Abstract
We previously reported that the novel Pseudomonas aeruginosa toxin Cif is capable of decreasing apical membrane expression of the cystic fibrosis transmembrane conductance regulator (CFTR). We further demonstrated that Cif is capable of degrading the synthetic epoxide hydrolase (EH) substrate S-NEPC [(2S,3S)-trans-3-phenyl-2-oxiranylmethyl 4-nitrophenol carbonate], suggesting that Cif may be reducing apical membrane expression of CFTR via its EH activity. Here we report that Cif is capable of degrading the xenobiotic epoxide epibromohydrin (EBH) to its vicinal diol 3-bromo-1,2-propanediol. We also demonstrate that this epoxide is a potent inducer of cif gene expression. We show that the predicted TetR family transcriptional repressor encoded by the PA2931 gene, which is immediately adjacent to and divergently transcribed from the cif-containing, three-gene operon, negatively regulates cif gene expression by binding to the promoter region immediately upstream of the cif-containing operon. Furthermore, this protein-DNA interaction is disrupted by the epoxide EBH in vitro, suggesting that the binding of EBH by the PA2931 protein product drives the disassociation from its DNA-binding site. Given its role as a repressor of cif gene expression, we have renamed PA2931 as CifR. Finally, we demonstrate that P. aeruginosa strains isolated from cystic fibrosis patient sputum with increased cif gene expression are impaired for the expression of the cifR gene.
Pseudomonas aeruginosa is a ubiquitous opportunistic pathogen known to infect a variety of organisms, including plants, nematodes, fruit flies, and humans (19, 30, 49). Disease in humans shows a relative lack of tropism, as infections caused by P. aeruginosa can be found in tissues as varied as the skin, ocular epithelia, and the lung (3, 28, 31). This pathogenic plasticity is largely due to the presence of a multiplicity of virulence factors which allow for colonization and the establishment of infection (1). This arsenal of virulence factors includes the well-characterized type three secretion system and its effector molecules, elastases, phospholipases, phenazines, and rhamnolipids (6, 9, 25, 27, 44, 47, 52). These virulence factors contribute to both acute and chronic infections associated with burn wounds, ocular infections, P. aeruginosa ventilator-associated pneumonia, and cystic fibrosis (CF).
The genetic disease CF is the result of heritable mutations within the cystic fibrosis transmembrane conductance regulator (CFTR), the most common of which is the deletion of the phenylalanine at position 508 (18, 37, 38, 48). In healthy airway epithelia cells, CFTR acts to directly regulate the flux of Cl− ions and, indirectly, the flux of Na+ ions and water across the apical membrane (22, 37). Airway epithelial cells in CF patients demonstrate decreased functional CFTR at their apical membrane; thus, there is altered Cl− and Na+ ion and water flux across this membrane, resulting in increased viscosity and decreased height of the periciliary fluid, which in turn leads to decreased ciliary beating and a subsequent loss of the mucociliary elevator, a key component of the innate immune system. This breach in mucociliary clearance allows for a number of different pathogens, including P. aeruginosa, to colonize the CF lung and establish chronic infections (5). Following initial colonization by environmental isolates, these bacteria undergo a series of phenotypic changes including the acquisition of auxotrophy for various nutrients, alterations in quorum-sensing machinery, and the overproduction of the exopolysaccharide alginate (11-14, 41). The overproduction of alginate results in the mucoid phenotype associated with P. aeruginosa sputum isolates from chronically infected CF patients. The mucoid phenotype has been well studied and is believed to be the result of selective pressures including exposure to antibiotics as well as the host innate immune system (12, 13, 35).
We previously described a novel P. aeruginosa toxin that is packaged into outer membrane vesicles and is capable of dramatically reducing apical membrane expression of CFTR in several different epithelial cell lines (29, 46). This toxin, known as Cif, was predicted to belong to the family of epoxide hydrolases (EHs) and was experimentally shown to degrade the synthetic EH substrate S-NEPC [(2S,3S)-trans-3-phenyl-2-oxiranylmethyl 4-nitrophenol carbonate]. While it is not entirely understood how this EH activity may regulate apical membrane expression of CFTR, epoxides and their metabolites have been shown to act as signaling molecules in both endothelial and epithelial cells, regulating processes as varied as vasodilation and Cl− ion transport (33, 42). We found that a subset of P. aeruginosa strains isolated from the CF airway demonstrated a marked increase in cif transcription relative to laboratory strains, suggesting a role for Cif in the pathophysiology of CF.
Our previous demonstration that cif was differentially expressed in mucoid versus nonmucoid CF clinical isolates (29) led us to investigate the regulation of cif gene expression. Here we show that the Cif protein is capable of degrading the epoxide epibromohydrin (EBH) and that this xenobiotic compound induces cif gene expression via its ability to alter DNA binding of the TetR family regulator CifR. Finally, we show that reduced cifR gene expression can account for the high level of cif gene expression observed in the nonmucoid P. aeruginosa CF sputum isolates.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
All of the bacterial strains and plasmids used in this study are shown in Table 1. All bacterial strains were grown in lysogeny broth (LB) unless otherwise noted (2). Growth media were supplemented with antibiotics at the following concentrations: gentamicin, 10 μg/ml (Escherichia coli) and 100 μg/ml (P. aeruginosa); and ampicillin, 150 μg/ml (E. coli) and 1.5 mg/ml (P. aeruginosa). All strains were grown at 37°C. Yeast cultures were grown in either rich (yeast extract-peptone-dextrose) or minimal (SD-Ura; Sunrise Science Products, San Diego, CA) media at 30°C. All restriction enzymes were purchased from New England Biolabs (Beverly, MA). All plasmids were constructed in E. coli Top10 cells using standard protocols or in Saccharomyces cerevisiae INVSc1 (Invitrogen, Carlsbad, CA) using in vivo recombination and electroporated or conjugated into P. aeruginosa strain PA14, as previously reported (8, 40).
TABLE 1.
Strains, plasmids, and primers used
| Strain, plasmid, or primer | Relevant characteristic | Reference or source |
|---|---|---|
| Strain | ||
| S. cerevisiae INVSc1 | MATahis3D1 leu2 trp1-289 ura3-52 MAT his3D1 leu2 trp1-289 ura3-52 | Invitrogen |
| E. coli Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) f80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| SMC3505 | E. coli Top10 + pDPM73 (Cif-His) | 29 |
| SMC3814 | E. coli Top10 + pDPM79 (CifR-His) | This study |
| P. aeruginosa PA14 | Wild type | |
| SMC1584 | P. aeruginosa clinical isolate | 29 |
| SMC1586 | P. aeruginosa clinical isolate | 29 |
| SMC1591 | P. aeruginosa clinical isolate | 29 |
| SMC1593 | P. aeruginosa clinical isolate | 29 |
| SMC3816 | PA14 ΔPA2931 + pDPM79 | This study |
| SMC3817 | SMC1584 + pMQ71 | This study |
| SMC3818 | SMC1584 + pDPM79 | This study |
| SMC3819 | SMC1586 + pMQ71 | This study |
| SMC3820 | SMC1586 + pDPM79 | This study |
| Plasmid | ||
| pMQ71 | Arabinose-inducible expression vector Cbr Apr URA3 | 40 |
| pMQ30 | Allelic replacement vector; Gmr | 40 |
| pDPM79 | PA2931 hexa-histidine fusion in pMQ71; Apr Cbr URA3 | This study |
| pDPM84 | PA2931 in-frame deletion plasmid; Gmr | This study |
| Primer | ||
| PA2934_rt_1 | 5′-CTCCTGGCCGGCATCGCCCTGACCTTCTCC-3′ | |
| PA2934_rt_2 | 5′-CCATTCGTACCAGGTCTGGCCGAAGCCGTGC-3′ | |
| RplU_rt_1 | 5′-GGTGGCAAGCAGCACAAAGTCACCG-3′ | |
| RplU_rt_2 | 5′-GCGGACCTTGTCGTGACGGCCGTGG-3′ | |
| PA2931_rt_for | 5′-CCAGGGCTTCGCGGAACAGCCCTTCC-3′ | |
| PA2931_rt_rev | 5′-GACACCGCCCTGCAGCGAGCCATGG-3′ | |
| Cif_northern_for | 5′-BT-GCCGCGGCCTCCTGGCCGGCATCGCCC-3′a | |
| Cif_northern_rev | 5′-BT-CGGCGACCGTTCGCCGCTATAGCCGG-3′ | |
| PA2931_ko_1 | 5′-CGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACG GCTGGAAGGCTCCAAGGTAGAATGGATGG-3′ | |
| PA2931_ko_2 | 5′-CAAGTAAATTTATAGTGATCGATACAAATATGAGCG GGTCGCATCAGGCCGCC-3′ | |
| PA2931_ko_3 | 5′-GGCGGCCTGATGCGACCCGCTCATATTTGTATC GATCACTATAAATTTACTTG-3′ | |
| PA2931_ko_4 | 5′-GTGAGCGGATAACAATTTCACACAGGAAACAGCTAT GACCGCAGGCGCTCGCGGAAGCCTTGC-3′ | |
| PA2931_His_for | 5′-GAAAATCTTCTCTCATCCGCCTCAATGATGATGATG ATGATGGGGCCAGGCGCGCAGCGCCCGTTCG-3′ | |
| PA2931_His_rev | 5′-CTCCATACCCGTTTTTTTGGGCTAGCCCAAGGA AGCACACATATGGCAACGCGAGGCAGGCCAC-3′ | |
| EMSA_5′bt_for | 5′-BT-CCTGACCTCCATTATTTGTATCGATCAC-3′ | |
| EMSA_5′bt_rev | 5′-BT-GAGCAGGTTGGACATGGTCTTTTCC-3′ | |
| EMSA_for | 5′-CCTGACCTCCATTATTTGTATCGATCAC-3′ | |
| EMSA_rev | 5′-GAGCAGGTTGGACATGGTCTTTTCC-3′ |
BT, biotinylated.
EBH degradation assays.
EBH degradation by Cif was performed as previously described (7). Briefly, 50 μg of purified Cif was incubated with EBH at a final concentration of 10 mM in 20 mM HEPES buffer (pH 7.4) and 500 mM NaCl for 30 min at 37°C. Sodium periodate in acetonitrile was added at final concentrations of 1.67 mM and 30%, respectively, and incubated at room temperature for 1 h. Epinephrine-HCl was then added to a final concentration of 1.5 mM. The reaction was allowed to continue for 15 min at room temperature prior to removal of the supernatant and subsequent detection at a wavelength of 490 nM with a Molecular Devices SpectrMax M2 plate reader. Degradation of EBH was demonstrated by a reduction in absorbance at 490 nM.
EBH induction assays.
Wild-type P. aeruginosa PA14 was grown overnight in LB at 37°C and subsequently diluted 1:100 in LB supplemented with EBH at the concentrations indicated for each experiment. Cultures were grown at 37°C with shaking to an optical density at 600 nm (OD600) of 1.0. Cultures (500 μl) were centrifuged and pellets were frozen at −80°C. Samples for protein analysis were boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer as previously described and subsequently resolved using SDS-PAGE. Cif protein expression was analyzed using standard Western blotting as previously described (29). RNA for semiquantitative reverse transcription-PCR (sqRT-PCR) was harvested as described below.
Detection of the morB-PA2933-cif transcript.
RNA for Northern blot analysis was harvested as described below. Samples were prepared, electrophoresed, and transferred as previously described(17). Briefly, 20 μg of RNA was denatured using formaldehyde and formamide in MOPS (morpholinepropanesulfonic acid) buffer at 65°C for 15 min. Gel loading buffer was added to the denatured RNA, which was subsequently electrophoresed in a MOPS-formaldehyde-agarose gel for 1 h at 80 V. RNA was transferred to a Biodyne B modified nylon membrane (Thermo Scientific, Rockford, IL) using a standard wicking transfer for 4 to 8 h. Following cross-linking and blocking, 500 ng of a biotinylated cif-specific probe was added to the blocking buffer and incubated at 65°C for 12 h. The cif probe was synthesized using the biotinylated primers Cif_northern_for and Cif_northern_rev (Table 1) in a standard PCR followed by gel purification. Membranes were washed with SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and SDS buffer and subsequently labeled with a steptavidin-horseradish peroxidase conjugate followed by detection using a LightShift chemiluminescence kit per the manufacturer's instructions (Pierce, Rockford, IL).
Plasmid construction.
Construction of the PA2931 deletion plasmid was performed as previously described (40) using primers PA2931_ko_1, PA2931_ko_2, PA2931_ko_3, and PA2931_ko_4. Amplicons were created using primer pair ko_1 and ko_2 and primer pair ko_3 and ko_4 and were recombined using S. cerevisiae INVSc1 (Invitrogen) into plasmid pMQ30 linearized with restriction enzymes EcoRI, HindIII, and BamHI. The resulting recombinants were lysed and plasmids recovered by using standard techniques. Plasmid pDPM84 was transformed into wild-type P. aeruginosa strain PA14 as previously described (8), and transformants were selected for on solid LB media supplemented with gentamicin. Merodiploids were resolved on solid LB media supplemented with 10% sucrose as previously described (45).
Plasmid pDPM79 containing the C-terminal, six-histidine-tagged variant of PA2931 was constructed using standard yeast recombination techniques. Briefly, the PA2931 open reading frame (ORF) was amplified using primers PA2931_His_for and PA2931_His_rev, which contain regions homologous to the PA2931 ORF and the plasmid pMQ71. The resulting amplicon was recombined into the plasmid pMQ71.
Purification of the Cif protein.
Purification of the histidine-tagged Cif protein was performed as previously described (29).
Purification of the CifR protein.
The hexa-histidine-tagged variant of the PA2931 protein was expressed from the arabinose-inducible pDPM79 plasmid in E. coli Top10 cells (Invitrogen, Carlsbad, CA). Cells containing the plasmid were grown overnight at 37°C in LB supplemented with ampicillin, diluted 1:100 in 1 liter of LB supplemented with ampicillin and 0.2% arabinose, and incubated with shaking at 37°C for 8 h. Cultures were centrifuged at 7,000 × g for 20 min. Cell pellets were then mechanically lysed via French pressure lysis. Lysates were centrifuged at 20,000 × g for 30 min to remove cellular debris. Supernatants from centrifuged lysates were fractionated utilizing an Amersham HisTrap FF 5-ml nickel affinity column. Protein was eluted over a 20 to 500 mM imidazole gradient, with the bulk of the purified PA2931 protein eluting at ∼100 mM. Fractions containing PA2931 were pooled and concentrated using Amicon Ultra-15 10-kDa centrifugation columns (Millipore, Billerica, MA). The protein solution was then dialyzed against 20 mM Tris-500 mM NaCl, pH 7.5. The protein concentration was determined by utilizing a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). Protein purity was determined using SDS-PAGE followed by staining with Coomassie blue.
EMSA.
Electromobility shift assays (EMSA) were performed using a LightShift chemiluminescence EMSA kit (Pierce, Rockford, IL) per the manufacturer's instructions. The PA2931-morB intergenic region was amplified using primers EMSA_5′bt_for, EMSA_5′bt_rev, EMSA_for, and EMSA_rev, resulting in either the biotinylated probe or the cold-competitor DNA. The biotinylated probe was then incubated with the purified PA2931 protein for 30 min. Samples were loaded onto 10× Tris-borate-EDTA-PAGE gels and electrophoresed. Following electrophoresis, samples were electro-transferred to a Biodyne nylon membrane (Rockford, IL) in 0.5 Tris-borate-EDTA. Samples were then UV cross-linked to the membrane and blocked according to the manufacturer's instructions. Labeling and subsequent detection were carried out using a streptavidin-horseradish peroxidase conjugate and chemiluminescence, as described by the manufacturer. When EBH was included in the EMSA experiments, it was added simultaneously with the DNA probe and protein prior to incubation at 37°C.
RNA purification and cDNA synthesis.
RNA purification and cDNA synthesis were performed as previously described (29) with the following modifications: strains for sqRT-PCR were grown overnight in LB medium and subsequently diluted 1:100 in LB medium and grown to an OD600 of 1.0. Cultures (500 μl) were harvested and centrifuged at 16,000 × g for 2 min, and the cell pellets were frozen at −80°C. Strains were grown in triplicate, and two samples were harvested per replicate.
RESULTS
Cif degrades the epoxide EBH.
We previously reported that the P. aeruginosa secreted toxin Cif is capable of degrading the generic EH substrate S-NEPC, suggesting that this protein may act as an EH (29). A previous report by Jacobs et al. identified a soil pseudomonad capable of surviving on and degrading the xenobiotic epoxide EBH (24). Further analysis demonstrated that the biochemical activity produced by this pseudomonad was the result of a secreted protein with a mass similar to that of the Cif protein. Functional analysis of the secreted protein demonstrated that it was an EH. The amino acid sequence of the N terminus of this EH and three CNBr-generated fragments of the protein shared up to 40% identity with the Cif protein. The work by Jacobs and colleagues, as well as our previous demonstration that Cif could degrade S-NEPC, led us to hypothesize that Cif may be capable of degrading the xenobiotic epoxide EBH.
To assay EH activity with EBH as the substrate, we monitored the production of the vicinal diol of EBH, 3-bromo-1,2-propanediol. This product is readily oxidized by periodate, producing aldehyde and ketone variants of 3-bromo-1,2-propanediol. The oxidation of 3-bromo-1,2-propanediol by periodate results in the reduction of periodate. The reduction of 3-bromo-1,2-propanediol by periodate can be detected using epinephrine, which itself is readily oxidized by periodate, resulting in a red color. The reduction of periodate results in an inability to oxidize epinephrine and hence no colorimetric change (7). Thus, the degradation of EBH by Cif can be readily assayed based on the fact that as more EBH is degraded, more periodate is reduced, and hence less epinephrine is oxidized (Fig. 1A).
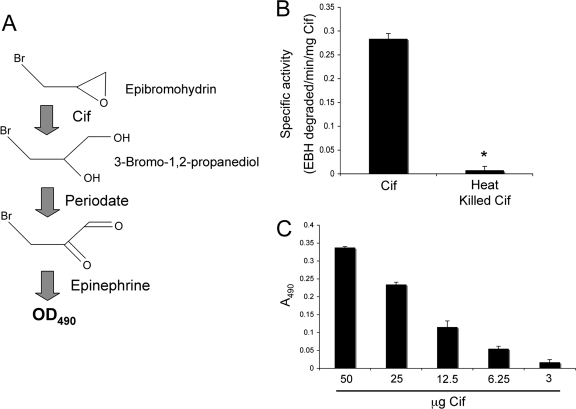
FIG. 1.
Cif degrades the epoxide EBH. The degradation of epoxides by EHs creates highly reactive vicinal diols. The colorimetric detection of these diols is outlined in panel A. Generation of the vicinal diol of EBH by Cif can be detected spectrophotometrically through the oxidation of 3-bromo-1,2-propanediol by periodate and subsequent analysis of the redox state of epinephrine added to the reaction. (B) Purified Cif (50 μg) was incubated with 10 mM EBH, and absorbance at 490 nm was monitored. Also shown is a heat-inactivated protein (heat kill) control. Data were normalized to samples containing buffer alone. *, P value of <0.05. Values are presented as specific activity (EBH degraded/min/mg Cif). (C) Dose response of EBH degradation by Cif. Bar 1 shows 10 mM EBH and 50 μg Cif; bar 2, EBH and 25 μg Cif; bar 3, EBH and 12.5 μg Cif; bar 4, EBH and 6.25 μg Cif; and bar 6, EBH and 3 μg Cif. Samples were incubated at 37°C for 15 min. Values are presented as the OD490, with the OD490 measurements of each condition subtracted from the control condition containing EBH alone.
When we incubated EBH with the buffer alone, we observed a red color detected at OD490, indicating oxidation of the epinephrine due to the presence of periodate (data not shown). However, when purified Cif was incubated with EBH, we observed an absorbance threefold less than what we observed in the no-protein control, consistent with our hypothesis that Cif is an EH and is capable of degrading EBH (Fig. 1B). Furthermore, when Cif was heated to 95°C for 20 min (heat kill), thus denaturing the protein, we saw a complete loss of the EH activity compared to activity with the non-heat-treated protein. As an additional control, supernatant from E. coli Top10 cells harboring the empty vector pMQ70 was fractionated using conditions similar to those used to purify the Cif protein. Neither the supernatant alone nor fractions derived from the supernatant of the empty vector strain demonstrated any degradation of EBH (data not shown).
To further test Cif's ability to degrade EBH, we performed a dose-response experiment, incubating EBH with decreasing amounts of purified Cif (Fig. 1C). Consistent with Cif's ability to degrade EBH, we observed decreasing EBH degradation with decreasing concentrations of the Cif protein.
Both cif gene and Cif protein expression are induced by EBH.
The expression of a variety of catabolic genes is induced in the presence of their cognate substrates. Typically, this increased gene expression is the result of a regulatory protein that either induces gene expression or derepresses expression in the presence of the cognate effector molecule. The observation that Cif is capable of degrading the epoxide EBH led us to hypothesize that cif gene expression may be positively regulated in the presence of this molecule.
To assess the response of cif gene expression to EBH, we grew wild-type P. aeruginosa PA14 in the presence of increasing concentrations of EBH and then performed sqRT-PCR. We found that the addition of EBH to P. aeruginosa cultures resulted in a dose-dependent increase in cif transcript levels, while the transcript levels of the control rplU transcript remained largely unchanged (Fig. 2A).
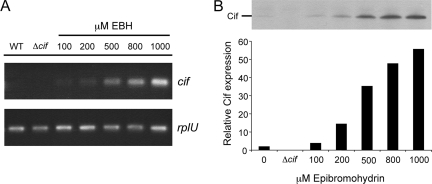
FIG. 2.
EBH induction of cif gene and Cif protein expression. Wild-type (WT) P. aeruginosa strain PA14 was grown in LB supplemented with increasing concentrations of the epoxide EBH. Expression of the cif gene and Cif protein in response to EBH was assayed using either sqRT-PCR (A) or Western blotting (B), respectively. The expression of rplU, a gene previously shown to be constitutively expressed under most laboratory conditions, served as a control (panel A). Samples for Western blotting were normalized to total protein content (panel B). The graphical interpretation of the Western blot data shown in panel B is the relative Cif expression normalized to the total protein electrophoresed.
To further confirm these results, wild-type P. aeruginosa cultures grown under identical conditions were assayed by Western blotting (Fig. 2B). We also found a dose-dependent increase in Cif protein levels. These results demonstrate that EBH is a potent inducer of cif gene expression, resulting in increased Cif protein levels.
cif is transcribed as part of a three-gene operon.
The observation that EBH induced expression of the cif gene led us to further investigate the regulation of this gene. The chromosomal organization of the cif gene predicts that it belongs to a three-gene operon with the morB and PA2933 genes (Fig. 3A). To determine if in fact cif was transcribed as part of a polycistronic mRNA, we performed Northern analysis using a biotinylated probe specific for the cif gene. RNA was isolated from wild-type PA14, the Δcif mutant, and the ΔPA2931 mutant and probed for cif gene expression.
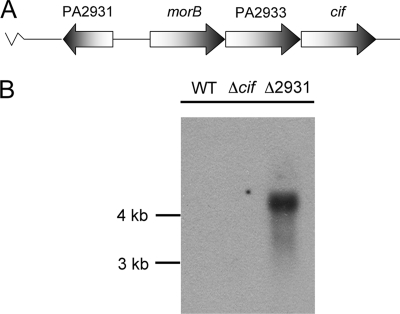
FIG. 3.
cif is cotranscribed with morB and PA2933. (A) The cif gene is predicted to belong to a three-gene operon including morB, PA2933, and cif. (B) RNA from the wild type (WT) and the Δcif and ΔPA2931 mutants was assayed using Northern blotting to determine the transcript size detected by the cif-specific probe. A single band at ∼4.2 kb was detected in the ΔPA2931 mutant.
A single band was detected at 4.2 kb, consistent with the prediction that cif is cotranscribed with the morB and PA2933 genes (Fig. 3B). Due to relatively low cif transcription levels, we were unable to detect the cif-containing operon transcript via Northern analysis in the wild-type genetic background. However, in a P. aeruginosa PA14 mutant that overexpresses cif as a result of a mutation in the PA2931 ORF (see below), we were able to detect the transcript of the appropriate size. These data suggest that all three genes are transcribed from the predicted promoter region immediately adjacent to the morB gene.
PA2931 negatively regulates cif gene expression.
With the demonstration that cif gene expression was variable and inducible, we next sought to identify genes that regulate cif gene expression. Divergently transcribed from the morB gene is the PA2931 gene, predicted to encode a TetR family DNA-binding protein. Members of this family are known to regulate gene expression by binding to the promoter region and occluding the RNA polymerase-binding site, thus inhibiting gene expression. Via in silico analysis, we found a predicted promoter region immediately adjacent to the morB-PA2933-cif operon. The relative proximity and orientation of the PA2931 gene led us to hypothesize that it negatively regulates cif gene expression.
To test this hypothesis, a clean deletion of the PA2931 gene was created in P. aeruginosa PA14. A hexa-histidine-tagged variant of the cloned PA2931 gene was created in the multicopy, arabinose-inducible plasmid pMQ71 (40) (designated pDPM79) to be used both in complementation studies here and for expression of the PA2931 protein for purification described below. RNA from these strains was isolated, and cif and rplU gene expression were assayed by using sqRT-PCR. These strains were also assayed for Cif protein expression by using Western blotting.
Deletion of the PA2931 gene resulted in a marked increase in cif gene expression (Fig. 3B and 4A). Introduction of the PA2931 histidine-tagged variant expressed from the plasmid pDPM79 into the PA2931 deletion strain resulted in a restoration of wild-type levels of cif transcript.
FIG. 4.
cifR negatively regulates cif gene expression. cif gene and Cif protein expression were assayed using either sqRT-PCR or Western blotting, respectively. (A) The wild type (WT), the Δcif mutant, and the ΔPA2931 mutant harboring the empty vector pMQ71 or expressing the PA2931 gene from the plasmid pDPM79 were assayed for cif and rplU gene expression. Deletion of the PA2931 gene resulted in higher levels of cif gene transcript, which was complemented by expression of the histidine-tagged variant of PA2931. (B) Western blot of the WT P. aeruginosa strain PA14, the ΔPA2931 mutant containing the empty vector pMQ71, and the ΔPA2931 mutant carrying a plasmid expressing a histidine-tagged variant of PA2931 from the arabinose-inducible expression plasmid pDPM79. Arabinose was added to a final concentration of 0.2% where indicated.
The marked increase in expression seen in the ΔPA2931 strain, compared to that in the wild-type strain, required the use of a low number of PCR cycles in order to demonstrate the difference in expression and to capture cif expression in the ΔPA2931 strain during the linear range of the PCR. These PCR conditions resulted in almost undetectable levels of the cif amplicon in the wild-type strain. The use of a higher number of cycles on identical samples demonstrated that the cif transcript was detectable in the wild-type and complemented strains but not in the Δcif mutant strain, as would be expected (data not shown).
These data were further confirmed by using Western blotting. Cif protein expression was increased in the PA2931 deletion strain carrying the empty vector pMQ71, while strains expressing the histidine-tagged variant of the PA2931 protein from the plasmid pDPM79 demonstrated reduced Cif protein expression compared to that of the vector control (Fig. 4B).
These data taken together demonstrate that the protein encoded by the PA2931 gene negatively regulates cif gene transcription, resulting in decreased Cif protein expression. Based on these data, we have renamed the PA2931 gene cifR, reflecting its ability to repress cif gene expression.
CifR shows EBH-dependent binding upstream of the morB-PA2933-cif operon.
The TetR family of repressors has previously been shown to bind directly to the promoter region upstream of the genes they regulate in the absence of their cognate effector molecules (21, 23, 36). As mentioned above, the CifR protein is predicted to be a TetR family member. Considering this data together with the demonstration that the cifR gene is important for repression of cif gene expression, we hypothesized that CifR is repressing cif gene expression by binding to the predicted promoter region directly upstream of the morB-PA2933-cif operon (Fig. 5A). We employed EMSA to study CifR-DNA interactions. If indeed CifR is capable of binding to the cifR-morB intergenic region, a shift in migration of the probe should be observed when CifR is incubated with the biotinylated probe.
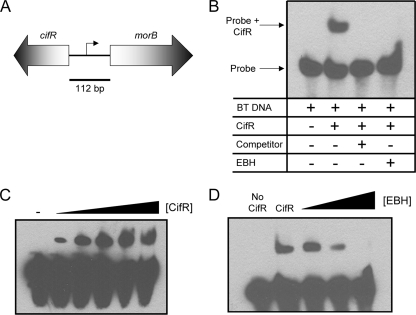
FIG. 5.
CifR binds to the intergenic region directly upstream of the cif-containing operon. (A) The intergenic region between the cifR and morB genes was amplified using biotinylated primers and used as a probe in EMSAs. (B) EMSA using the morB-cifR intergenic region as a probe and purified CifR. Lane 1, labeled probe; lane 2, probe and 1.15 nM CifR; lane 3, probe, 1.15 nM CifR, and a 20× molar excess of the unlabeled cold competitor; and lane 4, probe, 1.15 nM CifR, and 1 mM EBH. BT, biotinylated. (C) EMSA CifR dose response. Lane 1, probe alone; lane 2, probe and 115 pM CifR; lane 3, probe and 287.5 pM CifR; lane 4, probe and 575 pM CifR; lane 5, probe and 862.5 pM CifR; and lane 6, probe and 1.15 nM CifR. (D) EMSA EBH dose response. Lane 1, probe alone; lane 2, probe and 1.15 nM CifR; lane 3, probe, 1.15 nM CifR, and 10 μM EBH; lane 4, probe, 1.15 nM CifR, and 100 μM EBH; and lane 5, probe, 1.15 nM CifR, and 1 mM EBH.
The incubation of the purified CifR protein with the biotinylated probe resulted in a substantial mobility shift consistent with the formation of a CifR-DNA complex (lane 2, Fig. 5B, lane 2), compared to the shift with probe alone (Fig. 5B, lane 1). This binding was shown to be sensitive to the addition of the unlabeled cold probe, suggesting specificity of CifR for the intergenic region (Fig. 5B, lane 3). As an additional control, we utilized a nonspecific cold competitor derived from the rplU gene from P. aeruginosa. Inclusion of the rplU amplicon at a 100× molar excess did not result in a decrease in CifR binding to the biotinylated probe (data not shown).
The binding of CifR to the cifR-morB intergenic region is concentration dependent; as CifR concentration increased, we observed an increase in the CifR-DNA complex (Fig. 5C). Interestingly, under the conditions assayed, binding was observed at molar DNA-to-protein ratios as low as 1:100 (Fig. 5C, lane 2). By using a best-fit curve to determine the half-maximal effective concentration (EC50) of CifR binding to the intergenic region assayed, we found that CifR has an EC50 of approximately 118 pM. As this system consists of only two components, we can assume that it is noncompetitive, and thus the EC50 value is approximately equal to the CifR/DNA binding constant.
Based on the finding that EBH stimulates cif gene expression in vivo, we tested whether EBH could impact the CifR-DNA complex in vitro. The CifR-DNA complex is disrupted by the addition of EBH (Fig. 5B, lane 4). This finding is in direct agreement both with the results showing increased cif gene transcription in the presence of EBH and with several reports in the literature demonstrating that the presence and binding of certain ligands to TetR family protein members results in disassociation of the protein from its cognate DNA-binding site (16, 26, 39). It should be noted that EBH does not generally disrupt DNA-protein complexes, as we have tested the effects of EBH on another DNA-binding protein using EMSA and found that there was no effect on binding (data not shown).
As we showed in Fig. 2A and B, induction of cif gene expression and Cif protein expression by EBH is concentration dependent between 10 μM and 1 mM. As shown in Fig. 5D, the effect of EBH on CifR DNA binding is also concentration dependent. Interestingly, inhibition of the formation of the DNA-protein complex occurs at concentrations similar to those observed for Cif induction by EBH in vivo (Fig. 2A and B).
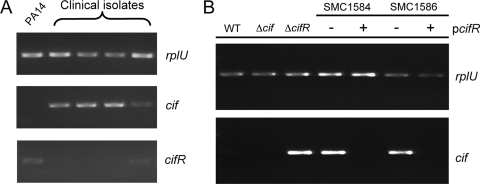
cif-overexpressing clinical isolates demonstrate altered cifR gene expression.
A previous report from our group demonstrated that nonmucoid P. aeruginosa strains isolated from CF sputum demonstrate relatively high levels of cif gene expression compared to levels associated with the lab strain P. aeruginosa PA14 (29). We also found that mucoid CF isolates, those producing high concentrations of the exopolysaccharide alginate, had relatively lower levels of cif gene expression compared to levels associated with PA14. The striking similarity between the nonmucoid cif-overexpressing strains and the ΔcifR strain led us to hypothesize that the increased cif expression observed in these nonmucoid clinical strains was due to decreased cifR gene expression.
Nonmucoid P. aeruginosa clinical isolates previously shown to overexpress cif were assayed using sqRT-PCR for rplU, cif, and cifR gene expression. All of the clinical isolates assayed demonstrated increased cif expression compared to that of the wild-type PA14 and compared to the rplU expression (Fig. 6A). Interestingly, all of the nonmucoid clinical isolates also demonstrated a marked decrease in cifR gene expression compared to that with the wild-type laboratory strain PA14. Furthermore, the one isolate that expressed slightly higher levels of cifR (lane 5) demonstrated a concomitant decrease in cif expression, suggesting an inverse relationship between cif and cifR expression in P. aeruginosa CF sputum isolates.
FIG. 6.
Clinical isolates overexpressing the cif gene demonstrate decreased cifR gene expression. (A) sqRT-PCR of wild-type P. aeruginosa PA14 and four nonmucoid P. aeruginosa CF sputum isolates. Expression of the cif, cifR, and rplU genes was assayed. (B) sqRT-PCR of wild-type (WT) P. aeruginosa, the Δcif and ΔcifR mutants, and two of the clinical isolates from panel A carrying either the empty vector pMQ71 (−) or the cifR-expressing plasmid pDPM79 (+).
To further demonstrate that the cif overexpression phenotype was due to a decrease in cifR expression, the plasmid pDPM79 expressing cifR was introduced into two of these strains. We found that the expression of cifR in the nonmucoid cif-overexpressing clinical strains results in a significant decrease in cif expression (Fig. 6B). Only two of four strains in Fig. 6A were assayed, as isolation of stable transformants of the other two strains was never achieved (data not shown).
The cif gene overexpression phenotypes appeared to be the result of heritable changes in gene expression, as these phenotypes were stable following serial passaging of the strains in both solid and liquid media (data not shown). We hypothesized that the loss of cifR expression in these clinical isolates was due to changes within the promoter region of the cifR gene. In order to identify any changes affecting cifR gene expression, we amplified the chromosomal region containing both the cifR promoter region and the ORF and sequenced the resulting amplicon. Surprisingly, there were no changes to the promoter region. While there were changes within the cifR ORF in these strains, none of these nucleotide changes were predicted to result in either missense or nonsense mutations.
DISCUSSION
We have previously demonstrated that P. aeruginosa produces a novel toxin, Cif, which is capable of decreasing apical membrane expression of CFTR in a variety of epithelial cell culture lines (29, 46). In silico analysis of the predicted amino acid sequence of this protein suggested that it may belong to the large family of α/β hydrolases and in particular to the family of EHs. Supporting this hypothesis, Cif was shown to degrade the general model EH substrate S-NEPC.
The ability to degrade S-NEPC, while suggestive, was not conclusive evidence that Cif is an EH. In order to better understand the biological activity of Cif, i.e., decreased apical membrane expression of CFTR, we sought to further characterize the biochemical activity of this protein. Previous work identified a soil pseudomonad capable of degrading the epoxide EBH (24). This previous study demonstrated that this EH activity was the result of a secreted protein with a mass and amino acid sequence similar to those to the Cif protein, leading us to hypothesize that Cif may be a homolog of the EH described previously that would thus be capable of degrading EBH. The finding that Cif could indeed degrade EBH supported this hypothesis. Furthermore, the demonstration that EBH could significantly induce cif gene expression supports the conclusion that this family of molecules can serve as substrates for Cif.
Epoxides have been previously shown to act as signaling molecules regulating processes as varied as Cl− ion transport in renal tubular epithelial cells, angiogenesis, and vasodilation mediated by endothelial cells (32, 33, 42). Furthermore, several studies have demonstrated the presence of high levels of leukotrienes within the CF airway. Leukotrienes are produced from the inactive epoxide molecule leukotriene A4, which is metabolized by the EH leukotriene hydrolase to the active, vicinal diol form, leukotriene B4. Leukotriene B4 has been shown to be a potent chemoattractant, specifically for neutrophils, and is generally considered a mediator of inflammation (10, 32, 34, 50, 51). It still remains unclear how Cif alters apical membrane expression of CFTR, but it may be through the metabolism of these epoxide signaling molecules. Thus, CifR may regulate cif gene expression, and hence Cif biosynthesis, by binding of native epoxide molecules in the CF lung. These molecules have yet to be identified. While we do not believe that EBH, an industrial pollutant, is found in the CF lung, or acts as a signaling molecule within the CF lung, it serves as a model compound for better understanding the regulation of cif gene expression by CifR.
The family of TetR repressors has been shown to regulate the expression of genes involved in a variety of pathways, from antibiotic resistance to carbon catabolism (20, 23, 36). Typically TetR family members have a DNA-binding helix-turn-helix (HTH) motif as well as an effector-binding domain. The protein acts to regulate gene expression by binding to the promoter region of its target gene and thus inhibiting transcription. The presence of an effector causes conformational shifts in the HTH domain, resulting in disassociation of the protein from the promoter and hence derepression of gene expression (16, 21, 26, 36). Comparison of the CifR amino acid sequence to known TetR family members showed a similar organization, including a HTH DNA-binding domain and a predicted effector-binding domain with no homology to any known TetR protein. We have shown here that the CifR protein is capable of binding to the predicted promoter region of the cif-containing operon and that this interaction is sensitive to the epoxide EBH (Fig. 7). These data readily explain the EBH-dependent induction in cif gene expression shown in Fig. 2A. Interestingly, we found that this disassociation was concentration dependent and that this phenomenon occurred at concentrations similar to those shown in Fig. 2A, indicating that the concentration of EBH used to disrupt the DNA-protein complex in the in vitro studies is physiologically relevant.
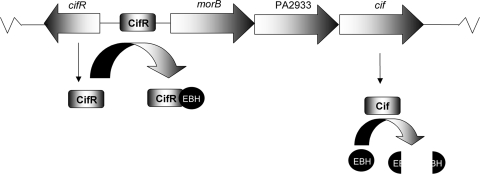
FIG. 7.
Model for cif gene expression. CifR represses cif gene expression by binding to the promoter region immediately adjacent to the cif-containing operon. The binding of EBH by CifR results in disassociation of the repressor from the promoter, resulting in increased cif gene expression and biosynthesis of the EH Cif, which in turn degrades EBH.
In silico analysis of the cifR-morB intergenic region has identified a putative CifR-binding site consisting of two palindromic sequences that overlap both the putative −10 and −35 sequences of the morB and cifR promoters. Work is ongoing to identify and characterize the CifR-binding site. Interestingly, the putative promoter regions of the morB and cifR genes overlap significantly, as does the putative CifR-binding site. Thus, most likely, CifR not only represses expression of the cif-containing operon but also negatively regulates its own expression in the absence of EBH.
We have previously demonstrated that nonmucoid P. aeruginosa strains isolated from CF sputum demonstrated significantly increased cif gene expression compared to that with the P. aeruginosa strain PA14 (29), while mucoid CF isolates displayed lower levels of cif gene expression. These observations led us to hypothesize that the differential cif gene expression was the result of changes in expression of CifR. In support of this hypothesis, we found that in several of the nonmucoid strains overexpressing cif, cifR gene expression was significantly lower than the expression seen in the laboratory strain PA14. Furthermore, when CifR was expressed from the plasmid pDPM79 in two of the cif-overexpressing clinical isolates, we observed a level of cif transcript that was significantly lower than that with the vector control strains. Interestingly, the cif overexpression phenotype of nonmucoid clinical isolates was stable, as repeated passaging of the strains in laboratory media did not effect cif gene expression, suggesting that these strains had acquired mutations either in the cifR promoter region or within the ORF itself. However, DNA sequence analysis of the cifR promoter region did not illuminate any mutations in the promoter. We did find several mutations within the ORF, although none of these are predicted to result in either missense or nonsense mutations. Several models for the decreased expression of cifR in these strains can be invoked, including changes in mRNA stability, structural changes in the mRNA, or changes in genes that regulate cifR expression. Currently, we believe the latter to be the case, and studies are ongoing to identify regulators of both cif and cifR gene expression.
It is generally believed that CF patients are initially colonized by nonmucoid environmental isolates, which eventually convert to the mucoid phenotype (4, 15, 35, 43). These data, taken together with our demonstration that nonmucoid clinical isolates show relatively high cif expression compared to that of mucoid clinical isolates, suggest that Cif may play a role in initial colonization of the CF lung and early infection. Classically, TetR family regulators have been shown to regulate gene expression as a response to environmental stimuli, as is the case with CifR and its response to EBH. We hypothesize that CifR may respond to the presence of an endogenous epoxide and regulate cif gene expression accordingly, thus affecting CFTR membrane expression through an as-yet-unidentified mechanism.
An alternative hypothesis is that there is a selective pressure for strains that overexpress cif in the context of the CF lung and that this pressure leads to a population that has increased cif expression as a result of decreases in cifR expression. The demonstration that the cif-overexpressing phenotype in clinical isolates was heritable and stable in these strains suggests that this phenotype is due to genotypic changes altering cifR gene expression. Thus, rather than sensing the environment and responding through classic regulatory pathways, it would appear that P. aeruginosa in the CF lung may simply remove the regulatory elements for some genes, thus altering their basal gene expression.
Acknowledgments
We thank Joseph Schwartzman at the Dartmouth Hitchcock Medical Center for providing clinical isolates and Robert M. Q. Shanks for critical review of the manuscript. We also acknowledge Charles Midgett for help in determining EC50 values for CifR/DNA binding. Additionally, we thank Christopher Bahl for providing purified Cif used in Fig. 1C.
This work was supported by an NIH training grant predoctoral fellowship (T32-DF007301) to D.P.M., a Rosalind Borrison Memorial Predoctoral Fellowship to D.P.M., and grant HL074175 to B.A.S. and G.A.O.
Editor: A. Camilli
Footnotes
Published ahead of print on 5 May 2008.
REFERENCES
- 1.Bals, R., D. J. Weiner, and J. M. Wilson. 1999. The innate immune system in cystic fibrosis lung disease. J. Clin. Investig. 103303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielecki, P., J. Glik, M. Kawecki, and V. A. Martins Dos Santos. 2008. Towards understanding Pseudomonas aeruginosa burn wound infections by profiling gene expression. Biotechnol. Lett. 30777-790. [DOI] [PubMed] [Google Scholar]
- 4.Boucher, J. C., J. Martinez-Salazar, M. J. Schurr, M. H. Mudd, H. Yu, and V. Deretic. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher, R. C. 2004. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 23146-158. [DOI] [PubMed] [Google Scholar]
- 6.Britigan, B. E., M. A. Railsback, and C. D. Cox. 1999. The Pseudomonas aeruginosa secretory product pyocyanin inactivates α1 protease inhibitor: implications for the pathogenesis of cystic fibrosis lung disease. Infect. Immun. 671207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cedrone, F., T. Bhatnagar, and J. C. Baratti. 2005. Colorimetric assays for quantitative analysis and screening of epoxide hydrolase activity. Biotechnol. Lett. 271921-1927. [DOI] [PubMed] [Google Scholar]
- 8.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64391-397. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54735-774. [DOI] [PubMed] [Google Scholar]
- 10.Cromwell, O., M. J. Walport, G. W. Taylor, H. R. Morris, B. R. O'Driscoll, and A. B. Kay. 1982. Identification of leukotrienes in the sputum of patients with cystic fibrosis. Adv. Prostaglandin Thromboxane Leukot. Res. 9251-257. [PubMed] [Google Scholar]
- 11.D'Argenio, D. A., M. Wu, L. R. Hoffman, H. D. Kulasekara, E. Deziel, E. E. Smith, H. Nguyen, R. K. Ernst, T. J. Larson Freeman, D. H. Spencer, M. Brittnacher, H. S. Hayden, S. Selgrade, M. Klausen, D. R. Goodlett, J. L. Burns, B. W. Ramsey, and S. I. Miller. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64512-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deretic, V., D. W. Martin, M. J. Schurr, M. H. Mudd, N. S. Hibler, R. Curcic, and J. C. Boucher. 1993. Conversion to mucoidy in Pseudomonas aeruginosa. Biotechnology (New York) 111133-1136. [DOI] [PubMed] [Google Scholar]
- 13.Deretic, V., M. J. Schurr, J. C. Boucher, and D. W. Martin. 1994. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J. Bacteriol. 1762773-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deretic, V., M. J. Schurr, and H. Yu. 1995. Pseudomonas aeruginosa, mucoidy and the chronic infection phenotype in cystic fibrosis. Trends Microbiol. 3351-356. [DOI] [PubMed] [Google Scholar]
- 15.Doggett, R. G. 1969. Incidence of mucoid Pseudomonas aeruginosa from clinical sources. Appl. Microbiol. 18936-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frenois, F., J. Engohang-Ndong, C. Locht, A. R. Baulard, and V. Villeret. 2004. Structure of EthR in a ligand bound conformation reveals therapeutic perspectives against tuberculosis. Mol. Cell 16301-307. [DOI] [PubMed] [Google Scholar]
- 17.Fu, Z., N. P. Donegan, G. Memmi, and A. L. Cheung. 2007. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J. Bacteriol. 1898871-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuller, C. M., and D. J. Benos. 1992. Cftr! Am. J. Physiol. 263C267-C286. [DOI] [PubMed] [Google Scholar]
- 19.Hendrickson, E. L., J. Plotnikova, S. Mahajan-Miklos, L. G. Rahme, and F. M. Ausubel. 2001. Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J. Bacteriol. 1837126-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hillerich, B., and J. Westpheling. 2008. A new TetR family transcriptional regulator required for morphogenesis in Streptomyces coelicolor. J. Bacteriol. 19061-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinrichs, W., C. Kisker, M. Duvel, A. Muller, K. Tovar, W. Hillen, and W. Saenger. 1994. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 264418-420. [DOI] [PubMed] [Google Scholar]
- 22.Howard, M., X. Jiang, D. B. Stolz, W. G. Hill, J. A. Johnson, S. C. Watkins, R. A. Frizzell, C. M. Bruton, P. D. Robbins, and O. A. Weisz. 2000. Forskolin-induced apical membrane insertion of virally expressed, epitope-tagged CFTR in polarized MDCK cells. Am. J. Physiol. Cell Physiol. 279C375-C382. [DOI] [PubMed] [Google Scholar]
- 23.Huffman, J. L., and R. G. Brennan. 2002. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 1298-106. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs, M. H., A. J. Van den Wijngaard, M. Pentenga, and D. B. Janssen. 1991. Characterization of the epoxide hydrolase from an epichlorohydrin-degrading Pseudomonas sp. Eur. J. Biochem. 2021217-1222. [DOI] [PubMed] [Google Scholar]
- 25.Jaffar-Bandjee, M. C., A. Lazdunski, M. Bally, J. Carrere, J. P. Chazalette, and C. Galabert. 1995. Production of elastase, exotoxin A, and alkaline protease in sputa during pulmonary exacerbation of cystic fibrosis in patients chronically infected by Pseudomonas aeruginosa. J. Clin. Microbiol. 33924-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kisker, C., W. Hinrichs, K. Tovar, W. Hillen, and W. Saenger. 1995. The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J. Mol. Biol. 247260-280. [DOI] [PubMed] [Google Scholar]
- 27.Kong, F., L. Young, Y. Chen, H. Ran, M. Meyers, P. Joseph, Y. H. Cho, D. J. Hassett, and G. W. Lau. 2006. Pseudomonas aeruginosa pyocyanin inactivates lung epithelial vacuolar ATPase-dependent cystic fibrosis transmembrane conductance regulator expression and localization. Cell. Microbiol. 81121-1133. [DOI] [PubMed] [Google Scholar]
- 28.Lau, G. W., D. J. Hassett, and B. E. Britigan. 2005. Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol. 13389-397. [DOI] [PubMed] [Google Scholar]
- 29.MacEachran, D. P., S. Ye, J. M. Bomberger, D. A. Hogan, A. Swiatecka-Urban, B. A. Stanton, and G. A. O'Toole. 2007. The Pseudomonas aeruginosa secreted protein PA2934 decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 753902-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahajan-Miklos, S., L. G. Rahme, and F. M. Ausubel. 2000. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol. Microbiol. 37981-988. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto, K. 2004. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol. Chem. 3851007-1016. [DOI] [PubMed] [Google Scholar]
- 32.Newman, J. W., C. Morisseau, and B. D. Hammock. 2005. Epoxide hydrolases: their roles and interactions with lipid metabolism. Prog. Lipid Res. 441-51. [DOI] [PubMed] [Google Scholar]
- 33.Nusing, R. M., H. Schweer, I. Fleming, D. C. Zeldin, and M. Wegmann. 2007. Epoxyeicosatrienoic acids affect electrolyte transport in renal tubular epithelial cells: dependence on cyclooxygenase and cell polarity. Am. J. Physiol. Renal Physiol. 293F288-F298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penning, T. D. 2001. Inhibitors of leukotriene A4 (LTA4) hydrolase as potential anti-inflammatory agents. Curr. Pharm. Des. 7163-179. [DOI] [PubMed] [Google Scholar]
- 35.Poschet, J. F., J. C. Boucher, A. M. Firoved, and V. Deretic. 2001. Conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Methods Enzymol. 33665-76. [DOI] [PubMed] [Google Scholar]
- 36.Ramos, J. L., M. Martinez-Bueno, A. J. Molina-Henares, W. Teran, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riordan, J. R., J. M. Rommens, B. Kerem, N. Alon, R. Rozmahel, Z. Grzelczak, J. Zielenski, S. Lok, N. Plavsic, J. L. Chou, et al. 1989. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 2451066-1073. [DOI] [PubMed] [Google Scholar]
- 38.Rommens, J. M., M. C. Iannuzzi, B. Kerem, M. L. Drumm, G. Melmer, M. Dean, R. Rozmahel, J. L. Cole, D. Kennedy, N. Hidaka, et al. 1989. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science 2451059-1065. [DOI] [PubMed] [Google Scholar]
- 39.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 2942158-2163. [DOI] [PubMed] [Google Scholar]
- 40.Shanks, R. M., N. C. Caiazza, S. M. Hinsa, C. M. Toutain, and G. A. O'Toole. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ Microbiol. 725027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 1038487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spector, A. A., and A. W. Norris. 2007. Action of epoxyeicosatrienoic acids on cellular function. Am. J. Physiol. Cell Physiol. 292C996-C1012. [DOI] [PubMed] [Google Scholar]
- 43.Speert, D. P., S. W. Farmer, M. E. Campbell, J. M. Musser, R. K. Selander, and S. Kuo. 1990. Conversion of Pseudomonas aeruginosa to the phenotype characteristic of strains from patients with cystic fibrosis. J. Clin. Microbiol. 28188-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stonehouse, M. J., A. Cota-Gomez, S. K. Parker, W. E. Martin, J. A. Hankin, R. C. Murphy, W. Chen, K. B. Lim, M. Hackett, A. I. Vasil, and M. L. Vasil. 2002. A novel class of microbial phosphocholine-specific phospholipases C. Mol. Microbiol. 46661-676. [DOI] [PubMed] [Google Scholar]
- 45.Straus, D., and F. M. Ausubel. 1990. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc. Natl. Acad. Sci. USA 871889-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swiatecka-Urban, A., S. Moreau-Marquis, D. P. Maceachran, J. P. Connolly, C. R. Stanton, J. R. Su, R. Barnaby, G. A. O'Toole, and B. A. Stanton. 2006. Pseudomonas aeruginosa inhibits endocytic recycling of CFTR in polarized human airway epithelial cells. Am. J. Physiol. Cell Physiol. 290C862-C872. [DOI] [PubMed] [Google Scholar]
- 47.Terada, L. S., K. A. Johansen, S. Nowbar, A. I. Vasil, and M. L. Vasil. 1999. Pseudomonas aeruginosa hemolytic phospholipase C suppresses neutrophil respiratory burst activity. Infect. Immun. 672371-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welsh, M. J., and A. E. Smith. 1993. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell 731251-1254. [DOI] [PubMed] [Google Scholar]
- 49.Yorgey, P., L. G. Rahme, M.-W. Tan, and F. M. Ausubel. 2001. The roles of mucD and alginate in the virulence of Pseudomonas aeruginosa in plants, nematodes and mice. Mol. Microbiol. 411063-1076. [DOI] [PubMed] [Google Scholar]
- 50.Zakrzewski, J. T., N. C. Barnes, J. F. Costello, and P. J. Piper. 1987. Lipid mediators in cystic fibrosis and chronic obstructive pulmonary disease. Am. Rev. Respir. Dis. 136779-782. [DOI] [PubMed] [Google Scholar]
- 51.Zakrzewski, J. T., N. C. Barnes, P. J. Piper, and J. F. Costello. 1987. Detection of sputum eicosanoids in cystic fibrosis and in normal saliva by bioassay and radioimmunoassay. Br. J. Clin. Pharmacol. 2319-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zulianello, L., C. Canard, T. Kohler, D. Caille, J. S. Lacroix, and P. Meda. 2006. Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect. Immun. 743134-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]