Abstract
Mycoplasma pneumoniae is a bacterial pathogen of the human respiratory tract that causes a wide range of airway diseases as well as extrapulmonary symptoms. It possesses a distinct, differentiated terminal structure, termed the attachment organelle, that mediates adherence to the host respiratory epithelium. Previously, we reported that surface-associated M. pneumoniae elongation factor Tu (EF-Tu, also called MPN665) serves as a fibronectin (Fn)-binding protein, facilitating interactions between mycoplasmas and extracellular matrix. In the present study, we determined that binding of M. pneumoniae EF-Tu to Fn is primarily mediated by the EF-Tu carboxyl region. A 179-amino-acid region spanning the carboxyl terminus (designated EC; amino acids 192 to 394) binds Fn in a dose-dependent manner. Further analysis of carboxyl constructs (ED3 and ED4) and their deletion truncations (ED3.1, ED3.2, and ED4.1) revealed that the carboxyl region possessed two distinct sites with different Fn-binding efficiencies. Immunogold electron microscopy using antibodies raised against recombinant ED3 and ED4 demonstrated the surface accessibility of the EF-Tu carboxyl region. Competitive binding assays using intact radiolabeled mycoplasmas and purified recombinant ED3 and ED4 proteins, along with antibody blocking assays, reinforced the role of the surface-exposed EF-Tu carboxyl region in Fn binding. Alkali and high-salt treatment of mycoplasma membranes and Triton X-114-partitioned mycoplasma fractions confirmed the stable association of EF-Tu within the mycoplasma membrane. These observations highlight the unique, multifaceted, and unpredictable role of the classically defined cytoplasmic protein EF-Tu relative to cellular function, compartmentalization, and topography.
Microbial pathogens possess multiple adherence mechanisms, providing biological versatility in their interactions with host targets. For successful colonization of the respiratory tract, Mycoplasma pneumoniae adheres to host respiratory epithelial cell receptors through tip-mediated protein adhesin molecules, designated P1 (24) and P30 (16). Interestingly, hemadsorption-negative mutants (22) of M. pneumoniae that fail to express these specific adhesins still exhibit low-level adherence to target cells (32). Therefore, classical tip-mediated cytadherence may not account for all adherence pathways. Consistent with this possibility, we demonstrated that surfaced localized elongation factor Tu (EF-Tu) and the pyruvate dehydrogenase E1β subunit of M. pneumoniae mediate mycoplasma binding to fibronectin (Fn) (17).
Fn is a high-molecular-weight glycoprotein, abundantly found in fibrillar form in extracellular matrix (ECM) and in soluble form in body fluids. While the primary role of Fn is to serve as a substrate for mammalian cell adhesion (55), a wide variety of microbial pathogens bind Fn for colonization purposes, which contributes to the infectious process. Since our initial observation describing the interaction of Treponema pallidum with Fn (43), many Fn-binding proteins (FnBPs) have been identified, like the MSCRAMM (microbial surface components recognizing adhesive matrix molecules) family of proteins that includes FnBPA and FnBPB from Staphylococcus aureus (27, 52) and SfbI/protein F1, protein F2 (23), SfbII (33), FnbA, and FnbB (37) from streptococcus. While the primary Fn-binding region for the MSCRAMM family of FnBPs is located on the N-terminal 30-kDa type 1 heparin-binding fragment of Fn, secondary binding sites have also been reported (35). Among mycoplasmas, the first Fn interaction was identified in Mycoplasma penetrans (18), followed by M. pneumoniae, where its EF-Tu unexpectedly bound Fn (17).
EF-Tu is the most abundant bacterial protein (25), constituting approximately 10% of M. pneumoniae total protein (47). It is responsible for critical steps in protein synthesis in its GTP-associated form (31). In addition to its role in protein synthesis, EF-Tu has been linked to many “moonlighting” roles, including protein disulfide activities (48), chaperone-like properties (10), initiation of Qβ phage replication (6), and upregulation in the presence of high concentrations of iron (54). EF-Tu forms filaments or bundles of filaments in vitro and is speculated to be a structural element in the “cytoskeletal web” (39). EF-1α, its eukaryotic counterpart, also demonstrates variable activities by binding to actin filaments and microtubules (20) and influencing the assembly and stability of cytoskeletal polymers (40).
The membrane location of EF-Tu has been observed in numerous prokaryotes and is attributed to posttranslational modifications. For example, in Escherichia coli, EF-Tu is transferred from cytoplasm to periplasm during osmotic shock (5) and to the membrane during starvation when subpopulations of EF-Tu are methylated (57). It has been identified as a major cell wall protein of Mycobacterium leprae (38) and is a periplasmic component in Neisseria gonorrhoeae (44). In M. pneumoniae, 17% of the total EF-Tu is observed in the membrane fraction (17). The ability of EF-Tu to function as an M. pneumoniae FnBP provided evidence of its biological versatility, which was subsequently reinforced by reports of EF-Tu membrane association in Lactobacillus johnsonii and Listeria monocytogenes, where it mediated binding to mucin (19) and fibrinogen (50), respectively. Recently, it has been reported that in Pseudomonas aeruginosa EF-Tu binds factor H and plasminogen (34). Thus, EF-Tu joins the group of enzymes, including enolase (4), glyceraldehyde-3-phosphate dehydrogenase (1, 41), and pyruvate dehydrogenase E1β subunit (17) that exhibit unexpected biological functions, in addition to their well-defined enzymatic activities.
In this study we identified specific regions of EF-Tu that are surface localized and mediate binding to Fn, further reinforcing the intricate and dynamic interplay between cytosolic enzymes and their moonlighting activities.
MATERIALS AND METHODS
Bacterial strains, plasmids, and DNA manipulations.
E. coli INVαF′ [F′endA1 rec1 hsdR17 supE44 gyrA96 lacZM15 (lacZYAargF)] (Invitrogen, Carlsbad, CA) and E. coli BL21(DE3) [F′ompT hsdSB (rB− mB−) gal dcm (DE3)] (Stratagene, La Jolla, CA) were grown in Luria-Bertani broth and used to clone, express, and purify M. pneumoniae EF-Tu full-length (FL) and its truncated forms. The following vectors and bacterial cells were used: pCR2.1 (TA cloning vector; Invitrogen) and E. coli INVαF′ for gene manipulations and pET-19b (N-terminal His10 tag; expression vector; Novagen/EMD Biosciences Inc., San Diego, CA) and E. coli BL21(DE3) for protein expression. An M. pneumoniae low-passage clinical isolate designated S1 was used for isolation of genomic DNA (16) in competitive enzyme-linked immunosorbent assays (ELISAs) and antibody blocking assays.
Mycoplasma culture conditions.
M. pneumoniae cells were grown to mid-log phase in SP-4 medium at 37°C for 72 h in 150-cm2 tissue culture flasks as earlier reported (17). Mycoplasmas were harvested by washing surface-adherent cells three times with phosphate-buffered saline (PBS) (150 mM NaCl, 10 mM sodium phosphate, pH 7.4) before cells were scraped and pelleted at 12,500 × g for 15 min at 4°C. For radiolabeling, M. pneumoniae cell pellets were resuspended in 1/10 of their original volume in Dulbecco's modified Eagle's medium without cysteine or methionine and supplemented with 10% fetal bovine serum. [35S]methionine (1 mCi; specific activity of 43.5 TBq/mmol; PerkinElmer Inc., Waltham, MA), was added, and mycoplasmas were incubated at 37°C for 4 h on a rocker, pelleted, and washed four times with PBS before use.
Cloning, expression, and purification of EF-Tu truncations.
M. pneumoniae EF-Tu FL and individual truncations (EN, EC, ED2, ED3, ED4, ED3.1, ED3.2, and ED4.1) (Table 1) were amplified, and the resultant DNA fragments were cloned into PCR2.1 vector (Invitrogen) and transformed into E. coli INVαF′ cells. PCR2.1 plasmids with inserts were digested with NdeI and BamHI, and excised fragments were cloned into pET-19b vector and transformed into E. coli INVαF′. pET-19b plasmids with inserts were isolated using Qiagen mini-prep kits according to the manufacturer's protocol, screened for individual inserts by PCR amplification using specific primers (Table 1), and sequenced. One positive clone representing each truncation was transformed into E. coli BL21(DE3) for expression studies. E. coli cells were grown to an optical density at 600 nm of 0.6 at 37°C in Luria-Bertani medium containing 100 μg/ml ampicillin (Sigma, St. Louis, MO). Induction of all recombinant proteins was accomplished with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma) for 3 h at 37°C with aeration at 220 rpm. Large-scale purification of recombinant proteins was performed under native conditions using columns packed with Ni-nitrilotriacetic acid Superflow (Qiagen, Valencia, CA), except for ED3.1, ED3.2, and ED4.1, which were purified under denatured conditions using 8 M urea. All proteins were desalted in 50 mM Tris-Cl (pH 7.4) and 5% glycerol, and protein concentrations were estimated by the bicinchoninic acid method (Pierce, Rockford, IL). Each purified protein was separated on Nu-PAGE (where PAGE is polyacrylamide gel electrophoresis) 4 to 12% gradient gels (Invitrogen) and visualized using Coomassie blue. Immunoblotting of parallel gels transferred onto membranes was performed with polyclonal anti-EF-Tu antibodies and anti-His monoclonal antibody (MAb) (Clontech/BD Biosciences, San Jose, CA). Purified recombinant proteins were also tested with anti-His MAb and anti-EF-Tu antiserum using an ELISA format (described below) except that increasing concentrations of recombinant proteins, instead of Fn, were coated onto wells. All recombinant proteins were reactive with anti-His MAb in a concentration-dependent manner.
TABLE 1.
Construction of M. pneumoniae EF-Tu and truncations
| Protein | PCR primer sequencea
|
Nucleotide positionb | No. of amino acids | Amino acid residue nos. | |
|---|---|---|---|---|---|
| Forward primer (5′→3′) | Reverse primer (5′→3′) | ||||
| MPN EF-Tu FL | GAGACGTAATTCAAACATATGGCAAGAGAG | GGCTTTCCTTGAGGATCCTAACAGAGTCAA | −18 to 1185 | 394 | 1-394 |
| EN | GAGACGTAATTCAAACATATGGCAAGAGAG | CGGCCAGGATCCGTCATGGTGTCTTAGATTGCC | −18 to 645 | 215 | 1-215 |
| EC | GCTAAGATCCATGATCATATGAATGCAGTTG | GTGCCTGGCTTTCCTTGAGGATCCTAACAGAGT | 577-1185 | 202 | 192-394 |
| ED1 | GAGACGTAATTCAAACATATGGCAAGAGAG | CCATGCGTGGATCCCCCACTTAGCGGGCCAA | −18 to 372 | 124 | 1-124 |
| ED2 | GACTACATTAAGCATATGATTACTGGTGCTGC | CGGCCAGGATCCGTCATGGTGTCTTAGATTGCC | 276-645 | 122 | 91-215 |
| ED3 | GCTAAGATCCATGATCATATGAATGCAGTTG | CCGGTGTGAGGATCCCCTTCTTACTTCTTTAAAGC | 577-942 | 122 | 192-314 |
| ED4 | GAACGTGGTCATATGTTAGCTAAACCAGG | GTGCCTGGCTTTCCTTGAGGATCCTAACAGAGT | 879-1185 | 102 | 292-394 |
| ED3.1 | GCTAAGATCCATGATCATATGAATGCAGTTG | GGATCCTTAGCTTTACACTTGACCACGTTCCACTTC | 577-876 | 100 | 192-292 |
| ED3.2 | CTTCTTGTTGCATATGGAAGACACCATGACG | GGATCCTTAGCTTTACACTTGACCACGTTCCACTTC | 646-876 | 77 | 215-292 |
| ED4.1 | GCTTTACATATGGAAGAAGGTGGTCGTCAC | GTGCCTGGCTTTCCTTGAGGATCCTAACAGAGT | 943-1185 | 80 | 314-394 |
Introduced BamHI and NdeI sites underlined, and introduced stop codons are in bold.
−, upstream position of nucleotide from the adenine nucleotide of the starting ATG codon.
Identification of EF-Tu truncations that mediate Fn binding.
Interactions between EF-Tu and Fn were measured by ELISA. In brief, individual wells of 96-well plates (Reacti-Bind amine-binding maleic anhydride-activated plates; Pierce) were coated overnight at 4°C with 100 μl of 100 ng/well Fn in PBS (pH 7.4). Wells were blocked with 200 μl of 0.1% bovine serum albumin (BSA; Sigma) in PBS for 2 h at room temperature (RT). Then, 100 μl of increasing concentrations (25 nM, 50 nM, 75 nM, and 100 nM in 0.1% BSA-PBS) of recombinant His-tagged EF-Tu FL and its truncations was individually added in separate wells and incubated for 2 h at room temperature. Wells were washed three times with PBS and further incubated with 100 μl of a 1:2,000 dilution of anti-His MAb in 1% BSA-PBS for 2 h. Wells were again washed three times with PBS and incubated with 100 μl of a 1:2,000 dilution of goat anti-mouse immunoglobulin G (IgG) alkaline phosphatase (AP) antibody (Invitrogen) in 1% BSA-PBS. Wells coated with 0.1% BSA alone served as negative controls. All assays were performed in triplicate and developed using p-nitrophenyl phosphate substrate (Sigma). Plates were read by an ELISA reader (Dynatech Laboratories, Chantilly, VA) at 405 nm.
Generation of mouse polyclonal antibodies.
Six-week-old BALB/c mice (n = 5 for each construct) were prebled and screened by immunoblotting for lack of reactivity against recombinant proteins EF-Tu FL, EN, ED3, and ED4. Then, individual mice were immunized intraperitoneally with recombinant proteins plus complete Freund's adjuvant and boosted as described earlier (29). Animals were screened after each boost by immunoblotting using recombinant proteins and whole M. pneumoniae cell lysates. All mice exhibited strong immunoreactivity to recombinant EF-Tu FL, EF-Tu in whole-cell mycoplasma lysates, and each specific recombinant fragment used for immunization.
Antibody blocking assay.
[35S]methionine-labeled viable mycoplasmas were treated with prebleed serum or anti-EN, anti-ED3, or anti-ED4 antiserum at 1:1,000 dilution for 1 h at RT before incubation with immobilized Fn (100 μl of 1 μg of Fn in PBS overnight) on ELISA plates for 1 h at 37°C. Individual wells were extensively rinsed with PBS, and radioactive counts were determined. Values obtained using M. pneumoniae incubated with individual prebled sera represented background and were subtracted from test values.
Competitive ELISA.
Purified recombinant ED3 and ED4 were used to compete with biosynthetically [35S]methionine-labeled M. pneumoniae cells for binding to Fn coated onto microtiter plates as already described. ED3 and ED4 at 75, 375, and 750 nM final concentrations were added separately to individual Fn-coated wells and incubated for 2 h at RT. Then, radiolabeled mycoplasmas were added and incubated for 1 h at 37°C. After extensive washing, M. pneumoniae binding to Fn was assessed by determining radioactive counts per well.
Phase partitioning of M. pneumoniae components using Triton X-114.
M. pneumoniae cells at the mid-log phase of growth were separated into Triton X-114 insoluble and aqueous fractions by slight modification of the method of Bordier (8). Washed M. pneumoniae cells were suspended in 1 ml of prewashed, condensed 1% (wt/vol) Triton X-114 in PBS, incubated on ice for 2 h, and centrifuged at 13,000 × g for 30 min at 4°C. The supernatant was incubated at 37°C for 5 min for rapid condensation of Triton X-114 and centrifuged at 13,000 × g for 10 min at RT to generate an aqueous upper phase and lower Triton phase; the latter was freed of the aqueous phase by extraction two more times. The final aqueous and Triton phases were ethanol precipitated, pellets were suspended in 1× PBS (pH 7.4), and soluble and insoluble protein fractions were quantified by the bicinchoninic acid method. Equal amounts of each fraction were loaded onto Nu-PAGE 4 to 12% gradient gels, and electrophoresis and protein transfers were performed prior to immunoblotting with mouse anti-EF-Tu (1:3,000) or rabbit anti-EF-G (1:3,000) antiserum in 3% milk in Tris-buffered saline ([TBS] Blotto) for 1 h at RT. After extensive washes with 1× TBS-0.05% Tween-20, membranes were incubated with goat anti-mouse IgG AP or goat anti-rabbit AP (1:2,000) in 3% Blotto for 1 h at RT. After multiple washes with 1× TBS-0.05% Tween-20, membranes were developed with nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolylphosphate (Roche Diagnostics, Indianapolis, IN).
Mycoplasma membrane purification.
Pelleted SP-4 grown, mid-log-phase M. pneumoniae cells were subjected to membrane isolation by osmotic lysis (17, 45). Individual sucrose gradient-purified mycoplasma membrane preparations were treated with 0.1 M Na2CO3 (pH 11.5) for 1 h at 37°C (2) or suspended in 2 mM Tris (pH 7.4) with 1 M NaCl and 3 M KCl (49) and incubated on ice for 1 h. Polar reagents such as alkali or high-salt concentrations dissociate peripherally associated proteins from the membrane. Treated samples were centrifuged at 13,000 × g for 30 min. Supernatants containing peripheral proteins and pellets containing integral membrane proteins were loaded onto Nu-PAGE 4 to 12% gels and transferred onto nitrocellulose membranes for immunoblotting with rabbit anti-EF-Tu antiserum (1:2,000) and goat anti-rabbit AP (1:2,000).
Immunogold electron microscopy.
Immunogold labeling of M. pneumoniae cells was performed as reported previously (17). After mycoplasma cells were washed with 100 mM Tris-HCl buffer (pH 7.5) and incubated with buffer A (100 mM Tris-HCl [pH 7.5] containing 1% BSA and supplemented with 1% heat-inactivated goat serum), intact viable M. pneumoniae cells were incubated with anti-EN, anti-ED3, or anti-ED4 mouse antiserum diluted 1:1,000 in buffer A for 120 min at 37°C. Then, mycoplasma cells were washed with buffer A, followed by incubation for 60 min at RT with goat anti-mouse IgG-gold (10-nm particles) complex diluted 1:20 in buffer B (PBS, pH 7.4, with 1% BSA). Sequential washes with buffer B, PBS, and deionized water were performed, and mycoplasmas were mounted on Formvar-coated nickel grids and fixed with 1% glutaraldehyde-4% formaldehyde for 20 min at RT. Individual grids were examined with a Philips 208S transmission electron microscope at 80 kV accelerating voltage after staining with 7% uranyl acetate followed by Reynolds lead citrate.
Interaction of EF-Tu FL, ED3, and ED4 with different Fn domains.
Equal amounts (3 μg) of commercially available Fn peptides representing a 30-kDa fibrin-heparin-binding domain (Sigma), 45-kDa gelatin-binding domain (Sigma), 40-kDa heparin-binding domain (Chemicon, Temecula, CA), and 120-kDa cell-binding domain (Chemicon) and of BSA as a control were electrophoresed in individual lanes of 4 to 12% Nu-PAGE gels and transferred onto nitrocellulose membranes. Individual nitrocellulose membranes were then incubated with 10 μg/ml recombinant EF-Tu FL, ED3, or ED4 proteins for 2 h at RT and immunoblotted using mouse anti-His MAb (1:5,000) followed by goat anti-mouse AP (1:2,000). The intensity of each band was analyzed using Kodak document imaging software (version 3.5.5B), and interactions were classified based on the mean intensity values as follows: 0 to 30, no binding; 31 to 60, weak; 61 to 90, intermediate; and >91, strong.
RESULTS
Identification of EF-Tu regions that interact with Fn.
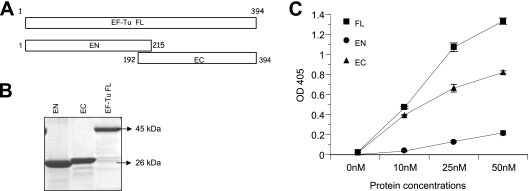
In an attempt to identify the regions of EF-Tu that interact with Fn, we generated two recombinant proteins, designated EF-Tu N-terminal (EN, amino acids 1 to 215) and C-terminal (EC, amino acids 192 to 394) regions, with an overlapping region of 24 amino acids (Fig. 1A). While the purified recombinant EN protein matched its predicted theoretical molecular mass of 26 kDa, recombinant EC ran higher than its predicted molecular mass of 24 kDa (Fig. 1B).
FIG. 1.
Identification of Fn-binding region of M. pneumoniae EF-Tu. (A) Schematic representation of EF-Tu amino and carboxyl termini. EF-Tu FL (amino acids 1 to 394) was divided into two overlapping amino-terminal (EN, residues 1 to 215) and carboxyl-terminal (EC, residues 192 to 394) constructs. (B) Purification of EF-Tu FL, EN, and EC proteins. Regions encoding EF-Tu FL, EN, and EC were cloned, expressed, and purified as described in Materials and Methods, separated on 4 to 12% Nu-PAGE gels, and stained with Coomassie blue. (C) Binding of recombinant EF-Tu FL, EN, and EC to immobilized Fn. Microtiter plates were coated with 0.1 μg of human Fn. Increasing concentrations of recombinant proteins were incubated in individual wells for 2 h at RT. Bound proteins were detected with mouse anti-His MAb and goat anti-mouse AP-conjugated antibodies, followed by color development with p-nitrophenyl phosphate substrate. Results are expressed as means ± standard deviations. Each sample point is based upon triplicate values. OD 405, optical density at 405 nm.
To determine which EF-Tu regions bound Fn, we added increasing concentrations (nM) of recombinant His-tagged EF-Tu FL, EN, and EC to immobilized Fn and monitored binding. While EN demonstrated markedly reduced binding to Fn, EC bound in a dose-dependent manner, like EF-Tu FL (Fig. 1C).
Two distinct regions of the carboxyl terminus bind to Fn.
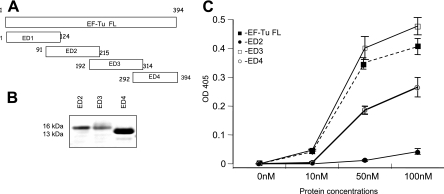
To further define the Fn-binding region of EC, we constructed two overlapping truncations, ED3 and ED4. We also attempted to construct two overlapping amino-terminal truncations of EN (ED1 and ED2) to further reinforce their limited role in Fn binding. Except for ED1, all constructs were expressed and purified under native conditions (Fig. 2A). Binding assays with ED2, ED3, ED4, and FL revealed that within the carboxyl terminus, ED3 exhibited higher binding for Fn than ED4 and EF-Tu FL (Fig. 2B). As expected, like EN, ED2 had minimal binding to Fn.
FIG. 2.
Characterization of minimal Fn-binding regions of M. pneumoniae EF-Tu. (A) Schematic representation of deletion constructs of amino and carboxyl regions of EF-Tu. EN was further divided into ED1 (residues 1 to 124) and ED2 (residues 91 to 215) with 23 overlapping amino acids while EC was divided into ED3 (residues 192 to 314) and ED4 (residues 292 to 394) with 22 overlapping amino acids. (B) Purification of ED2, ED3, and ED4. All recombinant proteins were purified as described in Materials and Methods, separated by electrophoresis with 4 to 12% Nu-PAGE gels, and stained with Coomassie blue. ED1 could not be expressed. (C) Dose-dependent binding of recombinant EF-Tu-related proteins to immobilized Fn. ELISAs were performed with recombinant ED2, ED3, and ED4 as described in the legend of Fig. 1C. OD 405, optical density at 405 nm.
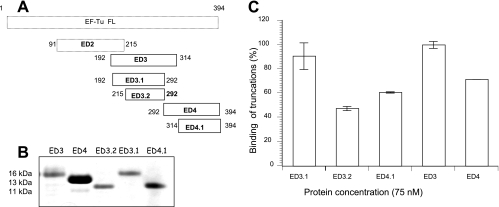
Since ED3 and ED4 possessed Fn-binding properties, we set out to determine the minimal EC-related Fn-interacting region(s) by generating additional truncations. ED3.1 was constructed by deleting the overlapping 22 amino acids shared by ED3 and ED4 (Fig. 3A). ED3.2 overlapped with ED3.1 but was missing 23 amino acids that overlapped with ED2. ED4.1 was missing the overlapping amino acids shared by ED3 and ED4 (Fig. 3A). Recombinant ED3 and ED3.1 (Fig. 3B) demonstrated the highest Fn-binding activity (Fig. 3C), and ED4.1 marginally differed from ED4 (Fig. 3C). Interestingly, ED3.2 showed a marked decrease in its Fn-binding ability, especially compared to ED3 and ED3.1.
FIG. 3.
Construction and Fn binding of overlapping carboxyl regions of M. pneumoniae EF-Tu. (A) Schematic representation of deletion constructs ED3.1, ED3.2, and ED4.1. ED3.1 and ED3.2 were constructed by deleting the overlapping regions of ED3 and ED4. ED3.2 additionally had a deletion of the overlapping region of ED3 and ED2. ED4.1 was constructed by deleting the ED3-overlapping region. (B) Purification of ED3.1, ED3.2, and ED4.1. ED3.1, ED3.2, and ED4.1 were purified under denatured conditions and then refolded in 50 mM Tris buffer. Purified proteins were separated by electrophoresis with 4 to 12% Nu-PAGE gels and stained with Coomassie blue. (C) Interaction of ED3.1, ED3.2, and ED4.1 with Fn. ELISAs were performed using 75 nM concentrations of recombinant proteins in triplicate as described in Materials and Methods. Values were determined, and binding percentages were calculated using ED3 binding values at 75 nM as 100%. Results are expressed as means ± standard deviations. Each sample point is based upon triplicate values.
EF-Tu FL, ED3, and ED4 interact with multiple Fn regions.
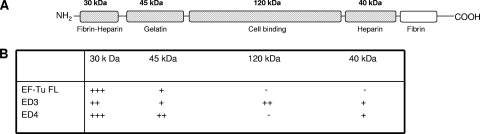
Ligand blotting was performed to determine the domains of Fn (Fig. 4A) that bound EF-Tu FL, ED3, and ED4. While EF-Tu FL interacted with the N-terminal 30-kDa fibrin-heparin and 45-kDa gelatin-binding domains, ED4 also interacted with the N-terminal 30- and 45-kDa domains plus the C-terminal 40-kDa heparin-binding domain. ED3 interacted with the N-terminal 30-kDa and 45-kDa and C-terminal 40-kDa domains and uniquely bound to the 120-kDa cell-binding domain (Fig. 4B). Thus, ED3 and ED4 possessed both common and distinct Fn-binding regions.
FIG. 4.
Interaction of M. pneumoniae EF-Tu FL and its truncations with different Fn domains. (A) Schematic representation of different domains of Fn. The figure was adapted from Virkola et al. (53). Fn domains used for the ligand blot analyses are indicated in cross-hatched boxes. (B) Interaction of EF-Tu FL, ED3, and ED4 with domains of Fn. Equal amounts of available Fn fragments were separated individually on 4 to 12% Nu-PAGE gels and transferred onto nitrocellulose membranes, which were incubated with 10 μg/ml recombinant EF-Tu proteins in 3% Blotto in 1× TBS for 2 h at RT (see Materials and Methods). Membranes were probed with mouse anti-His MAb (1:5,000) and subsequently with goat anti-mouse AP (1:2,000) reagent. Based on the mean intensities of His tag antibody recognition of recombinant EF-Tu proteins, interactions were classified as demonstrating weak (+), intermediate (++), strong (+++) or no (−) binding.
M. pneumoniae EF-Tu subpopulation is associated with the Triton X-114-enriched and membrane fractions.
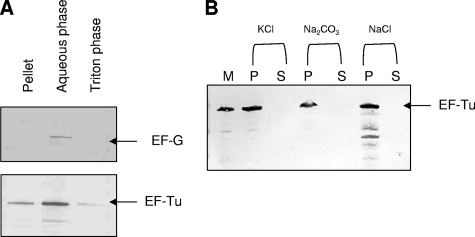
Earlier, we reported the surface accessibility of M. pneumoniae EF-Tu by whole-cell radioimmunoprecipitation, membrane fractionation, trypsin sensitivity, antibody-blocking assays, and immunogold labeling (17). Here, we further characterized the mem-brane association of M. pneumoniae EF-Tu by subjecting intact mycoplasmas to Triton X-114 phase partitioning. Although most of the EF-Tu partitioned to the aqueous cytosolic fraction, we detected a subpopulation of EF-Tu in Triton X-114 soluble and insoluble fractions (Fig. 5A). Antibody reactive against EF-G (a gift from R. Herrmann), a protein limited to the cytoplasm, was used as control for cytosolic contamination of Triton-soluble fractions.
FIG. 5.
M. pneumoniae EF-Tu association with mycoplasma membrane. (A) Triton X-114 phase partitioning of intact M. pneumoniae cells. Equal amounts of sample were separated on Nu-PAGE gels under reducing conditions and transferred onto nitrocellulose membranes. Immunoblotting was performed with mouse anti-EF-Tu (1:3,000) or rabbit anti-EF-G (1:3,000) antiserum in 3% Blotto for 1 h at RT. (B) Alkali and high-salt treatment of M. pneumoniae membranes. M. pneumoniae S1 membranes (M) were treated with 3 M KCl, 0.1 M Na2CO3, and 1 M NaCl. Supernatant (S) and pellet (P) fractions were subjected to Nu-PAGE under reducing conditions, transferred onto nitrocellulose membranes, and immunoblotted with anti-EF-Tu antiserum as described in Materials and Methods.
The primary structure of EF-Tu indicates two putative transmembrane domains (amino acids 28 to 35 and 101 to 108) by the dense alignment surface method (14), and therefore EF-Tu could be associated with the mycoplasma membrane. Treatment of mycoplasma membrane preparations with alkali (0.1 M Na2CO3 [pH 11.5]) and high salts (3 M KCl and 1 M NaCl), which aids the dissociation of peripherally associated proteins, did not completely release membrane-associated EF-Tu (Fig. 5B).
Surface accessibility of the M. pneumoniae EF-Tu carboxyl regions and its interaction with Fn.
Since the carboxyl terminus of M. pneumoniae EF-Tu binds Fn, it was likely that this region was surface exposed in intact mycoplasmas. Therefore, soluble recombinant ED3 and ED4 fragments were used in competitive Fn-binding ELISAs with viable M. pneumoniae cells. Both fragments competed effectively as recombinant ED3 blocked M. pneumoniae cell binding at 75, 375, and 750 nM by 80, 85, and 90%, respectively, and ED4 at similar concentrations blocked binding by 60, 75, and 80%, respectively.
Preincubation of viable M. pneumoniae cells with antibodies raised against purified recombinant fragments ED3 and ED4 reduced mycoplasma binding by 80 and 75%, respectively. In contrast, antiserum against EN demonstrated 20% blocking. Preimmune sera possessed no blocking capabilities.
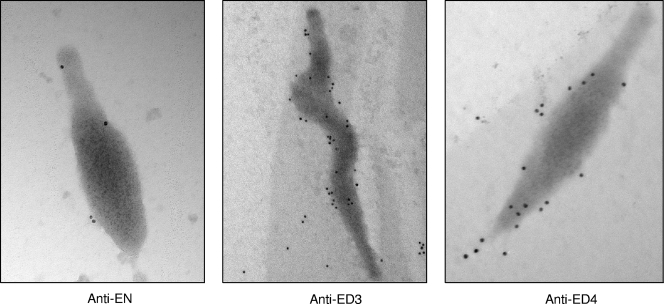
The surface accessibility of the Fn-interacting carboxyl region of EF-Tu was further confirmed by immunoelectron microscopy. Surface immunogold labeling of M. pneumoniae cells occurred consistently with antisera generated against ED3 and ED4, while antiserum to EN revealed limited labeling (Fig. 6). Exposure of mycoplasmas to preimmune sera resulted in no labeling (data not shown).
FIG. 6.
Immunogold labeling of EF-Tu on intact M. pneumoniae cells. Mycoplasmas were incubated with antisera (1:1,000) generated against EN, ED3, or ED4 truncations followed by anti-mouse IgG gold complex (10 nm). Mycoplasma membrane-associated gold labeling of EF-Tu is readily observed with ED3 and ED4 antisera and much less so with EN antiserum. EN magnification, ×71,000; ED3 magnification, ×44,000; and ED4 magnification, ×71,000.
DISCUSSION
In this study we characterized the regions of M. pneumoniae EF-Tu that interact with Fn. We determined that Fn binding by EF-Tu is primarily mediated by the carboxyl terminus (amino acids 192 to 394; recombinant peptides ED3 and ED4) (Fig. 2), which includes the well-described EF-Tu domains 2 and 3 (amino acids 209 to 394) involved in interactions with macromolecular ligands during the translation process (31). In contrast, EN (amino acids 1 to 215) and its truncation ED2 (Fig. 2), which correspond to domain 1 (amino acids 1 to 200) and are involved in binding GTP (31), exhibited minimal Fn-binding properties. Based on our experimental data using deletion truncations, EF-Tu possessed two Fn-interacting regions in the carboxyl terminus with different binding properties. One Fn-binding region included amino acids 192 to 292 (Fig. 3, ED3.1), and the other Fn-binding region included amino acids 314 to 394 (Fig. 3, ED4.1). Amino acids 192 to 214 contributed to maximum Fn binding, as observed by the deletion of these 23 amino acids from ED3.1 (i.e., truncation ED3.2) (Fig. 3A) which decreased binding to Fn by ∼50% (Fig. 3C). However, these 23 amino acids by themselves do not appear to directly mediate Fn interactions, since ED2 alone (amino acids 91 to 215) (Fig. 3A) exhibited minimal Fn binding (Fig. 2). Of interest are the amino acids EDT (216 to 218) that follow the 23 amino acids in the carboxyl terminus. Amino acids EDT belong to a larger motif that mediates Fn binding in several MSCRAMM FnBPs (13, 51). Interestingly, ED3, ED3.1, and ED3.2 possess the EDT motif, although ED3.2 exhibits reduced Fn binding. In ED3 and ED3.1, EDT is preceded by 192 to 215 amino acids, and Fn binding is enhanced. In contrast, ED2, regardless of possessing these 23 amino acids, lacks EDT and exhibits negligible Fn-binding ability. Hence, the 23 amino acids plus the EDT motif appear essential for efficient EF-Tu-Fn interactions.
The second carboxyl-terminal Fn-interacting region of EF-Tu is located within ED4 (amino acids 292 to 394) and ED4.1 (amino acids 314 to 394) (Fig. 3). This region revealed no significant homologies to any conserved motifs identified in other FnBPs. Several sequence and organizational variations have been reported as motifs in Fn-binding bacterial proteins. For example, in staphylococci and streptococci, the primary Fn-binding site is localized to unstructured repeats of 35 to 40 residues, and a secondary Fn-binding site lies outside of the carboxyl-terminal repeats (51). Further, FnBPs like the Fn attachment protein family in mycobacteria consist of two noncontiguous regions of four amino acids (58), and Campylobacter jejuni CadF consists of a single Fn-binding domain of four amino acids (30). Overall, M. pneumoniae EF-Tu lacks the recognizable motifs and repeats associated with other FnBPs, indicating the existence of alternative sequence-independent and conformational properties that mediate Fn interactions.
The existence of two Fn-binding regions within the carboxyl terminus of M. pneumoniae EF-Tu was unexpected and led us to predict that more than one recognition site on Fn served as a target for EF-Tu. Like a majority of FnBPs (26, 42), EF-Tu FL and its carboxyl truncations exhibited significant interaction with the N-terminal 30-kDa type I fibrin-heparin-binding region (Fig. 4A and B). Also, the carboxyl region of EF-Tu interacted with the C-terminal 40-kDa type II heparin-binding domain. Similar interactions have been observed with some FnBPs (9, 36). ED3 interacted specifically with the 120-kDa cell-binding domain. Thus, M. pneumoniae EF-Tu has at least two Fn-interacting regions, and these two regions recognize common as well as specific domains on Fn. Interestingly, EF-Tu FL did not recognize the 40- or 120-kDa Fn domains (Fig. 4B), suggesting that in recombinant EF-Tu FL, the ED3 and ED4 sites are unavailable for Fn binding. Clearly, the contribution of EF-Tu and its potential impact on mycoplasma airway colonization and internalization via Fn-binding dynamics need to be further investigated.
Using Triton X-114, we demonstrated the presence of EF-Tu in all mycoplasma fractions (Fig. 5A). Similar studies with Triton X-100 identified M. pneumoniae EF-Tu as a major component of the insoluble cytoskeleton fraction (46). The inability to remove membrane-associated EF-Tu with alkali or high-salt treatments (Fig. 5B) lends credence to its strong membrane association. While M. pneumoniae membrane-localized EF-Tu was reported to be trypsin sensitive (17), competitive ELISAs using recombinant ED3 and ED4 and antibody blocking assays with antisera generated against ED3 and ED4 proved conclusively that the Fn-interacting carboxyl terminus is surface exposed. Antiserum generated against EF-Tu FL exhibited blocking consistent with our previous report (17). Immunoelectron microscopy results further supported the surface accessibility of the EF-Tu carboxyl terminus (Fig. 6).
Despite the classification of EF-Tu as a cytosolic protein, we have shown that EF-Tu can relocate to the mycoplasma membrane surface with an exposed carboxyl terminus to facilitate Fn binding. While there are other reports that document the presence of cytosolic proteins, including EF-Tu, on surfaces of different bacteria, important questions regarding how these proteins translocate to the surface remain unanswered, especially since conventional secretion or anchoring signals are absent. Physiological factors, including osmotic stress, have been implicated in E. coli EF-Tu association with the periplasm, and other possible mechanisms for export of cytosolic proteins have been described (50), including the impact of posttranslational modifications (7).
Attachment and colonization of target tissues by M. pneumoniae and other microbial pathogens are prerequisites for successful infection and disease progression. While adherence of virulent M. pneumoniae is primarily mediated by adhesins, like P1 (24), the absence of this protein (21, 32) still permits weak adherence. Furthermore, monospecific antibodies against P1 inhibit attachment of virulent M. pneumoniae to the respiratory epithelium by a maximum of 75% (3), further suggesting the involvement of other adhesin-like proteins, as well as alternate mechanisms of mycoplasma binding to host cells. Fn is an abundant and available pathogen target, as earlier noted; it exists in soluble form in blood fluids and plasma and in fibrillar form in ECM. As ECM underlies the epithelial and endothelial cells and surrounds connective tissue, mycoplasmas could readily access ECM following epithelial damage. Events following M. pneumoniae cytadherence have been studied in infected hamster tracheal organ cultures and include inhibition of respiratory cell macromolecular synthesis, followed by characteristic ciliostasis, cytoplasmic and nuclear vacuolization, and extensive epithelial cell fragmentation and sloughing (11, 12). Recently, we described the role of a unique ADP-ribosylating and vacuolating M. pneumoniae toxin (annotated MPN372) in eliciting the characteristic cytopathology associated with mycoplasma infection (28). Interestingly, this toxin is surface associated and binds selectively to human lung surfactant protein A (29). Thus, in association with toxin-mediated epithelial cell damage, M. pneumoniae could also directly access subepithelial tissue targets and ECM by multiple pathways, including EF-Tu interactions with Fn. Further, these distinct pathogenic pathways may also contribute to the ability of M. pneumoniae to invade and establish intracellular and perinuclear residence (15, 56). Taken together, these findings indicate that M. pneumoniae is a sophisticated and versatile bacterial pathogen with wide-ranging colonization strategies to overcome host defenses and trigger acute and chronic airway and extrapulmonary diseases.
Acknowledgments
This work was supported by Public Health Service grants AI070412 and AI045737 from the National Institutes of Health.
We thank Marianna Cagle for generating mouse polyclonal antibodies. We thank Rose Garza for her assistance with finalizing the manuscript.
Editor: A. Camilli
Footnotes
Published ahead of print on 14 April 2008.
REFERENCES
- 1.Alvarez, R. A., M. W. Blaylock, and J. B. Baseman. 2003. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol. Microbiol. 481417-1425. [DOI] [PubMed] [Google Scholar]
- 2.Balish, M. F., T. W. Hahn, P. L. Popham, and D. C. Krause. 2001. Stability of Mycoplasma pneumoniae cytadherence-accessory protein HMW1 correlates with its association with the triton shell. J. Bacteriol. 1833680-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baseman, J. B., R. M. Cole, D. C. Krause, and D. K. Leith. 1982. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J. Bacteriol. 1511514-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. Alpha-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 401273-1287. [DOI] [PubMed] [Google Scholar]
- 5.Berrier, C., A. Garrigues, G. Richarme, and A. Ghazi. 2000. Elongation factor Tu and DnaK are transferred from the cytoplasm to the periplasm of Escherichia coli during osmotic downshock presumably via the mechanosensitive channel MscL. J. Bacteriol. 182248-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumenthal, T. 1980. Interaction of host-coded and virus-coded polypeptides in RNA phage replication. Proc. R. Soc. Lond. B 210321-335. [DOI] [PubMed] [Google Scholar]
- 7.Boel, G., V. Pichereau, I. Mijakovic, A. Maze, S. Poncet, S. Gillet, J. C. Giard, A. Hartke, Y. Auffray, and J. Deutscher. 2004. Is 2-phosphoglycerate-dependent automodification of bacterial enolases implicated in their export? J. Mol. Biol. 337485-496. [DOI] [PubMed] [Google Scholar]
- 8.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 2561604-1607. [PubMed] [Google Scholar]
- 9.Bozzini, S., L. Visai, P. Pignatti, T. E. Petersen, and P. Speziale. 1992. Multiple binding sites in fibronectin and the staphylococcal fibronectin receptor. Eur. J. Biochem. 207327-333. [DOI] [PubMed] [Google Scholar]
- 10.Caldas, T. D., A. El Yaagoubi, and G. Richarme. 1998. Chaperone properties of bacterial elongation factor EF-Tu. J. Biol. Chem. 27311478-11482. [DOI] [PubMed] [Google Scholar]
- 11.Collier, A. M., and W. A. Clyde. 1971. Relationships between Mycoplasma pneumoniae and human respiratory epithelium. Infect. Immun. 3694-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collier, A. M., W. A. Clyde, Jr., and F. W. Denny. 1969. Biologic effects of Mycoplasma pneumoniae and other mycoplasmas from man on hamster tracheal organ culture. Proc. Soc. Exp. Biol. Med. 1321153-1158. [DOI] [PubMed] [Google Scholar]
- 13.Courtney, H. S., D. L. Hasty, and J. B. Dale. 2002. Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann. Med. 3477-87. [DOI] [PubMed] [Google Scholar]
- 14.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10673-676. [DOI] [PubMed] [Google Scholar]
- 15.Dallo, S. F., and J. B. Baseman. 2000. Intracellular DNA replication and long-term survival of pathogenic mycoplasmas. Microb. Pathog. 29301-309. [DOI] [PubMed] [Google Scholar]
- 16.Dallo, S. F., A. Chavoya, and J. B. Baseman. 1990. Characterization of the gene for a 30-kilodalton adhesion-related protein of Mycoplasma pneumoniae. Infect. Immun. 584163-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dallo, S. F., T. R. Kannan, M. W. Blaylock, and J. B. Baseman. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 461041-1051. [DOI] [PubMed] [Google Scholar]
- 18.Giron, J. A., M. Lange, and J. B. Baseman. 1996. Adherence, fibronectin binding, and induction of cytoskeleton reorganization in cultured human cells by Mycoplasma penetrans. Infect. Immun. 64197-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granato, D., G. E. Bergonzelli, R. D. Pridmore, L. Marvin, M. Rouvet, and I. E. Corthesy-Theulaz. 2004. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 722160-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross, S. R., and T. G. Kinzy. 2005. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 12772-778. [DOI] [PubMed] [Google Scholar]
- 21.Hansen, E. J., R. M. Wilson, and J. B. Baseman. 1979. Isolation of mutants of Mycoplasma pneumoniae defective in hemadsorption. Infect. Immun. 23903-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen, E. J., R. M. Wilson, W. A. Clyde, Jr., and J. B. Baseman. 1981. Characterization of hemadsorption-negative mutants of Mycoplasma pneumoniae. Infect. Immun. 32127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanski, E., J. Jaffe, and V. Ozeri. 1996. Proteins F1 and F2 of Streptococcus pyogenes. Properties of fibronectin binding. Adv. Exp. Med. Biol. 408141-150. [DOI] [PubMed] [Google Scholar]
- 24.Hu, P. C., A. M. Collier, and J. B. Baseman. 1977. Surface parasitism by Mycoplasma pneumoniae of respiratory epithelium. J. Exp. Med. 1451328-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson, G. R., and J. P. Rosenbusch. 1976. Abundance and membrane association of elongation factor Tu in E. coli. Nature 26123-26. [DOI] [PubMed] [Google Scholar]
- 26.Joh, D., E. R. Wann, B. Kreikemeyer, P. Speziale, and M. Hook. 1999. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18211-223. [DOI] [PubMed] [Google Scholar]
- 27.Jonsson, K., C. Signas, H. P. Muller, and M. Lindberg. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 2021041-1048. [DOI] [PubMed] [Google Scholar]
- 28.Kannan, T. R., and J. B. Baseman. 2006. ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. Proc. Natl. Acad. Sci. USA 1036724-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kannan, T. R., D. Provenzano, J. R. Wright, and J. B. Baseman. 2005. Identification and characterization of human surfactant protein A binding protein of Mycoplasma pneumoniae. Infect. Immun. 732828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konkel, M. E., J. E. Christensen, A. M. Keech, M. R. Monteville, J. D. Klena, and S. G. Garvis. 2005. Identification of a fibronectin-binding domain within the Campylobacter jejuni CadF protein. Mol. Microbiol. 571022-1035. [DOI] [PubMed] [Google Scholar]
- 31.Krab, I. M., and A. Parmeggiani. 1998. EF-Tu, a GTPase odyssey. Biochim. Biophys. Acta 14431-22. [DOI] [PubMed] [Google Scholar]
- 32.Krause, D. C., D. K. Leith, R. M. Wilson, and J. B. Baseman. 1982. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun. 35809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreikemeyer, B., D. R. Martin, and G. S. Chhatwal. 1999. SfbII protein, a fibronectin binding surface protein of group A streptococci, is a serum opacity factor with high serotype-specific apolipoproteinase activity. FEMS Microbiol. Lett. 178305-311. [DOI] [PubMed] [Google Scholar]
- 34.Kunert, A., J. Losse, C. Gruszin, M. Huhn, K. Kaendler, S. Mikkat, D. Volke, R. Hoffmann, T. S. Jokiranta, H. Seeberger, U. Moellmann, J. Hellwage, and P. F. Zipfel. 2007. Immune evasion of the human pathogen Pseudomonas aeruginosa: elongation factor Tuf is a factor H and plasminogen binding protein. J. Immunol. 1792979-2988. [DOI] [PubMed] [Google Scholar]
- 35.Kuusela, P., T. Vartio, M. Vuento, and E. B. Myhre. 1985. Attachment of staphylococci and streptococci on fibronectin, fibronectin fragments, and fibrinogen bound to a solid phase. Infect. Immun. 5077-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuusela, P., T. Vartio, M. Vuento, and E. B. Myhre. 1984. Binding sites for streptococci and staphylococci in fibronectin. Infect. Immun. 45433-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindgren, P. E., P. Speziale, M. McGavin, H. J. Monstein, M. Hook, L. Visai, T. Kostiainen, S. Bozzini, and M. Lindberg. 1992. Cloning and expression of two different genes from Streptococcus dysgalactiae encoding fibronectin receptors. J. Biol. Chem. 2671924-1931. [PubMed] [Google Scholar]
- 38.Marques, M. A., S. Chitale, P. J. Brennan, and M. C. Pessolani. 1998. Mapping and identification of the major cell wall-associated components of Mycobacterium leprae. Infect. Immun. 662625-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer, F. 2003. Cytoskeletons in prokaryotes. Cell Biol. Int. 27429-438. [DOI] [PubMed] [Google Scholar]
- 40.Moore, R. C., N. A. Durso, and R. J. Cyr. 1998. Elongation factor-1α stabilizes microtubules in a calcium/calmodulin-dependent manner. Cell Motil. Cytoskeleton 41168-180. [DOI] [PubMed] [Google Scholar]
- 41.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48585-617. [DOI] [PubMed] [Google Scholar]
- 43.Peterson, K. M., J. B. Baseman, and J. F. Alderete. 1983. Treponema pallidum receptor binding proteins interact with fibronectin. J. Exp. Med. 1571958-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porcella, S. F., R. J. Belland, and R. C. Judd. 1996. Identification of an EF-Tu protein that is periplasm-associated and processed in Neisseria gonorrhoeae. Microbiology 1422481-2489. [DOI] [PubMed] [Google Scholar]
- 45.Razin, S. 1983. Cell lysis and isolation of membranes, vol. 1. Academic Press, Inc., New York, NY.
- 46.Regula, J. T., G. Boguth, A. Gorg, J. Hegermann, F. Mayer, R. Frank, and R. Herrmann. 2001. Defining the mycoplasma “cytoskeleton”: the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology 1471045-1057. [DOI] [PubMed] [Google Scholar]
- 47.Regula, J. T., B. Ueberle, G. Boguth, A. Gorg, M. Schnolzer, R. Herrmann, and R. Frank. 2000. Towards a two-dimensional proteome map of Mycoplasma pneumoniae. Electrophoresis 213765-3780. [DOI] [PubMed] [Google Scholar]
- 48.Richarme, G. 1998. Protein-disulfide isomerase activity of elongation factor EF-Tu. Biochem. Biophys. Res. Commun. 252156-161. [DOI] [PubMed] [Google Scholar]
- 49.Riethman, H. C., M. J. Boyer, and K. S. Wise. 1987. Triton X-114 phase fractionation of an integral membrane surface protein mediating monoclonal antibody killing of Mycoplasma hyorhinis. Infect. Immun. 551094-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaumburg, J., O. Diekmann, P. Hagendorff, S. Bergmann, M. Rohde, S. Hammerschmidt, L. Jansch, J. Wehland, and U. Karst. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 42991-3006. [DOI] [PubMed] [Google Scholar]
- 51.Schwarz-Linek, U., M. Hook, and J. R. Potts. 2004. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52631-641. [DOI] [PubMed] [Google Scholar]
- 52.Signas, C., G. Raucci, K. Jonsson, P. E. Lindgren, G. M. Anantharamaiah, M. Hook, and M. Lindberg. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. USA 86699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virkola, R., M. Brummer, H. Rauvala, L. van Alphen, and T. K. Korhonen. 2000. Interaction of fimbriae of Haemophilus influenzae type B with heparin-binding extracellular matrix proteins. Infect. Immun. 685696-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong, D. K., B. Y. Lee, M. A. Horwitz, and B. W. Gibson. 1999. Identification of fur, aconitase, and other proteins expressed by Mycobacterium tuberculosis under conditions of low and high concentrations of iron by combined two-dimensional gel electrophoresis and mass spectrometry. Infect. Immun. 67327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamada, K. M., L. H. Hahn, and K. Olden. 1980. Structure and function of the fibronectins. Prog. Clin. Biol. Res. 41797-819. [PubMed] [Google Scholar]
- 56.Yavlovich, A., M. Tarshis, and S. Rottem. 2004. Internalization and intracellular survival of Mycoplasma pneumoniae by non-phagocytic cells. FEMS Microbiol. Lett. 233241-246. [DOI] [PubMed] [Google Scholar]
- 57.Young, C. C., and Bernlohr. 1991. Elongation factor Tu is methylated in response to nutrient deprivation in Escherichia coli. J. Bacteriol. 1733096-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao, W., J. S. Schorey, R. Groger, P. M. Allen, E. J. Brown, and T. L. Ratliff. 1999. Characterization of the fibronectin binding motif for a unique mycobacterial fibronectin attachment protein, FAP. J. Biol. Chem. 2744521-4526. [DOI] [PubMed] [Google Scholar]