Abstract
The SH2 domain-containing inositol 5′-phosphatase, SHIP, negatively regulates various hematopoietic cell functions and is critical for maintaining immune homeostasis. However, whether SHIP plays a role in controlling bacterial infections in vivo remains unknown. Salmonella enterica causes human salmonellosis, a disease that ranges in severity from mild gastroenteritis to severe systemic illness, resulting in significant morbidity and mortality worldwide. The susceptibility of ship+/+and ship−/− mice and bone marrow-derived macrophages to S. enterica serovar Typhimurium infection was compared. ship−/− mice displayed an increased susceptibility to both oral and intraperitoneal serovar Typhimurium infection and had significantly higher bacterial loads in intestinal and systemic sites than ship+/+mice, indicating a role for SHIP in the gut-associated and systemic pathogenesis of serovar Typhimurium in vivo. Cytokine analysis of serum from orally infected mice showed that ship−/− mice produce lower levels of Th1 cytokines than do ship+/+ animals at 2 days postinfection, and in vitro analysis of supernatants taken from infected bone marrow-derived macrophages derived to mimic the in vivo ship−/− alternatively activated (M2) macrophage phenotype correlated with these data. M2 macrophages were the predominant population in vivo in both oral and intraperitoneal infections, since tissue macrophages within the small intestine and peritoneal macrophages from ship−/− mice showed elevated levels of the M2 macrophage markers Ym1 and Arginase 1 compared to ship+/+ cells. Based on these data, we propose that M2 macrophage skewing in ship−/− mice contributes to ineffective clearance of Salmonella in vivo.
Lipid phosphatases, including the Src homology 2 domain-containing inositol 5′-phosphatase, SHIP, play critical roles in balancing immune cell signaling cascades (19). SHIP is a 145-kDa molecule that is restricted to hematopoietic cells and negatively regulates the phosphatidylinositol 3-kinase (PI3K) pathway by hydrolyzing the critical second messenger phosphatidylinositol 3,4,5-triphosphate [PtdIns(3,4,5)P3] to PtdIns(3,4)P2. By downregulating PI3K activity, SHIP restrains a wide array of cellular processes such as migration, proliferation, and survival (19). Furthermore, in many cells of both the innate and adaptive immune system, including macrophages, mast cells, and B cells, SHIP negatively regulates proinflammatory responses and, without this activity, immune homeostasis is lost (4, 22, 37).
The biological importance of SHIP is exemplified in the ship−/− mouse model. Although these mice are viable, they suffer from various maladies, including shortened life span; overproduction of granulocytes, macrophages, and myeloid suppressor cells; extramedullary hematopoiesis; aberrant natural killer (NK) cell development; ineffective allograft rejection; and osteoporosis (10, 12, 30, 32, 42, 44). In addition, bone marrow-derived macrophages (BMDMs) and monocytes derived from ship−/− mice under standard in vitro conditions are hyper-responsive to cytokines and growth factors such as interleukin-3 (IL-3) and granulocyte-macrophage colony-stimulating factor, and SHIP is known to negatively regulate mast cell degranulation (16, 18). Furthermore, as a result of SHIP deficiency, peritoneal and alveolar macrophages from ship−/− mice display an M2 or, alternatively activated, phenotype (35). These cells differ from classically activated, or M1, macrophages in that they produce much lower levels of inflammatory mediators, such as nitric oxide, tumor necrosis factor alpha (TNF-α), IL-12p70, and IL-23, after stimulation by pattern recognition receptor ligation and thus are ineffective in combating pathogens (11, 21). For example, it has recently been shown that M2 macrophages are impaired in their ability to limit the growth of intracellular Mycobacterium tuberculosis due to reduced nitric oxide production and increased iron levels within the phagosome, and they are also associated with murine susceptibility to cutaneous leishmaniasis (14, 15). However, M2 macrophages do play a vital role in the resolution of immune responses and are essential for promoting tissue healing and repair (11). Thus, SHIP is intimately involved in maintaining the delicate balance of macrophage differentiation, maturation, and phenotype that ultimately has a dramatic effect on ensuring an appropriate response by the immune system.
However, although SHIP is an indispensable regulator of immune homeostasis, it is unknown how SHIP functions during an immune response after infection in vivo. Recent studies suggest that SHIP may direct the outcome of pathogenesis; for example, in vitro, SHIP regulates both the macrophage proinflammatory response to the intracellular pathogen, “Francisella tularensis subsp. novicida” (33), and phagosome maturation (17). In addition, SHIP is essential for maintaining endotoxin tolerance in both mice and macrophages (40). Because many bacterial pathogens initially stimulate innate immune cells such as macrophages through lipopolysaccharide (LPS) and an appropriate inflammatory program is required to control infections, these studies suggest that an aberrant immune response may be mounted against such invaders in ship−/− mice (31).
To address whether SHIP plays a role in preventing bacterial infections, we established a Salmonella enterica serovar Typhimurium infection model system in ship+/+ and ship−/− mice and BMDMs. Serovar Typhimurium is a facultative intracellular pathogen that causes gastroenteritis in humans, and infection in mice mimics the more severe and systemic human disease, typhoid fever (38). Infecting mice orally with serovar Typhimurium provides a model for the natural route of infection for this pathogen, allowing us to study the role of SHIP in both colonization of the gut as well as migration of bacteria to systemic organs. In contrast, intraperitoneal (i.p.) infection bypasses the gut phase of pathogenesis, directing focus to the role of SHIP in systemic disease (38). In both oral and i.p. serovar Typhimurium infections, the macrophage provides a protective niche where intracellular Salmonella survive, replicate, and spread throughout the body to cause systemic illness (31). Because SHIP plays important roles in regulating macrophage behavior, we chose to focus our attention on the differences in disease susceptibility between ship+/+ and ship−/− mice and on the impact SHIP has on the macrophage-dependent innate immune response to Salmonella infection.
Our results indicate that SHIP does indeed play a crucial role in modulating the immune response during Salmonella pathogenesis both in vivo and in vitro. These data show for the first time that SHIP regulates innate immune responses necessary for the control of bacterial infections in vivo. Our data also suggest that SHIP may have a significant impact on Salmonella pathogenesis during the establishment of infection in the gut, as well as the spread of infection to foci in systemic organs. Furthermore, we propose that increased susceptibility to Salmonella infection in ship−/− mice may be due to a lack of effector M1 macrophages that are required to control disease.
MATERIALS AND METHODS
Reagents.
Serovar Typhimurium LPS was obtained from Sigma Chemical Company (St. Louis, MO). For mouse genotyping, the primers for SHIP A (sense oligonucleotide; 5′-TCTGTGCAGCTCAGTTTCCTCT-3′), SHIP B (antisense oligonucleotide; 5′-CGTCCCACCATCCTATGACATAA-3′), and TK promoter (antisense oligonucleotide; 5′-CTGCATCTGCGTGTTCGAATT-3′) were obtained from Sigma Genosys (Oakville, Ontario, Canada). TNF-α, gamma interferon (IFN-γ), IL-12p70, IL-6, and IL-10 enzyme-linked immunosorbent assay (ELISA) kits and a cytometric bead array (CBA) kit were from BD Biosciences (Mississauga, Ontario, Canada). Cell culture reagents, including fetal bovine serum (FBS), 1 M sodium pyruvate, 200 mM l-glutamine, and premixed penicillin (10,000 U/ml)-streptomycin (10,000 μg/ml), were from Gibco (Burlington, Ontario, Canada). Dulbecco modified Eagle medium (DMEM) and phosphate-buffered saline with 0.901 mM CaCl2 (PBS+/+) or without calcium (PBS−/−) were from HyClone (Mississauga, Ontario, Canada). The antibodies used in the Western blot analyses included mouse monoclonal P1C1 anti-SHIP sc-8425 from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); polyclonal rabbit anti-Ym1 from StemCell Technologies (Vancouver, British Columbia, Canada); monoclonal mouse anti-Arg1 from BD Transduction (Lexington, KY); and anti-GAPDH from Research Diagnostics (Flanders, NJ). The antibodies used in immunohistochemistry included polyclonal rabbit anti-Ym1 from StemCell Technologies; monoclonal mouse anti-Arg1 from BD Transduction; polyclonal rat anti-F4/80 from ADB Serotec (Hornby, Ontario, Canada); and goat anti-rat Alexa Fluor 568, goat anti-rabbit Alexa Fluor 488, and goat anti-mouse Alexa Fluor 488 from Invitrogen (Burlington, Ontario, Canada). Monoclonal antibodies and dyes used in flow cytometry included biotin rat anti-mouse Ly-6G and GR-1, allophycocyanin rat anti-mouse Mac-1, phycoerythrin (PE) Ar. hamster anti-mouse CD11c, PE Ar. hamster anti-mouse CD-3, rat anti-mouse B220, and streptavidin-fluorescein isothiocyanate (SA-FITC) from BD Biosciences; polyclonal biotin rat anti-F4/80 from ADB Serotec; and 7-amino-actinomycin D (7AAD) from BD Biosciences.
Tissue culture of BMDMs.
Bone marrow cells were obtained by flushing tibiae and femora from uninfected ship+/+ and ship−/− mice. A total of 5 × 106 cells were first suspended in 10 ml of DMEM supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml)-streptomycin (100 μg/ml), and 30% conditioned medium from L929 cells as a source of macrophage colony stimulating factor. M2 inducing medium was further supplemented with 2% mouse serum taken from ship+/− mice. After the cells were allowed to adhere to non-tissue-culture-treated petri plates for 3 h at 37°C and 5% CO2, nonadherent cells were transferred to fresh petri plates for differentiation at 37°C in 5% CO2 for 10 days, with complete medium changes on adherent cells at days 4 and 7. This procedure results in a population of cells that is 95% positive for the macrophage markers F4/80 and Mac-1 (40). RAW 264.7 macrophages were grown in culture in DMEM-10% FBS and skewed to an M2 phenotype 24 h prior to infection by using 100 ng of recombinant mouse IL-4 (R&D Systems, Minneapolis, MN)/ml.
Bacterial culture and preparation.
Serovar Typhimurium SL1344 and Citrobacter rodentium DBS100 were grown overnight in 3 and 5 ml of Luria broth (LB), respectively, at 37°C with shaking. For in vitro infections, serovar Typhimurium SL1344 was opsonized by a wash with 100 μl of overnight culture and resuspension in 100 μl of 30% mouse serum in DMEM, followed by incubation at 37°C for 25 min. Opsonized bacteria were then diluted 1:10 for infection of BMDMs. To heat kill the bacteria, 100 μl of overnight culture of serovar Typhimurium SL1344 was washed and resuspended in 100 μl of PBS+/+, followed by incubation at 80°C for 30 min. For UV killing, 100 μl of overnight culture of serovar Typhimurium SL1344 was washed and resuspended in 100 μl of PBS+/+ and diluted 1:10 in PBS+/+, followed by incubation under a 254-nm UV light for 24 h. No heat-killed or UV-killed bacteria were viable after 48 h of incubation on LB agar at 37°C. Prior to infection, heat-killed and UV-killed bacteria were centrifuged at 13,200 rpm for 4 min in an Eppendorf Mini-Spin benchtop centrifuge (catalog no. F45-12-11 rotor) and diluted 1:10 in 1 ml of DMEM for infection of the BMDMs. To enumerate bacteria from overnight cultures, serial dilutions were prepared in sterile PBS+/+, plated on LB agar supplemented with 100 μg of streptomycin/ml, and incubated for 24 h at 37°C. Approximately 3 × 109 bacteria were present in each 3-ml overnight culture.
In vivo infections.
Six- to eight-week-old 129/SvJ × C57/BL6 F2 ship+/+ and ship−/− mice were obtained from G. Krystal at the BC Cancer Research Centre and then infected orally with a dose of 106 live serovar Typhimurium SL1344 or 108 C. rodentium or intraperitoneally (i.p.) with 102 serovar Typhimurium SL1344 for survival, bacterial load enumeration, cytokine analysis, and immunohistochemistry experiments. For heat kill survival experiments, mice were infected either orally or i.p. with 108 heat-killed serovar Typhimurium SL1344. Oral inocula were diluted from overnight cultures in sterile HEPES buffer (pH 8.0; Gibco), and i.p. inocula were diluted in sterile Hanks balanced salt solution (Sigma). For serovar Typhimurium infections, mice were sacrificed immediately when moribund (for survival experiments) and at 2 or 5 days postinfection (for bacterial load determination and cytokine analyses). For C. rodentium experiments, mice were sacrificed at 7 days postinfection for bacterial load determination. For all in vivo infection data except the flow cytometry experiments, three independent experiments were performed with four each of the ship+/+ and ship−/− mice, for a total of twelve animals for each mouse type. All in vivo experiments were performed in accordance with the protocols and guidelines provided by the Animal Care Committee at the University of British Columbia.
Enumeration of bacterial load from infected mice.
Mice were sacrificed 2 or 5 days postinfection for oral or i.p. serovar Typhimurium infections, respectively, and colons, small intestines, livers, mesenteric lymph nodes (MLN), and spleens were harvested into 1 ml of sterile, cold PBS+/+. For C. rodentium infections, mice were sacrificed 7 days postinfection, and colons were harvested into 1 ml of sterile, cold PBS+/+. Organs were homogenized, and serial dilutions of homogenate were prepared in sterile PBS+/+ for plating. Bacteria were grown on LB agar plus 100 μg of streptomycin (Sigma)/ml for 24 h at 37°C and subsequently counted.
In vitro serovar Typhimurium infections.
BMDMs or RAW 264.7 macrophages were removed from petri plates using nonenzymatic cell dissociation buffer (Gibco) and washed twice in 10 ml of DMEM to remove antibiotics. BMDMs and RAW 264.7 macrophages were seeded at 105 cells/well in 1 ml of DMEM plus 10% FBS, 2 mM l-glutamine, and 1 mM sodium pyruvate (BMDM medium) or DMEM plus 10% FBS plus 100 ng of recombinant mouse IL-4/ml (raw medium; R&D Systems) in 24-well tissue culture plates 12 h prior to infection. Cells were infected at a multiplicity of infection (MOI) of 10 with either opsonized SL1344 or heat-killed or UV-killed serovar Typhimurium SL1344 and then centrifuged at 1,000 rpm in a Beckman GS-6R benchtop centrifuge at 23°C for 5 min to synchronize infection, followed by incubation at 37°C in 5% CO2 for 15 min. To remove extracellular bacteria and prevent reinfection, cells were washed three times in sterile PBS+/+ and supplied with 1 ml of BMDM or raw medium plus 50 μg of gentamicin (Sigma)/ml for 2 h. Cell supernatants were then replaced with 1 ml of BMDM or raw medium plus 10 μg of gentamicin/ml. LPS-treated control cells were given BMDM medium plus gentamicin plus 100 ng of serovar Typhimurium LPS/ml for the time periods indicated. All in vitro experiments were performed a total of three times from independent derivations of BMDMs or RAW 264.7 cell passages, and each manipulation of cells was performed in triplicate in each experiment.
Cytokine analyses.
At designated time points postinfection, 1 ml of supernatant was removed from each well of infected or control BMDMs, divided into aliquots, and frozen at −80°C until assayed for cytokines by using mouse TNF-α, IL-10, IL-6, and IL-12p70 ELISA kits. For analysis of mouse serum cytokine levels, blood was obtained by cardiac puncture, incubated at 37°C for 1 h, and then spun at 13,200 rpm in an Eppendorf 5415D benchtop centrifuge for 10 min to separate the serum. The serum was divided into aliquots, frozen at −80°C, and analyzed by using a mouse inflammation CBA assay kit from BD Biosciences. Briefly, cytokine levels were analyzed in a multiplex fashion, whereby single samples of mouse serum were incubated with a combination of antibody-coated beads specific for IL-12p70, IL-6, IL-10, TNF-α, and IFN-γ and a PE detector solution. Samples were assessed by flow cytometry on a BD FACScalibur flow cytometer (BD Biosciences), and cytokine levels were then analyzed by using the kit-associated CBA software.
Intracellular replication assays.
BMDMs and RAW 264.7 macrophages were infected with serovar Typhimurium SL1344 as described above, and supernatants were removed at 2 and 24 h postinfection. After three washes in PBS+/+, the cells were lysed in 250 μl of PBS+/+, 1% Triton X-100, and 0.1% sodium dodecyl sulfate. Lysates were serially diluted and plated onto LB agar plus 100 μg of streptomycin (Sigma)/ml for 24 h at 37°C and subsequently counted. Fold replication numbers were generated by dividing the bacterial loads enumerated from the 24-h lysates by those from the 2-h lysates.
Cell death assays.
BMDMs were infected with live, heat-killed, or UV-killed serovar Typhimurium SL1344 or treated with LPS, and the supernatants were removed at 8 and 24 h postinfection. After three washes with PBS+/+, the cells were removed by using a rubber scraper from the wells and placed into 300 μl of PBS−/−-2% FBS-0.5% NaN3 (fluorescence-activated cell sorting [FACS] buffer). Cells were transferred to 96-well round-bottom tissue culture plates and stained for necrosis by using 7AAD (1:250). 7AAD positivity was assessed by flow cytometry using CellQuest Pro software on a FACScalibur flow cytometer, and samples were analyzed by using FlowJo flow cytometry software (Tree Star, Ashland, OR).
Western blotting.
Peritoneal macrophages were obtained from infected and uninfected mice by lavage with 10 ml of DMEM-10% FBS. Cells were washed once in DMEM, counted, spun at 1,000 rpm in a Beckman-Coulter Allegra X-12R benchtop centrifuge at 23°C for 5 min, and lysed directly into 1× Laemmli Western sample buffer. Samples were boiled for 3 min and spun at 13,200 rpm in an Eppendorf 5415D benchtop centrifuge for 30 s before being loaded onto SDS-10% polyacrylamide gels. Western analysis for Ym1, Arg1, SHIP, and GAPDH was performed as described previously (35).
Immunohistochemistry.
Tissues were removed from mice and immediately fixed in 10% neutral buffered formalin, incubated at 23°C for 24 h, and then transferred into 70% ethanol. Fixed tissues were embedded in paraffin, cut into 5-μm sections, and stained with hematoxylin and eosin using standard techniques by Wax-it Histology Services (Vancouver, British Columbia, Canada). Prior to staining, slides with 5-μm sections of tissue were deparaffinized and rehydrated. Antigen retrieval was carried out by digesting tissues in 20 μg of proteinase K (Sigma)/ml in TE buffer (50 mM Tris base, 1 mM EDTA [pH 8.0]) for 15 min. Tissues were then immunostained by using the following primary antibodies: rabbit anti-Ym-1 (1:500), mouse anti-Arg1 (1:100), and rat anti-F4/80 (1:1,000). The secondary antibodies used included goat anti-rat Alexa Fluor 468 (1:500), goat anti-mouse Alexa Flour 488 (1:500), and goat anti-rabbit Alexa Fluor 488 (1:1,000). After staining, tissues were mounted using ProLong Gold Antifade reagent (Invitrogen) containing DAPI (4′,6′-diamidino-2-phenylindole) for DNA staining. Images were obtained by using a Zeiss AxioImager microscope equipped with an AxioCam HRm camera operating using AxioVision software (Carl Zeiss, Ltd., Toronto, Ontario, Canada).
Flow cytometry.
For flow cytometric analysis of lymphoid cell populations, the spleens and MLN from orally infected mice were pooled and harvested into 5 ml of DMEM, 1% HEPES buffer (Gibco), and 0.01 mg of Vibrio alginolyticus collagenase (Roche Diagnostics, Indianapolis, IN)/ml and then minced by using scissors and forceps, followed by incubation at 37°C for 1 h. Cell suspensions were separated by drawing them through an 18-gauge needle three times with a 5-ml syringe. Cells were washed in FACS buffer, centrifuged at 1,200 rpm in a Beckman-Coulter Allegra X-12R benchtop centrifuge at 23°C for 5 min, and resuspended in 10 ml of FACS buffer for flow cytometry staining. Approximately 106 cells were stained per well in 96-well round-bottom tissue culture plates using the following primary antibodies and dilutions: PE Ar. hamster anti-mouse CD-3 (1:400) plus biotinylated rat anti-mouse B220 (1:200), allophycocyanin rat anti-mouse Mac-1 (1:100) plus biotin rat anti-mouse F4/80 (1:100), and PE Ar. hamster anti-mouse CD11c (1:200) plus biotin rat anti-mouse GR-1 (1:100). All wells were also stained with SA-FITC (1:400) as a secondary conjugate for biotinylated primary antibodies. The percent positive cells was assessed by using CellQuest Pro software on a FACScalibur flow cytometer and analyzed by using FlowJo flow cytometry software.
Statistical analyses.
For in vivo time-of-death experiments, log-rank statistical analyses for survival data were performed on curves generated by GraphPad Prism 4.0 (MacKiev Software). For enumeration of the bacterial loads from infected mice, as well as replication assays, flow cytometry, ELISA, and CBA data, statistical analyses were performed by using two-tailed, unpaired Student t tests with a 95% confidence interval in GraphPad Prism 4.0. In the figures the error bars represent the standard error of the mean. Statistical significance based on P values of <0.05, <0.01, and < 0.001 are represented in the figure graphs by one, two, and three asterisks, respectively.
RESULTS
SHIP is required to control Salmonella infection in vivo.
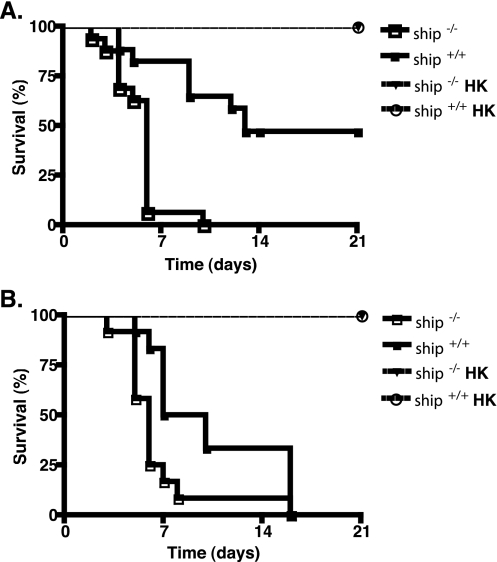
Because endotoxin tolerance is associated with resistance to Salmonella infection (20) and this response is defective in ship−/− mice (40), we hypothesized that these animals would be more susceptible to gram-negative bacterial infection. To test this, we infected ship+/+ and ship−/− mice orally or i.p. with serovar Typhimurium and monitored their survival. We found that ship−/− mice were significantly more susceptible to oral Salmonella infection than were ship+/+ mice (P < 0.001, Fig. 1A). Even with a low dose of 106 bacteria, ship−/− mice died as early as day 2 postinfection and no animals survived longer than 10 days, whereas 47% of ship+/+ mice survived to at least 21 days postinfection. This phenotype was not specific to oral Salmonella infection, since ship−/− mice were significantly more susceptible to Salmonella after i.p. infection with 102 bacteria than ship+/+ mice (P = 0.0119, Fig. 1B).
FIG. 1.
SHIP controls susceptibility to Salmonella in vivo. (A) ship+/+and ship−/− mice were infected orally with 106 live or 106 and 108 heat-killed (HK) serovar Typhimurium SL13344, and the time of death was assessed over a 3-week period. (B) ship+/+and ship−/− mice were infected i.p. with 102 live or 102 and 108 heat-killed (HK) serovar Typhimurium SL13344, and the time of death was assessed over a 3-week period. For both panels, three independent experiments were performed with a total of 12 animals for both ship+/+and ship−/− mice.
Inoculation with LPS is lethal to ship−/− mice within 54 h (40). Therefore, we wanted to examine the possibility that death seen at early time points after oral or i.p. infection in ship−/− mice was due to endotoxic shock from Salmonella LPS in the infection inoculum. To do this, ship+/+ and ship−/− mice were infected orally or i.p. with a dose of 106 or 102 heat-killed salmonellae, respectively, and the survival was monitored. Our results show that 100% of both ship+/+ and ship−/− mice survived these treatments (Fig. 1). In addition, 100% of mice infected either orally or i.p. with a high dose of 108 heat-killed Salmonella, which more closely mimicked bacterial load levels seen when mice were moribund, survived infection (Fig. 2A and B). Furthermore, ship−/− mice are not susceptible to sepsis induced by LPS found on replicating C. rodentium (Fig. 2C). Although we cannot completely exclude the possibility that LPS may still play some role in mortality in ship−/− mice, these data suggest that whereas ship−/− mice cannot control Salmonella replication, levels of LPS present during early and later time points of infection are not sufficient to cause mortality.
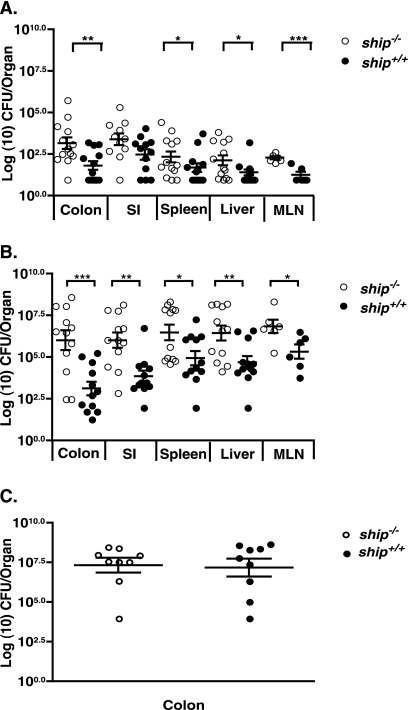
FIG. 2.
SHIP is required to control Salmonella but not Citrobacter replication in vivo. (A) ship+/+and ship−/− mice were infected orally with 106 serovar Typhimurium SL13344 and sacrificed at 2 days postinfection. Colons, small intestines (SI), livers, MLN, and spleens were harvested from the mice, homogenized, and plated to enumerate the bacterial load. (B) ship+/+and ship−/− mice were infected i.p. with 102 serovar Typhimurium SL13344 and sacrificed at 5 days postinfection. Colons, small intestines, livers, MLN, and spleens were harvested from the mice, homogenized, and plated to enumerate bacterial load. (C) ship+/+and ship−/− mice were infected orally with 108 C. rodentium DBS100 and sacrificed at 7 days postinfection. Colons were harvested from the mice, homogenized, and plated to enumerate the bacterial load. For all figures, three independent experiments were performed with a total of 12 animals for both ship+/+and ship−/− mice.
Higher bacterial burdens in the organs of infected mice corresponded to increased susceptibility to infection. We found that bacterial loads were significantly higher at 2 days postinfection in all organs (spleen, P = 0.0245; liver, P = 0.0459; MLN, P = 0.0006; and colon, P = 0.005), except for the small intestines, of orally infected ship−/− mice compared to ship+/+ mice (Fig. 2A). Colony counts were also significantly higher at 5 days postinfection in all organs (spleen, P = 0.0230; liver, P = 0.0068; MLN, P = 0.0290; small intestines, P = 0.0011; and colon, P = 0.005) of ship−/− mice compared to ship+/+mice infected i.p. with serovar Typhimurium (Fig. 2B).
Importantly, we found that when mice were challenged with C. rodentium, an extracellular attaching-and-effacing pathogen that does not cause systemic disease (26), there was no difference in colonization of the colon between ship+/+ and ship−/− mice (Fig. 2C), and infection in either strain did not lead to morbidity or mortality in mice used in CFU experiments after 7 days. These data suggest that the outcome of infection in ship−/− mice may be dependent on the intracellular or extracellular nature of the pathogen and highlight the role SHIP may play in preventing systemic infection.
ship−/− mice produce levels of inflammatory cytokines typical of M2 macrophages during Salmonella infection.
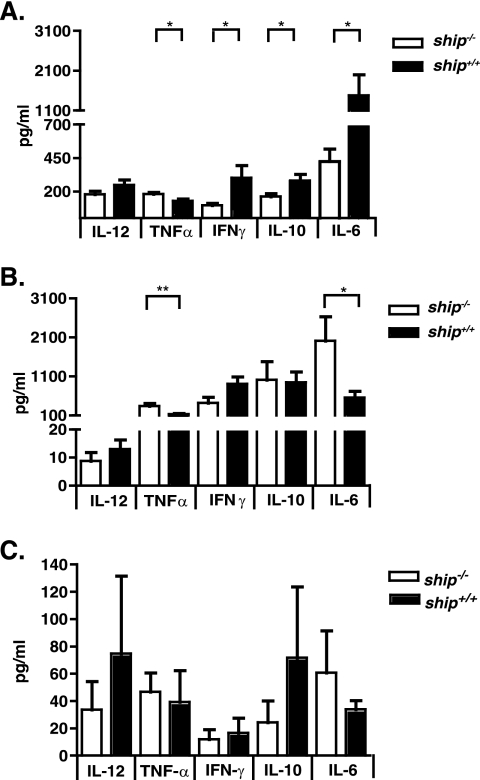
IL-12 and IFN-γ comprise the central axis of Th1 cytokines that are known to drive the immune response against Salmonella (5, 23). Thus, we sought to determine whether the increased susceptibility to Salmonella in ship−/− mice was associated with lower levels of these cytokines during infection. Mice were infected orally or i.p. with 106 or 102 serovar Typhimurium SL1344, respectively, and blood was taken from the mice at 2 days after oral infection or 5 days after i.p. infection for cytokine analyses. These were the same animals used to generate bacterial load data shown in Fig. 2. We found that, in response to oral infection, ship−/− mice produced significantly lower levels of IFN-γ (P = 0.0377), IL-6 (P = 0.0259), and IL-10 (P = 0.0301) than did ship+/+ mice, and the levels of IL-12p70 were also decreased, albeit not significantly (Fig. 3A). During an i.p. infection, a similar trend was observed, with the exception of IL-6. ship−/− mice produced significantly higher levels of IL-6 (P = 0.0299) but lower levels of IL-12p70 and IFN-γ than ship+/+ mice; however, these differences were not significant (Fig. 3B). In both oral and i.p. infections the TNF-α levels were significantly higher in ship−/− mice than in ship+/+ mice (P = 0.0173 and P = 0.0085, respectively). Importantly, no significant differences were found between cytokine levels in uninfected ship−/− and ship+/+ mice (Fig. 3C). Trends toward low IL-12 and IFN-γ levels produced in ship−/− mice upon Salmonella infection suggest a cytokine profile characteristic of M2 macrophages; therefore, association between increased susceptibility to Salmonella and SHIP deficiency could be due to a lack of M1 macrophages that produce the Th1 cytokines required to prevent disease.
FIG. 3.
SHIP deficiency leads to altered levels of inflammatory cytokine production after Salmonella infection in vivo. ship+/+and ship−/− mice were infected orally with 106 serovar Typhimurium SL1344 (A), i.p. with 102 serovar Typhimurium SL1344 (B), or left uninfected (C) and sacrificed at 2 days postinfection or 5 days postinfection, respectively. Blood samples were obtained via cardiac puncture, and serum was separated for cytokine analysis. Cytokines were analyzed by using the flow cytometry-based CBA mouse inflammation kit. For panels A and B, three independent experiments were performed with a total of 12 animals for both ship+/+and ship−/− mice.
M2 macrophage skewing is associated with increased susceptibility to Salmonella in ship−/− mice.
Previous work has shown that SHIP deficiency skews the macrophage phenotype in vivo toward M2 in the lung and peritoneal cavity (35) and that M2 macrophages are ineffective at mounting immune responses against pathogens, especially those requiring Th1 cytokines for clearance (21, 33). Therefore, we suspected that increased susceptibility to Salmonella infection in ship−/− mice could be due, in part, to a lack of M1 effector macrophages. To address whether macrophages in ship−/− mice were M2, we looked for the presence of two M2 macrophage markers, Ym1 and Arginase 1 (Arg1), in tissue sections and peritoneal macrophages isolated from both uninfected mice and those infected orally or i.p. with serovar Typhimurium SL1344. We found that histological sections of the small intestine of orally infected ship−/− mice showed a large amount of inflammatory cells, many of which were Ym1- and Arg1-positive macrophages, in the submucosa, whereas infected ship+/+ mice showed no inflammatory infiltration, and few were Ym1 or Arg1 positive (Fig. 4A to C). In addition, we found that peritoneal macrophages from i.p.-infected ship−/− mice showed elevated levels of Ym1 and Arg1 compared to macrophages from uninfected and infected ship+/+ mice (Fig. 4D). Taken together, these results suggest that macrophages in the gut and peritoneal cavity in ship−/− mice are heavily skewed to an M2 phenotype; thus, these sites may be less protected by effector cells during Salmonella infection.
FIG. 4.
M2 macrophage markers are found in the guts and peritoneal cavities of ship−/− mice. (A and B) Sections of small intestine were taken from uninfected (ship+/+ and ship−/−) and orally infected ship+/+and ship−/− mice (ship+/+ SL1344 and ship−/− SL1344) at day 2 postinfection and stained for the M2 macrophage markers YM1 or Arg1 (green) or and F4/80 (red) as a macrophage control. (C) Sections of small intestine were taken from uninfected ship+/+ and ship−/− mice (−SL1344) and orally infected ship+/+and ship−/− mice (+SL1344) at day 2 postinfection and stained with hematoxylin and eosin to show the pathology. All photographs were taken at ×40 magnification. (D) Peritoneal macrophages were obtained from uninfected (−SL1344) or i.p.-infected (+SL1344) ship+/+and ship−/− mice at 2 days postinfection. Cells were washed, counted, and lysed directly into Western sample buffer for protein analysis. Both bands are specific for Arg1 (35). For panels A to D, the results of representative experiments are shown.
Because many other cell types besides macrophages are key in clearing Salmonella infections and because there are known differences in various immune cell populations between uninfected ship+/+ and ship−/− mice (2, 12, 39), we questioned whether Salmonella infection affected the distribution of T cells, B cells, dendritic cells, macrophages, or neutrophils found in the spleens and MLN of ship+/+ versus ship−/− mice. Interestingly, the distribution of none of these cells in the spleens and MLN of ship+/+ versus ship−/− mice was significantly altered upon Salmonella infection (data not shown). Thus, while inherent differences in lymphocyte populations may contribute to susceptibility to Salmonella infection, these phenotypes are not exaggerated in infected mice.
M2 skewing of BMDMs provides a model for macrophage function in oral Salmonella infection in ship−/− mice.
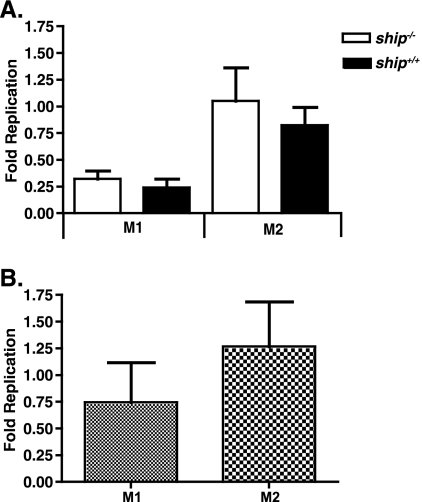
Classically activated macrophages are the primary reservoirs for Salmonella in vivo and modulate bacterial clearance via the production of cytokines such as IL-12p70 and IFN-γ (31). To investigate the role of the macrophage in the ship−/− response to Salmonella infection, we derived macrophages from the bone marrow of ship+/+ and ship−/− mice under M1 or M2 derivation conditions and compared their responses to Salmonella infection. To assess the capability of M1 versus M2 macrophages to prevent intracellular Salmonella replication, numbers of viable Salmonella were assessed by using a standard 24-h gentamicin protection assay. We found that replication was slightly higher in BMDMs from ship+/+ and ship−/− mice derived under M2 inducing conditions, as well as in RAW 264.7 cells skewed to an M2 phenotype using IL-4; however, these differences were not significant (Fig. 5). Importantly, there was no significant difference in intracellular Salmonella replication between M1-derived ship+/+ and ship−/− BMDMs or between M2-derived ship+/+ and ship−/− BMDMs (Fig. 5A).
FIG. 5.
Fold replication of serovar Typhimurium in M1 versus M2 macrophages. (A) BMDMs were obtained from ship+/+ and ship−/− mice and derived in the presence of FBS alone (M1) or FBS plus 2% mouse serum (M2) and infected with serovar Typhimurium SL1344. (B) RAW 264.7 macrophages were grown in either DMEM plus 10% FBS (M1) or skewed to an M2 phenotype by the addition of 100 ng of IL-4/ml (M2) 24 h prior to infection. For both panels, the cells were seeded and infected with serovar Typhimurium SL1344 at an MOI of 10 in a gentamicin protection assay, and the bacteria were enumerated at 2 and 24 h postinfection. The graphs represent the fold replication of intracellular serovar Typhimurium by dividing the CFU values obtained at 24 h by those obtained at 2 h. For both panels, three independent experiments were performed, with each treatment being performed in triplicate, for a total nine wells for each treatment.
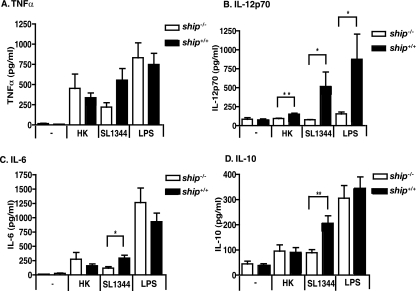
Differences in cytokine profiles produced by M1 or M2 BMDMs during infection with serovar Typhimurium SL1344 were more apparent and dependent upon SHIP genotype. Interestingly, we found that, as in orally infected ship−/− mice, Salmonella-infected M2-derived ship−/− BMDMs produced significantly lower levels of IL-12p70 (P = 0.0201), IL-6 (P = 0.017), and IL-10 (P = 0.0027) than did ship+/+ cells (Fig. 6). Interestingly, these differences could only be seen under M2 derivation conditions; M1-derived macrophages showed significantly higher levels of IL-12p70 (P = 0.0326), IL-6 (P = 0.0017), and IL-10 (P = 0. 0143) when infected with Salmonella (data not shown). Under M2 derivation conditions, significant differences were not found in TNF-α production from Salmonella-infected ship+/+ and ship−/− cells or in IL-6 and IL-10 production from ship+/+ and ship−/− cells stimulated with LPS or heat-killed bacteria (Fig. 6A, C, and D). IL-12p70 production by ship+/+ cells was significantly greater than by ship−/− cells with LPS (P = 0.0465) and heat-killed bacteria (P = 0.0052, Fig. 6B). No significant differences were found in the production of any cytokine examined when cells were infected with UV-killed bacteria (data not shown). Importantly, there were no significant differences seen in cell death in ship+/+ versus ship−/− BMDMs caused by infection with Salmonella or treatment with heat-killed bacteria or LPS (Fig. 7), suggesting that the lower cytokine production seen in Salmonella-infected ship−/− BMDMs is not a result of increased susceptibility to necrosis in our model. Taken together, these results suggest that, whereas M2-derived BMDMs from ship−/− mice may not be less effective in preventing intracellular Salmonella replication than ship+/+ cells, they do produce very different cytokine profiles that closely mimic those seen in ship−/− mice infected with Salmonella.
FIG. 6.
Salmonella-infected BMDMs from ship−/− mice derived under M2 inducing conditions show decreased levels of inflammatory cytokines compared to ship+/+ cells. (A to D) BMDMs were obtained from ship+/+and ship−/− mice and derived in the presence of FBS plus 2% mouse serum for 10 days. Cells were seeded and either left untreated (−), infected with serovar Typhimurium SL1344 (SL1344) or heat-killed serovar Typhimurium SL1344 (HK) at an MOI of 10, or treated with 100 ng of serovar Typhimurium LPS (LPS)/ml for 8 h, and supernatants were collected. Cytokine analysis was performed by using ELISAs. For all four panels, three independent experiments were performed with each treatment being performed in triplicate for a total nine wells for each treatment.
FIG. 7.
ship−/− BMDMs are not more susceptible to death in vitro upon infection with Salmonella. BMDMs were obtained from ship+/+and ship−/− mice and derived in the presence of FBS plus 2% mouse serum for 10 days. Cells were seeded and either left untreated (−), infected with serovar Typhimurium SL1344 (SL1344) or heat-killed serovar Typhimurium SL1344 (HK) at an MOI of 10, or treated with 100 ng of serovar Typhimurium LPS/ml (LPS) for 8 h (A) or 24 h (B). Cells were collected and stained with the cell death marker 7AAD and analyzed via flow cytometry. Three independent experiments were performed, with each treatment being performed in triplicate for a total nine wells for each treatment.
DISCUSSION
Salmonella species pose a global threat to human health. Throughout the world there are an estimated 20 million cases of typhoid fever each year, and nontyphoidal enterocolitis is the second most common cause of food poisoning and the most common cause of death from food-borne illnesses in developed nations (3). Research using the serovar Typhimurium mouse model of systemic salmonellosis has provided many insights into the behavior of this pathogen and the nature of immune responses required to clear intracellular infection. However, despite this progress, the ultimate cause of mortality in mice remains unknown.
One possibility is that negative regulation of immune responses during bacterial infections ultimately decides the fate of the host. For example, endotoxin tolerance is required for resistance to serovar Typhimurium infections (6, 20, 46). In endotoxin-tolerant mice, protection against infection is mediated primarily by increased efficiency of innate immune effector cells (20), and both dissemination and proliferation of serovar Typhimurium are controlled more efficiently than in LPS-insensitive strains (46). In addition, regulation of proinflammatory pathways by enzymes such as PI3K has also been shown to play a critical role in determining the outcome of infections (7). For example, PI3K−/− mice show increased susceptibility to nematode infection and gram-negative induced septic peritonitis (9, 13) but higher resistance to Toxoplasma and Leishmania spp. due to PI3K-dependent skewing of a Th1 immune response (8).
SHIP modulates immune homeostasis, endotoxin tolerance, PI3K signaling, and macrophage inflammatory responses in vitro, but its role in the immune response to in vivo infection was undefined prior to this study. Because SHIP suppresses PI3K and PI3K has been shown to affect outcomes of various pathogenic infections, it is probable that SHIP is an important mediator in this regulatory cascade. Furthermore, since ship−/− mice do not display endotoxin tolerance (36, 40) and ship−/− macrophages cannot respond to “Francisella tularensis subsp. novicida” infection (33), we hypothesized that SHIP deficiency might affect the innate immune response to gram-negative pathogens. Here, we report that indeed SHIP-dependent regulation of innate immune responses is critical for the control of intracellular bacterial infections in vivo. Furthermore, our results suggest that an excess of M2 macrophages in ship−/− mice may exacerbate Salmonella pathogenesis.
Th1-mediated immunity is essential for final clearance of Salmonella infection both in vivo and in vitro (5). For example, neutralization of IFN-γ and IL-12 increases murine susceptibility to Salmonella infection, whereas exogenous addition of these cytokines increases host survival, and patients able to clear gastroenteric Salmonella infection have higher serum levels of these cytokines (1, 28, 41). Consistent with this, we found slightly lower levels of IL-12p70 and significantly lower levels of IFN-γ in ship−/− mice during oral Salmonella infection (Fig. 3A), and a similar trend was observed during i.p. infections as well (Fig. 3B). Importantly, this phenotype was associated with a significant increase in susceptibility to disease (Fig. 1 and 2).
In addition, the levels of other innate immune cytokines, such as IL-6 and IL-10, were altered in infected ship−/− mice. However, the role for these in Salmonella infection is less clear. The fact that IL-6 is upregulated during Salmonella infections in vivo and regulates PMN killing of Salmonella in vitro (5, 27) suggests that it plays a protective role against disease. Interestingly, we found IL-6 levels were lower in ship−/− mice orally infected with Salmonella and higher in i.p.-infected mice, suggesting that IL-6 modulation in ship−/− mice likely plays a critical role in controlling Salmonella in ship−/− mice independently of the other Th1 cytokines examined and may be heavily dependent on the route of infection. This is supported by experiments that plotted IL-6 production over the duration of oral versus i.p. infections (data not shown), where we found that ship−/− mice produce lower levels of IL-6 at early time points during both oral and i.p. infections, but not later. Therefore, we believe that SHIP may play an important role in IL-6 regulation in both oral and i.p. infections and at distinct times during Salmonella pathogenesis.
In contrast to IL-6, it has been suggested that IL-10 may be antiprotective against Salmonella infection (34), due to its classical role as an anti-inflammatory cytokine that suppresses the functions of macrophages, dendritic cells, NK cells, and T cells (25). However, more recently it has been shown that adequate IL-10 production by Th1 cells is an essential component of the immune response against intracellular pathogens such as Leishmania and Toxoplasma spp. (29, 43). Thus, the lower levels of IL-10 produced by Salmonella-infected ship−/− mice may also exacerbate disease.
The production of Th1 cytokines and the subsequent recruitment and activation of phagocytes within Salmonella-infected tissues is heavily dependent on M1 macrophages; thus, these cells are essential in the fight against this intracellular pathogen (11, 21). In contrast, M2 macrophages are incapable of controlling other intracellular pathogens like M. tuberculosis and Leishmania, both of which require strong Th1 immunity for clearance (14, 15). Interestingly, in the case of the ship−/− mouse, there is a skewing of macrophages in the peritoneal cavity and lungs to an M2 phenotype, and it has been shown that BMDMs from ship−/− mice derived in FBS with added mouse serum are M2 (35). Rauh et al. hypothesized that this is due to uncontrolled PI3K signaling in the absence of SHIP (35), and recent evidence has shown that the Src family kinases, Lyn and Hck, are important for this phenotype (45). Based our data showing decreased Th1 responses in infected ship−/− mice, we suspected that M2 skewing was contributing to increased susceptibility to Salmonella infection in vivo. Consistent with this, we found that peritoneal macrophages from i.p.-infected ship−/− animals showed a strong M2 phenotype (Fig. 4D). During this type of infection, macrophages within the peritoneal cavity are the first innate immune cells to encounter Salmonella and are responsible for front line defense to prevent further spread to the blood and systemic sites (23). During oral Salmonella infection, however, resident tissue macrophages, as well as dendritic cells present in the Peyer's patches of the small intestine are the cells that first interact with bacteria colonizing gut tissues (24). Our positive staining for Ym1 and Arg1 in these areas further supported that M2 macrophages are indeed poised at critical sites during both oral and i.p. Salmonella infection in ship−/− mice (Fig. 4A and B).
Interestingly, when we attempted to extrapolate our in vivo data to ship−/− macrophages in vitro, we found that M2 ship−/− BMDMs infected with Salmonella produced a cytokine profile that most closely paralleled the one seen in orally infected ship−/− mice. For example, M2 ship−/− BMDMs produced significantly lower levels of IL-12p70, IL-10, and IL-6 upon Salmonella infection. Lower, but not significant, production of TNF-α was also a hallmark sign of an M2 phenotype (Fig. 6). Importantly, reduction in cytokines was not attributed to increased cell death in ship−/− BMDM (Fig. 7). In contrast, our data using conventional derivation conditions that are known to induce an M1 phenotype (35) showed that Salmonella-infected ship−/− M1 BMDMs produced a Th1 cytokine profile that was opposite to the one seen in in vivo oral Salmonella infections, with significantly higher levels of IL-12p20, IL-10, and IL-6 being produced. Thus, studying the behavior of M2 macrophages in vitro can provide insight into how these cells behave in a natural, oral route of Salmonella infection in ship−/− mice.
M2 macrophages do not produce high levels of bactericidal mediators such as reactive nitrogen or oxygen intermediates that play a role in controlling intracellular replication of Salmonella. Indeed, we found that cells derived under M2 inducing conditions using mouse serum or IL-4 allowed slightly higher Salmonella replication in a 24-h period (Fig. 5). However, this difference was independent of SHIP genotype in BMDMs. In addition, consistent with other unpublished work from our laboratory, Salmonella did not replicate well in BMDMs from either ship+/+ or ship−/− mice (Fig. 5A). Overall, these results suggest that the cytokine response of the M2 macrophage may be a more robust measure of how these cells modulate of the outcome of Salmonella infection rather than how they prevent bacterial replication.
Our data provide the first evidence that the restriction of M2 macrophage generation by SHIP may play a critical role in the prevention of bacterial infections in vivo. First and foremost, ship−/− mice are more susceptible to both oral and i.p. Salmonella infection, but not to increased colonization with C. rodentium, indicating that SHIP is pivotal in controlling the ability of Salmonella to establish infection in the gut, as well as to migrate to, and replicate in, systemic sites of infection. Furthermore, morbidity is associated with lower levels of hallmark M1 cytokines in the blood, and primary macrophages are M2 skewed during infection. These results emphasize the importance of negative immune regulators during Salmonella infection and open the door to future investigations of the role of SHIP and alternatively activated macrophages in other infection models.
Acknowledgments
Work in our laboratory is supported by operating grants from the Canadian Institutes of Health Research (CIHR), the Howard Hughes Medical Institute (HHMI), Genome Canada, and the Foundation for National Institutes of Health. B.B.F. is a CIHR Distinguished Investigator, an HHMI International Research Scholar, and the UBC Peter Wall Distinguished Professor.
We thank Guntram Grassl and Erin Boyle for immunohistochemistry protocols, technical support, and helpful advice.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 21 April 2008.
REFERENCES
- 1.Bao, S., K. W. Beagley, M. P. France, J. Shen, and A. J. Husband. 2000. Interferon-gamma plays a critical role in intestinal immunity against Salmonella typhimurium infection. Immunology 99464-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brauweiler, A. M., I. Tamir, and J. C. Cambier. 2000. Bilevel control of B-cell activation by the inositol 5-phosphatase SHIP. Immunol. Rev. 17669-74. [DOI] [PubMed] [Google Scholar]
- 3.CDC. 2005. Salmonella surveillance: annual summary, 2004. U.S. Department of Health and Human Services, CDC, Atlanta, GA.
- 4.Damen, J. E., L. Liu, P. Rosten, R. K. Humphries, A. B. Jefferson, P. W. Majerus, and G. Krystal. 1996. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5-triphosphate 5-phosphatase. Proc. Natl. Acad. Sci. USA 931689-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckmann, L., and M. F. Kagnoff. 2001. Cytokines in host defense against Salmonella. Microbes Infect./Inst. Pasteur 31191-1200. [DOI] [PubMed] [Google Scholar]
- 6.Freudenberg, M. A., T. Merlin, M. Gumenscheimer, C. Kalis, R. Landmann, and C. Galanos. 2001. Role of lipopolysaccharide susceptibility in the innate immune response to Salmonella typhimurium infection: LPS, a primary target for recognition of gram-negative bacteria. Microbes Infect./Inst. Pasteur 31213-1222. [DOI] [PubMed] [Google Scholar]
- 7.Fukao, T., and S. Koyasu. 2003. PI3K and negative regulation of TLR signaling. Trends Immunol. 24358-363. [DOI] [PubMed] [Google Scholar]
- 8.Fukao, T., M. Tanabe, Y. Terauchi, T. Ota, S. Matsuda, T. Asano, T. Kadowaki, T. Takeuchi, and S. Koyasu. 2002. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat. Immunol. 3875-881. [DOI] [PubMed] [Google Scholar]
- 9.Fukao, T., T. Yamada, M. Tanabe, Y. Terauchi, T. Ota, T. Takayama, T. Asano, T. Takeuchi, T. Kadowaki, J. Hata Ji, and S. Koyasu. 2002. Selective loss of gastrointestinal mast cells and impaired immunity in PI3K-deficient mice. Nat. Immunol. 3295-304. [DOI] [PubMed] [Google Scholar]
- 10.Ghansah, T., K. H. Paraiso, S. Highfill, C. Desponts, S. May, J. K. McIntosh, J. W. Wang, J. Ninos, J. Brayer, F. Cheng, E. Sotomayor, and W. G. Kerr. 2004. Expansion of myeloid suppressor cells in SHIP-deficient mice represses allogeneic T-cell responses. J. Immunol. 1737324-7330. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, S., and P. R. Taylor. 2005. Monocyte and macrophage heterogeneity. Nat. Rev. 5953-964. [DOI] [PubMed] [Google Scholar]
- 12.Helgason, C. D., J. E. Damen, P. Rosten, R. Grewal, P. Sorensen, S. M. Chappel, A. Borowski, F. Jirik, G. Krystal, and R. K. Humphries. 1998. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 121610-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsch, E., V. L. Katanaev, C. Garlanda, O. Azzolino, L. Pirola, L. Silengo, S. Sozzani, A. Mantovani, F. Altruda, and M. P. Wymann. 2000. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science 2871049-1053. [DOI] [PubMed] [Google Scholar]
- 14.Holscher, C., B. Arendse, A. Schwegmann, E. Myburgh, and F. Brombacher. 2006. Impairment of alternative macrophage activation delays cutaneous leishmaniasis in nonhealing BALB/c mice. J. Immunol. 1761115-1121. [DOI] [PubMed] [Google Scholar]
- 15.Kahnert, A., P. Seiler, M. Stein, S. Bandermann, K. Hahnke, H. Mollenkopf, and S. H. Kaufmann. 2006. Alternative activation deprives macrophages of a coordinated defense program to Mycobacterium tuberculosis. Eur. J. Immunol. 36631-647. [DOI] [PubMed] [Google Scholar]
- 16.Kalesnikoff, J., V. Lam, and G. Krystal. 2002. SHIP represses mast cell activation and reveals that IgE alone triggers signaling pathways which enhance normal mast cell survival. Mol. Immunol. 381201-1206. [DOI] [PubMed] [Google Scholar]
- 17.Kamen, L. A., J. Levinsohn, and J. A. Swanson. 2007. Differential association of phosphatidylinositol 3-kinase, SHIP-1, and PTEN with forming phagosomes. Mol. Biol. Cell 182463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, C. H., G. Hangoc, S. Cooper, C. D. Helgason, S. Yew, R. K. Humphries, G. Krystal, and H. E. Broxmeyer. 1999. Altered responsiveness to chemokines due to targeted disruption of SHIP. J. Clin. Investig. 1041751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krystal, G. 2000. Lipid phosphatases in the immune system. Semin. Immunol. 12397-403. [DOI] [PubMed] [Google Scholar]
- 20.Lehner, M. D., J. Ittner, D. S. Bundschuh, N. van Rooijen, A. Wendel, and T. Hartung. 2001. Improved innate immunity of endotoxin-tolerant mice increases resistance to Salmonella enterica serovar Typhimurium infection despite attenuated cytokine response. Infect. Immun. 69463-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantovani, A., A. Sica, and M. Locati. 2007. New vistas on macrophage differentiation and activation. Eur. J. Immunol. 3714-16. [DOI] [PubMed] [Google Scholar]
- 22.March, M. E., and K. Ravichandran. 2002. Regulation of the immune response by SHIP. Semin. Immunol. 1437-47. [DOI] [PubMed] [Google Scholar]
- 23.Mastroeni, P. 2002. Immunity to systemic Salmonella infections. Curr. Mol. Med. 2393-406. [DOI] [PubMed] [Google Scholar]
- 24.McSorley, S. J., S. Asch, M. Costalonga, R. L. Reinhardt, and M. K. Jenkins. 2002. Tracking salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity 16365-377. [DOI] [PubMed] [Google Scholar]
- 25.Moore, K. W., R. de Waal Malefyt, R. L. Coffman, and A. O'Garra. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19683-765. [DOI] [PubMed] [Google Scholar]
- 26.Mundy, R., T. T. MacDonald, G. Dougan, G. Frankel, and S. Wiles. 2005. Citrobacter rodentium of mice and man. Cell. Microbiol. 71697-1706. [DOI] [PubMed] [Google Scholar]
- 27.Nadeau, W. J., T. G. Pistole, and B. A. McCormick. 2002. Polymorphonuclear leukocyte migration across model intestinal epithelia enhances Salmonella typhimurium killing via the epithelial derived cytokine, IL-6. Microbes Infect./Institut Pasteur 41379-1387. [DOI] [PubMed] [Google Scholar]
- 28.Nauciel, C., and F. Espinasse-Maes. 1992. Role of gamma interferon and tumor necrosis factor alpha in resistance to Salmonella typhimurium infection. Infect. Immun. 60450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Garra, A., and P. Vieira. 2007. T(H)1 cells control themselves by producing interleukin-10. Nat. Rev. 7425-428. [DOI] [PubMed] [Google Scholar]
- 30.Oh, S. Y., T. Zheng, M. L. Bailey, D. L. Barber, J. T. Schroeder, Y. K. Kim, and Z. Zhu. 2007. Src homology 2 domain-containing inositol 5-phosphatase 1 deficiency leads to a spontaneous allergic inflammation in the murine lung. J. Allergy Clin. Immunol. 119123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohl, M. E., and S. I. Miller. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52259-274. [DOI] [PubMed] [Google Scholar]
- 32.Paraiso, K. H., T. Ghansah, A. Costello, R. W. Engelman, and W. G. Kerr. 2007. Induced SHIP deficiency expands myeloid regulatory cells and abrogates graft-versus-host disease. J. Immunol. 1782893-2900. [DOI] [PubMed] [Google Scholar]
- 33.Parsa, K. V., L. P. Ganesan, M. V. Rajaram, M. A. Gavrilin, A. Balagopal, N. P. Mohapatra, M. D. Wewers, L. S. Schlesinger, J. S. Gunn, and S. Tridandapani. 2006. Macrophage pro-inflammatory response to Francisella novicida infection is regulated by SHIP. PLoS Pathog. 2e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pie, S., P. Matsiota-Bernard, P. Truffa-Bachi, and C. Nauciel. 1996. Gamma interferon and interleukin-10 gene expression in innately susceptible and resistant mice during the early phase of Salmonella typhimurium infection. Infect. Immun. 64849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rauh, M. J., V. Ho, C. Pereira, A. Sham, L. M. Sly, V. Lam, L. Huxham, A. I. Minchinton, A. Mui, and G. Krystal. 2005. SHIP represses the generation of alternatively activated macrophages. Immunity 23361-374. [DOI] [PubMed] [Google Scholar]
- 36.Rauh, M. J., L. M. Sly, J. Kalesnikoff, M. R. Hughes, L. P. Cao, V. Lam, and G. Krystal. 2004. The role of SHIP1 in macrophage programming and activation. Biochem. Soc. Trans. 32785-788. [DOI] [PubMed] [Google Scholar]
- 37.Rohrschneider, L. R., J. F. Fuller, I. Wolf, Y. Liu, and D. M. Lucas. 2000. Structure, function, and biology of SHIP proteins. Genes Dev. 14505-520. [PubMed] [Google Scholar]
- 38.Santos, R. L., S. Zhang, R. M. Tsolis, R. A. Kingsley, L. G. Adams, and A. J. Baumler. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 31335-1344. [DOI] [PubMed] [Google Scholar]
- 39.Sly, L. M., V. Ho, F. Antignano, J. Ruschmann, M. Hamilton, V. Lam, M. J. Rauh, and G. Krystal. 2007. The role of SHIP in macrophages. Front. Biosci. 122836-2848. [DOI] [PubMed] [Google Scholar]
- 40.Sly, L. M., M. J. Rauh, J. Kalesnikoff, C. H. Song, and G. Krystal. 2004. LPS-induced upregulation of SHIP is essential for endotoxin tolerance. Immunity 21227-239. [DOI] [PubMed] [Google Scholar]
- 41.Stoycheva, M., and M. Murdjeva. 2005. Serum levels of interferon-gamma, interleukin-12, tumour necrosis factor-alpha, and interleukin-10, and bacterial clearance in patients with gastroenteric Salmonella infection. Scand. J. Infect. Dis. 3711-14. [DOI] [PubMed] [Google Scholar]
- 42.Takeshita, S., N. Namba, J. J. Zhao, Y. Jiang, H. K. Genant, M. J. Silva, M. D. Brodt, C. D. Helgason, J. Kalesnikoff, M. J. Rauh, R. K. Humphries, G. Krystal, S. L. Teitelbaum, and F. P. Ross. 2002. SHIP-deficient mice are severely osteoporotic due to increased numbers of hyper-resorptive osteoclasts. Nat. Med. 8943-949. [DOI] [PubMed] [Google Scholar]
- 43.Trinchieri, G. 2007. Interleukin-10 production by effector T cells: Th1 cells show self control. J. Exp. Med. 204239-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, J. W., J. M. Howson, T. Ghansah, C. Desponts, J. M. Ninos, S. L. May, K. H. Nguyen, N. Toyama-Sorimachi, and W. G. Kerr. 2002. Influence of SHIP on the NK repertoire and allogeneic bone marrow transplantation. Science 2952094-2097. [DOI] [PubMed] [Google Scholar]
- 45.Xiao, W., H. Hong, Y. Kawakami, C. A. Lowell, and T. Kawakami. 2008. Regulation of myeloproliferation and M2 macrophage programming in mice by Lyn/Hck, SHIP, and Stat5. J. Clin. Investig. 118924-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, H. R., and H. S. Hsu. 1992. Dissemination and proliferation of Salmonella typhimurium in genetically resistant and susceptible mice. J. Med. Microbiol. 36377-381. [DOI] [PubMed] [Google Scholar]