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sokurenko, E. V., H. S. Courtney, D. E. Ohman, P. Klemm, and D. L. Hasty. 1994. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J. Bacteriol. 176748-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Struve, C., and K. A. Krogfelt. 1999. In vivo detection of Escherichia coli type 1 fimbrial expression and phase variation during experimental urinary tract infection. Microbiology 1452683-2690. [DOI] [PubMed] [Google Scholar]
- 59.Svanborg Eden, C., L. Hagberg, L. A. Hanson, S. Hull, R. Hull, U. Jodal, H.Leffler, H. Lomberg, and E. Straube. 1983. Bacterial adherence—a pathogenetic mechanism in urinary tract infections caused by Escherichia coli. Prog. Allergy 33175-188. [PubMed] [Google Scholar]
- 60.Tarkkanen, A. M., B. L. Allen, B. Westerlund, H. Holthofer, P. Kuusela, L. Risteli, S. Clegg, and T. K. Korhonen. 1990. Type V collagen as the target for type-3 fimbriae, enterobacterial adherence organelles. Mol. Microbiol. 41353-1361. [DOI] [PubMed] [Google Scholar]
- 61.Wellens, A., C. Garofalo, H. Nguyen, N. Van Gerven, R. Slättegård, J. P. Hernalsteens, L. Wyns, S. Oscarson, H. De Greve, S. J. Hultgren, and J. Bouckaert. 2008. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS One 3e2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wright, K. J., P. C. Seed, and S. J. Hultgren. 2007. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell. Microbiol. 92230-2241. [DOI] [PubMed] [Google Scholar]
- 63.Wu, X. R., T. T. Sun, and J. J. Medina. 1996. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc. Natl. Acad. Sci. USA 939630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]