Abstract
Francisella tularensis, a gram-negative facultative intracellular bacterial pathogen, causes disseminating infections in humans and other mammalian hosts. Macrophages and other monocytes have long been considered the primary site of F. tularensis replication in infected animals. However, recently it was reported that F. tularensis also invades and replicates within alveolar epithelial cells following inhalation in a mouse model of tularemia. TC-1 cells, a mouse lung epithelial cell line, were used to study the process of F. tularensis invasion and intracellular trafficking within nonphagocytic cells. Live and paraformaldehyde-fixed F. tularensis live vaccine strain organisms associated with, and were internalized by, TC-1 cells at similar frequencies and with indistinguishable differences in kinetics. Inhibitors of microfilament and microtubule activity resulted in significantly decreased F. tularensis invasion, as did inhibitors of phosphatidylinositol 3-kinase and tyrosine kinase activity. Collectively, these results suggest that F. tularensis epithelial cell invasion is mediated by a preformed ligand on the bacterial surface and driven entirely by host cell processes. Once internalized, F. tularensis-containing endosomes associated with early endosome antigen 1 (EEA1) followed by lysosome-associated membrane protein 1 (LAMP-1), with peak coassociation frequencies occurring at 30 and 120 min postinoculation, respectively. By 2 h postinoculation, 70.0% (± 5.5%) of intracellular bacteria were accessible to antibody delivered to the cytoplasm, indicating vacuolar breakdown and escape into the cytoplasm.
Francisella tularensis, the causative agent of the disease tularemia, infects a wide range of animal hosts. Humans can be infected by a variety of routes, including physical contact with infected animals, insect bites, ingestion of contaminated food or water, and inhalation of organisms (13, 52). Disease severity is affected by both the route of inoculation and the bacterial subtype (13, 48, 52). F. tularensis strains are subdivided into two groups, A and B; the more severe form of tularemia in humans is caused by type A strains (13, 48, 52). Type A strain F. tularensis subspecies tularensis is found almost exclusively in North America, whereas type B strain F. tularensis subspecies holarctica is found throughout Europe, as well as in North America (35). There is a 5% to 15% mortality rate associated with untreated human tularemia caused by type A strains; however, that rate reaches 30% to 60% for untreated pneumonic and typhoid forms of the disease (11). The live vaccine strain (LVS) is an attenuated type B strain that causes a tularemia-like disease in mice and is used as a model organism to study F. tularensis pathogenesis. There has been heightened interest in the study of this organism in recent years due to its history of weaponization and potential for use as an agent of biological warfare. F. tularensis is a category A select agent on the CDC's bioterrorism agent list, which includes organisms with the potential to cause high numbers of casualties if disseminated in an aerosol form (26).
Much of the Francisella pathogenesis research has focused on the survival and replication of F. tularensis in macrophages and dendritic cells, and many of the genes identified to date that are required for full virulence contribute to survival or replication in the macrophage (1, 6, 17, 20, 27, 30, 32, 37, 42). We recently demonstrated that F. tularensis also invades and replicates in alveolar type II (ATII) epithelial cells of infected mice (21). ATII cells have a number of biological functions, including the production, secretion, and recycling of surfactant; proliferation to produce additional type II cells as well as transdifferentiation into type I cells; maintenance of alveolar fluid balance; and production of antimicrobial and anti-inflammatory substances (29). Due to their close proximity to endothelial cells and their role in gas exchange and fluid balance, these cells are ideally located to provide a portal through which bacteria could disseminate to distal organs.
F. tularensis attaches to and invades nonphagocytic cells, including the ATII cell lines A549 and MLE 12 and the lung epithelial cell line TC-1 (21, 28). Melillo et al. also reported that Escherichia coli expressing the F. tularensis surface protein FsaP binds to A549 cells (31), identifying a surface protein that may play a role in ATII cell association.
A primary function of monocytes is to engulf bacterial cells and other foreign particles (24, 44). However, the invasion of epithelial and other nonphagocytic host tissue cells requires bacterially mediated exploitation of host cell functions to gain entry (10, 15, 16). Alveolar epithelial cells provide a site where F. tularensis can replicate in the infected host (21), and as such it is important to understand how bacterial interaction with these cells may differ from interaction with macrophages. Herein we describe our efforts to understand how F. tularensis invades lung epithelial cells.
MATERIALS AND METHODS
Bacterial strains.
Francisella tularensis LVS was obtained from the CDC, Atlanta, GA. LVSgfp was constructed using the pKK214GFP plasmid (a gift of Mats Forsman). F. tularensis LVS and LVSgfp were propagated on chocolate agar supplemented with 1% IsoVitaleX (Becton-Dickinson). Listeria monocytogenes EGD1/2a, Salmonella enterica serovar Typhimurium, Yersinia pseudotuberculosis, and Campylobacter jejuni strain 81-176 were gifts from Paul Orndorff, Craig Altier, Ralph Isberg, and Deborah Threadgill, respectively. All bacterial strains were grown on LB agar, with the exception of C. jejuni, which was grown on Mueller-Hinton agar with 5% CO2, and F. tularensis LVS, which was grown on chocolate agar as described above. All bacterial strains were grown at 37°C. Salmonella enterica was grown overnight in LB broth under static conditions for invasion assays.
Cell culture.
TC-1 (ATCC CRL-2785) is a tumor cell line derived from primary lung epithelial cells of C57BL/6 mice. The cells were immortalized with human papillomavirus type 16 E6 and E7 and transformed with the c-H-ras oncogene. These cells were grown in RPMI 1640 supplemented with 2 mM l-glutamine, 1.5 g/liter sodium bicarbonate, 10 mM HEPES, 1.0 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 10% fetal bovine serum. Cell cultures were maintained at 37°C and 5% CO2.
Attachment, invasion, and vacuolar escape assay.
To evaluate the percentage of TC-1 cells with associated bacteria, synchronized infections were carried out by adding F. tularensis LVS expressing green fluorescent protein (GFP) (LVSgfp) at a multiplicity of infection (MOI) of 100 to TC-1 cells chilled to 4°C. Bacterial suspensions were made in tissue culture media from bacteria grown as described above. Bacterial concentration of suspensions was determined using a Klett meter. Dilutions of suspensions were plated to verify MOI. Plates were centrifuged at 300 × g for 5 min at 4°C and then rapidly warmed by placing in a 37°C water bath for 2 min before transferring to a 37°C, 5% CO2 incubator. For experiments using killed LVS, organisms were treated with 4% paraformaldehyde (PFA) for 10 min. There was no growth from a PFA-treated bacterial aliquot that was plated on chocolate agar, demonstrating that PFA treatment killed all organisms. At 10 min postinoculation, samples were washed with phosphate-buffered saline (PBS) to remove unattached bacteria, and prewarmed medium was added to monolayers. Cells were collected by trypsinization at 10, 20, 30, and 60 min postinoculation and samples were processed at 4°C. Cells to be analyzed for bacteria association were fixed in 4% PFA for 10 min and analyzed by flow cytometry.
Parallel wells were analyzed for intracellular bacteria by collecting samples as described above. Extracellular bacteria were labeled using anti-F. tularensis lipopolysaccharide (LPS) antibody (USBiological) conjugated to Pacific Blue (Molecular Probes), 1:1,000, for 30 min at 4°C. Cells were then lysed with water and centrifuged at 300 × g to removed eukaryotic cell debris. Supernatant containing bacteria was centrifuged at 16,000 × g, fixed with 4% PFA, and analyzed by flow cytometry to differentiate intracellular (GFP only) from extracellular (GFP and Pacific Blue) bacteria.
To evaluate bacterial escape into the cytoplasm, synchronized inoculations of TC-1 cells and staining of extracellular bacteria were done as described above. Cells were collected by trypsinization at 10, 20, 30, 60, and 120 min postinoculation. TC-1 cells were treated with 50 μg/ml digitonin in KHM buffer (110 mM potassium acetate-20 mM HEPES-2 mM MgCl2, pH 7.3) for 1 min, washed in KHM buffer, and incubated with anti-F. tularensis LPS antibody conjugated to Alexa Fluor 647 (Molecular Probes), 1:1,000, for 30 min at 4°C. Cells were then washed in KHM buffer, lysed with water, and centrifuged at 300 × g to remove cellular debris. Supernatants were centrifuged at 16,000 × g and bacterial pellets were fixed in 4% PFA and analyzed by flow cytometry to differentiate extracellular (GFP, Pacific Blue, and Alexa Fluor 647), cytoplasmic (GFP and Alexa Fluor 647), and vacuolar (GFP-only) bacteria.
Flow cytometry of whole cells and bacteria was performed using a CyAn ADP flow cytometer (Dako Cytomation). Data were analyzed using Summit Software (Dako), with gating for single events for whole-cell samples and GFP-positive single events for bacterial samples.
Inhibitor assays.
Inhibitor assays were carried out with cytochalasin D, colchicine, wortmannin, and genistein. Cytochalasin D, an actin polymerization inhibitor, was used at 0.5 and 1.0 μM concentrations. Colchicine, a microtubule polymerization inhibitor, was used at 0.1 and 0.25 μM concentrations. Wortmannin, an inhibitor of phosphatidylinositol 3-kinase (PI 3-kinase) activity, was used at 100 and 200 nM concentrations. Genistein, an inhibitor of tyrosine kinase activity, was used at 50 and 100 μM concentrations. Monolayers were preincubated for 1 hour with escalating concentrations of inhibitor, and then bacteria suspended in media containing inhibitor were added at MOI of 100 for LVS or 25 to 50 for other bacteria. Samples without inhibitor included bacteria suspended in tissue culture media alone or in media containing the carrier in which the inhibitor was reconstituted where appropriate. After 4 hours, cells were washed with PBS to remove inhibitor. Extracellular bacteria were killed by adding media containing 25 μg/ml of gentamicin and incubated for 2 h before monolayers were washed with PBS and scraped from the plate using sterile applicator sticks. Serial dilutions were plated on chocolate agar to quantify CFU of intracellular bacteria. Inhibitors did not affect LVS viability, as demonstrated by incubating bacteria in tissue culture media containing inhibitor for 4 hours, followed by plating onto chocolate agar to quantify CFU. Inhibitors did not affect eukaryotic cell viability at the concentrations used, as determined by trypan blue exclusion. The effect of inhibitors on bacterial attachment to cells was determined as described above for invasion, except that gentamicin was not added, allowing bacteria attached to cells but not internalized to be included in the CFU recovered. Inhibitors had no effect on LVS attachment to TC-1 cells. Results are expressed as the percentage of LVS or control organism that survived gentamicin treatment, relative to that for the sample without inhibitor inoculated with the same organism. Invasion without inhibitor is defined as 100% invasion. Data presented are the results of assays done in triplicate and are representative of multiple repetitions of each experiment. Data were analyzed for statistical significance by paired two-tailed t test and were considered significantly different from the untreated control when the P value was <0.01.
Fluorescence microscopy.
Cells were grown on poly-l-lysine-coated coverslips in 24-well cell culture plates to ∼90% confluence. LVSgfp was added at an MOI of 100 to prechilled TC-1 plates and centrifuged at 300 × g for 3 minutes to synchronize infection as described above. At 10, 20, 30, 60, 120, and 180 min postinoculation, monolayers were washed with PBS to remove unattached bacteria and fixed using 4% PFA. Samples were blocked with PBS-Fc block-5% donkey serum and incubated with anti-F. tularensis LPS diluted 1:1,000 for 30 min at 4°C followed by donkey anti-mouse AMCA (7-amino-4-methylcoumarin-3-acetic acid; Jackson ImmunoResearch) diluted 1:100 for 30 min at 4°C to label extracellular bacteria. For early endosome antigen 1 (EEA1) or lysosome-associated membrane protein 1 (LAMP-1), staining samples were blocked using PBS-0.1% saponin-5% serum of host species of secondary antibody followed by goat polyclonal anti-EEA1 (N-19; Santa Cruz Biotechnologies) diluted 1:200 in blocking solution or rat monoclonal anti-mouse LAMP-1 (1D4B; developed by J. T. August, obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242) diluted 1:200 in blocking solution. Secondary antibodies used were donkey anti-goat Cy5 (Jackson ImmunoResearch) or goat anti-rat Alexa Fluor 647 (Molecular Probes) diluted 1:500 in PBS-0.1% saponin. Samples were examined using a Zeiss Axioplan 2 epifluorescence microscope and SlideBook digital deconvolution software (Intelligent Imaging Innovations). One hundred intracellular bacteria were counted for each time point and condition. Images of sequential vertical planes were acquired to determine the location of bacteria within EEA1- or LAMP-1-containing vacuoles. Data presented are the results of three independent experiments.
Electron microscopy.
Cell monolayers grown on polystyrene plates were rinsed with PBS or serum-free medium and fixed in 3% glutaraldehyde-0.15 M sodium phosphate, pH 7.4. Following three rinses with sodium phosphate buffer, the monolayers were postfixed for 1 h in 1% osmium tetroxide-1.25% potassium ferrocyanide-0.15 M sodium phosphate buffer, rinsed in deionized water, dehydrated using increasing concentrations of ethanol (30%, 50%, 75%, 100%, 100%; 10 min each), and embedded in Polybed 812 epoxy resin (Polysciences, Inc., Warrington, PA). The embedded samples were sectioned parallel and perpendicular to the substrate at 70 nm using a diamond knife. Ultrathin sections were collected on 200-mesh copper grids and stained with 4% aqueous uranyl acetate for 15 min, followed by Reynolds’ lead citrate for 7 min. Sections were observed using a Leo EM910 transmission electron microscope at 80 kV (Leo Electron Microscopy, Thornwood, NY) and photographed using a Gatan Bioscan digital camera (Gatan, Inc., Pleasanton, CA).
RESULTS
Kinetics of F. tularensis epithelial cell attachment and invasion.
Francisella tularensis LVS invades and replicates within TC-1, MLE 12, and A549 lung epithelial cell lines (21). While the intracellular replication rates within these lines are indistinguishable, TC-1 cells support the highest initial invasion frequency (21). In addition, TC-1 cells are a mouse cell line, and LVS infection causes a tularemia-like disease in mice similar to that seen with virulent strains in humans. We therefore used TC-1 cells to examine the initial stages of F. tularensis invasion.
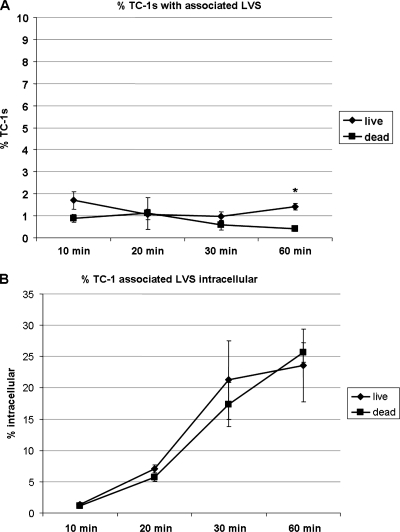
To determine the kinetics of lung cell attachment and invasion by F. tularensis, we synchronized the infection of TC-1 cells at an MOI of 100 with LVSgfp. At 10, 20, 30, and 60 min postinoculation, cells were washed to remove nonadherent bacteria and collected to quantify TC-1 cells associated with GFP-expressing bacteria by flow cytometry (Fig. 1A). At these same time points, in parallel samples, extracellular bacteria were stained using a Pacific Blue-labeled antibody to F. tularensis LPS and TC-1 cells were lysed to recover cell-associated bacteria. GFP-positive bacteria recovered from cells were analyzed by flow cytometry to determine intracellular (GFP alone) versus extracellular (GFP and Pacific Blue) localization (Fig. 1B). At 10 min, 1.7% (± 0.4%) of TC-1 cells had associated bacteria, of which 1.4% (± 0.1%) of TC-1 cell-associated bacteria were intracellular. At 20 min postinoculation, 1.1% (± 0.2%) of TC-1 cells had cell-associated bacteria, of which 7.0% (± 0.6%) were internal. At 30 min postinoculation, 1.0% (± 0.2%) of TC-1 cells had cell-associated bacteria, of which 21.3% (± 6.3%) were internal. At 60 min postinoculation, 1.4% (± 0.1%) of TC-1 cells had cell-associated bacteria, of which 23.6% (± 5.8%) were intracellular.
FIG. 1.
F. tularensis LVS association with and internalization by TC-1 lung epithelial cells. (A) Percentage of TC-1 cells with associated live or PFA-fixed LVS at designated times postinoculation. *, Data are significantly different from untreated control data (P < 0.01 by paired two-tailed t test). (B) Percentages of TC-1 cell-associated live or PFA-fixed LVS that are intracellular at the designated times postinoculation.
To determine if LVS entry into lung epithelial cells required viable bacteria, we repeated the above-described experiments using killed LVSgfp. At 10 min, 0.9% (± 0.2%) of TC-1 cells had dead bacteria associated with them, of which 1.2% (± 0.4%) of TC-1 cell-associated bacteria were intracellular. At 20 min postinoculation, 1.1% (± 0.7%) of TC-1cells had cell-associated dead bacteria, of which 5.7% (± 0.6%) were internal. At 30 min postinoculation, 0.6% (± 0.2%) of TC-1 cells had cell-associated dead bacteria, of which 17.3% (± 3.5%) were internal. At 60 min postinoculation, 0.4% (± 0.1%) of cells had dead bacteria associated with them, of which 25.7% (± 1.6%) were intracellular. These results indicated that both live and dead LVS attached to and invaded lung epithelial cells. Further, invasion frequency and kinetics were not significantly different between live and dead bacteria (P < 0.01), except for cell association at 60 min (Fig. 1). The ability of nonviable F. tularensis to invade lung epithelial cells suggested that a preformed ligand on the bacterial surface may interact with the host cell.
Effects of cytoskeleton and signaling pathway inhibitors on F. tularensis invasion of epithelial cells.
Inhibitors of eukaryotic cell function were used to determine the contribution of host cell signaling and cytoskeleton rearrangement to LVS invasion of lung epithelial cells. Bacterial entry into host cells generally requires rearrangement of cytoskeletal structures; either microfilaments alone, as is the case with Salmonella, Shigella, Listeria, and Yersinia spp. (12), or both microfilaments and microtubules, as is the case with Neisseria gonorrhoeae and Campylobacter jejuni (4, 43).
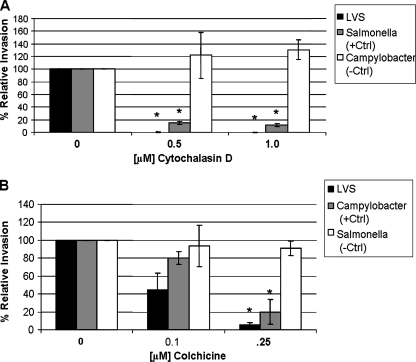
To determine the contribution of actin polymerization to LVS invasion, we incubated TC-1 lung epithelial cells with cytochalasin D, an actin polymerization inhibitor that disrupts microfilaments, and measured bacterial internalization by gentamicin protection assay. Treatment of TC-1 cells with 0.5 and 1.0 μM cytochalasin D decreased LVS invasion more than 99% (± 0.1%) (Fig. 2A), whereas Salmonella enterica invasion, which is known to be actin dependent (36), decreased by 84.6% (± 2.3%) and 88.1% (± 2.5%) at the same inhibitor concentrations, respectively. TC-1 cell invasion by Campylobacter jejuni strain 81-176, a strain which does not require actin for invasion of intestinal epithelial cells (34), was not decreased by actin inhibition.
FIG. 2.
The effect of actin and microtubule polymerization on F. tularensis LVS invasion of lung epithelial cells. TC-1 cells were treated with designated concentrations of cytochalasin D (A) or colchicine (B). Results are expressed as the percentage of LVS, Salmonella, or Campylobacter organisms that survived gentamicin treatment relative to that for the sample without inhibitor inoculated with the same organism (defined as 100% invasion). *, Data are significantly different from untreated control data (P < 0.01 by paired two-tailed t test).
Microtubules are responsible for the cytoplasmic organization of eukaryotic cells and the control of organelle transport and are a primary component of cilia and flagella (55). Microtubule polymerization has been shown to contribute to the epithelial cell invasion of Neisseria gonorrhoeae and Campylobacter jejuni (4, 43). To determine the contribution of microtubules to LVS invasion, we treated lung epithelial cells with colchicine, which binds tubulin and inhibits microtubule polymerization, and determined invasion by gentamicin protection assay. Treatment of TC-1 cells with colchicine decreased LVS invasion by 55.2% (± 18.3%) and 94.2% (± 2.4%) at 0.1 and 0.25 μM concentrations, respectively (Fig. 2B). TC-1 cell invasion by Campylobacter jejuni strain 81-176, a strain for which invasion is blocked by microtubule depolymerization in intestinal epithelial cells (34), was decreased significantly at the higher colchicine concentration. Salmonella enterica invasion, which is not considered microtubule dependent (4), did not demonstrate a statistically significant decrease in invasion at these concentrations.
Signaling pathways are frequently manipulated by bacteria to cause the cytoskeletal rearrangement necessary to gain entry into nonphagocytic cells (10, 15, 16). PI 3-kinase and tyrosine kinase signaling are exploited by other pathogens for invasion (25, 45). These pathways were examined for their contribution to Francisella tularensis entry into lung epithelial cells.
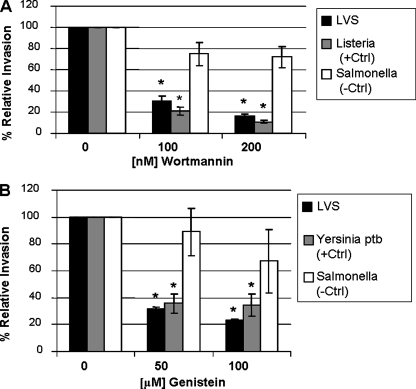
PI 3-kinases phosphorylate inositol phospholipids, forming lipid products that are in turn involved in cellular functions such as cell growth, actin rearrangement, and vesicular trafficking (51). To determine the importance of PI 3-kinase signaling to F. tularensis invasion, lung epithelial cells were treated with wortmannin, an inhibitor of PI 3-kinase activity, and invasion was assessed by gentamicin protection assay. Wortmannin decreased LVS invasion of TC-1 cells by 69.4% (± 4.6%) and 84.1% (± 1.8%) when cells were exposed to 100 and 200 nM concentrations, respectively (Fig. 3A). Invasion by Listeria monocytogenes, which is PI 3-kinase dependent (25), was significantly decreased. Salmonella enterica invasion, which is not PI 3-kinase dependent (49), was not significantly decreased at these concentrations in TC-1 cells.
FIG. 3.
The effect of PI 3-kinase and tyrosine kinase activity on LVS invasion of lung epithelial cells. Wortmannin (A) and genistein (B) were added to TC-1 cells at the indicated concentrations. Results are expressed as the percentage of LVS or control organisms that survived gentamicin treatment relative to that for the sample without inhibitor inoculated with the same organism (defined as 100% invasion). *, Data are significantly different from untreated control data (P < 0.01 by paired two-tailed t test).
Cells monitor and respond to their external environment via receptors that lead to intracellular signaling events (53). Activation of receptor tyrosine kinases can lead to receptor internalization as well as the initiation of a cascade of downstream signaling events (53). Some organisms, such as Yersinia pseudotuberculosis, exploit signaling through host cell receptors to gain entry into cells, and this entry can be blocked by tyrosine kinase inhibitors (45). To determine the contribution of tyrosine kinase signaling to LVS invasion, the inhibitor genistein was added to lung epithelial cells and the number of intracellular organisms measured by gentamicin protection assay. Genistein decreased LVS invasion of TC-1 cells by 68.1% (± 1.1%) and 77.1% (± 1.1%) at 50 and 100 μM concentrations, respectively (Fig. 3B). Invasion by Yersinia pseudotuberculosis was significantly reduced at these concentrations, while Salmonella enterica invasion, which is tyrosine kinase independent (45), was not significantly decreased in TC-1 cells.
Collectively, these results indicate that actin and microtubule rearrangement both contribute to the entry of F. tularensis into lung epithelial cells. PI 3-kinase and tyrosine kinase, proteins which regulate cytoskeletal rearrangement, both impact F. tularensis invasion of lung epithelial cells.
LVS traffics along the endocytic pathway in lung epithelial cells.
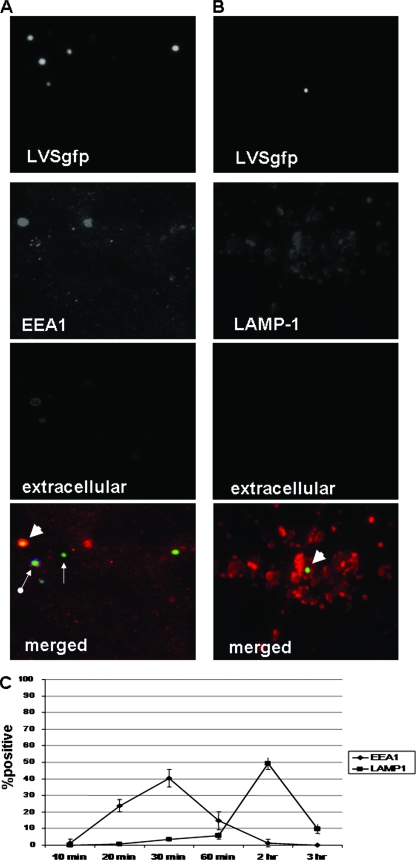
To characterize the trafficking of F. tularensis LVS along the endocytic pathway in lung epithelial cells, we synchronized the addition of LVSgfp to TC-1 cells and evaluated these cells microscopically for the presence of bacteria inside vacuoles containing the early endosomal marker EEA1 or the late endosomal/lysosomal marker LAMP-1 (Fig. 4A and B). LVS association with EEA1-containing phagosomes increased from 1.4% (± 2.4%) at 10 min to 40.4% (± 5.3%) at 30 min before decreasing (Fig. 4C). LVS association with LAMP-1-containing vacuoles peaked 2 hours postinoculation at 49.3% (± 3.5%) (Fig. 4C). These results indicate that LVS traffics along the endocytic pathway in lung epithelial cells, associating first with early endosomes before progressing to late endosomes/lysosomes.
FIG. 4.
Representative fluorescence microscopy images demonstrating LVSgfp localization within EEA1- or LAMP-1-containing endosomes in TC-1 cells. Images depict TC-1 cells stained for EEA1 30 min after inoculation with LVSgfp (A) or 2 h postinoculation stained for LAMP-1 (B). Single-color images: LVSgfp images depict bacteria alone, EEA1 and LAMP-1 images depict staining for EEA1 or LAMP-1 only, and extracellular images depict staining of extracellular bacteria (no extracellular bacteria were present in panel B). The merged color images depict LVSgfp (green), vacuoles labeled with anti-EEA1 antibody (N19) (red) or anti-LAMP-1 (1D4B) (red), or extracellular LVS labeled with anti-F. tularensis LPS antibody conjugated to Pacific Blue (blue). Extracellular LVSgfp (arrow with round end), intracellular but not EEA1-associated LVSgfp (small arrow), and LVS associated with EEA1-containing vacuoles (large arrowhead). (C) Trafficking of LVSgfp in TC-1 cells. One hundred intracellular bacteria were counted for each condition and scored for association with EEA1- and LAMP-1-containing vacuoles. Three replicates were examined for each time point and condition.
LVS escapes the phagosome and replicates in the cytoplasm of lung epithelial cells.
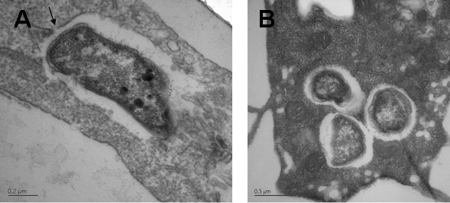
Once inside host cells, invasive bacteria either replicate within the endosome or escape the vacuole and replicate in the cytoplasm. To determine the intracellular location of bacteria, TC-1 cells were inoculated with LVS at an MOI of 100 and examined at 1 and 24 h by transmission electron microscopy. At 1 h postinoculation, LVS were contained within TC-1 cells in a membrane-bound vacuole (Fig. 5A). The membrane was easily visualized and in some cases was beginning to degrade (Fig. 5A). By 24 h, all observed intracellular bacteria were no longer contained within a visible membrane and appeared to be free in the cytoplasm (Fig. 5B), though they were surrounded by an electron-lucent zone that has been noted by other researchers (8, 19).
FIG. 5.
Transmission electron micrographs of TC-1 infected with F. tularensis LVS. (A) TC-1 cells 1 h postinoculation showing LVS in a membrane-bound vacuole that in some cases appeared to be degrading (arrow). (B) LVS cells 24 h postinoculation were free in the cytoplasm.
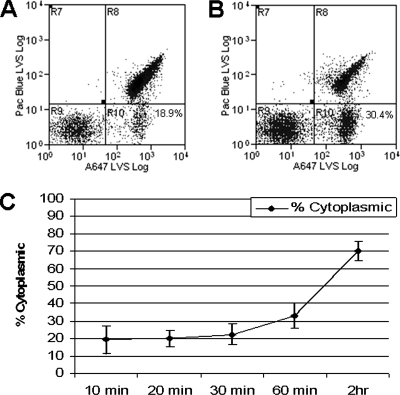
To determine the kinetics of LVS escape from the endocytic pathway into the cytoplasm of lung epithelial cells, we differentially labeled extracellular, cytoplasmic, and vacuolar LVSgfp and analyzed recovered bacteria by flow cytometry (modified from the technique of Checroun et al. [reference 7]). We first labeled bacteria that were extracellular and attached to TC-1 cells by use of Pacific Blue-conjugated anti-F. tularensis LPS. We then permeabilized the cytoplasmic membrane of TC-1 cells by use of digitonin, allowing Alexa Fluor 647-conjugated anti-F. tularensis LPS to access cytoplasmic bacteria, but not those enclosed in intact vacuoles, which are digitonin impermeable. TC-1 cells were lysed and cell-associated bacteria recovered and analyzed by flow cytometry, gating on GFP to differentiate bacteria from residual TC-1 debris. The resulting three populations of bacteria were analyzed: extracellular (Pacific Blue, Alexa Fluor 647, and GFP), cytoplasmic (Alexa Fluor 647 and GFP), and vacuolar (GFP only) (Fig. 6A and B). At 10 min postinoculation, 19.3% (± 8.1%) of intracellular bacteria were accessible to antibody delivered to the cytoplasm and therefore were considered cytoplasmic (Fig. 6C). This number increased to 70.0% (± 5.5%) at 2 h postinoculation, indicating that the majority of intracellular bacteria were no longer contained within intact endocytic vacuoles. It should be noted that once the vacuolar membrane begins to degrade, bacteria are accessible to antibody. As a result, bacteria that were associated with LAMP-1-staining vacuoles at 2 h postinoculation (Fig. 4C) would be identified as cytoplasmic by this assay if the vacuolar membrane was no longer intact. When TC-1 cells were treated with saponin, which permeabilizes both vacuolar and cytoplasmic membranes, greater than 99% of bacteria were accessible to antibody labeling (data not shown), demonstrating that a population of bacteria were protected from staining when only the cytoplasmic membrane was permeabilized. Microscopic examination of saponin-treated TC-1 cells revealed that anti-F. tularensis antibody was able to access bacteria in EEA1-containing vacuoles, while antibody was excluded from these vacuoles in digitonin-treated cells (data not shown). When TC-1 cells were inoculated with PFA-killed bacteria, >90% of organisms were vacuolar at 60 min postinoculation, indicating that killed bacteria did not escape into the cytoplasm (data not shown). Thus, bacterial viability was required for escape from the epithelial cell endosome.
FIG. 6.
LVSgfp was analyzed by flow cytometry for escape from vacuoles into the cytoplasm of TC-1 cells. Extracellular bacteria were labeled with anti-F. tularensis LPS antibody conjugated to Pacific Blue (region R8). Cytoplasmic bacteria were identified by labeling with anti-F. tularensis LPS conjugated to Alexa Fluor 647 after digitonin permeabilization of the cytoplasmic membrane (region R10). Vacuolar bacteria were inaccessible to antibody and therefore GFP positive only (region R9). Representative flow cytometry data of bacteria recovered 10 min (A) or 60 min (B) postinoculation. The value shown in region R10 represents the percentage of intracellular bacteria that are cytoplasmic. (C) Percentages of intracellular bacteria present in the cytoplasm at designated times postinoculation.
DISCUSSION
The ability of many facultative intracellular bacterial pathogens to cause disease is dependent upon their ability to invade and replicate within various host cells (14, 22, 41). F. tularensis survival and replication within macrophages and dendritic cells has been well described (1, 5, 6, 8, 33). We previously demonstrated that F. tularensis LVS also localizes to and replicates within ATII epithelial cells following inhalation of organisms in a mouse model of infection (21), prompting us to investigate lung epithelial cell invasion by this organism.
ATII cells account for about 12% of the total cells, and 2% of the surface area, of the alveolar region in mice (50). Approximately 98% of the surface area, and 10% of the total cell number, is comprised of type I cells, which provide structure and are the site of gas exchange (50). Type II cells have a number of biological functions, including the production, secretion, and recycling of surfactant; proliferation to produce additional type II cells as well as transdifferentiation into type I cells; maintenance of alveolar fluid balance; and production of antimicrobial and anti-inflammatory substances (29). Due to their close proximity to endothelial cells and their role in gas exchange and fluid balance, these cells are ideally located to provide a portal through which bacteria could disseminate to distal organs.
To characterize the interactions of F. tularensis with lung epithelial cells, we investigated the initial interactions of LVS with the lung epithelial cell line TC-1. We determined that bacteria associated with and were internalized by lung epithelial cells within 10 min of inoculation and that viable bacteria were not necessary for gaining entry into these cells. We previously found that the frequency with which F. tularensis invaded lung epithelial cell lines was low (21). The low invasion frequency may be due to the inherent inability of a transformed cell line grown in a nonpolarized fashion to recapitulate the complex environment within the host lung. These cells may also intermittently express a relevant receptor or other process that facilitates F. tularensis invasion in vivo. A system using primary cells grown in a polarized manner may more realistically reproduce the environment in the lung (18). Given the quantity of cells required for studies, cell culture lines provide a reasonable place to begin the investigation of the interaction of bacteria with epithelial cells and have been described as a model for such interactions (28).
Pathogens have developed numerous means of exploiting host cell functions to gain entry into nonphagocytic cells (10, 15). These invasion strategies are typically classified as either a zipper mechanism, which is utilized by pathogens such as Yersinia pseudotuberculosis and Listeria monocytogenes, or a trigger mechanism, which is utilized by organisms such as Salmonella enterica serovar Typhimurium and Shigella flexneri. The zipper mechanism is characterized by bacterial surface proteins binding to host cell receptors, leading to internalization. The trigger mechanism is characterized by the injection of bacterial effector proteins into the host cell via a type III secretion system, resulting in bacterial engulfment via induced macropinocytosis. Entry by either of these mechanisms requires the manipulation of host cell cytoskeletal components and signaling pathways. A number of host cell receptors play a role in F. tularensis entry into macrophages, including Fcγ, complement receptor 3, macrophage mannose receptor, and class A scavenger receptors (3, 38, 47). However, very little is known about how this organism is able to access cells that are not considered to be professional phagocytes.
To better understand the process of F. tularensis entry into lung epithelial cells, we investigated cytoskeletal components and signaling pathways that are involved in epithelial invasion by other bacterial pathogens for their contribution to internalization of LVS by a lung epithelial cell line. Lindemann et al. previously demonstrated that actin and microtubules are necessary for F. tularensis invasion of the human epithelial cell line HEp-2 (28). We determined that both actin and microtubules contribute to F. tularensis invasion of lung epithelial cells, as do PI 3-kinases and tyrosine kinases, both of which can control signaling events leading to cytoskeletal rearrangement. We showed that F. tularensis internalization by lung epithelial cells is dependent upon a variety of host cell mechanisms and that interruption of any of these mechanisms interferes with bacterial invasion.
We were unable to identify the characteristic membrane ruffling that is seen with Salmonella invasion either by phalloidin staining or field emission scanning electron microscopy under conditions where we were able to clearly identify membrane ruffling with Salmonella (data not shown). While this points toward F. tularensis not causing massive actin reorganization at the site of entry, it is possible that membrane ruffling occurs but was not detected by us. Listeria monocytogenes, another organism that is able to invade epithelial cells, also requires PI 3-kinase and tyrosine kinase function for invasion, in a cell-type-dependent manner (39, 40). This organism has two invasion proteins, InlA and InlB, which interact with different host receptors, leading to different signaling events. It is possible that F. tularensis also uses multiple receptors for entry into lung epithelial cells and that interruption of various signaling pathways could decrease uptake via various receptors. Toll-like receptors (TLRs) may play a role in the modulation of the immune response to F. tularensis (9, 23, 54), and functional TLRs are present on ATII cells (2). However, TLRs have not been implicated among macrophage receptors for F. tularensis internalization identified to date (3, 38, 47). Melillo et al. have reported that E. coli expressing the F. tularensis surface protein FsaP was able to bind A549 cells (31). We have also demonstrated that killed F. tularensis was taken up by lung epithelial cells. This information, taken together with the absence of genes predicted to encode a type III secretion system, points toward the presence of a preformed ligand receptor interaction, as with the zipper mechanism of uptake, rather than the injection into the host cell of effector proteins, as with organisms that gain entry via the trigger mechanism. Further studies are needed to identify receptors necessary for the uptake of F. tularensis by nonphagocytic cells.
To investigate F. tularensis trafficking along the endocytic pathway in lung epithelial cells, we examined the association of LVS with EEA1- and LAMP-1-containing vacuoles as well as escape into the cytoplasm. F. tularensis traffics along the endocytic pathway in macrophages before escaping to the cytoplasm, where replication occurs. The timing of this escape seems to be dependent upon the Francisella species and host cell type tested. “F. tularensis subsp. novicida” begins to disrupt the phagosomal membrane of quiescent human macrophages at 4 h and is free in the cytoplasm by 12 h postinfection (46). LVS and clinical isolates of F. tularensis associate with EEA1- and then LAMP-1-containing vacuoles in mouse bone marrow-derived macrophages, human macrophages, and mouse and human macrophage-like cell lines before degrading the phagosomal membrane and beginning to escape into the cytoplasm between 1 and 2 hours postinoculation (7, 8, 19). We determined that LVS was initially associated with EEA1-containing vacuoles, and then with LAMP-1-containing vacuoles, before being found free in the cytoplasm of lung epithelial cells in a manner and kinetic consistent with that seen for macrophages.
F. tularensis LVS is found in ATII cells in the lungs of C57BL/6 mice 1, 3, and 7 days after intranasal inoculation and replicates in these cells as disease progresses. By day 7 organisms are widespread in the alveolar epithelium (21). These observations demonstrate that that ATII cell invasion and replication is a part of the F. tularensis disease process. Understanding how this bacterium gains access to these cells, and just as importantly, how it replicates and establishes a stronghold in the lung epithelium, is necessary to understanding the progression of respiratory tularemia, as well as potentially providing insight into methods that may be used to block bacterial uptake or replication and thus prevent disease.
Acknowledgments
We gratefully acknowledge Victoria Madden of the Microscopy Services Laboratory and Larry Arnold of the Flow Cytometry Facility at UNC-CH for their expertise. We thank Richard Cheney and Jean Celli for helpful advice. We also thank Robert Fulcher and Todd Kijek for editing assistance and general advisement.
This work was supported by a Southeast Regional Center of Excellence in Biodefense and Emerging Infections grant (NIH/NIAID U54-AI057157) and by the National Institutes of Health (R21-AI053399).
Editor: J. N. Weiser
Footnotes
Published ahead of print on 21 April 2008.
REFERENCES
- 1.Anthony, L. D., R. D. Burke, and F. E. Nano. 1991. Growth of Francisella spp. in rodent macrophages. Infect. Immun. 593291-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, L., A. R. Medford, K. M. Uppington, J. Robertson, I. R. Witherden, T. D. Tetley, and A. B. Millar. 2004. Expression of functional toll-like receptor-2 and -4 on alveolar epithelial cells. Am. J. Respir. Cell Mol. Biol. 31241-245. [DOI] [PubMed] [Google Scholar]
- 3.Balagopal, A., A. S. MacFarlane, N. Mohapatra, S. Soni, J. S. Gunn, and L. S. Schlesinger. 2006. Characterization of the receptor-ligand pathways important for entry and survival of Francisella tularensis in human macrophages. Infect. Immun. 745114-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, D., K. Itoh, and C. Sasakawa. 2003. Role of microfilaments and microtubules in the invasion of INT-407 cells by Campylobacter jejuni. Microbiol. Immunol. 47469-473. [DOI] [PubMed] [Google Scholar]
- 5.Bolger, C. E., C. A. Forestal, J. K. Italo, J. L. Benach, and M. B. Furie. 2005. The live vaccine strain of Francisella tularensis replicates in human and murine macrophages but induces only the human cells to secrete proinflammatory cytokines. J. Leukoc. Biol. 77893-897. [DOI] [PubMed] [Google Scholar]
- 6.Bosio, C. M., and S. W. Dow. 2005. Francisella tularensis induces aberrant activation of pulmonary dendritic cells. J. Immunol. 1756792-6801. [DOI] [PubMed] [Google Scholar]
- 7.Checroun, C., T. D. Wehrly, E. R. Fischer, S. F. Hayes, and J. Celli. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. USA 10314578-14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 723204-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, L. E., K. A. Shirey, E. Barry, A. Santiago, P. Rallabhandi, K. L. Elkins, A. C. Puche, S. M. Michalek, and S. N. Vogel. 2007. Toll-like receptor 2-mediated signaling requirements for Francisella tularensis live vaccine strain infection of murine macrophages. Infect. Immun. 754127-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cossart, P., and P. J. Sansonetti. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304242-248. [DOI] [PubMed] [Google Scholar]
- 11.Dennis, D. T., T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, M. Layton, S. R. Lillibridge, J. E. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 2852763-2773. [DOI] [PubMed] [Google Scholar]
- 12.Dramsi, S., and P. Cossart. 1998. Intracellular pathogens and the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14137-166. [DOI] [PubMed] [Google Scholar]
- 13.Feldman, K. A., R. E. Enscore, S. L. Lathrop, B. T. Matyas, M. McGuill, M. E. Schriefer, D. Stiles-Enos, D. T. Dennis, L. R. Petersen, and E. B. Hayes. 2001. An outbreak of primary pneumonic tularemia on Martha's Vineyard. N. Engl. J. Med. 3451601-1606. [DOI] [PubMed] [Google Scholar]
- 14.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 835189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finlay, B. B., and P. Cossart. 1997. Exploitation of mammalian host cell functions by bacterial pathogens. Science 276718-725. [DOI] [PubMed] [Google Scholar]
- 16.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol Rev. 61136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forslund, A. L., K. Kuoppa, K. Svensson, E. Salomonsson, A. Johansson, M. Bystrom, P. C. Oyston, S. L. Michell, R. W. Titball, L. Noppa, E. Frithz-Lindsten, M. Forsman, and A. Forsberg. 2006. Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol. Microbiol. 591818-1830. [DOI] [PubMed] [Google Scholar]
- 18.Gentry, M., J. Taormina, R. B. Pyles, L. Yeager, M. Kirtley, V. L. Popov, G. Klimpel, and T. Eaves-Pyles. 2007. Role of primary human alveolar epithelial cells in host defense against Francisella tularensis infection. Infect. Immun. 753969-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golovliov, I., V. Baranov, Z. Krocova, H. Kovarova, and A. Sjostedt. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 715940-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golovliov, I., A. Sjostedt, A. Mokrievich, and V. Pavlov. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222273-280. [DOI] [PubMed] [Google Scholar]
- 21.Hall, J. D., R. R. Craven, J. R. Fuller, R. J. Pickles, and T. H. Kawula. 2006. Francisella tularensis replicates within alveolar type II epithelial cells in vitro and in vivo following inhalation. Infect. Immun. 751034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284967-970. [DOI] [PubMed] [Google Scholar]
- 23.Hong, K. J., J. R. Wickstrum, H. W. Yeh, and M. J. Parmely. 2007. Toll-like receptor 2 controls the gamma interferon response to Francisella tularensis by mouse liver lymphocytes. Infect. Immun. 755338-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hume, D. A., I. L. Ross, S. R. Himes, R. T. Sasmono, C. A. Wells, and T. Ravasi. 2002. The mononuclear phagocyte system revisited. J. Leukoc. Biol. 72621-627. [PubMed] [Google Scholar]
- 25.Ireton, K., B. Payrastre, H. Chap, W. Ogawa, H. Sakaue, M. Kasuga, and P. Cossart. 1996. A role for phosphoinositide 3-kinase in bacterial invasion. Science 274780-782. [DOI] [PubMed] [Google Scholar]
- 26.Khan, A. S., S. Morse, and S. Lillibridge. 2000. Public-health preparedness for biological terrorism in the USA. Lancet 3561179-1182. [DOI] [PubMed] [Google Scholar]
- 27.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. USA 1014246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindemann, S. R., M. K. McLendon, M. A. Apicella, and B. D. Jones. 2007. An in vitro model system used to study adherence and invasion of Francisella tularensis live vaccine strain in nonphagocytic cells. Infect. Immun. 753178-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason, R. J. 2006. Biology of alveolar type II cells. Respirology 11(Suppl.)S12-S15. [DOI] [PubMed] [Google Scholar]
- 30.Meibom, K. L., I. Dubail, M. Dupuis, M. Barel, J. Lenco, J. Stulik, I. Golovliov, A. Sjostedt, and A. Charbit. 2008. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol. Microbiol. 671384-1401. [DOI] [PubMed] [Google Scholar]
- 31.Melillo, A., D. D. Sledjeski, S. Lipski, R. M. Wooten, V. Basrur, and E. R. Lafontaine. 2006. Identification of a Francisella tularensis LVS outer membrane protein that confers adherence to A549 human lung cells. FEMS Microbiol. Lett. 263102-108. [DOI] [PubMed] [Google Scholar]
- 32.Nano, F. E., N. Zhang, S. C. Cowley, K. E. Klose, K. K. Cheung, M. J. Roberts, J. S. Ludu, G. W. Letendre, A. I. Meierovics, G. Stephens, and K. L. Elkins. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 1866430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nutter, J. E., and Q. N. Myrvik. 1966. In vitro interactions between rabbit alveolar macrophages and Pasteurella tularensis. J. Bacteriol. 92645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oelschlaeger, T. A., P. Guerry, and D. J. Kopecko. 1993. Unusual microtubule-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc. Natl. Acad. Sci. USA 906884-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oyston, P. C., A. Sjostedt, and R. W. Titball. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2967-978. [DOI] [PubMed] [Google Scholar]
- 36.Patel, J. C., and J. E. Galan. 2005. Manipulation of the host actin cytoskeleton by Salmonella—all in the name of entry. Curr. Opin. Microbiol. 810-15. [DOI] [PubMed] [Google Scholar]
- 37.Pechous, R., J. Celli, R. Penoske, S. F. Hayes, D. W. Frank, and T. C. Zahrt. 2006. Construction and characterization of an attenuated purine auxotroph in a Francisella tularensis live vaccine strain. Infect. Immun. 744452-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierini, L. M. 2006. Uptake of serum-opsonized Francisella tularensis by macrophages can be mediated by class A scavenger receptors. Cell. Microbiol. 81361-1370. [DOI] [PubMed] [Google Scholar]
- 39.Pizarro-Cerda, J., and P. Cossart. 2006. Subversion of cellular functions by Listeria monocytogenes. J. Pathol. 208215-223. [DOI] [PubMed] [Google Scholar]
- 40.Pizarro-Cerda, J., and P. Cossart. 2004. Subversion of phosphoinositide metabolism by intracellular bacterial pathogens. Nat. Cell Biol. 61026-1033. [DOI] [PubMed] [Google Scholar]
- 41.Qin, A., and B. J. Mann. 2006. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raynaud, C., K. L. Meibom, M. A. Lety, I. Dubail, T. Candela, E. Frapy, and A. Charbit. 2007. Role of the wbt locus of Francisella tularensis in lipopolysaccharide O-antigen biogenesis and pathogenicity. Infect. Immun. 75536-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson, W. P., and J. C. Sadoff. 1988. Induced engulfment of Neisseria gonorrhoeae by tissue culture cells. Infect. Immun. 562512-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenberger, C. M., and B. B. Finlay. 2003. Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat. Rev. Mol. Cell Biol. 4385-396. [DOI] [PubMed] [Google Scholar]
- 45.Rosenshine, I., V. Duronio, and B. B. Finlay. 1992. Tyrosine protein kinase inhibitors block invasin-promoted bacterial uptake by epithelial cells. Infect. Immun. 602211-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santic, M., M. Molmeret, and Y. Abu Kwaik. 2005. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell. Microbiol. 7957-967. [DOI] [PubMed] [Google Scholar]
- 47.Schulert, G. S., and L. A. Allen. 2006. Differential infection of mononuclear phagocytes by Francisella tularensis: role of the macrophage mannose receptor. J. Leukoc. Biol. 80563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staples, J. E., K. A. Kubota, L. G. Chalcraft, P. S. Mead, and J. M. Petersen. 2006. Epidemiologic and molecular analysis of human tularemia, United States, 1964-2004. Emerg. Infect. Dis. 121113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steele-Mortimer, O., J. H. Brumell, L. A. Knodler, S. Meresse, A. Lopez, and B. B. Finlay. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell. Microbiol. 443-54. [DOI] [PubMed] [Google Scholar]
- 50.Stone, K. C., R. R. Mercer, P. Gehr, B. Stockstill, and J. D. Crapo. 1992. Allometric relationships of cell numbers and size in the mammalian lung. Am. J. Respir. Cell Mol. Biol. 6235-243. [DOI] [PubMed] [Google Scholar]
- 51.Takenawa, T., and T. Itoh. 2001. Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim. Biophys. Acta 1533190-206. [DOI] [PubMed] [Google Scholar]
- 52.Tarnvik, A., H. S. Priebe, and R. Grunow. 2004. Tularaemia in Europe: an epidemiological overview. Scand. J. Infect. Dis. 36350-355. [DOI] [PubMed] [Google Scholar]
- 53.Teis, D., and L. A. Huber. 2003. The odd couple: signal transduction and endocytosis. Cell. Mol. Life Sci. 602020-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thakran, S., H. Li, C. L. Lavine, M. A. Miller, J. E. Bina, X. R. Bina, and F. Re. 2008. Identification of Francisella tularensis lipoproteins that stimulate the Toll-like receptor (TLR) 2/TLR1 heterodimer. J. Biol. Chem. 2833751-3760. [DOI] [PubMed] [Google Scholar]
- 55.Valiron, O., N. Caudron, and D. Job. 2001. Microtubule dynamics. Cell. Mol. Life Sci. 582069-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]