Abstract
The importance of glucokinase (GK; EC 2.7.1.12) in glucose homeostasis has been demonstrated by the association of GK mutations with diabetes mellitus in humans and by alterations in glucose metabolism in transgenic and gene knockout mice. Liver GK activity in humans and rodents is allosterically inhibited by GK regulatory protein (GKRP). To further understand the role of GKRP in GK regulation, the mouse GKRP gene was inactivated. With the knockout of the GKRP gene, there was a parallel loss of GK protein and activity in mutant mouse liver. The loss was primarily because of posttranscriptional regulation of GK, indicating a positive regulatory role for GKRP in maintaining GK levels and activity. As in rat hepatocytes, both GK and GKRP were localized in the nuclei of mouse hepatocytes cultured in low-glucose-containing medium. In the presence of fructose or high concentrations of glucose, conditions known to relieve GK inhibition by GKRP in vitro, only GK was translocated into the cytoplasm. In the GKRP-mutant hepatocytes, GK was not found in the nucleus under any tested conditions. We propose that GKRP functions as an anchor to sequester and inhibit GK in the hepatocyte nucleus, where it is protected from degradation. This ensures that glucose phosphorylation is minimal when the liver is in the fasting, glucose-producing phase. This also enables the hepatocytes to rapidly mobilize GK into the cytoplasm to phosphorylate and store or metabolize glucose after the ingestion of dietary glucose. In GKRP-mutant mice, the disruption of this regulation and the subsequent decrease in GK activity leads to altered glucose metabolism and impaired glycemic control.
Glucokinase (GK; EC 2.7.1.12), the principal hexokinase in liver parenchymal cells and in pancreatic β cells, is a critical component of the physiological glucose-sensing apparatus (1). In humans, GK heterozygous mutations lead to the autosomal, dominant, maturity-onset diabetes of the young (MODY) (2, 3) phenotype. In mice, changes in GK activity by gene knockout and by overexpression resulted in altered glucose homeostasis and demonstrated that modest changes in liver GK activity alone were sufficient to cause significant alterations in blood glucose levels (4–9). Liver GK activity has been reported to be lower in some obese humans with type 2 diabetes mellitus when compared with nondiabetic normal weight or obese individuals, suggesting a role for liver GK in glucose homeostasis in type 2 diabetes (10).
In the liver, GK activity is allosterically inhibited by GK regulatory protein (GKRP) (11–13). This protein, found in the livers of all animal species where GK is present, shows no inhibitory effect on other known hexokinases (11–13). In rodents, GKRP inhibition of GK is relieved by high concentrations of glucose and by fructose 1-phosphate, and is potentiated by fructose 6-phosphate (11–13). Recent studies have demonstrated that both GK and GKRP are in the nucleus of rat hepatocytes cultured in a medium containing 5.5 mM glucose (14, 15). When the growth medium was supplemented with higher concentrations of glucose or 0.5 mM fructose, GK alone translocated into the cytoplasm (14). The physiological significance of the GK/GKRP subcellular localization and the glucose- and fructose-regulated intracellular translocation is unclear. Given the importance of hepatic GK activity in glucose homeostasis, the physiological consequences of decreasing or eliminating the protein regulator was investigated in mice by genetic ablation.
Materials and Methods
Gene Targeting.
An 8-kb genomic DNA fragment containing 1.7 kb of the promoter region, exon 1, intron 1, exon 2, and part of intron 2, was isolated from a λ-129/sv mouse genomic library (CLONTECH) by hybridization to an oligonucleotide probe corresponding to the 5′ end of the rat GKRP cDNA. In the positive/negative selection pPNT-based gene targeting vector (16), GKRP exons 1 and 2 were replaced by a phosphoglycerokinase promoter-driven, neomycin-resistance gene cassette in the opposite transcription orientation, so that neomycin was flanked by 1.6 kb and 3.8 kb of genomic sequence, 5′ and 3′, respectively. Mouse 129/sv embryonal stem cells (ES cells, CJ7) were electroporated with linearized targeting vector, and resistant colonies were isolated as described by Hogan et al. (17). Southern blot hybridization analysis of BamHI-digested genomic DNA to a random-primed, 500-bp 32P-labeled EcoRV/PstI promoter region probe identified five ES cell clones with disruption in one GKRP allele. These clonal cells were microinjected into or fused with blastocysts and implanted into pseudopregnant ICR foster females. Three chimeras capable of germ-line transmission were crossed to ICR and C57BL/6 mice, and the heterozygous mutant mice were interbred to generate GKRP wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice. The results presented are from 129sv/ICR animals except for experiments determining the effect of a high-sucrose/high-fat diet, for which mice that were mostly C57BL/6 (N3) were used. The C57BL/6 genetic background was chosen because mice in this genetic background have been shown to respond to high-sucrose/high-fat dietary conditions and to develop diabetes. The mice were housed in a 12-hr light/dark cycle and fed ad lib.
Northern Blot Hybridization Analysis.
Ad-lib fed or 16-hr fasted, 5- to 6-week-old mice were sacrificed; their livers were harvested and frozen in liquid N2, and total RNA was prepared by using TRIzol reagent (GIBCO/BRL). RNA (20 μg) was separated on a 1% agarose-formaldehyde gel, transferred onto nylon membranes (Gene Screen, DuPont/NEN), and hybridized to 32P-labeled, random-primed rat GKRP, human GK, rat phosphoenolpyruvate carboxykinase (PEPCK), and mouse rpL30 cDNA probes (18, 19). The radioactivity in the hybridized bands was quantitated in a β counter (Betagen, Waltham, MA), and mRNA values were normalized to rpL30 mRNA.
Western Blot Analysis.
Liver extracts (100,000 × g supernatants) were prepared as described previously (7). Ten-microgram protein samples were separated on a 4–20% Mini-SepraGel SDS/PAGE (Integrated Separation Systems, Hyde Park, MA) (20, 21) and electrotransferred onto NitroScreen membranes (DuPont/NEN) according to manufacturers’ instructions. GKRP and GK were detected with affinity-purified primary antibodies to rat GKRP or human GK raised in rabbits and with an alkaline phosphatase-conjugated secondary antibody and BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) color development or Immunostar chemiluminescence (Bio-Rad).
GK Activity Determination.
Liver extracts (100,000 × g supernatants) were assayed for GK activity as described previously (7). GK activity (milliunits/mg of protein) was calculated as the difference in rates at 0.5 mM and 50 mM glucose.
Immunocytochemical Analysis of Primary Hepatocytes.
Hepatocytes were isolated from livers of 8- to 10-week-old, ad-lib fed mice, cultured, and probed with anti-GKRP or GK antibodies as detailed earlier (14). Low-glucose medium contained 5.5 mM glucose, high-glucose medium contained 27.5 mM glucose, and low-fructose + glucose medium contained 0.5 mM fructose and 5.5 mM glucose. Whole-cell extracts (100,000 × g supernatants) were also prepared from cells cultured in the presence or absence of 50 μM cycloheximide and probed for GK protein by Western blot analysis (7, 14).
Plasma Chemistry Analysis.
About 200 μl of tail vein blood from 16-hr fasted mice were collected in EDTA-coated tubes. Plasma was separated, and glucose, triglycerides, lactate, and nonesterified fatty acid values were determined as described previously (7). Insulin levels were quantitated by radioimmunoassay, using rat insulin antibodies and rat insulin standard (Linco Research Immunoassay, St. Charles, MO).
Liver Glycogen Analysis.
Livers weighing ≈1 g were homogenized in 3 M perchloric acid. After complete digestion, a portion of the liver homogenates was used for glycogen measurement (22).
Glucose Tolerance Test.
Mice were fasted for 16 hr and d-glucose (1 g/kg of body weight) in saline was injected intraperitoneally in conscious and restrained mice. Glucose levels were determined in ≈20-μl blood samples collected from the tail vein before the injection and at 30, 60, 90, 120, 150, and 180 min after the injection with glucose strips and a glucometer (Bayer, West Haven, CT). For β cell efficiency, mice were sacrificed by decapitation 30 min after glucose injection, and plasma insulin levels were quantitated by radioimmunoassay.
Mouse Diets.
Mice were fed ad lib regular laboratory chow or research diet 12327, which contains 40% sucrose and 40% fat (Research Diets, New Brunswick, NJ).
Insulin Injection.
Mice that were fasted for 7 hr were given an intraperitoneal injection of bovine insulin (5.0 units/kg; GIBCO/BRL) and d-glucose (1 g/kg) in saline. Glucose was included to prevent the animals from succumbing to hypoglycemia. Groups of mice were sacrificed just before and 2, 4, 8, and 24 hr after insulin injection. Livers were harvested and frozen in liquid N2, and hepatic GK mRNA and protein levels were determined.
Statistical Analysis.
Independent, two-tailed Student’s t tests were performed for comparisons between groups. Differences were considered significant at P < 0.05.
Results
Generation of GKRP-Mutant Mice.
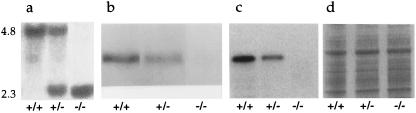
The GKRP gene was mutated by homologous recombination in mouse ES cells with a targeting vector in which exons 1 and 2 were replaced with a neomycin resistance gene cassette. Southern hybridization analysis of genomic DNA from neomycin and FIAU [1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-5-iodouracil]-resistant clones, with probes specific for DNA regions outside the targeted locus and not included in the targeting vector, identified five independent ES cell clones with one disrupted GKRP allele. These were expanded and microinjected into or fused with blastocysts derived from ICR mice. Three male chimeras were generated that were capable of germ-line transmission of the disrupted GKRP allele when bred with either ICR or C57BL/6 females. Breeding of +/− mice produced +/+, +/−, and −/− offspring (Fig. 1a) in the expected Mendelian ratio. Both male and female GKRP mutants showed no physical abnormalities, were fertile, and developed normally. Northern hybridization analysis of liver RNA detected the expected 2.4-kb GKRP mRNA in +/+ mice, with the levels reduced in +/− mice and undetectable in −/− mice (Fig. 1b). Similarly, Western blot analysis of liver extracts with GKRP polyclonal antibodies detected the ≈65-kDa GKRP band in +/+ mice, with decreased levels in +/− mice and a complete absence in −/− mice (Fig. 1 c and d). The GKRP expression data demonstrate that there was no compensation from the wild-type allele in +/− mice as well as the true null nature of the mutation in −/− mice.
Figure 1.
GKRP gene. (a) Southern hybridization analysis of BamHI-digested DNA with promoter region probe identifying the normal (4.8-kb) and the mutant (2.3-kb) alleles. (b) Northern hybridization analysis of liver RNA showing the lack of compensation from the normal allele in +/− mice and the absence of a GKRP transcript in −/− mice. (c) Western blot analysis of liver extracts showing the ≈65-kDa GKRP in +/+ and +/− mice, decreased levels of GKRP in +/− mice, and the complete absence of GKRP in −/− mice. (d) Coomassie blue-stained SDS/PAGE gel showing equivalent amounts of protein loaded.
Reduced Levels of GK in GKRP-Mutant Mouse Liver.
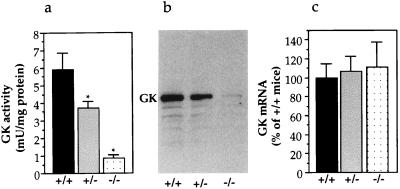
Because GKRP is an allosteric inhibitor of GK, a decrease or complete absence of GKRP was predicted to result in higher GK enzymatic activity in the liver. To test this prediction, GK activity levels were determined in liver samples obtained from 16-hr fasted and ad-lib fed mice. Surprisingly, and contrary to the prediction, GK activity was found to be dramatically diminished in the GKRP-mutant mouse liver. The fasting liver GK activity was decreased in +/− mice to 62% and in −/− mice to 15% of that in +/+ mice (Fig. 2a). The rate of liver glucose phosphorylation at 0.5 mM glucose remained unchanged (data not shown). Because glucose phosphorylation depends on non-GK hexokinases at 0.5 mM glucose, this result suggests the absence of compensation from other hexokinases. Confirming low GK activity, Western blot analysis of liver extracts with GK polyclonal antibodies detected reduced levels of GK in +/− mice and dramatically reduced levels of GK in −/− mice (Fig. 2b). The reduction in GK protein and activity in mutant mice was not because of diminished steady-state GK mRNA levels, because the liver GK mRNA levels in +/− and −/− mice remained comparable to +/+ mice (Fig. 2c). Lower GK activity was also obtained in the liver of ad-lib fed mice, 85% in +/− mice and 48% in −/− mice as compared with +/+ mice (data not shown). The GK data suggest a positive regulatory role for GKRP in the posttranscriptional modulation of GK levels and activity in the liver. In the mutant mouse liver, this regulation is disrupted, resulting in constitutively diminished GK activity.
Figure 2.
GK gene expression in GKRP-mutant liver. (a) Glucose phosphorylation in liver extracts of fasted mice, showing reduced GK activity in GKRP mutants. mU, milliunits. (+/+, n = 8; +/−, n = 7; −/−, n = 7). *, P ≤ 0.05, +/+ versus +/− or −/−. (b) Representative Western blot analysis of liver extracts with GK antibodies showing the GK protein and decreased levels in +/− and in −/− livers. (c) Liver RNA analysis showing the absence of significant changes in GK mRNA levels in GKRP mutants. Data are presented as percentage of +/+ mice liver with +/+ taken as 100%. (+/+, n = 3; +/−, n = 3; −/−, n = 3). Mean ± SEM.
Posttranscriptional Expression of GK Is Compromised in GKRP-Mutant Mouse Liver.
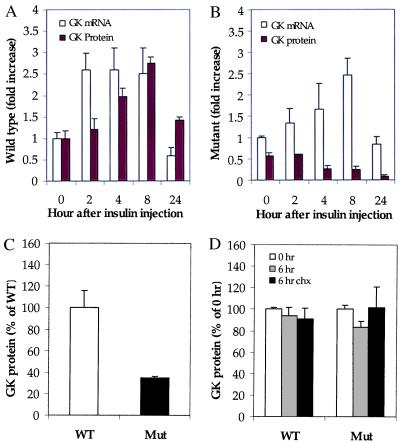
To explore the mechanism of loss of GK protein in the GKRP-mutant mouse liver, animals were fasted for 7 hr and then given an intraperitoneal injection of insulin and glucose. It has been previously demonstrated in rats that liver GK gene transcription can be transiently induced by insulin (23, 24). The short duration of fasting and the coadministration of glucose with the insulin were preferred in this experiment to prevent animals from succumbing to hypoglycemia. Mice were sacrificed before and 2, 4, 8, and 24 hr after insulin injection, and livers were excised and analyzed for GK mRNA and protein. In +/+ mice, insulin treatment stimulated liver GK mRNA levels >2.5-fold over control, preinjection levels by 2 hr. The levels returned to baseline by 24 hr (Fig. 3A). The increase in GK message also resulted in a time-delayed increase in GK protein in +/+ liver (Fig. 3A). GKRP mRNA and protein levels remained unchanged in the +/+ liver samples, showing that GKRP gene expression is not regulated by insulin (data not shown). Confirming this observation, no significant effect on GKRP mRNA levels was observed in hepatocytes in culture after exposure to increasing concentrations of insulin (data not shown). In −/− mouse liver, the increase in GK mRNA levels was delayed but reached +/+ liver levels by 8 hr (Fig. 3B). However, the GK protein levels in liver were never increased (Fig. 3B). Blood glucose concentration, monitored at each time point, showed an essentially identical decrease in both +/+ and −/− mice and, therefore, was not an independent factor that affected the stability of GK protein (data not shown).
Figure 3.
Posttranscriptional expression of GK gene is compromised in GKRP-mutant mouse liver. (A) GK expression in wild-type liver; GK mRNA (empty bars) and protein (filled bars) levels are enhanced by insulin. (B) GK expression in mutant mouse liver; GK mRNA (empty bars) is increased by insulin, whereas GK protein (filled bars) is not. Data are presented as -fold over time-0 wild-type mouse liver, with wild-type value taken as 1.0. Mean ± SEM, n = 3 for wild type and n = 3 for mutant per each time point. (C) Densitometric quantitation of the Western blot of extracts prepared from primary hepatocytes cultured in media containing 5.5 mM glucose, showing decreased levels of GK in homozygous mutant (Mut) hepatocytes. Data are presented as percentage of wild-type (WT) hepatocytes. (D) Densitometric quantitation of the Western blot of extracts from cells grown for an additional 6 hr in the presence or absence of cycloheximide showing absence of altered levels of GK degradation in Mut hepatocytes. GK values in WT and Mut cells, cultured in the absence of cycloheximide at time 0, were both considered as 100%, and data are presented as a percentage of these values.
To determine whether GK in GKRP-mutant mouse liver was susceptible to higher levels of proteolytic degradation, primary hepatocytes were prepared from ad-lib fed +/+ and −/− mouse liver and cultured in low-glucose-containing medium (5.5 mM) in the presence or absence of the protein synthesis inhibitor cycloheximide. After 6 hr, cells were lysed and GK protein levels were quantitated. Western blot analysis of whole-cell extracts from hepatocytes before cyclohexamide treatment showed that GK protein in the −/− cells was only 35% of the amount in +/+ cells (Fig. 3C). In the +/− cells the GK protein level was 52% of the amount in +/+ cells (data not shown). However, the relative levels of GK in +/+ and −/− hepatocytes before and after cycloheximide treatment were not changed, suggesting that GK in −/− cells was not susceptible to higher levels of degradation within the time frame and nature of the experimental protocol (Fig. 3D). Taken together, these data show that GKRP plays a major regulatory role in the posttranscriptional expression of the GK gene, but the exact site of the regulation is unclear at this time.
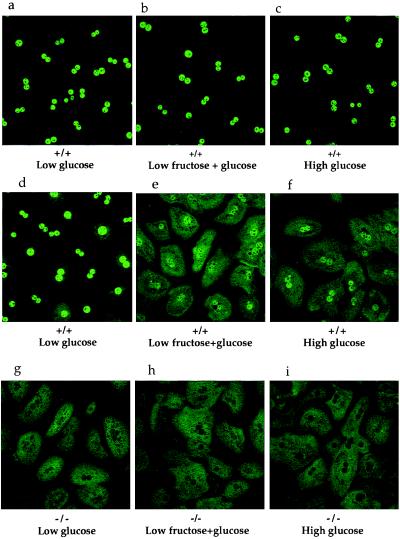
Absence of GKRP in Hepatocytes Leads to Exclusion of GK from the Nucleus.
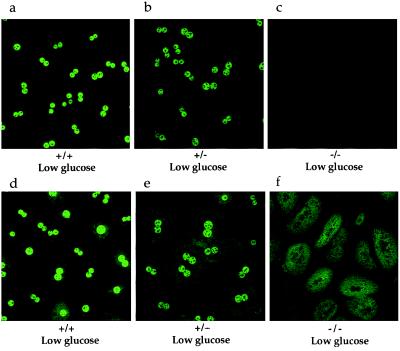
GK and GKRP have been previously reported to be localized mainly in the nucleus of rat primary hepatocytes cultured in low-glucose-containing medium (14). GK, but not GKRP, was able to translocate into the cytoplasm when the hepatocytes were cultured in high-glucose- or low-fructose + low-glucose-containing media (14). However, the physiological relevance of this intracellular compartmentalization and glucose- and fructose-induced translocation remained unclear. GKRP-mutant mice offered an opportunity to investigate this regulatory phenomenon and determine what effect the absence of GKRP would have on the intracellular localization of GK and on the response to glucose and fructose. Primary hepatocytes were isolated and cultured from ad-lib fed +/+, +/−, and −/− mice and were independently probed with anti-GKRP and anti-GK antibodies. In the hepatocytes cultured in low-glucose-containing medium (5.5 mM, equivalent to normal fasting glucose levels in rodents and humans), the GKRP antibody staining was primarily localized in the nucleus (Fig. 4a). As anticipated, GKRP antibody staining was decreased in +/− and absent in −/− hepatocyte nuclei (Fig. 4 b and c). Under these culture conditions, GK antibody staining was also almost exclusively localized in the nucleus of +/+ cells (Fig. 4d). The nuclear staining was diminished in +/− hepatocytes and undetectable in −/− hepatocytes (Fig. 4 e and f), respectively. As shown above, the +/− and −/− cells contained 52% and 35% GK protein, respectively, as compared with +/+ cells under these culture conditions. Cytoplasmic staining was higher in both +/− and −/− hepatocytes, more so in −/− hepatocytes, as compared with +/+ hepatocytes (Fig. 4 d–f).
Figure 4.
Subcellular localization of GKRP and GK in mouse primary hepatocytes cultured in low-glucose-containing medium. (a–c) Immunocytochemical analysis of hepatocytes with anti-GKRP showing nuclear localization of GKRP in +/+ and +/− cells, with antibody staining decreased in +/− cells (b) and absent in −/− cells (c). (d–f) Immunocytochemical analysis of hepatocytes with anti-GK antibodies showing nuclear localization of GK in +/+ cells and +/− cells, with antibody staining reduced in +/− cells (e) and the absence of GK antibody staining in −/− cells’ nuclei, in which the staining is increased in the cytoplasm (f).
GKRP remained in the nucleus of +/+ hepatocytes cultured in high-glucose (27.5 mM)-containing or in low-fructose (0.5 mM) + low-glucose (5.5 mM)-containing media (Fig. 5a–c). However, in +/+ hepatocytes cultured in high-glucose-containing media or in low-fructose + low-glucose-containing media, the GK antibody staining was substantially diminished in the nuclei, and there was a large increase in cytoplasmic staining (Fig. 5 d–f). The exclusive cytoplasmic localization of GK staining in −/− hepatocytes remained unaltered under these conditions (Fig. 5 g–i). There is no effect of insulin on either GK or GKRP intracellular localization in rat hepatocytes under these culture conditions (data not shown). The GKRP and GK immunolocalization studies show that GKRP is a nuclear protein and that GK remained sequestered in the nucleus, probably bound to GKRP, when the hepatocytes were cultured in low-glucose-containing media. When this association was lost in the mutant mice or disrupted in the presence of high levels of glucose, GK was principally found in the cytoplasm.
Figure 5.
Subcellular localization of GKRP and GK in mouse primary hepatocytes. Immunocytochemical analysis with anti-GKRP and anti-GK antibodies. (a–c) GKRP continued to remain in the nucleus of +/+ hepatocytes cultured in low-glucose- (a), low-fructose + low-glucose- (b), or in high-glucose-containing media (c). (d–f) GK is present in the nucleus of +/+ hepatocytes cultured in low-glucose-containing medium (d), translocated into the cytoplasm of +/+ hepatocytes cultured in low-fructose + low-glucose- (e) or in high-glucose-containing media (f). (g–i) GK present in the cytoplasm of −/− hepatocytes cultured in low-glucose medium (g) continued to stay in the cytoplasm in low-fructose + glucose- (h) or in high-glucose-containing medium (i).
Impaired Glycemic Control in GKRP-Mutant Mice.
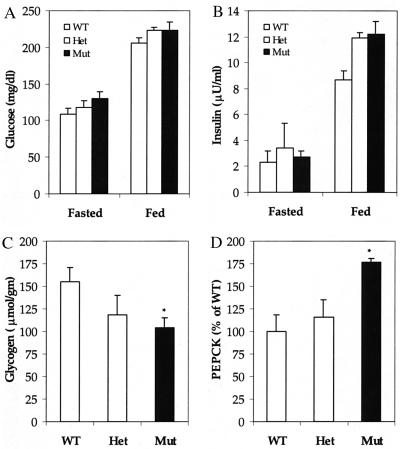
To determine the physiological effects caused by the absence of GKRP in the liver, plasma metabolite levels were determined in GKRP-mutant mice that were fed a normal laboratory chow. Surprisingly, both fasting and fed glucose and insulin levels in +/− and −/− mice remained relatively unchanged as compared with age-matched +/+ mice (Fig. 6 A and B). Other plasma chemistry parameters, including lactate, triglycerides, and nonesterified fatty acids, also were not significantly different in −/− mice (data not shown). However, the total liver glycogen concentration was 24% lower in +/− mice (not statistically significant) and 33% lower in −/− mice than in +/+ mice, indicating impaired glucose metabolism (Fig. 6C). This could be because of lower rates of glucose phosphorylation and/or reduced glycogen synthase activity in the liver, because glucose-6-PO4 is an allosteric activator of this enzyme (25). The mRNA levels of PEPCK, a pivotal enzyme in the gluconeogenesis pathway, were elevated in the mutant mouse liver (Fig. 6D). As PEPCK gene expression in rodent liver has been shown to be negatively regulated by insulin, an increase in mRNA level in the mutant mouse liver under normal insulin conditions suggests the development of insulin resistance (26).
Figure 6.
Plasma glucose, insulin, liver glycogen, and PEPCK mRNA levels. (A and B) Eight-week-old male mice were fasted for 16 hr or fed ad lib, and glucose (A) and insulin (B) levels were determined. μU, microunit. Fasted: n = 5 for wild type (WT); n = 6 for heterozygous mutant (Het); n = 6 for homozygous mutant (Mut). Fed: n = 6 for WT; n = 5 for Het; n = 5 for Mut. (C) Eight-week-old male, ad-lib fed mice were sacrificed, livers were harvested, and glycogen levels were quantitated. n = 3 for WT; n = 3 for Het; n = 3 for Mut. (D) PEPCK mRNA levels were quantitated by Northern blot analysis of total RNA prepared from fasted mouse liver. n = 3 for WT; n = 3 for Het; n = 3 for Mut. The mRNA level in WT is taken as 100%. Data are presented as mean ± SEM. *, Differences were considered statistically significant at P < 0.05.
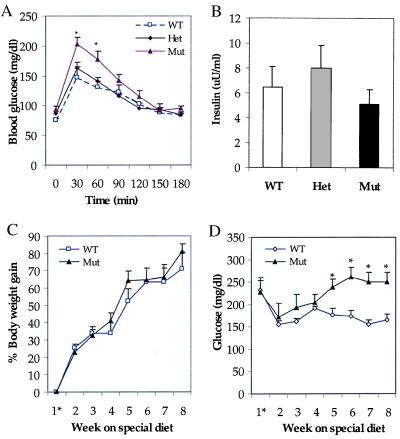
Abnormalities in glucose utilization in GKRP-mutant mice were further investigated by challenging them with an acute glucose load. Fasted mice were given an intraperitoneal injection of glucose, and changes in blood glucose levels were monitored over a period of 3 hr. Blood glucose levels in +/− mice were not significantly different from those in +/+ mice at any time point tested (Fig. 7A). In contrast, the blood glucose levels at 30 and 60 min were higher in −/− mice, as compared with +/+ mice, suggesting impaired glucose disposal (Fig. 7A). It is unlikely that this result was caused by inefficient pancreatic β cell insulin secretory response to glucose in −/− mice, because no significant differences in insulin levels were seen in −/− mice as compared with +/+ mice when the plasma insulin levels were determined 30 min after the glucose load (Fig. 7B).
Figure 7.
Glucose homeostasis in GKRP-mutant mice. (A) Glucose tolerance test showing elevated blood glucose levels in Mut mice as compared with WT and Het mice. n = 7 for WT; n = 7 for Het; n = 13 for Mut. (B) Plasma insulin levels in mice 30 min after glucose injection, showing the absence of significant changes. n = 4 for WT; n = 4 for Het; n = 4 for Mut. (C and D) Effect of high-sucrose + high-fat feeding on body weight and plasma glucose levels. Although both groups gained body weight at similar rates (C), the plasma glucose levels are significantly elevated in Mut mice as compared with WT mice (D). n = 5 for WT; n = 5 for Mut. 1* indicates after weaning. Data represent mean ± SEM. *, Differences were considered statistically significant at P < 0.05.
To further investigate glucose disposal in GKRP-mutant mice, the effects of chronic high-sucrose/high-fat feeding on plasma glucose levels were determined. For this, the +/+ and −/− mice were maintained from the time of weaning on a special diet containing 40% sucrose + 40% fat for 8 weeks and monitored for body weight and changes in plasma chemistry. The two groups of mice gained body weight at similar rates (Fig. 7C). However, in the −/− mice, the fasting plasma glucose levels were higher than those in +/+ mice with the progression of the experiment and became significantly elevated by week 5 (Fig. 7D). As shown in Table 1, at week 7 the fasting plasma glucose levels in −/− mice were increased by 60%. The fasting insulin and lactate levels were nearly doubled, whereas triglycerides and nonesterified fatty acid values did not show any differences in −/− mice as compared with +/+ mice. As anticipated, the plasma glucose levels in +/+ and −/− mice that were maintained on a normal chow diet during this experimental period showed no significant differences (data not shown). The elevated levels of lactate in −/− mice were likely because of increased glucose usage by skeletal muscle in response to increased plasma glucose and insulin levels. The higher plasma glucose levels in −/− mice in the presence of nearly double the concentration of insulin demonstrates impaired glucose homeostasis and suggests there was development of insulin resistance. Thus, the mild phenotype shown by −/− mice on normal chow was considerably enhanced when stressed with a high-sucrose/high-fat-containing diet; these mice became hyperglycemic and hyperinsulinemic. It is likely that the −/− mouse liver, with no GKRP regulation of GK and with reduced GK activity, is unable to effectively take up glucose and/or to regulate glucose production.
Table 1.
Plasma chemistry parameters in GKRP-mutant mice fed a high-sucrose/high-fat diet
| Plasma analysis | Wild type | Mutant |
|---|---|---|
| Glucose, mg/dl | 151.0 ± 10.0 | 232.0 ± 25.0* |
| Insulin, μU/ml | 10.8 ± 1.8 | 20.8 ± 3.6* |
| Lactate, mg/dl | 21.4 ± 1.8 | 41.7 ± 5.1* |
| NEFA, meq/1 | 1.2 ± 0.1 | 1.2 ± 0.1 |
| Triglycerides, mg/dl | 169.0 ± 15.3 | 161.0 ± 15.0 |
Eleven-week-old male mice, at week 7 on a high-sucrose/high-fat diet, were fasted for 16 hr. Data are presented as mean ± SEM. n = 5 for wild type; n = 5 for mutant. μU, microunit; NEFA, nonesterified fatty acid. *, Differences from wild type were considered statistically significant at P < 0.05.
Discussion
It has been speculated that GKRP is a candidate type 2 diabetes mellitus gene because gain of function mutations could potentially lead to enhanced liver GK inhibition and the development of MODY phenotype (27, 28). Loss of function mutations in GKRP would be predicted to result in higher GK activity. Contrary to this prediction, the liver GK activity was diminished in GKRP-mutant mice. In the +/− mouse liver, with only about half the level of GKRP present as compared with +/+, there was a significant loss of GK protein and activity. In the −/− mouse liver, the loss was more severe with only 15% and 48% GK activity present in the fasting and fed stages, respectively, as compared with +/+ mouse liver. The relatively modest loss of GK activity in fed −/− mouse liver as compared with the fasted −/− mouse liver is likely because of glucose stabilization of GK in the hepatocytes. Glucose stabilization of GK protein has been reported for the β cell isoform in cultured islets (29). The molecular cause for the loss of GK protein is presently unclear. GK protein turnover, at least as measured in isolated hepatocyte cultures, was not elevated. It is unknown whether GK protein turnover is regulated similarly in vivo. A small but detectable decrease from the basal GK protein values was observed in −/− mouse liver after insulin administration. This is probably because of the glucose-lowering effect of insulin that was observed in both wild-type and mutant mice. The GKRP-mutant mice, therefore, show that in addition to the accepted role of acute control of GK activity through direct inhibition to match changing glucose levels, GKRP plays a broader regulatory role in maintaining liver GK levels and activity.
Based on the data presented here, we propose that GKRP functions as an anchor protein for GK in the hepatocyte nucleus and helps to maintain a reserve pool of GK during the fasting state. GK in −/− mouse hepatocytes, although lower than in +/+ cells, was essentially completely excluded from the nucleus, and GK antibody staining was largely limited to the nucleus in +/+ hepatocytes under low-glucose conditions. Although GKRP remained in +/+ mouse hepatocyte nucleus at all conditions tested, there was a substantial decrease in GK nuclear staining and a corresponding increase in the cytoplasmic staining under high-glucose concentrations in the medium or in the presence of 0.5 mM fructose in low-glucose-containing medium. Both of these conditions have been previously demonstrated to relieve GKRP inhibition of GK in vitro (11–14). Thus, a physical association probably exists between GKRP and GK in the hepatocyte nucleus. When this association is absent, as in the mutant mice, or disrupted by fructose or high levels of glucose, GK is functionally excluded from the nucleus. It is unclear at present whether this phenomenon is active in β cells of the pancreas, in which GKRP protein is present in barely detectable levels.
Physiologically, GK nuclear localization may determine glucose phosphorylating capability of hepatocytes. For example, after a meal or when challenged with a glucose load, the GK/GKRP association is disrupted, and GK from the nuclear pool is rapidly released and mobilized into the cytoplasm to provide glucose phosphorylation activity. After glucose levels return to fasting levels, GK translocates back into the nucleus, sequestered away and available for immediate release. In the absence of GKRP, the total liver GK activity is diminished and the hepatocytes are unable to mobilize GK into the cytoplasm to meet changing glucose levels. Loss of this level of regulation leads to impaired glycemic control as evidenced by lower glycogen levels, elevated gluconeogenic enzyme PEPCK gene expression, and higher glucose levels after a glucose challenge. Decreased glycogen content in GKRP-mutant mouse liver is consistent with impaired liver glycogen synthesis observed in MODY 2 patients with defective GK activity (30). A defect in liver glycogen synthesis may also play a role in the mild hyperglycemia in mutant mice. The impairment in glycemic control in GKRP-mutant mice on a regular chow diet is further exacerbated by a high-fat/high-sucrose diet.
In rodents and humans, the level of hepatic glucose output is regulated by the relative rates of glucose phosphorylation and dephosphorylation by GK and glucose-6-phosphatase, respectively (25). The rate of liver glucose production is inversely correlated to the rate of glucose phosphorylation by GK (25). Therefore, it is significant that an additional level of regulation in the form of GKRP has evolved to regulate liver GK activity. Thus, the evolutionarily conserved GKRP/GK subcellular distribution and the response to glucose and fructose offer a rapid on/off switch for glucose sensing by GK in the liver.
The data presented here represent an example in which sequestration of a metabolic enzyme in the nucleus is used as a mechanism to “buffer” its activity and rapidly respond to the metabolic demand for its activity. Another metabolic enzyme, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) has been shown to be present in both the cytoplasm and nucleus (31). It was suggested that GAPDH shuttles between the two compartments and performs an additional nonglycolytic function in RNA biogenesis (31). Further characterization of the GK/GKRP regulatory mechanism may reveal additional non-glucose-phosphorylation-related roles for these proteins. In conclusion, the GKRP-mutant mice show that the GK/GKRP system represents an important regulatory mechanism for glucose homeostasis in the liver.
Acknowledgments
We thank Drs. Todd Kirchgessner and Gordon Robinson for critically reading the manuscript, as well as the staff of Veterinary Sciences and the microinjection facility for their technical support.
Abbreviations
- GK
glucokinase
- GKRP
glucokinase regulatory protein
- MODY
maturity-onset diabetes of the young
- ES cell
mouse embryonal stem cell
- PEPCK
phosphoenolpyruvate carboxykinase
- +/+
wild type
- +/−
heterozygous knockout
- −/−
homozygous knockout
References
- 1.Matschinsky F A. Diabetes. 1996;45:223–241. doi: 10.2337/diab.45.2.223. [DOI] [PubMed] [Google Scholar]
- 2.Froguel P, Zouali H, Vionnet N, Velho G, Vaxillaire M, Sun F, Lesage S, Stoffel M, Takeda J, Passa P, et al. N Engl J Med. 1993;328:697–702. doi: 10.1056/NEJM199303113281005. [DOI] [PubMed] [Google Scholar]
- 3.Gidh-Jain M, Takeda J, Wu L, Lange A, Vionnet N, Stoffel M, Velho G, Sun F, Cohen D, Froguel P, et al. Proc Natl Acad Sci USA. 1993;90:1932–1936. doi: 10.1073/pnas.90.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bali D, Svetlanov A, Lee H-W, Fusco-DeMane D, Leiser M, Li B, Barzilai N, Surana M, Hou H, Fleischer N, et al. J Biol Chem. 1995;270:21464–21467. doi: 10.1074/jbc.270.37.21464. [DOI] [PubMed] [Google Scholar]
- 5.Grupe A, Hultgren B, Ryan A, Ma Y, Bauer M, Stewart T. Cell. 1995;83:69–78. doi: 10.1016/0092-8674(95)90235-x. [DOI] [PubMed] [Google Scholar]
- 6.Efrat S, Leiser M, Wu Y-J, Fusco-DeMane D, Emran O, Surana M, Jetton T, Magnuson M, Weir G, Fleischer N. Proc Natl Acad Sci USA. 1994;91:2051–2055. doi: 10.1073/pnas.91.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hariharan N, Farrelly D, Hagan D, Hillyer D, Arbeeny C, Sabrah T, Treloar A, Brown K, Kalinowski K, Mookhtiar K. Diabetes. 1997;46:11–16. doi: 10.2337/diab.46.1.11. [DOI] [PubMed] [Google Scholar]
- 8.Ferre T, Pujol A, Riu E, Bosch F, Valera A. Proc Natl Acad Sci USA. 1996;93:7225–7230. doi: 10.1073/pnas.93.14.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Postic C, Shiota M, Niswender K, Jetton T, Chen Y, Moates J, Shelton K, Linder J, Cherrington A, Magnuson A. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 10.Caro J, Triester S, Patel V, Tapscott E, Frazier N, Dohm G. Horm Metab Res. 1995;27:19–22. doi: 10.1055/s-2007-979899. [DOI] [PubMed] [Google Scholar]
- 11.Vandercammen A, Van Schaftingen E. Biochem J. 1993;294:551–556. doi: 10.1042/bj2940551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veiga-da-Cunha M, Detheux M, Watelet N, Van Schaftingen E. Eur J Biochem. 1994;225:43–51. doi: 10.1111/j.1432-1033.1994.00043.x. [DOI] [PubMed] [Google Scholar]
- 13.Veiga-da-Cunha M, Courtois S, Michel A, Gosselain E, Van Schaftingen E. J Biol Chem. 1996;271:6292–6297. doi: 10.1074/jbc.271.11.6292. [DOI] [PubMed] [Google Scholar]
- 14.Brown K, Kalinowski S, Megill J, Durham S, Mookhtiar K. Diabetes. 1996;46:179–186. doi: 10.2337/diab.46.2.179. [DOI] [PubMed] [Google Scholar]
- 15.Toyoda Y, Miwa I, Satake S, Anai M, Oka Y. Biochem Biophys Res Commun. 1995;215:467–473. doi: 10.1006/bbrc.1995.2488. [DOI] [PubMed] [Google Scholar]
- 16.Tybulewicz V, Crawford C, Jackson P, Bronson R, Mulligan R. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 17.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Hagan D, Kienzle B, Jamil H, Hariharan N. J Biol Chem. 1994;269:28737–28744. [PubMed] [Google Scholar]
- 20.Markwell M, Hass S, Bieber L, Tolbert N. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Passonneau V, Lauderdale R. Anal Biochem. 1974;60:405–412. doi: 10.1016/0003-2697(74)90248-6. [DOI] [PubMed] [Google Scholar]
- 23.Iynedjian P, Pilot P, Nouspikel T, Milburn J, Quadde C, Hughes S, Ucla C, Newgard C. Proc Natl Acad Sci USA. 1989;86:7838–7842. doi: 10.1073/pnas.86.20.7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iynedjian P, Gjinovci A, Renold A. J Biol Chem. 1988;263:740–744. [PubMed] [Google Scholar]
- 25.Davis S N, Pagliassotti M J. In: The Role of the Liver in Maintaining Glucose Homeostasis. Pagliassotti M J, Davis S N, Cherrington A D, editors. Austin, TX: Landes; 1994. pp. 85–107. [Google Scholar]
- 26.Kimbal S, Vary T, Jefferson L. Annu Rev Physiol. 1994;56:321–348. doi: 10.1146/annurev.ph.56.030194.001541. [DOI] [PubMed] [Google Scholar]
- 27.Warner J, Leek J, Intody S, Markham A, Bonthron D. Mamm Genome. 1995;6:532–536. doi: 10.1007/BF00356171. [DOI] [PubMed] [Google Scholar]
- 28.Vionnet N, Hani E, Lesage S, Philippi A, Hager J, Varret M, Stoffel M, Tanizawa Y, Chiu K, Glaser B, et al. Diabetes. 1997;46:1062–1068. doi: 10.2337/diab.46.6.1062. [DOI] [PubMed] [Google Scholar]
- 29.Liang Y, Najafi H, Smith R, Zimmerman E, Magnuson M, Tal M, Matschinsky M. Diabetes. 1992;41:792–806. doi: 10.2337/diab.41.7.792. [DOI] [PubMed] [Google Scholar]
- 30.Velho G, Petersen K, Perseghin G, Hwang J-H, Rothman D, Pueyo M, Cline G, Froguel P, Shulman G. J Clin Invest. 1996;98:1755–1761. doi: 10.1172/JCI118974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh R, Green M. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]