Abstract
The human parasite Plasmodium falciparum has the potential to express a vast repertoire of variant proteins on the surface of the infected red blood cell (iRBC). Variation in the expression pattern of these proteins is linked to antigenic variation and thereby evasion of host antibody-mediated immunity. The genes in the stevor multigene family code for small variant antigens that are expressed in blood-stage parasites where they can be detected in membranous structures called Maurer's clefts (MC). Some studies have indicated that STEVOR protein may also be trafficked to the iRBC membrane. To address the location of STEVOR protein in more detail, we have analyzed expression in several cultured parasite lines and in parasites obtained directly from patients. We detected STEVOR expression in a higher proportion of parasites recently isolated from patients than in cultured parasite lines and show that STEVOR is trafficked in schizont-stage parasites from the MC to the RBC cytosol and the iRBC membrane. Furthermore, STEVOR protein is also detected at the apical end of merozoites. Importantly, we show that culture-adapted parasites do not require STEVOR for survival. These findings provide new insights into the role of the stevor multigene family during both the schizont and merozoite stages of the parasite and highlight the importance of studying freshly isolated parasites, rather than parasite lines maintained in culture, when investigating potential mediators of host-parasite interactions.
Modification of the surface of the infected erythrocyte by the human malaria parasite Plasmodium falciparum is linked to immune evasion and pathology (17). The genes of var, rif, and stevor multigene families encoded on the parasite genome are thought to undergo switching and clonal variation, resulting in antigenic variation and thus prolonging the chronic infectivity of the parasite (7, 15, 17, 19, 21, 23, 24, 31, 32). The protein products of both the rif and var families have been shown to be transported to the surface of the infected red blood cell (iRBC) (10, 17, 19, 23), and there is evidence that STEVOR proteins may also be located there (21). Whether STEVOR proteins also play a role in adhesion and sequestration of the iRBCs in different organs of the host, thus contributing to disease, or whether they play a role in immune evasion through antigenic variation is not yet known.
The genes of the stevor multigene family along with those of the var and rif multigene families are located in the subtelomeric regions of P. falciparum chromosomes (7, 12). Transcription of the genes in these families is tightly regulated with var genes being transcribed ahead of rif genes in early and late ring stages, while stevor genes are transcribed later at the trophozoite stage, 22 h after invasion (15, 18). Transcription of var genes is regulated epigenetically, ensuring that only a single functional gene is transcribed in each cell (9, 28). In contrast, little is known about the transcriptional regulation of either rif or stevor, but it is apparent that stevor transcription is not coregulated with var despite similar locations of the genes (20, 32).
Genes of the rif and stevor gene families encode proteins with a predicted two-transmembrane-spanning structure (21, 30). These proteins are thought to be located on the iRBC surface, but no binding domains (similar to the Duffy binding ligand region of P. falciparum erythrocyte membrane protein 1 [PfEMP1]) have been identified. Antibodies in hyperimmune sera from adults recognize RIFIN proteins, indicating that these proteins are immunogenic and induce malaria-specific immunoglobulin G (IgG) antibodies (1-3). Another small family of proteins, SURFIN proteins encoded by the 10-gene surf family, are expressed on the iRBC surface of mature blood-stage parasites and are also expressed at the apical tips of merozoites (36).
While some recent studies have shown that transcription of stevor is upregulated in some field isolates, the pattern of stevor transcription in different parasite lines and most importantly in different field isolates is unclear. Our current understanding of the biological role of STEVOR protein is based on relatively few studies including immunofluorescence-based localization studies using peptide-specific antibodies in multiple life stages and on tracking of epitope-tagged STEVOR or green fluorescent protein-chimeric STEVOR constructs expressed under the control of constitutive promoters (15, 21, 22, 27). These studies have produced conflicting results. Using either episomally expressed green fluorescent protein-STEVOR chimeras or specific antibodies, STEVOR protein was located in Maurer's clefts (MC) in blood-stage trophozoites and in the plasma membranes of gametocytes (15, 22, 27) of the 3D7 parasite line. However, studies using epitope-tagged STEVOR protein expressed in the parasite isolate NF54 provided evidence that STEVOR moves beyond MC to the iRBC membrane (21). None of the studies has addressed whether native, untagged STEVOR protein remains in the MC or is relocated to the surfaces of RBCs infected with asexual stage parasites, in the same way as PfEMP1 is.
To determine the location of STEVOR protein in blood-stage parasites, we have investigated the expression of STEVOR in both a number of laboratory-adapted parasite lines and in parasites freshly isolated from patients. These studies confirm that STEVOR is initially located in the MC, but in late-stage schizonts, the protein is relocated to the periphery of the iRBC. They show that STEVOR is expressed at a much greater level in recently isolated parasites than in long-term cultured parasites. In addition, we provide evidence that the stevor gene is transcribed in merozoites and that STEVOR is detectable at the apical end of the merozoite. Taken together, these findings indicate that STEVOR has multiple distinct functions within the blood-stage parasite and plays an important role in parasite survival in its host.
MATERIALS AND METHODS
Parasite culture.
The parasites used in this study were either well-characterized laboratory lines or were isolated from blood samples obtained at the KEMRI-Wellcome Trust, Kilifi, Kenya. This study was approved by the Institutional Ethical Review Board of Nanyang Technological University, Singapore, Singapore, and by the Kenyan Medical Research Institute National Ethics Committee. Long-term parasite lines were cultured in fresh human RBCs in RPMI 1640 complete medium supplemented with 10% Albumax as described previously (35). Parasites obtained from blood samples from patients in Kilifi, Kenya, were transferred to RPMI 1640 medium supplemented with 10% human AB serum; no fresh RBCs were added, and the parasites were maintained in culture for only approximately 30 h, or allowing for delayed growth, until they reached late trophozoite/schizont stages. Parasitemia was monitored by microscopy; thin blood films were fixed in methanol onto glass slides, and stained with Giemsa reagent. Purified merozoites were prepared as previously described (4, 11).
RNA extraction and reverse transcription.
For RNA extraction, iRBC pellets (approximately 100 μl) were lysed in 1 ml prewarmed (37°C) Trizol LS reagent (Gibco Invitrogen) and then stored at −80°C in screw top cryovials. RNA was extracted as described previously (18), using DNase I digestion (Invitrogen) to remove contaminating DNA. RNA was then reverse transcribed using Superscript II that was primed with random hexamer primers (Invitrogen). A negative control lacking reverse transcriptase was used to confirm that amplification was solely from RNA. The stevor gene was amplified using stevor-specific PCR primers as described previously (15), using PCR with genomic DNA from all samples to ensure that all the primers worked efficiently.
Preparation of recombinant STEVOR proteins.
The DNA sequence from stevor gene PF10_0395 coding for an N-terminal fragment was PCR amplified and cloned into the pET-24a (+) vector (Novagen). The fragment corresponds to amino acids 27 to 180 in the native molecule, with an expected molecular mass of 18.5 kDa. Recombinant STEVOR protein was expressed as a fusion protein with a His tag in Escherichia coli BL21-CodonPlus (DE3)-RIL competent cells. The fusion protein was purified via the His tag by affinity chromatography on a Ni-nitrilotriacetic acid agarose (Qiagen) column, and further purified by fast protein liquid chromatography (AKTAPrime system; Amersham, United Kingdom) over a 16/60 Superdex 75 preparative grade column with tissue culture grade phosphate-buffered saline (PBS) (pH 7.2) as the mobile phase. Proteins were extensively dialyzed against PBS before further use, and protein concentration was measured using the bicinchoninic acid reagent (Pierce, Rockford, IL).
Antisera and antibody preparation.
Antipeptide sera were produced as previously described (15). Antibody was affinity purified using the STEVOR peptides covalently coupled to N-hydroxysuccinimide-activated Sepharose high-performance HiTRAP columns per the manufacturer's recommendations.
BALB/c mice were immunized intraperitoneally with 50 μg recombinant protein in 200 μl (100 μl in each of two sites) PBS containing the MPL plus TDM adjuvant system (Sigma) followed by three boosts, at intervals of ∼2 weeks. Rabbit antisera were prepared by ProSci Incorporated, using standard protocols.
All antisera were tested by enzyme-linked immunosorbent assays for either the presence of peptide- or recombinant-protein-specific antibodies. Rabbit sera were also tested for reactivity with a negative-control peptide and keyhole limpet hemocyanin- or His-tagged positive-control proteins (Calbiochem-Behring).
Western blotting.
Parasitized RBCs or uninfected RBC pellets (1 × 107 cells) were fractionated by the method described previously (25). Proteins were resolved on NuPAGE 12% bis-Tris gels (Invitrogen) under reducing conditions in NuPAGE 1× morpholineethanesulfonic acid-sodium dodecyl sulfate buffer (Invitrogen) per the manufacturer's directions. Proteins were run alongside 1× SeeBlue Plus2-prestained standard (Invitrogen). An identically loaded gel was stained first with Coomassie brilliant blue R-250 (Bio-Rad), before destaining in Coomassie R-250 1× solution (Bio-Rad), per the manufacturer's instructions, until the proteins became visible. The proteins were electrophoretically transferred to Hybond C membrane (Amersham Biosciences) in NuPAGE 1× transfer buffer (Invitrogen) containing 10% methanol and NuPAGE antioxidant (Invitrogen). Specific proteins were detected using rabbit anti-S1 serum: bound antibodies were detected using secondary horseradish peroxidase-conjugated swine anti-rabbit IgG (Dako), used at 1 in 2,000 dilution. The peroxidase was detected using Pierce Supersignal West Pico chemiluminescent substrate according to the manufacturer's protocol.
Immunofluorescence assays.
Thin blood smears were made following the development of the parasite to the late trophozoite/schizont stage in cultures. Slides were fixed at 4°C in acetone for 5 min. Slides were allowed to reach room temperature, blocked with PBS containing 2% bovine serum albumin, and then incubated with primary antibodies including the following: rabbit anti-S1 (1:200) and anti-peptide 1 (1:80) antibodies, mouse (B28) anti-P. falciparum skeleton binding protein 1 (anti-PfSBP1) (1:200) (kind gift of Catherine Braun-Breton) (5), rabbit 1E1 anti-MSP119 (1 in 500), and mouse polyclonal anti-P. falciparum reticulocyte binding protein homologue 1 (anti-PfRH1) (1 in 200) antibody (11a). This was followed by secondary antibodies: swine anti-rabbit IgG (labeled with fluorescein isothiocyanate) (1:200) (Dako) or goat anti-mouse IgG (labeled with tetramethyl rhodamine isocyanate) (1:200) (Southern Biotech).
Percentage measurement from immunofluorescence assay (IFA) slides of STEVOR-expressing iRBCs.
For determination of STEVOR-positive iRBCs, thin blood smears were stained with both anti-STEVOR antibodies as well as the DNA stain 4′,6′-diamidino-2-phenylindole (DAPI). As parasite maturation was asynchronous and varied in the different patient isolates, mature schizont-stage parasites as established by nuclear staining were counted in multiple fields. At the same time, the number of schizonts expressing STEVOR protein was determined by counting the number of fluorescein isothiocyanate-positive cells in the fields analyzed. A minimum of 100 fields were analyzed. The percentage of parasites expressing STEVOR was determined with the total number of DAPI-positive schizonts as the denominator.
Microarray and quantitative real-time PCR analysis.
Merozoite RNA and RNA obtained from different time points of synchronized P. falciparum 3D7 intracellular blood-stage parasites were hybridized against a 3D7 reference pool as described previously (6) using a P. falciparum long oligonucleotide microarray (13). Microarray hybridizations were incubated for 14 to 16 h using the Maui hybridization system (Bio Micro Systems). Data were acquired and analyzed by GenePix Pro 3 (Axon Instruments). Array data were stored and normalized using the NOMAD microarray database (http://ucsf-nomad.sourceforge.net/). The array features were unflagged, and the array data that have median intensities greater than the local background plus one times the standard deviation of the background were extracted from the database. Quantitative real-time PCR was performed using 3D7 RNA from the different developmental stages and the published primers as previously described (32). The gene for the housekeeping protein actin was used to normalize the data (14).
RESULTS
Transcription of stevor in different laboratory lines and patient isolates.
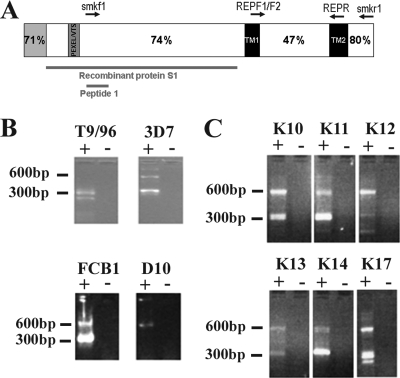
stevor transcription in five laboratory lines and in 25 different patients' samples was analyzed by reverse transcription-PCR (RT-PCR) (Fig. 1B and C and data not shown). In line with previous results obtained with P. falciparum 3D7 (15), several RT-PCR products of the approximate expected size were observed after gel electrophoresis (Fig. 1B and C), indicating that more than one stevor transcript is present in both laboratory and field parasite populations at one time. stevor transcripts were detected in all 25 samples from patients in Kilifi, Kenya, tested, but no stevor RT-PCR products could be detected in the A4 laboratory line. PCR using genomic DNA from the A4 clone gave the expected stevor products, indicating that the PCR primers worked efficiently and supporting the conclusion that this parasite line does not transcribe stevor genes. Sequencing of the RT-PCR products obtained from the different parasite samples confirmed that the stevor sequences were amplified.
FIG. 1.
(A) Schematic representation of the STEVOR protein. The N-terminal signal sequence (gray) including a canonical signal sequence (encoded by exon 1) and the Plasmodium exported element/vacuolar transport signal (PEXEL/VTS) site are shown. Predicted transmembrane domains are shown in black (TM1 and TM2). Percentage similarity data between different STEVOR regions are taken from reference 17. The locations of peptide 1 and recombinant proteins are marked with red bars. (B and C) Gel electrophoresis of nested stevor RT-PCR products from RNA extracted from four P. falciparum laboratory clones (3D7, T9/96, FcB1, and D10) (B) or six representative P. falciparum isolates from patients in Kilifi, Kenya (C). Five microliters of each product was loaded onto a 3% Metaphor agarose gel. Results show two amplicons, products carried over from the external PCR (approximately 600 bp) with primers smf1 and smr1, and internal PCR products (approximately 300 bp) from primers RepF1/2 and RepR. Some sample contained reverse transcriptase (+). The negative control (−) contained sterile water in place of reverse transcriptase enzyme.
STEVOR expression is not essential for parasite survival in culture.
In contrast to previous studies that have clearly shown that the P. falciparum A4 clone transcribes rif and expresses RIFIN proteins (18, 19), our RT-PCR data indicated that STEVOR protein is not expressed in this parasite. To confirm this finding, Western blot analysis was performed on extracts from P. falciparum 3D7 and A4 lines using polyclonal rabbit serum raised against the more conserved N-terminal region of the STEVOR protein (Fig. 1A). This anti-STEVOR recombinant protein (anti-S1) serum was tested using extracts of P. falciparum schizonts (Fig. 2). Since STEVOR is an integral membrane protein (27), it was expected that STEVOR proteins would be detected in the membrane pellet fraction following sequential protein extraction (25).
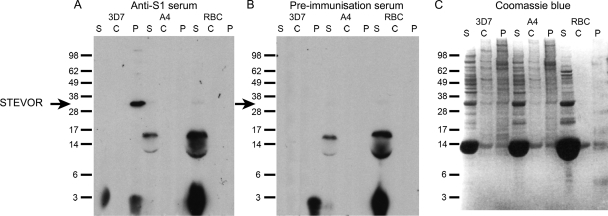
FIG. 2.
(A and B) P. falciparum 3D7 (STEVOR-positive) and A4 (STEVOR-negative) clones were fractionated into three protein populations: soluble proteins (S), carbonate-extracted peripheral membrane-associated proteins (C), and carbonate-insoluble pellet integral membrane proteins (P). An uninfected RBC control was treated identically. The location of the approximately 35-kDa STEVOR protein detected by anti-S1 rabbit serum is indicated by the black arrow to the left of the gel in panels A and B. (B) Rabbit preimmunization serum did not detect STEVOR. A horseradish peroxidase-conjugated secondary swine anti-rabbit IgG antibody and chemiluminescence was used to visualize the result on photographic film. An identical protein gel was stained with Coomassie blue to allow comparison of protein sample loading (C). Proteins were separated on 12% bis-Tris gels in morpholineethanesulfonic acid-sodium dodecyl sulfate running buffer under reducing conditions. The positions of SeeBlue molecular markers used for protein size comparisons (in kilodaltons) are shown to the left of the gels.
Anti-S1 serum detected a protein of the expected molecular mass for STEVOR in the pellet fraction of P. falciparum 3D7 iRBCs but not in this fraction of A4 parasites. A single band was observed in 3D7 at approximately 35 kDa (Fig. 2A), corresponding to the predicted size of STEVOR. This band was not present in the equivalent pellet fraction from clone A4 or in the uninfected RBC control. Western blots with the preimmunization rabbit serum (Fig. 2B) and the Coomassie blue-stained gel (Fig. 2C) are shown as controls.
STEVOR is translocated out of Maurer's clefts in late-stage schizonts.
The recent findings that STEVOR protein expressed from an episome can be detected at the RBC membrane of the P. falciparum NF54 parasite line contrasts with results of other earlier studies showing that STEVOR was trafficked to MC (15, 21, 27). To investigate whether this discrepancy reflected a true difference in the different parasite lines or represents shortcomings in the initial studies, we determined the location of native STEVOR expression in the P. falciparum laboratory lines D10 (Fig. 3A), 3D7 (data not shown), and a representative field isolate (K1657) from Kilifi, Kenya (Fig. 3B) using affinity-purified rabbit anti-peptide 1 immunoglobulin (15). IFAs showed that this anti-STEVOR antibody gave a punctate staining pattern throughout the iRBC, present from the late trophozoite stage and throughout early/mid-stage schizont development in 3D7, D10, and K1657 (Fig. 3 and data not shown). However, in the highly developed segmented schizont, the staining pattern of STEVOR was more diffuse both around the parasite nuclei and throughout the iRBC cytosol, with some potentially associated with the iRBC surface membrane.
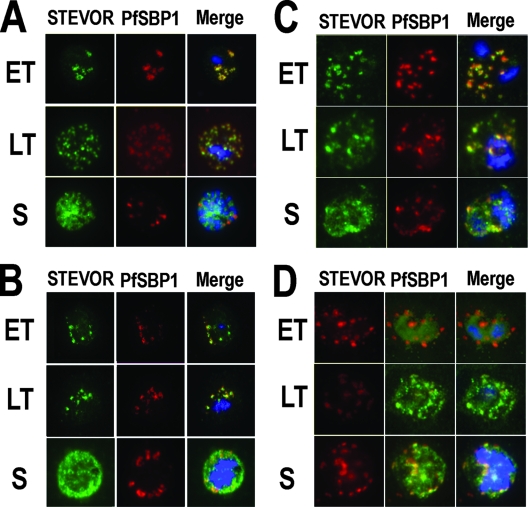
FIG. 3.
STEVOR-specific immunofluorescence staining of mature (>24 h after red blood cell invasion) blood-stage P. falciparum D10 (A and C) and representative field isolates K1657 and K1640 from patients in Kilifi, Kenya (B and D), respectively. Parasites were stained using anti-STEVOR peptide 1 antibodies (A and B) or anti-S1 serum (C and D). Early trophozoite (ET), late trophozoite (LT), and schizont (S) stage parasites are shown. All samples were stained with either rabbit anti-STEVOR peptide 1 affinity-purified antibodies (Stevor) or with anti-S1 rabbit serum (STEVOR), as well as the Maurer's cleft specific anti-PfSBP1 mouse serum (PfSBP1) and the DNA-specific nuclear stain DAPI (at 1 μg/ml). The individual stains and the merged image (Merge) are shown.
In both the laboratory lines and in the K1657 field isolate, STEVOR-specific antibodies and antibodies against the MC-specific marker PfSBP1 colocalized at late trophozoite and early schizont stages, but at the late schizont stage, the staining patterns of each were distinct, suggesting that STEVOR proteins traffic to, and thus may be functional at the erythrocyte surface. MC staining by PfSBP1 antibodies was punctate throughout the iRBC cytoplasm as described previously (5), although the majority of MC were often found in the periphery of the iRBC cytosol during the late schizont stage, compared with earlier trophozoites (Fig. 3A, compare LT with S). This pattern and distribution of MC were consistent, regardless of whether parasites had been cultured or were freshly derived from a field isolate.
For all parasite lines and isolates, control staining using normal rabbit immunoglobulin or secondary antibody reagents alone was negative (data not shown). The positive-control anti-MSP119-specific antibody showed characteristic staining of the merozoite's surface and did not colocalize with the MC-specific anti-PfSBP1 (data not shown).
To confirm the findings seen with the STEVOR antipeptide antibodies, anti-S1 rabbit serum was used to determine STEVOR expression in the P. falciparum laboratory lines D10 (Fig. 3C), cell lines 3D7 and A4 (data not shown), and two representative field isolates from patients in Kilifi, Kenya: K1640 (Fig. 3D) and K1489 (data not shown). As described for the antipeptide antibodies, the anti-S1 serum also gave a punctate staining pattern in late trophozoites, early and late schizonts, and again was more diffuse in the late schizonts (Fig. 3), a distribution similar to that seen with the affinity-purified anti-peptide 1 antibodies. Costaining using antibodies specific for PfSBP1 and the anti-STEVOR S1 antibodies showed that the STEVOR proteins colocalized with MC in the late trophozoite through to the schizont stages and verified that the punctuate STEVOR staining was within the MC. Localization of STEVOR protein changed during parasite development, from early trophozoites through to late schizont stages and from within the MC to a more diffuse staining throughout the parasitophorous vacuole and iRBC cytosol in the late schizonts. Analysis of bright field images showed that staining of STEVOR was closely associated with the iRBC surface membrane (data not shown).
Prior to localization within the MC, STEVOR is thought to travel via the parasite endoplasmic reticulum across the parasite plasma membrane and into the lumen of the parasitophorous vacuole (27). Further evidence for this trafficking is provided here using anti-STEVOR S1 antibodies, as STEVOR staining was also observed around the parasite early in trophozoite development (Fig. 3D, ET stages). No staining of any P. falciparum A4 schizonts with any anti-STEVOR antibodies was observed, as this clone does not transcribe or express STEVOR proteins (as shown from RT-PCR [Fig. 1B] and Western blotting [Fig. 2]).
Differences in STEVOR expression in field isolates compared with cultured parasites.
In line with previous observations (15), only a relatively small percentage of STEVOR-positive parasites was detected by IFA in the culture-adapted laboratory lines, with less than 30% of schizonts being positive in either P. falciparum 3D7 or D10. The intensity and pattern of STEVOR expression are identical in the two parasite lines (data not shown). On the other hand, the number of STEVOR-positive schizonts detected by IFA in a number of different field isolates is greater than 90% (Table 1). Similar to the laboratory-adapted parasites, there was little difference in the intensity and pattern of STEVOR expression for the different field isolates (Fig. 3). In contrast, there was an apparent increase in the intensity of STEVOR-specific IFA signal for the field isolates compared to the culture-adapted parasites, and the amount of STEVOR associated with the RBC membrane appeared to be higher in the field isolates (compare Fig. 3A and C with Fig. 3B and D). However, this observation will need to be confirmed using a more quantitative approach. The low proportion of STEVOR-positive iRBCs in clone 3D7 detected with the antiserum specific for a STEVOR sequence present in this parasite clone, together with the observation that the same antiserum stains a greater proportion of parasites within the field isolates often with greater intensity suggest that differences in expression levels represent true differences rather than variation in detection.
TABLE 1.
Infected RBCs that are positive for STEVOR expression
| P. falciparum clone or Kilifi field isolatea | % of infected RBCs that are STEVOR-expressing schizonts |
|---|---|
| P. falciparum clones | |
| 3D7 | <30 |
| D10 | <30 |
| A4 | 0 |
| Kilifi field isolates | |
| K1657 | >90 |
| K1903 | >90 |
| K1640 | >90 |
| K1127 | >90 |
Field isolate from patients in Kilifi, Kenya.
Differential trafficking of STEVOR to the apical ends of merozoites.
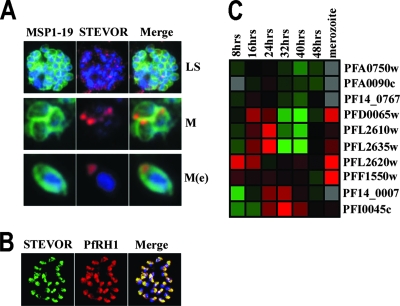
IFAs were carried out on schizont-stage iRBCs of the laboratory line 3D7 that had been allowed to mature further than previously to allow distinction of individual merozoites (Fig. 4). STEVOR staining could be clearly detected associated with individual merozoites as determined by costaining with anti-MSP119 specific antibodies both while still inside the iRBC (Fig. 4A) and after release (Fig. 4B). Closer examination of individual merozoites stained with both anti-MSP119 and anti-STEVOR S1 serum revealed that STEVOR was mainly found at a single end of each merozoite compared to the overall merozoite surface staining seen with antibodies to MSP1 (Fig. 4A). To define further the location of STEVOR, double staining with antibodies raised against a protein expressed at the apical ends of merozoites, PfRH1 (11a, 29, 34) was performed. STEVOR staining colocalized with anti-PfRH1 antibody staining, which is restricted to the apical tip of each merozoite (Fig. 4B). This suggests that STEVOR may be present at the apical ends of the merozoites either in the rhoptries or micronemes.
FIG. 4.
(A) STEVOR-specific immunofluorescence staining of P. falciparum 3D7: intact fully segmented schizonts (LS) and free merozoites (M) using STEVOR anti-S1 serum (STEVOR) and the merozoite surface-specific MSP119 antiserum (MSP1-19). In addition, samples were also stained with the DNA-specific nuclear stain DAPI (blue). Merged images of all stains (Merge) are shown. A single merozoite with digitally enhanced magnification is also shown [M(e)]. (B) STEVOR-specific immunofluorescence staining of P. falciparum 3D7 using STEVOR anti-S1 serum (STEVOR) and the apical end-specific anti-PfRH1 antiserum (PfRH1) are shown. (C) Microarray analysis of stevor transcribed during the asexual blood stage (8 to 48 hours after invasion) and in purified merozoites. Red indicates an elevated level of transcripts in relation to a pool of RNA, while green reflects a reduced level (6). Accession numbers for individual stevor genes (http://plasmodb.org/plasmol) that show regulated transcription are given to the right.
To investigate whether the merozoite-expressed STEVOR is due to additional transcription of stevor in late-stage schizonts/merozoites or possibly represents the translocation of already existing STEVOR into the merozoites from the MC and RBC cytosol, RNA was extracted from both purified merozoites and different time points of intracellular parasite development in the iRBCs. Microarray analysis indicated that there is a second peak of stevor transcription in merozoites (Fig. 4C), and this was confirmed by quantitative real-time RT-PCR analysis (see supplemental material).
DISCUSSION
To date, the biological role of STEVOR protein is still not clear, though the presence of a hypervariable region and expression switching in the blood-stage parasite is evidence suggesting that this protein plays a role in antigenic variation. In addition, the expression of STEVOR in other life cycle stages, including gametocytes and sporozoites, hints at a more complex array of functions. There is also still uncertainty about the final location of STEVOR in iRBCs, with different studies indicating a final location in Maurer's clefts (15, 27) or in one study, at the RBC membrane (21). As these studies used different techniques or parasite lines, it has not been clear whether these results reflect true differences between parasite lines or variation in the methodology used. Furthermore, the low expression level of STEVOR in the blood stage of culture-adapted parasites has raised the question of whether the expression observed in these parasites truly reflects what is happening in a natural infection. In an attempt to resolve these issues and obtain a more precise location of STEVOR in the P. falciparum-infected RBCs, we have investigated the expression of STEVOR in a number of laboratory-adapted parasite lines and parasites directly isolated from patients.
stevor transcripts could be detected in all 25 parasite samples directly obtained from patients and in all but one (A4 clone) culture-adapted parasite line. In line with previous studies (15), multiple distinct transcripts were detected by sequencing in all samples, indicating that multiple stevor genes are transcriptionally active in a parasite population. There was no apparent difference in terms of transcription between the culture-adapted parasites and the field isolates used here. The absence of STEVOR protein in the A4 line was confirmed by Western blot analysis and IFA and indicates that STEVOR is not required for parasite survival in culture. It is also a clear indication that RIFIN and STEVOR proteins are regulated independently, as this parasite line has previously been shown to transcribe and express RIFIN effectively (18, 19).
Two important differences are observed when comparing the expression of STEVOR protein from laboratory lines and field isolates. The frequency of iRBCs from fresh field isolates in which STEVOR was detected is significantly higher, with >90% of all schizonts being specifically detected in IFA compared to <30% in culture-adapted parasites. Moreover, the expression levels as measured by fluorescence intensity observed in the IFA were higher in the patient isolates. This is in agreement with recent microarray studies using field isolates that have shown increased transcription of parasite surface proteins, including STEVOR proteins in these parasites (8). Due to the fact that our antisera against STEVOR most likely do not recognize all STEVOR variants expressed in the patient isolates, the overall frequency of iRBCs expressing STEVOR could be even higher than the more than 90% observed here. Whether the observed difference in STEVOR expression reflects a difference at the transcriptional level or at the control of expression is not yet clear. The much higher frequency of stevor transcript detected in single micromanipulated parasites compared to IFA-positive parasites (15) would indicate that the difference may reside at the level of expression control. The loss or reduction of STEVOR expression in culture-adapted parasite lines is consistent with a role of STEVOR in host-parasite interaction and immune evasion and highlights the importance of studying parasites that have been obtained directly from the host.
We provide convincing evidence that both in laboratory and field-derived parasites, STEVOR protein moves from MC to the RBC cytosol and possibly to the membrane of the iRBC. The amount of STEVOR that moves out to the RBC membrane appears to be higher for the field isolates, and this is consistent with the overall downregulation of STEVOR expression in culture-adapted parasites. This redistribution occurs during schizogony and is most apparent in late-stage schizonts. The association of STEVOR with the iRBC membrane is in line with the recent findings by Lavazec et al. (21), who showed association of an episomally expressed tagged STEVOR with the RBC membrane. In our study, it was not possible to determine unambiguously whether STEVOR is exposed on the outside of the iRBC, due to the low frequency of STEVOR-positive parasites in culture-adapted lines, the limited amount of available parasite material from patient samples, and the potential limitations of the antibody reagents available.
We have demonstrated that STEVOR is also located at the apical end of the merozoite, similar to the RIFIN and SURFIN proteins (26, 36). It is feasible that STEVOR, after it is released from the MC into the RBC cytosol, is somehow translocated to the apical tip of the merozoite. However, our data showing colocalization with an internal apical organelle-specific marker make this difficult to imagine. Importantly, our observation that a second peak of stevor transcription is observed in merozoites supports the hypothesis that a new set of STEVOR protein is expressed in merozoites and transported to the apical end. A recent study has shown that distinct subtypes of RIFIN are associated with different cellular locations inside the parasite (26), and this could also be the case for STEVOR. A more detailed analysis of the gene structure and sequence of stevor could shed some light on this.
We have shown here that native STEVOR is differentially trafficked to the RBC membranes of iRBC and to the apical ends of merozoites (Fig. 5) and provide some preliminary evidence that this is underpinned by a subset of STEVOR proteins. Moreover, we show that STEVOR expression seems to be required only in parasites directly exposed to the host, as STEVOR expression is reduced or completely absent in culture-adapted parasites. A role of STEVOR in immune evasion by iRBCs and merozoites is clearly a likely possibility, though it raises the question why different sets of RIFIN are also expressed in similar locations. Understanding the role STEVOR plays at the surface of the iRBC and at the apical end of the merozoite is now a key question.
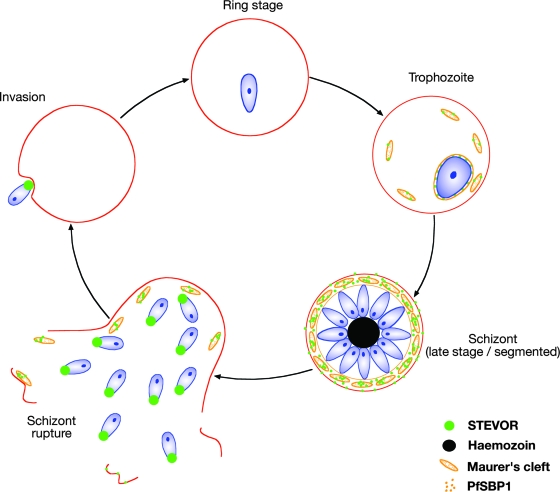
FIG. 5.
Model of STEVOR expression during asexual blood stage development, showing localization of STEVOR protein during the erythrocytic cycle. At the trophozoite stage, STEVOR is detected in both the parasite cytosol and Maurer's cleft. In late schizonts, STEVOR also can be detected in the RBC cytosol and associated with the erythrocyte membrane. In ruptured schizonts and free merozoites, STEVOR is also detected at the apical ends of merozoites.
Supplementary Material
Acknowledgments
We are grateful to Sue Kyes for parasite clone A4. We thank Catherine Braun-Breton for the kind gift of the PfSBP1 antisera.
J.E.B. was in receipt of a studentship from the Medical Research Council (United Kingdom). This work was supported by a Biomedical Research Council (BMRC) Singapore grant (04/1/22/19/364), and the BioMalPar European Network of Excellence was supported by a European grant (LSHP-CT-2004-503578) from Priority 1 “Life Sciences, Genomics and Biotechnology for Health” in the 6th Framework Programme.
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 12 May 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abdel-Latif, M. S., G. Cabrera, C. Kohler, P. G. Kremsner, and A. J. Luty. 2004. Antibodies to rifin: a component of naturally acquired responses to Plasmodium falciparum variant surface antigens on infected erythrocytes. Am. J. Trop. Med. Hyg. 71179-186. [PubMed] [Google Scholar]
- 2.Abdel-Latif, M. S., K. Dietz, S. Issifou, P. G. Kremsner, and M. Q. Klinkert. 2003. Antibodies to Plasmodium falciparum rifin proteins are associated with rapid parasite clearance and asymptomatic infections. Infect. Immun. 716229-6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdel-Latif, M. S., A. Khattab, C. Lindenthal, P. G. Kremsner, and M. Q. Klinkert. 2002. Recognition of variant Rifin antigens by human antibodies induced during natural Plasmodium falciparum infections. Infect. Immun. 707013-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackman, M. J. 1994. Purification of Plasmodium falciparum merozoites for analysis of the processing of merozoite surface protein-1. Methods Cell Biol. 45213-220. [DOI] [PubMed] [Google Scholar]
- 5.Blisnick, T., M. E. Morales Betoulle, J. C. Barale, P. Uzureau, L. Berry, S. Desroses, H. Fujioka, D. Mattei, and C. Braun Breton. 2000. Pfsbp1, a Maurer's cleft Plasmodium falciparum protein, is associated with the erythrocyte skeleton. Mol. Biochem. Parasitol. 111107-121. [DOI] [PubMed] [Google Scholar]
- 6.Bozdech, Z., M. Llinas, B. L. Pulliam, E. D. Wong, J. Zhu, and J. L. DeRisi. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, Q., N. Cloonan, K. Fischer, J. Thompson, G. Waine, M. Lanzer, and A. Saul. 1998. stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol. Biochem. Parasitol. 97161-176. [DOI] [PubMed] [Google Scholar]
- 8.Daily, J. P., K. G. Le Roch, O. Sarr, D. Ndiaye, A. Lukens, Y. Zhou, O. Ndir, S. Mboup, A. Sultan, E. A. Winzeler, and D. F. Wirth. 2005. In vivo transcriptome of Plasmodium falciparum reveals overexpression of transcripts that encode surface proteins. J. Infect. Dis. 1911196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duraisingh, M. T., T. S. Voss, A. J. Marty, M. F. Duffy, R. T. Good, J. K. Thompson, L. H. Freitas, Jr., A. Scherf, B. S. Crabb, and A. F. Cowman. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 12113-24. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez, V., M. Hommel, Q. Chen, P. Hagblom, and M. Wahlgren. 1999. Small, clonally variant antigens expressed on the surface of the Plasmodium falciparum-infected erythrocyte are encoded by the rif gene family and are the target of human immune responses. J. Exp. Med. 1901393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florens, L., M. P. Washburn, J. D. Raine, R. M. Anthony, M. Grainger, J. D. Haynes, J. K. Moch, N. Muster, J. B. Sacci, D. L. Tabb, A. A. Witney, D. Wolters, Y. Wu, M. J. Gardner, A. A. Holder, R. E. Sinden, J. R. Yates, and D. J. Carucci. 2002. A proteomic view of the Plasmodium falciparum life cycle. Nature 419520-526. [DOI] [PubMed] [Google Scholar]
- 11a.Gao, X., K. P. Yeo, S. K. Aw, C. Kuss, J. K. Iyer, S. Ganesan, R. Rajamanonmani, J. Lescar, Z. Bozdech, and P. R. Preiser. 2008. Antibodies targeting the PfRH1 binding domain inhibit invasion of Plasmodium falciparum merozoites. PLoS Pathog., in press. [DOI] [PMC free article] [PubMed]
- 12.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, G., M. Llinas, J. Li, P. R. Preiser, and Z. Bozdech. 2007. Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy. BMC Bioinform. 8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer, J. K., A. Amaladoss, S. Ganesan, and P. R. Preiser. 2007. Variable expression of the 235 kDa rhoptry protein of Plasmodium yoelii mediate host cell adaptation and immune evasion. Mol. Microbiol. 65333-346. [DOI] [PubMed] [Google Scholar]
- 15.Kaviratne, M., S. M. Khan, W. Jarra, and P. R. Preiser. 2002. Small variant STEVOR antigen is uniquely located within Maurer's clefts in Plasmodium falciparum-infected red blood cells. Eukaryot. Cell 1926-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Kyes, S., P. Horrocks, and C. Newbold. 2001. Antigenic variation at the infected red cell surface in malaria. Annu. Rev. Microbiol. 55673-707. [DOI] [PubMed] [Google Scholar]
- 18.Kyes, S., R. Pinches, and C. Newbold. 2000. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol. Biochem. Parasitol. 105311-315. [DOI] [PubMed] [Google Scholar]
- 19.Kyes, S. A., J. A. Rowe, N. Kriek, and C. I. Newbold. 1999. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 969333-9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavazec, C., S. Sanyal, and T. J. Templeton. 2007. Expression switching in the stevor and Pfmc-2TM superfamilies in Plasmodium falciparum. Mol. Microbiol. 641621-1634. [DOI] [PubMed] [Google Scholar]
- 21.Lavazec, C., S. Sanyal, and T. J. Templeton. 2006. Hypervariability within the Rifin, Stevor and Pfmc-2TM superfamilies in Plasmodium falciparum. Nucleic Acids Res. 346696-6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McRobert, L., P. Preiser, S. Sharp, W. Jarra, M. Kaviratne, M. C. Taylor, L. Renia, and C. J. Sutherland. 2004. Distinct trafficking and localization of STEVOR proteins in three stages of the Plasmodium falciparum life cycle. Infect. Immun. 726597-6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newbold, C. I., A. G. Craig, S. Kyes, A. R. Berendt, R. W. Snow, N. Peshu, and K. Marsh. 1997. PfEMP1, polymorphism and pathogenesis. Ann. Trop. Med. Parasitol. 91551-557. [DOI] [PubMed] [Google Scholar]
- 24.Noviyanti, R., and G. V. Brown. 2003. Phenotypic switching and var gene transcription in Plasmodium falciparum. Adv. Exp. Med. Biol. 531149-159. [DOI] [PubMed] [Google Scholar]
- 25.Papakrivos, J., C. I. Newbold, and K. Lingelbach. 2005. A potential novel mechanism for the insertion of a membrane protein revealed by a biochemical analysis of the Plasmodium falciparum cytoadherence molecule PfEMP-1. Mol. Microbiol. 551272-1284. [DOI] [PubMed] [Google Scholar]
- 26.Petter, M., M. Haeggstrom, A. Khattab, V. Fernandez, M. Q. Klinkert, and M. Wahlgren. 2007. Variant proteins of the Plasmodium falciparum RIFIN family show distinct subcellular localization and developmental expression patterns. Mol. Biochem. Parasitol. 15651-61. [DOI] [PubMed] [Google Scholar]
- 27.Przyborski, J. M., S. K. Miller, J. M. Pfahler, P. P. Henrich, P. Rohrbach, B. S. Crabb, and M. Lanzer. 2005. Trafficking of STEVOR to the Maurer's clefts in Plasmodium falciparum-infected erythrocytes. EMBO J. 242306-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ralph, S. A., and A. Scherf. 2005. The epigenetic control of antigenic variation in Plasmodium falciparum. Curr. Opin. Microbiol. 8434-440. [DOI] [PubMed] [Google Scholar]
- 29.Rayner, J. C., E. Vargas-Serrato, C. S. Huber, M. R. Galinski, and J. W. Barnwell. 2001. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J. Exp. Med. 1941571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sam-Yellowe, T. Y., L. Florens, J. R. Johnson, T. Wang, J. A. Drazba, K. G. Le Roch, Y. Zhou, S. Batalov, D. J. Carucci, E. A. Winzeler, and J. R. Yates III. 2004. A Plasmodium gene family encoding Maurer's cleft membrane proteins: structural properties and expression profiling. Genome Res. 141052-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scherf, A., R. Hernandez-Rivas, P. Buffet, E. Bottius, C. Benatar, B. Pouvelle, J. Gysin, and M. Lanzer. 1998. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 175418-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp, S., T. Lavstsen, Q. L. Fivelman, M. Saeed, L. McRobert, T. J. Templeton, A. T. Jensen, D. A. Baker, T. G. Theander, and C. J. Sutherland. 2006. Programmed transcription of the var gene family, but not of stevor, in Plasmodium falciparum gametocytes. Eukaryot. Cell 51206-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Taylor, H. M., M. Grainger, and A. A. Holder. 2002. Variation in the expression of a Plasmodium falciparum protein family implicated in erythrocyte invasion. Infect. Immun. 705779-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trager, W., and J. B. Jensen. 1976. Human malaria parasites in continuous culture. Science 193673-675. [DOI] [PubMed] [Google Scholar]
- 36.Winter, G., S. Kawai, M. Haeggstrom, O. Kaneko, A. von Euler, S. Kawazu, D. Palm, V. Fernandez, and M. Wahlgren. 2005. SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J. Exp. Med. 2011853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.