Abstract
SdrG is a surface-associated fibrinogen binding protein present in most strains of Staphylococcus epidermidis. Surface expression of SdrG was not detected by flow cytometry or immunofluorescence microscopy on S. epidermidis 0-47 grown in nutrient broth or in the presence of human serum. sdrG transcript levels increased 1 hour following a shift from growth in nutrient broth to growth in the bloodstream of a mouse and resulted in a concomitant increase in protein levels as detected by immunofluorescence microscopy. The environmental signal(s) resulting in the increase in expression is elusive, as growth under conditions known to mimic in vivo conditions (elevated CO2, iron limitation, human serum, and citrated human blood) did not affect expression of SdrG. Immunizing mice with either the N1N2N3 (amino acids 50 to 597) or N2N3 (amino acids 273 to 597) subdomain of the N-terminal A domain of recombinant SdrG (rSdrG) elicited a robust antibody response; however, only mice vaccinated with rSdrGN23 exhibited a significant reduction in 0-47 recovered after experimental infection. Since SdrG is expressed early during infection in response to specific host environmental cues present in the bloodstream and since antibodies to it are effective in reducing bacteremia, SdrG possesses attributes of a vaccine component effective against the pathogenic form of the ubiquitous human commensal S. epidermidis.
Staphylococcus epidermidis is a ubiquitous human commensal, but it is also the leading cause of nosocomial bacteremia infections. These infections typically accompany placement of indwelling devices such as venous catheters, prosthetic heart valves, or prosthetic joints (15, 32). An implanted device is readily coated with host proteins, and the staphylococci likely bind to these molecules via bacterial surface adhesins such as SdrG (Fbe), GehD, EmbP, and AtlE, which bind fibrinogen, collagen, fibronectin, and vitronectin, respectively (2, 4, 11, 14, 21, 34). It has been hypothesized that bacterial surface proteins function early during an infection to facilitate colonization (6, 7, 9, 23), although expression of these proteins during infection has not been examined. Bacteria attached to an implanted device continue to divide and associate into a three-dimensional structure or biofilm (11). These infections are difficult to treat not only because bacteria within a biofilm are recalcitrant to antibiotic therapy but also because the occurrence of antibiotic resistance in S. epidermidis clinical isolates is increasing (5, 11, 16, 26, 35). Treatment often requires costly and invasive surgical procedures to remove the prosthetic device (29). The additional morbidity imposed by this process underscores the medical need for vaccination to prevent these infections.
A limited number of published studies have examined potential vaccine targets for prevention of S. epidermidis infection. Vaccination with the biofilm-associated polysaccharide poly-N-acetylglucosamine reduced the level of infection in a rabbit model of infective endocarditis (30). In a proteomic approach to identify surface antigens, five antigens were each shown to reduce bacterial numbers in a murine infection model (28). Recently, antibodies directed against AtlE (amidase or repeat sequence) and ScaB were shown to be opsonic in vitro (25). Finally, incubation of S. epidermidis with anti-Fbe (anti-SdrG) immune serum prior to challenge reduced the bacteria recovered in a mouse model of infection (27). While the last study indicates that passive administration of antibodies to Fbe (SdrG) is effective in limiting infection in the murine model, it does not answer the question of whether active immunization can elicit a response sufficient to reduce bacteremia.
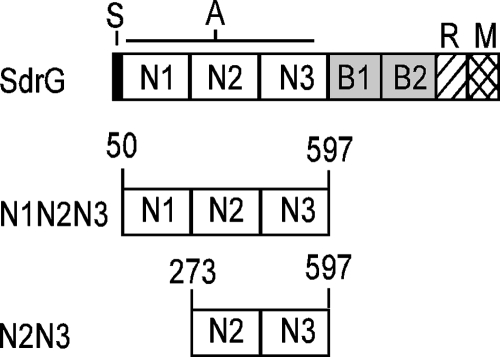
SdrG is a 119-kDa surface protein on S. epidermidis that mediates adhesion to fibrinogen and belongs to the class of bacterial adhesins termed MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) (4, 22, 23). MSCRAMMs are bacterial surface proteins that bind to host proteins and share a similar domain structure, with the ligand binding activity localized to the N-terminal A domain (Fig. 1), which facilitates a primary attachment during an infection. The A domain of SdrG includes the N1, N2, and N3 subdomains (amino acids 50 to 597), and binding of fibrinogen has been localized to a cleft between N2 and N3 (4, 12, 24). SdrG interacts with the Bβ chain of fibrinogen by a “dock, lock, and latch” mechanism with a calculated KD (equilibrium dissociation constant) of 1.4 × 10−7 M (4, 24). SdrG-mediated binding to fibrinogen is likely important for the staphylococci to interact with an indwelling device and initiate an infection. The presence of antibodies against SdrG in a vaccinated patient has the potential of preventing disease by being effective in both reducing attachment to a prosthetic device and mediating opsonophagocytic killing.
FIG. 1.
Schematic representation of SdrG. S, signal peptide; A, fibrinogen binding domain; B, repeats; R, serine-aspartate repeat; W, cell wall-spanning domain; M, membrane-spanning domain; C, cytoplasmic tail. N1N2N3 and N2N3 represent the truncated versions of rSdrG used in this study.
SdrG expression has been detected in some isolates of S. epidermidis using Western blot analysis or fluorescence-activated cell sorting (FACS); however, expression of SdrG during the course of infection has not been examined (1, 10, 12). We provide direct evidence that SdrG expression is increased in the early stages of a murine infection model compared to the undetectable level of SdrG expression occurring during in vitro culture. The data demonstrate that expression of sdrG by S. epidermidis 0-47 and by four different clinical isolates of methicillin-resistant S. epidermidis (MRSE) is induced within the first 60 min of a murine bloodstream infection. These results support the hypothesis that SdrG is an antigen whose expression increases following exposure to specific, but uncharacterized, host environmental signals present during the early stages of infection. Although two different recombinant forms of the SdrG A domain (His-tagged N123 [amino acids 50 to 597] and N23 [amino acids 273 to 597] [Fig. 1]) are highly immunogenic in mice, only rSdrGN23 reduces the CFU recovered in a murine infection model.
MATERIALS AND METHODS
Bacterial strains and chemicals.
The S. epidermidis strains utilized in this study are 0-47 (13) and four MRSE strains, GAR8896, GAR8933, GAR9155, and GAR9657, cultured from patients during a phase III antibiotic clinical trial conducted by Wyeth (3). Strains GAR8933, GAR9155, and GAR9657 are bloodstream isolates, whereas GAR8896 is a sputum isolate. GAR9155, GAR8933, and GAR8896 have distinct ribotoypes. Although isolated 5 months apart from distinct patients, GAR8933 and GAR9657 have the same ribotype and are possibly clonal. Bacterial cultures were started by diluting an overnight culture of S. epidermidis 0-47 grown in tryptic soy broth (TSB) or TSB supplemented with 70% human serum to an optical density at 600 nm of ∼0.1 in either TSB or TSB supplemented with 70% human serum. The bacteria were then grown at 37°C with shaking (200 rpm) and harvested at different phases of growth. All chemicals were obtained from Sigma Chemical Co. unless otherwise noted. rHis-SdrGN123 (SdrG-A) was provided by Inhibitex, Inc. (4, 31).
Expression and purification of rSdrG.
The gene encoding rSdrGN23 was amplified from a cloned His-SdrGN123 (SdrG-A) (4, 31) by PCR (forward primer, 5′GGAATTCCCATATGGAACAAGGTTCGAATGTTAATC3′; reverse primer, 5′GGAATTCCAAGCTTTTATTTTTCAGGAGGCAAGTCAC3′) and ligated into pET27b(+) at the NdeI and HindIII sites, thereby excluding all coding sequence from the vector except for a single methionine at the amino terminus. The 3′ primer was designed to include a stop codon before the HindIII site. The gene was cloned in Escherichia coli BL21(DE3) (Novagen) and confirmed by DNA sequencing. Protein was expressed by growing the bacteria in HySoy broth (28) supplemented with 25 μg/ml kanamycin at 37°C in a 10-liter fermentor (Braun Biotech International) until the optical density at 600 nm reached ∼1.0. Protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 3 h, the bacteria were harvested by centrifugation. The bacteria were resuspended in Tris-buffered saline (Bio-Rad) (pH 8.0) and lysed using a model 110Y microfluidizer (Microfluidics Corporation). The cell debris was removed by centrifugation, and the supernatant was fractionated with 80% ammonium sulfate. Material precipitated with ammonium sulfate was collected by centrifugation, dissolved in 20 mM Tris (pH 8.0)-25 mM NaCl, and dialyzed against the same buffer. Dialyzed material was applied to a TMAE FractoGel anion-exchange column (EM Separations), and bound protein was eluted with a 25 to 500 mM linear gradient of NaCl in 20 mM Tris (pH 8.0). Fractions containing rSdrGN23 were pooled and dialyzed against phosphate-buffered saline (PBS) (pH 7.4). The identity of isolated rSdrGN23 was confirmed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and a single NH2-terminal sequence starting at MEQGSNVNHL was displayed.
Animal studies.
All animal use protocols were reviewed and approved by the Pearl River Animal Care and Use Committee at Wyeth.
Generation of anti-rSdrG immune sera.
To produce anti-rSdrGN23 hyperimmune sera, five New Zealand White rabbits (2.5 to 3.5 kg; Charles River Laboratories) were vaccinated with 10 μg of rSdrGN23 mixed with 20 μg of the adjuvant QS21 (Antigenics) on weeks 0, 3, and 6. On week 8 the animals were exsanguinated, and the sera were collected and analyzed by antigen-specific enzyme-linked immunosorbent assay (ELISA). The serum most reactive with rSdrGN23 was used for flow cytometry and immunofluorescence microscopy (IF). Preimmune serum was collected from the rabbits on week 0 prior to the first vaccination to be used as a negative control.
Flow cytometry.
Preimmune and immune sera from rabbits immunized with rSdrGN23 or infected with S. epidermidis 0-47 (28) were used as primary antibodies for flow cytometry. Bacteria were incubated with preimmune or immune sera (1:100) in staining buffer (Hanks’ balanced salt solution [Mediatech, Inc.] with 10% goat serum) for 30 min on ice. Cells were then washed and stained with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch) on ice for 30 min. Bacteria were washed with staining buffer and fixed with 2% paraformaldehyde, and data were acquired and analyzed using a FACSCaliber flow cytometer and Cell Quest software (Becton Dickinson and Co.). A total of 30,000 events were collected for each sample.
Transcript analysis.
Ten female 12-week-old BALB/c mice (Charles River) were infected by intraperitoneal injection of ∼5 × 108 S. epidermidis 0-47 grown to late log phase in TSB. The bacterial challenge stock was diluted into 2 volumes of RNAlater (Ambion, Inc.) and stored at 4°C (T0 sample). Five animals were injected with sterile TSB as a mock-infected control, and blood was harvested after 1 hour as described below. At 1 and 3 hours postchallenge, five of the infected mice were sacrificed; 200 μl of blood from each was pooled and diluted into 4 ml RNAprotect (Qiagen), and the bacteria were pelleted, resuspended in RLT buffer (Qiagen), and transferred to a FastProtein Blue tube (MP Biomedical) for lysis in a FastPrep disruptor (QBIOgene). The RNA was purified using the RNeasy minikit (Qiagen) and treated with Turbo DNase (Ambion). The RNA concentration was measured in a ND-1000 spectrophotometer (NanoDrop Technologies) and its integrity assessed in a Bioanalyzer 2100 (Agilent Technologies). The RNA was reverse transcribed into cDNA using the RETROscript kit (Ambion) with random decamer primers.
Oligonucleotide primers were designed using Primer Express 2.0 (Applied Biosystems). cDNA samples were assayed in duplicate for specific transcripts, with 16S rRNA as a control, using Sybr Green PCR master mix (Applied Biosystems) and a standard cycle protocol.
IF.
Infected mice (as described above) were exsanguinated after 3 h, and the blood from five mice was pooled into ice-cold sodium citrate (pH 7.0) (final concentration, 0.4%). The eukaryotic cells were lysed with 1% NP-40 (Pierce Biotechnology). The bacteria were washed with PBS and incubated overnight at 4°C with rabbit anti-rSdrGN23 (1:100), rabbit anti-rSdrGN23 (1:100) pretreated for 3 h with 5 μg rSdrGN23, or preimmune serum (1:100) and detected with Alexa488-conjugated goat anti-rabbit antibody (1:250; Invitrogen). The labeled bacteria were dried on a microscope slide, and a coverslip was mounted with Vectashield HardSet medium (Vector Laboratories, Inc.). Images were obtained with a Leica TCS SL spectral confocal microscope (Leica Microsystems).
Vaccination and challenge of mice.
Groups of 4-week-old female BALB/c mice (Charles River) were vaccinated by subcutaneous injection on weeks 0, 3, and 6 with 10 μg rSdrGN23 or rHis-SdrGN123 mixed with 20 μg of the adjuvant QS21 and bled on weeks 0 and 8. The mice were then challenged with ∼5 × 108 CFU of S. epidermidis 0-47 as described above. After 24 hours the mice were sacrificed, and the bacteria present in the spleen were enumerated. Bacterial reduction was determined compared to a control group receiving QS21 in saline. Data were analyzed using JMP statistical software (SAS Institute).
SdrG-specific antigen ELISA.
Ninety-six-well Immunoplates (Nunc Nalgene International) were coated with 100 μl of 1-μg/ml rSdrGN23 or rHis-SdrGN123 in PBS (pH 7.5) overnight at 4°C. The plates were washed with 1× PBS-0.1% Tween 20 (PBST) and blocked with 1% (wt/vol) nonfat milk in PBS-0.05% Tween 20. Plates were washed with PBST, 100-μl portions of serially diluted (threefold) antisera were added to the plates, and the plates were incubated overnight at 4°C. The plates were washed, and bound primary antibodies were detected with biotin-conjugated goat anti-mouse immunoglobulin G (1:15,000 dilution) (Southern Biotech) in PBST followed by streptavidin-horseradish peroxidase conjugate (Symed; 1:5,000 in PBST) and developed with ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] (KPL, Inc.). Absorbance was measured at 405 nm in a plate reader (Molecular Devices). Antibody titers are expressed as the reciprocal of the highest serum dilution with an absorbance value of 0.1.
RESULTS
SdrG is not detected on the surface of S. epidermidis 0-47 in vitro.
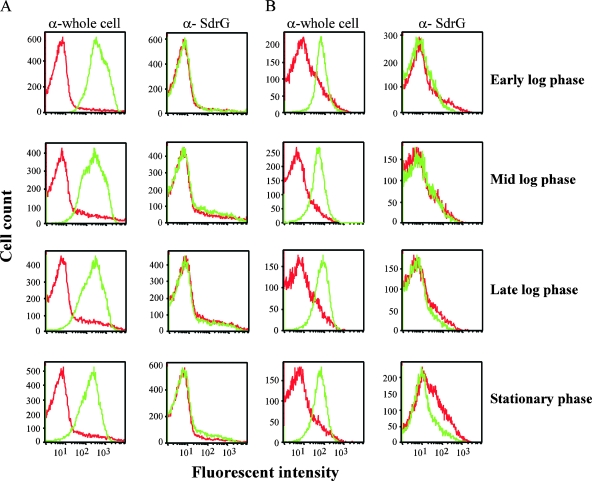
As an initial step in the characterization of SdrG expression, S. epidermidis 0-47 was harvested from TSB cultures at different growth phases and SdrG surface expression was assessed by flow cytometry. As a control to ensure the presence of S. epidermidis, the bacteria were stained with immune serum generated in rabbits challenged with live S. epidermidis 0-47 (28). Preimmune rabbit serum was included as a specificity control. SdrG staining was not detected at any of the growth phases tested (Fig. 2), whereas anti-whole-cell S. epidermidis 0-47 sera stained the bacteria at all growth phases (Fig. 2). These results indicate that surface expression of SdrG is not induced at any point in the growth phase to permit immunofluorescent staining with immune sera specifically reactive to SdrG. A lack of SdrG surface expression was confirmed by IF (see Fig. 4, and data not shown). Growth of S. epidermidis 0-47 in 70% serum to mimic a host environment alters the expression of surface antigens and might cause it to resemble that occurring during bacteremia (28). To determine the effect that growth in serum has on SdrG expression, we also analyzed surface expression of SdrG following growth in 70% human serum. Staining of bacteria grown in the presence of human serum was still at background levels (Fig. 2). Positive control sera against S. epidermidis 0-47 again stained bacteria in all samples (Fig. 2). Lack of anti-SdrG staining was also seen by IF and FACS following growth of 0-47 under several conditions that mimic specific in vivo signals, including growth under iron-depleted conditions, growth in 5% CO2, or growth in citrated human whole blood (data not shown). Under all the in vitro growth conditions tested, expression of SdrG was not detected on the surface of S. epidermidis 0-47.
FIG. 2.
Surface expression of SdrG on S. epidermidis 0-47. The bacteria were grown in either TSB (A) or 70% human serum (B) and harvested at different growth phases. They were then stained with immune serum (green) generated against either live whole S. epidermidis 0-47 or rSdrGN23 and compared to staining with the corresponding preimmune control serum (red). A total of 30,000 events were collected for each sample. These data are representative of three independent experiments.
FIG. 4.
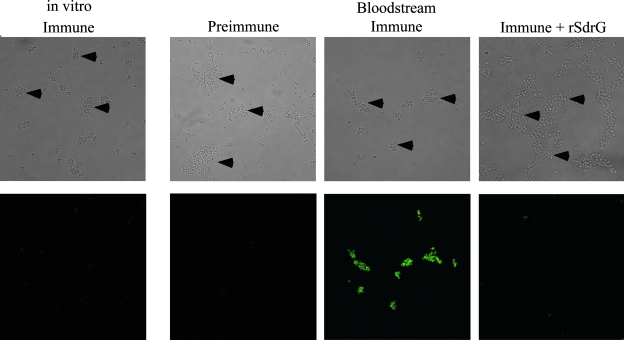
SdrG expression on S. epidermidis 0-47 in a murine model of bacteremia. A group of five mice was infected by intraperitoneal injection of 5 × 108 CFU 0-47. A 3 hours postinfection the blood from the mice was pooled and the bacteria isolated. The bacteria were then stained with rabbit anti-SdrGN23 immune serum and visualized with a confocal fluorescence microscope. In vitro-grown bacteria at the time of challenge were treated similarly and stained for comparison to the bacteria isolated from the bloodstreams of infected mice. Preincubation of the immune serum with rSdrG and staining with preimmune serum was included as a specificity control. Arrowheads point to the bacteria in the bright-field images. These data are representative of three independent experiments.
SdrG expression is increased in a murine infection model.
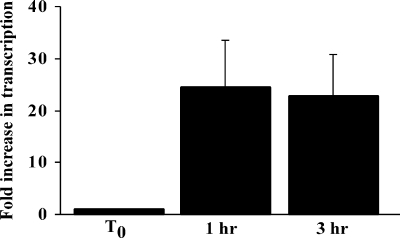
Although SdrG was not detected on S. epidermidis 0-47 grown in vitro, its role as a fibrinogen binding protein suggests that it functions in vivo and the environmental signals required to induce surface expression should be present during an infection. To test this hypothesis, mice were challenged by intraperitoneal injection of 0-47, and the bacteria were harvested from the bloodstream at 1 and 3 h postinfection to measure both sdrG mRNA and surface expression. sdrG transcript levels were measured in bacteria prior to challenge and then in bacteria isolated from bacteremic mice at 1 h (7.25 ± 0.5 log10 CFU/ml) and 3 h (7.33 ± 0.04 log10 CFU/ml) postchallenge. Following transition to the in vivo environment, transcription of sdrG mRNA increased to levels 24-fold and 23-fold greater at 1 and 3 h, respectively, than the transcript levels detected in the bacteria at the time of challenge (Fig. 3). Increased transcription of sdrG mRNA was specific to the bacteria in the infected mice, since amplification was not detected in mRNA purified from the blood of mock-infected mice (data not shown). To determine if an increase in mRNA results in a concomitant increase in surface expression, SdrG expression by 0-47 was assessed by IF at the time of challenge and on bacteria isolated from the blood of mice at 3 h postinfection. As expected, SdrG expression was below the limit of detection in the challenge dose of bacteria grown in TSB (late log phase). Within 3 h of exposure to the in vivo environment, SdrG surface expression increased substantially (Fig. 4), whereas neither preimmune serum nor immune serum blocked with competing purified rSdrGN23 protein stained the cells.
FIG. 3.
sdrG transcript levels in S. epidermidis in vivo versus in vitro. Groups of five BALB/c mice were infected with S. epidermidis 0-47 by intraperitoneal injection. At 1 and 3 hours postinfection, the blood from five mice was pooled and the RNA isolated. The sdrG transcript level in the bacteria was measured by quantitative reverse transcription-PCR following in vitro growth at the time of challenge (T0) and 1 h and 3 h after infection. Gene expression was assessed by the 2−ΔΔCT method (17) using 16S rRNA to normalize. Values for fold increase in transcription represent the means from three independent experiments ± standard errors of the means.
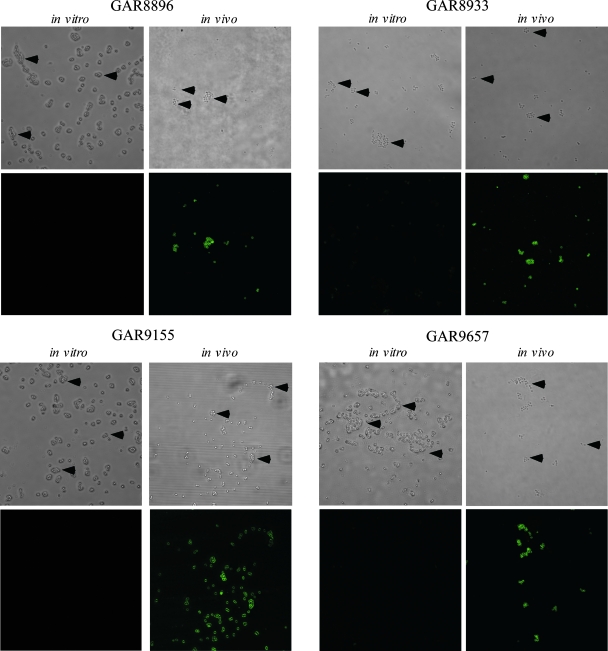
To determine if this type of regulation of SdrG is unique to 0-47, we tested the surface expression of SdrG on four MRSE clinical isolates using bacteria recovered from the murine bacteremia model. In all four isolates, surface expression of SdrG was detectable only during bacteremia and not on the in vitro-grown cells used for the challenge dose (Fig. 5).
FIG. 5.
SdrG expression on S. epidermidis clinical isolates. Four groups of five mice each were infected with one of four MRSE clinical isolates (GAR8896, GAR8933, GAR9155, and GAR9657). At 3 hours postinfection, the blood from the mice was pooled and the bacteria isolated. The bacteria at the time of challenge (in vitro) and following isolation from the bloodstream (in vivo) were then stained and visualized as for Fig. 4. Arrowheads point to the bacteria in the bright-field images. Preimmune serum did not react with any of the bacteria grown in vitro or in vivo (data not shown). These pictures are representatives of three independent experiments.
SdrG vaccination studies.
The evidence for SdrG surface expression during a murine bacteremia, as detected by antibody reactivity to the bacterial cell surface, supports the potential of SdrG as a prophylactic vaccine against S. epidermidis infection. To test this in an S. epidermidis murine infection model, mice were vaccinated with either of two different recombinant forms of the fibrinogen binding A domain of SdrG (rHis-SdrGN123 or rSdrGN23 [Fig. 1]). Both forms of rSdrG elicited high antigen-specific antibody titers (Table 1). Following challenge with 0-47, only the animals vaccinated with rSdrGN23 exhibited a consistent and statistically significant reduction in the number of bacteria recovered at 24 h postinfection (Table 1).
TABLE 1.
SdrG vaccination and S. epidermidis challengea
| Protein | Expt | nb | Antigen | CFU/spleen | Pc | Geometric mean titerd |
|---|---|---|---|---|---|---|
| SdrGN123 | 1 | 9 | Saline | 5.43 ± 0.97 | <0.04 | <100 |
| 10 | SdrGN23 | 4.30 ± 1.56 | 1.4 × 106 | |||
| 2 | 20 | Saline | 5.06 ± 0.93 | <0.01 | <100 | |
| SdrGN23 | 4.15 ± 0.79 | 1.5 × 106 | ||||
| 3 | 15 | Saline | 5.40 ± 0.89 | <0.04 | <100 | |
| SdrGN23 | 4.74 ± 0.69 | 1.0 × 106 | ||||
| rHis-SdrGN123 | 1 | 10 | Saline | 5.62 ± 1.19 | NS | <100 |
| rHis-SdrGN123 | 4.87 ± 1.42 | 1.2 × 106 | ||||
| 2 | 15 | Saline | 4.44 ± 1.83 | NS | <100 | |
| rHis-SdrGN123 | 5.52 ± 0.95 | 1.4 × 106 | ||||
| 3 | 15 | Saline | 4.24 ± 1.08 | NS | <100 | |
| rHis-SdrGN123 | 4.48 ± 0.89 | 6.8 × 106 |
Groups of female (4-week-old) BALB/c mice were vaccinated by subcutaneous injection with either saline, 10 μg rSdrGN23, or 10 μg rHis-SdrGN123 in 20 μg QS21 as adjuvant. At 2 weeks following the final vaccination, the animals were challenged by intraperitoneal injection of ∼5 × 108 CFU of S. epidermidis 0-47. At 24 hours postchallenge, the mice were sacrificed and the bacteria in the spleen enumerated.
Number of animals/group.
Statistical significance was determined using Student's t test. NS, not significant.
Serum antibody titers measured by ELISA.
DISCUSSION
Staphylococcus epidermidis is both a ubiquitous human commensal and an opportunistic pathogen responsible for the majority of nosocomial bloodstream infections (32, 33). Upon initiation of an infection, S. epidermidis likely undergoes global changes in gene expression that potentiate the transition from a commensal present on the skin to survival within the bloodstream of the host. In keeping with other models of bacterial pathogenesis, S. epidermidis expresses numerous surface adhesins that specifically bind host proteins and are likely crucial to the establishment of an infection. One such protein is the fibrinogen binding MSCRAMM SdrG. Detection of fluorescent staining of S. epidermidis with SdrG-specific immune serum by flow cytometry and IF showed that SdrG was not detected on the surface of S. epidermidis 0-47 grown under various in vitro conditions, including growth in TSB (Fig. 2A and 4), or in vitro conditions that may mimic some of the environmental signals encountered during a bacteremic infection, such as growth in human serum (Fig. 2B), iron-depleted conditions, 5 to 10% CO2, or citrated human whole blood (data not shown). However, following a shift of 0-47 from growth in TSB to survival within the bloodstream, sdrG transcript levels increased 24-fold within the first hour of infection and remained high for at least 3 hours (Fig. 3). This increase in transcript resulted in a corresponding increase in surface expression as detected by IF. Since a precise correlation of mRNA and protein levels does not always exist (8), it is important (particularly for vaccine development) to establish the relationship of surface expression of a potential vaccine target with mRNA levels, since it is not always possible to assay infected tissue samples directly by immunofluorescence assays. Our results demonstrate that the increased level of sdrG mRNA does result in a concomitant increase in surface protein as detected by IF on bacteria isolated during a murine bacteremia (Fig. 4). This fluorescent staining was specific for SdrG, since staining of the bacteria is blocked by preincuabtion of the immune serum with competing rSdrGN23. Therefore, the environmental signal or combination of signals required to upregulate SdrG expression is present in vivo but absent from all of the in vitro growth conditions tested (70% human serum, iron depletion, 5 to 10% CO2, or citrated human blood). A more detailed analysis of the sdrG promoter, in combination with the types of global analyses offered by microarray studies, presents an exciting opportunity to decipher some of the environmental cues involved in the regulation of SdrG and might provide valuable information into the understanding of the mechanisms by which a commensal transitions into a pathogen.
To determine the prevalence of the in vivo expression profile of SdrG in other S. epidermidis isolates, we tested the expression of SdrG on four different clinical isolates of MRSE representing three distinct ribotypes. In all four isolates, SdrG expression was increased in vivo relative to the inoculum, indicating that the same mode of regulation is common and possibly widespread among S. epidermidis clinical isolates. These data differ from previously published results showing that SdrG could be detected on the surface of in vitro-grown S. epidermidis HB and F40802 by FACS analysis (10) and by Western analysis of RP62A and HB (1, 12). The ability to demonstrate in vitro expression in these studies may be related to the sensitivity of the immunological reagents and assays, higher levels of expression, or increased accessibility of SdrG to antibody in the different isolates, but the studies did not directly compare in vitro versus in vivo conditions.
An antigen expressed early in an infection (e.g., SdrG) is an ideal target for a prophylactic vaccine. S. epidermidis infections are typically associated with indwelling prosthetic devices (e.g., venous catheter, heart valve, or prosthetic joint). Early in infection the bacteria bind specifically to host molecules (e.g., fibrinogen, vitronectin, or fibronectin) coating the implanted device. The bacteria then begin to divide and form a biofilm in which the bacteria are enmeshed in a complicated three-dimensional structure. At this point, the bacteria are likely refractory to both immune intervention and antimicrobial therapy. It is therefore necessary to target the bacteria prior to attachment and biofilm formation. The data presented here provide evidence for the hypothesis that SdrG is expressed early in infection and that an antibody response directed against this antigen could help to reduce infection, perhaps as a consequence of reducing binding to host components and enhancing opsonization of the bacteria.
Vaccination of mice with either form of the rSdrG (rSdrGN123 or rSdrGN23) resulted in high protein-specific antibody titers. However, only the animals vaccinated with rSdrGN23 exhibited a reduced level of 0-47 in a murine bacteremia model. Although there is no indwelling device in this model, the results demonstrate that immunization with rSdrGN23 elicits an antibody response that is capable of reducing bacterial burden and infection. It is unclear why the two forms of the rSdrG A domain behave differently as protective antigens, as the only differences between the proteins are the presence of a N-terminal hexahistidine tag and the N1 subdomain on the rHis-SdrGN123 protein. Although we have not ruled out any contribution of the His tag, it is more reasonable to suspect that the N1 plays a role in this difference. To date no function has been attributed to this subdomain, but it is possible that the presence of N1 could alter the immune response or mask a protective epitope(s) such that vaccination with rHis-SdrGN123 is not protective. Although there is some evidence indicating that the N1 domain in some MSCRAMMs is susceptible to proteolysis (19), further experimentation is required to understand the difference between the two forms of rSdrG.
Taken together, the above results indicate that SdrG is a surface antigen that is differentially regulated and expressed early during infection in response to host-specific signals in 0-47 and four other clinical isolates. In a recent study, the majority of serum samples from patients convalescing from S. epidermidis infections contained antibodies directed against SdrG, whereas sera from healthy individuals did not (1). These data indicate that the immune systems of healthy individuals colonized with S. epidermidis do not recognize SdrG, whereas after infection most patients have mounted an immune response against SdrG. These results are relevant in the strategy of developing vaccines against pathogenic forms of common human commensal bacteria where the unlikely event of inducing a sterilizing immunity may be perceived as a potential safety issue. Although there are no data regarding the expression of SdrG in S. epidermidis as it resides on human skin and mucosa, the serological data do support the hypothesis that SdrG is expressed during infection. Until data regarding expression of SdrG during the commensal state become available, it is tempting to speculate that the differential expression of SdrG in vitro versus in vivo represents one marker for the commensal versus pathogenic state of S. epidermidis. The fact that SdrG expression was detected only during bacteremia illustrates the value in identifying vaccine candidates that are uniquely expressed during an infection and reminds us that the common practice of using in vitro-cultured organisms to monitor the effectiveness of a vaccine (e.g., by whole-cell ELISA, FACS, or bactericidal or opsonic activity) cannot be the sole means on which to base a decision on vaccine potential. By examining SdrG expression in vivo, we demonstrate that it is an antigen that should be given serious consideration as a component in a prophylactic vaccine to protect against S. epidermidis infection.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 21 April 2008.
REFERENCES
- 1.Bowden, M. G., W. Chen, J. Singvall, Y. Xu, S. J. Peacock, V. Valtulina, P. Speziale, and M. Hook. 2005. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 1511453-1464. [DOI] [PubMed] [Google Scholar]
- 2.Bowden, M. G., L. Visai, C. M. Longshaw, K. T. Holland, P. Speziale, and M. Hook. 2002. Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J. Biol. Chem. 27743017-43023. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, P. A., D. T. Weaver-Sands, and P. J. Petersen. 2005. In vitro activity of tigecycline against isolates from patients enrolled in phase 3 clinical trials of treatment for complicated skin and skin-structure infections and complicated intra-abdominal infections. Clin. Infect. Dis. 41(Suppl. 5)S315-S332. [DOI] [PubMed] [Google Scholar]
- 4.Davis, S. L., S. Gurusiddappa, K. W. McCrea, S. Perkins, and M. Hook. 2001. SdrG, a fibrinogen-binding bacterial adhesin of the microbial surface components recognizing adhesive matrix molecules subfamily from Staphylococcus epidermidis, targets the thrombin cleavage site in the Bbeta chain. J. Biol. Chem. 27627799-27805. [DOI] [PubMed] [Google Scholar]
- 5.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, and M. Beach. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32S114-S132. [DOI] [PubMed] [Google Scholar]
- 6.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6484-488. [DOI] [PubMed] [Google Scholar]
- 7.Gillaspy, A. F., J. M. Patti, and M. S. Smeltzer. 1997. Transcriptional regulation of the Staphylococcus aureus collagen adhesion gene, cna. Infect. Immun. 651536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham, M. R., K. Virtaneva, S. F. Porcella, D. J. Gardner, R. D. Long, D. M. Welty, W. T. Barry, C. A. Johnson, L. D. Parkins, F. A. Wright, and J. M. Musser. 2006. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am. J. Pathol. 169927-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, B., X. Zhao, Y. Shi, D. Zhu, and Y. Zhang. 2007. Pathogenic implication of a fibrinogen-binding protein of Staphylococcus epidermidis in a rat model of intravascular-catheter-associated infection. Infect. Immun. 752991-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, A. E., P. R. Patel, P. J. Domanski, B. D. Prater, E. L. Gorovits, P. J. Syribeys, J. H. Vernachio, J. M. Patti, and J. T. Hutchins. 2007. A panel of monoclonal antibodies recognizing the Staphylococcus epidermidis fibrinogen-binding MSCRAMM SdrG. Hybridoma 2628-34. [DOI] [PubMed] [Google Scholar]
- 11.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev. Microbiol. 295-108. [DOI] [PubMed] [Google Scholar]
- 12.Hartford, O., L. O'Brien, K. Schofield, J. Wells, and T. J. Foster. 2001. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology 1472545-2552. [DOI] [PubMed] [Google Scholar]
- 13.Heilmann, C., C. Gerke, F. Perdreau-Remington, and F. Gotz. 1996. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 241013-1024. [DOI] [PubMed] [Google Scholar]
- 15.Heilmann, C., and G. Peters. 2000. Biology and pathogenicity of Staphylococcus epidermidis, p. 442-449. In V. A. Fischetti (ed.), Gram-positive pathogens. Americon Society for Microbiology, Washington, DC.
- 16.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.McAleese, F. M., E. J. Walsh, M. Sieprawska, J. Potempa, and T. J. Foster. 2001. Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. J. Biol. Chem. 27629969-29978. [DOI] [PubMed] [Google Scholar]
- 20.Reference deleted.
- 21.Nilsson, M., L. Frykberg, J. I. Flock, L. Pei, M. Lindberg, and B. Guss. 1998. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect. Immun. 662666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48585-617. [DOI] [PubMed] [Google Scholar]
- 23.Patti, J. M., and M. Hook. 1994. Microbial adhesins recognizing extracellular matrix macromolecules. Curr. Opin. Cell Biol. 6752-758. [DOI] [PubMed] [Google Scholar]
- 24.Ponnuraj, K., M. G. Bowden, S. Davis, S. Gurusiddappa, D. Moore, D. Choe, Y. Xu, M. Hook, and S. V. Narayana. 2003. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115217-228. [DOI] [PubMed] [Google Scholar]
- 25.Pourmand, M. R., S. R. Clarke, R. F. Schuman, J. J. Mond, and S. J. Foster. 2006. Identification of antigenic components of Staphylococcus epidermidis expressed during human infection. Infect. Immun. 744644-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raad, I., A. Alrahwan, and K. Rolston. 1998. Staphylococcus epidermidis: emerging resistance and need for alternative agents. Clin. Infect. Dis. 261182-1187. [DOI] [PubMed] [Google Scholar]
- 27.Rennermalm, A., M. Nilsson, and J. I. Flock. 2004. The fibrinogen binding protein of Staphylococcus epidermidis is a target for opsonic antibodies. Infect. Immun. 723081-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sellman, B. R., A. P. Howell, C. Kelly-Boyd, and S. M. Baker. 2005. Identification of immunogenic and serum binding proteins of Staphylococcus epidermidis. Infect. Immun. 736591-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steckelberg, J. M., and D. R. Osmon. 2000. Prosthetic joint infections, 3rd ed. ASM Press, Washington, DC.
- 30.Takeda, S., G. B. Pier, Y. Kojima, M. Tojo, E. Muller, T. Tosteson, and D. A. Goldmann. 1991. Protection against endocarditis due to Staphylococcus epidermidis by immunization with capsular polysaccharide/adhesin. Circulation 842539-2546. [DOI] [PubMed] [Google Scholar]
- 31.Vernachio, J. H., A. S. Bayer, B. Ames, D. Bryant, B. D. Prater, P. J. Syribeys, E. L. Gorovits, and J. M. Patti. 2006. Human immunoglobulin G recognizing fibrinogen-binding surface proteins is protective against both Staphylococcus aureus and Staphylococcus epidermidis infections in vivo. Antimicrob. Agents Chemother. 50511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2677-685. [DOI] [PubMed] [Google Scholar]
- 33.Vuong, C., and M. Otto. 2002. Staphylococcus epidermidis infections. Microbes Infect. 4481-489. [DOI] [PubMed] [Google Scholar]
- 34.Williams, R. J., B. Henderson, L. J. Sharp, and S. P. Nair. 2002. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 706805-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziebuhr, W., S. Hennig, M. Eckart, H. Kranzler, C. Batzilla, and S. Kozitskaya. 2006. Nosocomial infections by Staphylococcus epidermidis: how a commensal bacterium turns into a pathogen. Int. J. Antimicrob. Agents 28S14-S20. [DOI] [PubMed] [Google Scholar]