Abstract
Recombinant attenuated Salmonella vaccines (RASVs) have been used extensively to express and deliver heterologous antigens to host mucosal tissues. Immune responses can be enhanced greatly when the antigen is secreted to the periplasm or extracellular compartment. The most common method for accomplishing this is by fusion of the antigen to a secretion signal sequence. Finding an optimal signal sequence is typically done empirically. To facilitate this process, we constructed a series of plasmid expression vectors, each containing a different type II signal sequence. We evaluated the utilities of these vectors by fusing two different antigens, the α-helix domains of pneumococcal surface protein A (PspA) and pneumococcal surface protein C (PspC), to the signal sequences of β-lactamase (bla SS), ompA, and phoA and the signal sequence and C-terminal peptide of β-lactamase (bla SS+CT) on Asd+ plasmids under the control of the Ptrc promoter. Strains were characterized for level of expression, subcellular antigen location, and the capacity to elicit antigen-specific immune responses and protection against challenge with Streptococcus pneumoniae in mice. The immune responses to each protein differed depending on the signal sequence used. Strains carrying the bla SS-pspA and bla SS+CT-pspC fusions yielded the largest amounts of secreted PspA and PspC, respectively, and induced the highest serum IgG titers, although all fusion proteins tested induced some level of antigen-specific IgG response. Consistent with the serum antibody responses, RASVs expressing the bla SS-pspA and bla SS+CT-pspC fusions induced the greatest protection against S. pneumoniae challenge.
Attenuated mutants of Salmonella enterica serovar Typhi and Salmonella enterica serovar Typhimurium have been extensively studied as multivalent vectors expressing more than 50 different bacterial, viral, and protozoan antigens in preclinical and clinical trials (14, 15, 19, 21, 58). Recombinant attenuated Salmonella vaccines (RASVs) administered orally can colonize the gut-associated lymphoid tissue (GALT) and the secondary lymphatic tissues, including the liver and spleen, and elicit mucosal, humoral, and cellular immune responses against S. enterica and heterologous antigens during infection of the mouse (14, 19).
A number of factors may affect the immune response to protective antigens, including the abilities of vaccine strains to invade and colonize the host GALT, the stability of the plasmid expression system, and the antigen subcellular location (14, 19, 38). High-level expression of protective antigens by RASV strains often imposes an energy demand that decreases growth, fitness, and the ability to colonize lymphoid tissues, resulting in further attenuation and a reduction in immunogenicity (7, 14, 51). Means, such as regulated delayed in vivo antigen synthesis, have been developed to enhance the abilities of vaccine strains to efficiently invade and colonize GALT after oral immunization (7, 18, 51, 64). Another strategy for improving the immune response is to deliver heterologous antigens either secreted into the extracellular environment or displayed on the vaccine carrier surface. Such approaches are based on observations that antigens localized on the surfaces of Salmonella cells or extracellularly secreted produce greatly enhanced immune responses and protection (30, 31, 38, 41). In addition, secretion of heterologous antigens by RASV may decrease the toxicity of the protein to the bacterial vector, facilitate bacterial growth and antigen uptake by antigen-presenting cells, and continuously stimulate the host immune system during the colonization of lymphatic tissues by S. enterica, so as to enhance the immune response against the heterologous antigens (25, 58).
Previous studies have shown that PspA fused to a β-lactamase signal sequence (bla SS-pspA) can be transferred to the periplasmic space via the type II secretion system (T2SS) and subsequently released to the outside medium. This elicited higher PspA-specific immune responses than expression of the protein without a signal sequence and resulted in protection against virulent pneumococcal challenge (38, 39). In gram-negative bacteria, signal peptides of the T2SS play an important role in protein translocation and secretion. The T2SS involves a two-step process in which a preprotein containing a signal sequence is exported via the Sec pathway and processed into the mature protein in the periplasm (9, 10). A number of signal sequences, including outer membrane protein A (OmpA) (62), alkaline phosphatase (PhoA) (42), and murein lipoprotein (Lpp), have been used for efficient production and secretion of recombinant proteins in Escherichia coli. OmpA is an outer membrane protein, and its signal sequence (ompA SS) has been successfully used to direct the secretion of many recombinant proteins, including staphylokinase, thermoalkaliphilic lipase, Manduca diuresin, scFv antibody, 20-kDa human growth hormone, and peptide (9, 47, 50, 62). Fusions to the signal sequence of the E. coli periplasmic protein PhoA (phoA SS) have been reported to direct the secretion of recombinant human C-reactive protein, mouse endostatin, and human cytochrome P4501A1 in E. coli (20, 36, 67). β-Lactamase, encoded by the ampicillin resistance gene bla, is a well-characterized periplasmic protein in gram-negative bacteria. The translocation of β-lactamase depends on the presence of the β-lactamase signal sequence (bla SS) composed of the N-terminal 23 amino acid (aa) residues (37). Evidence obtained from previous studies confirms that the signal sequence plus an additional 12 aa of the mature β-lactamase is required to translocate β-lactamase through the cytoplasmic membrane of gram-negative bacteria (60). It has also been reported that, in addition to the N-terminal sequence, the C-terminal 21 aa residues of mature β-lactamase are important for efficient periplasmic secretion (45).
Streptococcus pneumoniae, a gram-positive human pathogen, causes serious health problems, including community-acquired pneumonia, otitis media, meningitis, and bacteremia, in persons of all ages. S. pneumoniae is a leading agent of childhood pneumonia worldwide, resulting in about 3 million deaths per year (28). Pneumococcal surface protein A (PspA) and pneumococcal surface protein C (PspC) have been considered pneumococcal subunit vaccine candidates. PspA and PspC/Hic are expressed in all clinically isolated pneumococcal strains (6, 32, 34). Immune responses to PspA and PspC can protect mice against virulent S. pneumoniae challenge (3-6, 38, 39, 48).
To date, there is no general rule for selecting the optimal signal sequence for a given protein antigen, as different signal sequences may differ in their efficiency at directing secretion of a given fusion protein. Finding the best signal sequence must be done empirically using a trial-and-error approach (9). In this paper, we describe four expression vectors, each encoding a different export signal sequence, and their use in constructing fusions to two antigens, PspA and PspC, expressed in an RASV engineered for delayed antigen expression (64). We evaluated each strain for level of antigen production, subcellular location, induction of immune responses, and protection in mice.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1 and Table 2, respectively. The amino acids and cleavage sites of signal sequences bla SS (37), ompA SS (62), phoA SS (42), and bla SS+CT (45) are listed in Table 3. E. coli and serovar Typhimurium cultures were grown at 37°C in LB broth or on LB agar plates (1). When required, antibiotics were added to culture media at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 12.5 μg/ml. Diaminopimelic acid (DAP) was added (50 μg/ml) for the growth of Asd− strains (24). Isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) and arabinose were added to media as indicated. S. pneumoniae strains WU2, D39, and L81905 were cultured on brain heart infusion agar containing 5% sheep blood or in Todd-Hewitt broth plus 0.5% yeast extract in an anaerobic container (5).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristic(s) | Derivation or reference |
|---|---|---|
| E. coli | ||
| χ6212 | F− λ− φ80 Δ(lacZYA-argF) endA1 recA1 hsdR17 deoR thi-1 glnV44 gyrA96 relA1 ΔasdA4 | 17 |
| χ289 | F− λ−glnV42 T3r | 13 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) dcm gal (DE3) | Novagen |
| S. enterica serovar Typhimurium | ||
| χ8914 | ΔpabA1516 ΔpabB232 ΔasdA16 | 69 |
| χ9241 | χ8914ΔrelA198::araC PBADlacI TT ΔaraBAD23 | This study |
| χ4700 | Δ(galE-uwrB)-1005 | 63 |
| S. pneumoniae | ||
| WU2 | Wild-type virulent, encapsulated type 3 | 5 |
| D39 | Wild-type virulent, encapsulated type 2 | 46 |
| L81905 | Wild-type virulent, encapsulated type 4 | 6 |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristic(s) | Derivation or reference |
|---|---|---|
| pIN-III-ompA | Secretion vector with ompA signal sequence | 50 |
| TOPO-TA | Commercial vector for TA cloning | Invitrogen |
| pYA3342 | Asd+ expression vector Ptrc promoter pBRori | 39 |
| pYA3493 | Asd+ vector bla SS-based secretion periplasmic plasmid pBRori | 39 |
| pYA3620 | Asd+ vector bla SS+CT-based secretion periplasmic plasmid pBRori | 14 |
| pYA3744 | 1.2-kb DNA encoding the α-helical region of PspC (aa 4-404) in pYA4106 | This study |
| pYA3802 | 0.8-kb DNA encoding the α-helical region of PspA (aa 3-285) in pYA3620 | This study |
| pYA4028 | 1.2-kb DNA encoding the α-helical region of PspC (aa 4-404) in pYA3493 | This study |
| pYA4088 | 0.8-kb DNA encoding the α-helical region of PspA (aa 3-285) in pYA3493 | This study |
| pYA4096 | His-tagged PspC (aa 4-404) in pET28a | This study |
| pYA4098 | 1.5-kb DNA encoding the codon-optimized α-helical and proline-rich regions of PspC (aa 4-477) in TOPO vector | This study |
| pYA4102 | Asd+ vector ompA SS-based plasmid pBRori | This study |
| pYA4106 | Asd+ vector phoA SS-based plasmid pBRori | This study |
| pYA4202 | 1.2-kb DNA encoding the α-helical region of PspC (aa 4-404) in pYA4102 | This study |
| pYA4266 | 0.8-kb DNA encoding the α-helical region of PspA (aa 3-285) in pYA4102 | This study |
| pYA4267 | 0.8-kb DNA encoding the α-helical region of PspA (aa 3-285) in pYA4106 | This study |
| pYA4269 | 1.2-kb DNA encoding the α-helical region of PspC (aa 4-404) in pYA3620 | This study |
| pYA4270 | 1.2-kb DNA encoding the α-helical region of PspC (aa 4-404) in pYA3342 | This study |
| UAB055 | α-helical region of PspA (aa 1-302) in pET20b | 3 |
TABLE 3.
Sequences for bla SS, bla SS-CT, ompA SS, and phoA SS and their fusions to pspA and pspC
| Signal sequence (no. of amino acids) | DNA sequence in expression plasmid or corresponding amino acid(s)a | Reference |
|---|---|---|
| bla SS (36) | ATG AGT ATT CAA CAT TTC CGT GTC GCC CTT ATT CCC TTT TTT GCG | 37 |
| M S I Q H F R V A L I P F F A | ||
| GCA TTT TGC CTT CCT GTT TTT GCT CAC CCA GAA ACG CTG GTG AAA | ||
| A F C L P V F A H P E T L V K | ||
| GTA AAA GAT GCT GAA TTC (EcoRI [pspA or pspC]) | ||
| V K D A ↑ E F | ||
| bla SS+CT (59) | ATG AGT ATT CAA CAT TTC CGT GTC GCC CTT ATT CCC TTT TTT GCG GCA TTT TGC | 45 |
| M S I Q H F R V A L I P F F A A F C | ||
| CTT CCT GTT TTT GCT CAC CCA GAA ACG CTG GTG AAA GTA AAA GAT GCT GAA | ||
| L P V F A H P E T L V K V K D A ↑ E | ||
| TTC (EcoRI [pspA or pspC]) | ||
| F | ||
| (PstI) CTG CAG GCA ACT ATG GAT GAA CGA AAT AGA CAG ATC GCT GAG ATA GGT GCC TCA | ||
| L R A T M D E R N R Q I A E I G A S | ||
| CTG ATT AAG CAT TGG TAA AAG CTT (HindIII) | ||
| L I K H W | ||
| ompA SS (24) | ATG AAA AAG ACA GCT ATC GCG ATT GCA GTG GCA CTG GCT GGT TTC GCT | 47 |
| M K K T A I A I A V A L A G F A | ||
| ACC GTA GCG CAG GCC GCG GAA TTC (EcoRI [pspA or pspC]) | ||
| T V A Q A A ↑ E F | ||
| phoA SS (28) | ATG AAA CAA AGC ACT ATT GCA CTG GCA CTG CTG CCG CTG CTG TTT | 40 |
| M K Q S T I A L A L L P L L F | ||
| ACC CCT GTG ACC AAA GCC CGT ACC CCA GAA ATG AAC CCG GGG ATC C (BamH [pspA or pspC]) | ||
| T P V T K A ↑ R T P E M N P G I |
The predicted cleavage site of each signal sequence is indicated by an arrowhead. Restriction sites (underlined) are defined in parentheses.
Vector construction.
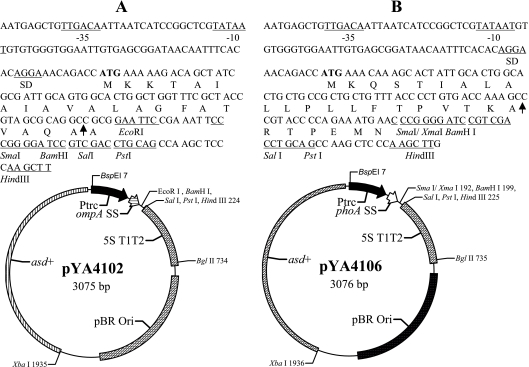
DNA manipulations were carried out as described by Sambrook et al. (53). Transformation of E. coli and S. enterica was done by electroporation (Bio-Rad, Hercules, CA). Transformants containing Asd+ plasmids were selected on LB agar plates without DAP. Only clones containing the recombinant plasmids were able to grow under these conditions (17, 24). The primers used in this paper are listed in Table 4. A 92-bp DNA fragment of the ompA SS gene was PCR amplified from the pIN-III-ompA plasmid template by using primers P1 and P2. The PCR-amplified fragment included the N terminus-encoding region of ompA from the ATG start codon, through the signal sequence (encoding 24 aa), to the region encoding the N-terminal end of mature OmpA (27). The 92-bp PCR product was digested with BspHI and EcoRI enzymes and cloned into the NcoI site (compatible with the BspHI site) and the EcoRI site of the Asd+ vector pYA3342, resulting in plasmid pYA4102 (Fig. 1). In a similar fashion, a 105-bp DNA fragment of the phoA signal sequence (40) was amplified by PCR from E. coli K-12 strain χ289, using primers P3 and P4. The PCR-amplified fragment included the N terminus-encoding region of phoA from the ATG start codon, through the signal sequence (encoding 28 aa), to the region encoding the N terminus end of mature PhoA. The PCR product was digested with BspHI and EcoRI enzymes and cloned into the NcoI site and the EcoRI site of the Asd+ vector pYA3342, resulting in plasmid pYA4106 (Fig. 1).
TABLE 4.
Primers used in this study
| Primer purpose and name | Gene | Sequence |
|---|---|---|
| Cloning ompA SS and phoA SS | ||
| P1 | ompA SS | 5′-TCATGAAAAAGACAGCTATCGCGATTGCA-3′ |
| P2 | ompA SS | 5′-ACGGAATTCAGCGGCCTGCGCTACGGTAGCGAAACC-3′ |
| P3 | phoA SS | 5′-TCATGAAACAAAGCACTATTGCACTGGCA-3′ |
| P4 | phoA SS | 5′-ACGGATCCCCGGGTTCATTTCTGGGGTACGGGC-3′ |
| Cloning pspA | ||
| P5 | pspA | 5′-GGAATTCTCTCCCGTAGCCAGTCAGTCT-3′ |
| P6 | pspA | 5′-TTCAAGCTTATTATGCTTTCTTAAGGTCAGCTT-3′ |
| P7 | pspA | 5′-TTCCTGCAGATTATGCTTTCTTAAGGTCAGCTT-3′ |
| P8 | pspA | 5′-CATGGATCCGTTCTCCCGTAGCCAGTCAGTCT-3′ |
| Cloning pspC | ||
| P9 | pspC | 5′-ACGAATTCGAAGGCCTGCCAAGTACCACTTCTTC-3′ |
| P10 | pspC | 5′-GCTGGTCGACCTATTATTTTTCTTTAACTTTATC-3′ |
| P11 | pspC | 5′-GCTCTGCAGTTTTTCTTTAACTTTATC-3′ |
| P12 | pspC | 5′-ATACCCGGGGGAAGGCCTGCCAAGTACCAC-3′ |
| P13 | pspC | 5′-GCTCTGCAGCTATTATTTTTTTCTTTAACTTTATC-3′ |
| P14 | pspC | 5′-GATGAATTCGAAGGCCTGCCAAGTACCACT-3′ |
| P15 | pspC | 5′-CGAGGATCCATTATTTTTCTTTAACTTTATCTTC-3′ |
| P16 | pspC | 5′-ATGCCATGGAAGGCCTGCCAAGTACCACTTCTTC-3′ |
FIG. 1.
Asd+ secretion vectors pYA4102 and pYA4106. The −35, −10, and Shine-Dalgarno (SD) Ptrc sequences are indicated, and the translation start codon is in boldface. An arrow within the sequence indicates the signal peptidase cleavage site. Unique restriction enzyme sites in the multicloning site are indicated. 5ST1T2 is a transcriptional terminator. (A) ompA SS vector pYA4102. The map of pYA4102 and the nucleotide sequences of the Ptrc promoter region and multicloning sites are shown. (B) phoA SS vector pYA4106. The map of pYA4106 and the nucleotide sequences of the Ptrc promoter region and multicloning sites are shown.
Construction of plasmids expressing PspA and PspC fusion proteins.
The template DNA for pspA cloning was plasmid pYA4088 (Table 2), which contains a copy of pspA encoding aa 3 to 285 of the mature PspA protein from S. pneumoniae RX1. Codons of the pspA gene have been optimized for expression in S. enterica, specifically those encoding aa 4 (CCC to CCG), aa 25 (GCG to GCT), aa 59 (CTA to CTG), aa 79 (CTA to CTG), aa 97 (ATA to ATC), aa 115 (CGA to CGT), aa 126 (GCT to GCG), aa 146 (CTA to CTG), aa 187 (AGA to CGT), aa 188 (CTA to CTG), and aa 223 (CTA to CTG). All pspA constructs were engineered to carry a TAA stop codon after codon 285 of the pspA coding sequence except bla SS+CT-pspA. PCR product 1 (primers P5 and P6), digested with EcoRI and HindIII enzymes, was cloned into the NcoI and HindIII sites of pYA4102, resulting in pYA4266. PCR product 2 (primers P5 and P7), digested with EcoRI and PstI enzymes, was cloned into the EcoRI and PstI sites of pYA3620, resulting in pYA3802. PCR product 3 (primers P8 and P6), digested with BamHI and HindIII enzymes, was cloned into the BamHI and HindIII sites of pYA4106, resulting in pYA4267.
A DNA fragment encoding aa 4 to 477 of the mature PspC protein from S. pneumoniae L81905 was PCR amplified from the bacterial genome and cloned into pTOPO-TA (Invitrogen, Carlsbad, CA). Codons of the pspC gene were optimized for expression in S. enterica to yield plasmid pYA4098 (Table 2), specifically those encoding aa 5 (GGA to GGC), aa 6 (CTA to CTG), aa 14 (AGG to CGC), aa 23 (GGA to GGC), aa 33 (CGA to CGC), aa 37 (AGG to CGC), aa 47 (ATA to ATC), aa 59 (CGA to CGC), aa 66 (CTA to CTG), aa 77 (ATA to ATC), aa 82 (ATA to ATC), aa 88 (CGA to CGC), aa 129 (GGA to GGC), aa 192 (CGA to CGC), aa 213 (AGG to CGC), aa 230 (CGA to CGC), aa 231 (AGA to CGC), aa 244 (CGG to CGC), aa 247 (CGA to CGC), aa 251(GGA to GGC), aa 253 (CTA to CTG), aa 337 (CTA to CTG), aa 346 (CGA to CGC), aa 367 (AGG to CGC), and aa 384 (CGA to CGC). A 1,203-bp DNA fragment encoding codon-optimized aa 4 to 404 of mature PspC was prepared and introduced into the signal sequence plasmids as follows. Note that during each PCR, a TAA stop codon was introduced into the DNA sequence after the region encoding aa 404, except with bla SS+CT-pspC. PCR product 4 (primers P9 and P10), digested with EcoRI and SalI enzymes, was cloned into the EcoRI and SalI sites of pYA3493, resulting in pYA4028. PCR product 5 (primers P9 and P11), digested with EcoRI and PstI enzymes, was cloned into the EcoRI and PstI sites of pYA3620, resulting in pYA4269. PCR product 6 (primers P12 and P13), digested with XmaI and PstI enzymes, was cloned into the XmaI and PstI sites of pYA4106, resulting in pYA3744. PCR product 7 (primers P14 and P15), digested with EcoRI and PstI enzymes, was cloned into the EcoRI and PstI sites of pYA4102, resulting in pYA4202. PCR product 8 (primers P16 and P13), digested with NcoI and PstI enzymes, was cloned into the NcoI and PstI sites of pYA3342, resulting in pYA4270. Nucleotide sequencing reactions were performed by the sequencing laboratory at Arizona State University, using ABI Prism fluorescent BigDye terminators according to the instructions of the manufacturer (PE Biosystems, Norwalk, CT).
SDS-PAGE and immunoblot analyses.
To evaluate PspA and PspC expression as a function of arabinose concentration, cells were grown in LB medium containing 0.05% or 0.2% added arabinose as indicated. Cells were grown in medium containing 1 mM IPTG as an additional control. When the cultures were grown to an optical density at 600 nm (OD600) of 0.8 (about 5 × 108 CFU/ml), 1 ml from each culture was centrifuged, suspended in 100 μl of phosphate-buffered saline (PBS; pH 7.4), and mixed with 100 μl 2× sodium dodecyl sulfate (SDS) loading buffer (39). Protein samples were boiled for 10 min, and then 10 μl was loaded onto a 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel and electrophoresed. Samples were transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk in PBS and incubated with rabbit polyclonal antibodies specific for PspA or PspC or antibody specific for GroEL (Sigma, St. Louis, MO) for 1 h at 37°C. Then, the plates were washed with PBS-Tween 20 three times. The PspA- and PspC-specific antibodies came from rabbits immunized with His-PspA derived from S. pneumoniae RX1 (3) or His-PspC derived from S. pneumoniae L81905 purified from pYA4096 expressed in BL21(DE3). Then, alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG) (Southern Biotech, Birmingham, AL) was added in PBS-milk. Immunoreactive bands were detected by the addition of nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (BCIP) (Sigma, St. Louis, MO). The reaction was stopped after 5 min by washing with several large volumes of deionized water.
Salmonella subcellular fractionation.
Periplasmic fractions were prepared by a modification of the lysozyme-osmotic shock method (23, 65) as previously described (39). Cultures were grown in LB to an OD600 of 0.6 and centrifuged. The supernatant fluid was saved for analysis of secreted proteins. Equal volumes of periplasmic, cytoplasmic, and supernatant fractions and total lysate samples were separated by SDS-PAGE for Western blot analysis. Salmonella outer membrane proteins (SOMPs) were prepared from serovar Typhimurium χ4700 cells (63) grown in LB broth without galactose for analysis by an enzyme-linked immunosorbent assay (ELISA). The use of SOMPs obtained from χ4700 grown in the absence of galactose precludes O-antigen contamination.
Immunization of mice.
Mice were kept 1 week after arrival to acclimate them to our animal facility before immunization. Each group of 8 or 10 inbred 7-week-old female BALB/c mice (Charles River Laboratories, Wilmington, MA) was deprived of food and water for 6 h before oral immunization. The recombinant serovar Typhimurium strains χ9241 (pYA4088), χ9241 (pYA3802), χ9241 (pYA4266), χ9241 (pYA4267), χ9241 (pYA4028), χ9241 (pYA4269), χ9241 (pYA3744), χ9241 (pYA4202), and χ9241 (pYA4270) were grown in LB with 0.2% arabinose to an OD600 of 0.8. The cultures were centrifuged at 4,000 × g at room temperature and suspended in buffered saline containing 0.01% gelatin (BSG) (12) to give a final concentration of 5 × 1010 CFU/ml. Twenty microliters (1 × 109 CFU) of the concentrated bacteria was orally administered. Serovar Typhimurium χ9241 (pYA3493) was used as the vector control. Food and water were returned to the mice 30 min after immunization. Blood was drawn by cheek pouch bleeding at 2-week intervals. Following centrifugation at 4,000 × g for 5 min, the serum was removed from the whole blood and stored at −70°C. Vaginal washes were collected in 50 μl BSG and stored at −20°C (71).
Pneumococcal challenge.
To assess the ability of the Salmonella PspA vaccine to protect immunized mice against S. pneumoniae, 2 × 104 CFU S. pneumoniae WU2 (100 50% lethal doses [LD50]) in 100 μl of BSG were administered by intraperitoneal (i.p.) injection 6 weeks after primary immunization (39, 48). To assess the protective efficacy of the Salmonella PspC vaccine, 4 × 103 CFU S. pneumoniae D39 (10 LD50) in 100 μl of BSG were administered i.p. 8 weeks after primary immunization (46).
ELISA.
IgG, IgG1, IgG2a, and IgA responses against SOMPs, PspA, and PspC in mouse sera were determined by ELISA (68). His-tagged PspA (3) or His-tagged PspC was purified from UAB055 or pYA4096 expressed in BL21 (DE3), respectively. Endotoxins were removed by Detoxi-Gel endotoxin removal columns (Pierce, Rockford, IL). Polystyrene 96-well flat-bottom microtiter plates (Dynatech Laboratories Inc., Chantilly, VA) were coated with 100 ng/well of each antigen, suspended in sodium carbonate-bicarbonate buffer (pH 9.6). The coated plates were incubated overnight at 4°C. Free binding sites were blocked with PBS-0.1% Tween 20 containing 1% bovine serum albumin. Immune mouse sera were serially diluted from an initial dilution of 1:1,000. The vaginal washes were serially diluted from an initial dilution of 1:10. A 100-μl volume of diluted sample was added to duplicate wells and incubated for 1 h at 37°C. Plates were treated with biotinylated goat anti-mouse IgG, IgG1, IgG2a, or IgA (Southern Biotechnology Inc., Birmingham, AL). Wells were developed with streptavidin-horseradish peroxidase conjugate (Invitrogen, Carlsbad, CA), followed by 2,2′-azino-bis-(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) (Sigma). Color development (absorbance) was recorded at 405 nm using an automated ELISA plate reader (model EL311SX; Biotek, Winooski, VT). Endpoint titers were expressed as the reciprocal log2 values of the last positive sample dilution. Absorbance readings two times higher than preimmune serum baseline values were considered indicative of positive reactions.
ELISPOT assay.
To assess the numbers of antigen-specific T-cell cytokines gamma interferon (IFN-γ) and interleukin 4 (IL-4), an enzyme-linked immunospot (ELISPOT) assay was performed (68). Spleen cells from two mice per group were pooled for this assay. Single-cell suspensions were prepared from the spleen by mechanical dissociation. Splenic mononuclear cells (1 × 107/ml) were resuspended in complete medium: RPMI 1640 (Invitrogen Life Technologies, Carlsbad, CA) containing 10% fetal bovine serum (Atlanta Biologicals), 10 mM HEPES buffer, 10 mM nonessential amino acids, 10 mM sodium pyruvate, and 100 U/ml penicillin plus 100 μg/ml streptomycin. Ninety-six-well nitrocellulose plates (Millititer HA; Millipore Corp., Bedford, MA) were used for the assay. The plates were coated with either anti-mouse IFN-γ or IL-4 and incubated overnight at 37°C in 5% CO2. The plates were washed and blocked with 10% fetal calf serum-RPMI 1640 for 1 h. The blocking solution was discarded. Then, 1 × 106 splenic cells in 10% fetal calf serum-RPMI 1640 were added to each well. Lymphocytes were stimulated with 5 μg/ml His-PspA or 5 μg/ml His-PspC in the presence of 10 U/ml mouse IL-2 (PeproTech) for 1 day at 37°C in 5% CO2. Concanavalin A (ConA) (5 μg/ml; Sigma) was used as a positive control for cytokine stimulation. Following overnight incubation, the plates were washed sequentially three times each with PBS and PBS-0.05% Tween 20. Goat-anti mouse IFN-γ and IL-4 were diluted in PBS-0.05% Tween 20 containing 1% bovine serum albumin and added to plates. The plates were treated with heavy-chain-specific, horseradish peroxidase-conjugated anti-goat antibodies (Southern Biotechnology Associates). The wells were washed three times and developed using an AEC (3-amino-9-ethylcarbazole) kit (Vector Laboratories). The plates were incubated at room temperature for 15 to 20 min and washed with water, and blots were counted by using an ELISPOT automatic plate counter (CTL Analyzers; Cellular Technology Ltd., Cleveland, OH).
Statistical analysis.
An analysis of variance (SPSS Software), followed by an application of Tukey's method, was used to evaluate differences in antibody titer and cytokine-forming cell (CFC) response, discerned to 95% confidence intervals. The Kaplan-Meier method (GraphPad Prism; GraphPad Software) was applied to obtain the survival fractions following i.p. challenge of orally immunized mice (68). By use of the Mantel-Haenszel log-rank test, the P value for statistical differences between groups surviving pneumococcal challenges and Salmonella-vaccinated groups or PBS controls was discerned at the 95% confidence interval.
RESULTS
Construction of expression plasmids with different signal sequences.
To evaluate the effects of the signal sequence on antigen expression and immunogenicity, we developed a set of four plasmid expression vectors from the same parent plasmid, pYA3342 (Table 2), so that all were identical except for the specific signal sequence. The four vector plasmids carry the Ptrc promoter, the asd+ gene, the 5ST1T2 transcriptional terminator, and the pBR origin of replication. Plasmids pYA3493 (bla SS) and pYA3620 (bla SS+CT) have been described by Kang et al. (39) and Curtiss et al. (14), respectively. Plasmids pYA4102 and pYA4106 carry the signal sequences from ompA (ompA SS) and phoA (phoA SS), respectively (Fig. 1). Both pYA4102 and pYA4106 were stably maintained for 50 or more generations in serovar Typhimurium asd hosts grown in the presence or absence of DAP.
Construction of plasmids carrying pspA and pspC fusions to bla SS, bla SS+CT, ompA SS, and phoA SS.
We are interested in developing vaccines for the prevention of diseases caused by S. pneumoniae. Therefore, we chose as test antigens the highly immunogenic α-helical region of pspA encoding aa residues 3 to 285 (849 bp; 283 aa) of the mature PspA/Rx1 protein (588 aa) and pspC sequences encoding aa residues 4 to 404 (1,203 bp; 401 aa) of the mature PspC/L81905 protein (516 aa). We constructed in-frame fusions of both antigens to all of the signal sequences described above. All of the fusions were confirmed by DNA sequencing.
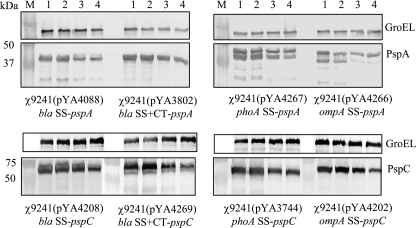
The amount of PspA antigen produced in serovar Typhimurium χ9241 harboring pYA4088, pYA3802, pYA4267, and pYA4266 was evaluated by Western blot analysis (Fig. 2A). Salmonella strains carrying the two bla SS fusions to pspA produced more antigen than the ompA and phoA fusions, although bla SS-pspA (pYA4088) produced slightly more than bla SS+CT (pYA3802). Two bands were observed in Western blots of protein from the strains carrying pYA4267 (phoA SS-pspA) and pYA4266 (ompA SS-pspA), indicating that the signal sequences were cleaved during secretion. There was no indication that either of the two bla fusion proteins was processed in this way.
FIG. 2.
Different PspA and PspC fusion proteins expressed in S. enterica. The pspA (A) and pspC (B) genes were fused to four different T2SS signal sequences (bla SS, bla SS+CT, ompA SS, and phoA SS) and transformed to serovar Typhimurium χ9241. Cells were grown in LB broth at 37°C to an OD600 of 0.8 and Western blot analyses performed on whole cells. Densitometry analyses of immunoreactive bands were evaluated by Quantity One software, and the relative density is shown below each blot. GroEL is used as a marker to indicate loading of the same amount of protein sample in each lane. These experiments were performed three times, with similar results.
The amount of PspC antigen produced in serovar Typhimurium χ9241 harboring pYA4028, pYA4269, pYA3744, pYA4202 and a control plasmid, pYA4270, which encodes cytoplasmically expressed PspC, was evaluated by Western blot analysis (Fig. 2B). Results showed that in this case, the strain carrying the bla SS+CT-pspC fusion protein (pYA4269) produced more antigen than the others, nearly twofold more than the bla SS-pspC fusion strain and threefold more than the ompA SS-pspC fusion strain. There was no indication that any of the signal sequences were cleaved. There was a small amount of a breakdown product in lanes 1, 2, and 3 in Fig. 2B, but those products are too small to represent the mature form of PspC. PspC without a signal sequence (pYA4270) was poorly expressed, producing nearly 20-fold less protein than the bla SS+CT-pspC fusion. Therefore, we conclude that for these two antigens, different signal sequences or the lack thereof (in the case of PspC) can influence the amount of a protein expressed by S. enterica.
Expression of PspA and PspC in Salmonella strain χ9241 is regulated by arabinose.
The attenuated serovar Typhimurium strain used in this study, χ9241, has been designed for delayed antigen expression (64). The construction and utility of this system will be described in a future publication (S. Wang, Y. Li, and R. Curtiss III, unpublished results). Briefly, this strain expresses LacI from the PBAD promoter such that LacI synthesis is regulated by arabinose availability (Table 1). The Ptrc promoter in our plasmid constructs is repressed by LacI. Therefore, when arabinose is added to the growth medium, LacI is produced, inhibiting transcription from the Ptrc promoter, leading to decreased antigen synthesis. Upon oral immunization of a mouse, the Salmonella vaccine colonizes the GALT, an environment with little or no arabinose (Wei Kong, unpublished results). As a result of growth in an arabinose-poor environment, LacI production will decrease and antigen expression will increase.
To evaluate PspA or PspC expression as a function of arabinose concentration, cells were grown in LB medium containing various concentrations of added arabinose. Note that there are trace amounts of arabinose in the yeast extract used to prepare the medium, approximately 0.0034% (Keith Ameiss, personal communication). As an additional control, cells were also grown in the presence of 1 mM IPTG. When IPTG is added to the medium, it diffuses into the cell, where it binds to the LacI repressor, causing a conformational change that decreases its affinity for lacO. Thus, PspA should be maximally expressed from the Ptrc promoter in the presence of IPTG, regardless of the arabinose concentration. Antigen levels in cells were evaluated by Western blot analysis. The results showed that the expression levels of all of the PspA and PspC fusions were repressed in the presence of added arabinose, as expected (Fig. 3). The levels of PspA and PspC expression were similar in LB broth with and without IPTG, indicating that the amount of arabinose normally present in LB was not sufficient to lead to Ptrc repression in our system. The PspA and PspC expression levels were similar for 0.2% and 0.05% arabinose in all groups, indicating that 0.05% arabinose was sufficient to cause the maximum level of Ptrc repression achievable in our system.
FIG. 3.
Expression levels of PspA and PspC fusion proteins are regulated by arabinose concentration. The pspA and pspC genes were fused to bla SS, bla SS+CT, ompA SS, and phoA SS and expressed in serovar Typhimurium χ9241. Serovar Typhimurium strains were cultured in LB broth with or without arabinose or IPTG. GroEL was used as a standardization marker. The lanes indicate media containing different concentrations of arabinose. Lane 1, LB Broth with 1 mM IPTG; lane 2, LB broth alone; lane 3, LB broth with 0.05% arabinose; lane 4, LB broth with 0.2% arabinose.
Subcellular localization of PspA and PspC in S. enterica.
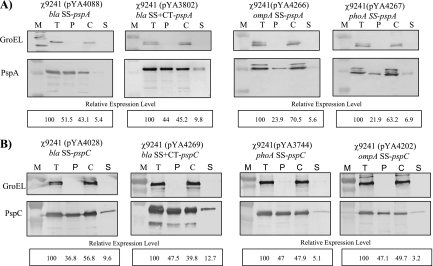
To examine the effects of signal sequences on the secretion of PspA and PspC, subcellular fractions from each strain, including cytoplasm, periplasm, and culture supernatant fractions, were prepared. PspA or PspC was detected in the periplasmic and culture supernatant fractions of all vaccine strains, indicating that all the signal sequences can facilitate the secretion of PspA and PspC protein (Fig. 4).
FIG. 4.
Subcellular location analyses of PspA and PspC fusion protein expressed in serovar Typhimurium. Total cell lysates and subcellular fractions, including cytoplasm, periplasm, and supernatant fractions, were prepared from serovar Typhimurium χ9241 harboring different SS-pspA (A) or SS-pspC (B) fusions. Cells were grown in LB broth at 37°C as described in Materials and Methods. Fractions equivalent to 25-μl volumes of the culture at an OD600 of 0.6 were evaluated by immunoblotting with PspA-specific (A) or PspC-specific (B) polyclonal rabbit antibody. GroEL was used as a cytoplasmic marker for fractionation. Standards are indicated to the left. M, protein ladders; T, total cell lysate; P, periplasm fraction; C, cytoplasm fraction; S, supernatant fraction. Densitometry analyses of immunoreactive bands were evaluated by Quantity One software, and the relative density is shown below each blot. Results are representative of three experiments.
Both bla fusions to PspA were more efficiently secreted to the periplasm than the ompA or phoA fusion (Fig. 4A), with the bla SS fusion being slightly more effective than the bla SS+CT fusion. In all cases, only minor amounts (5 to 10%) of PspA fusion protein were secreted into the supernatant. As mentioned above, the ompA and phoA fusion proteins appear as two bands in whole-cell extracts, suggesting signal sequence cleavage during secretion. This appears to be the case, as only the lower-molecular-weight band is present in the periplasm and supernatant fractions. It is interesting to note that these two proteins, while cleaved, were not as efficiently secreted as the uncleaved bla fusions.
In contrast to the PspA results, bla SS was the least efficient at directing PspC secretion to the periplasm (Fig. 4B), although this was compensated for somewhat by the amount of antigen directed to the supernatant. The bla SS+CT sequence was the most efficient overall, as there was a greater percentage of antigen directed to the supernatant with this fusion, resulting in 60% of the protein being secreted out of the cytoplasm. The other two signal sequences had roughly the same efficiency, with approximately 50% of the fusion protein secreted. Without any signal sequence, PspC was found only in the cytoplasm (data not shown).
GroEL, a cytoplasmic protein, was used as a marker for cell lysis (2, 29). We did not see any leakage of GroEL out of the cytoplasm, indicating that the PspA and PspC proteins detected in the periplasmic fraction and culture supernatant fluid were actively secreted and not present as a result of nonspecific membrane leaking or cell lysis.
Immune responses in mice after oral immunization with the recombinant serovar Typhimurium vaccines.
To investigate the influences of the different T2SSs delivered by RASV on the immunogenicities of PspA and PspC, we orally inoculated groups of BALB/c mice with a single dose of serovar Typhimurium χ9241 carrying one of the fusion protein plasmids. We did not observe any signs of disease in the immunized mice during the entire experimental period, confirming that the vaccine strains are avirulent.
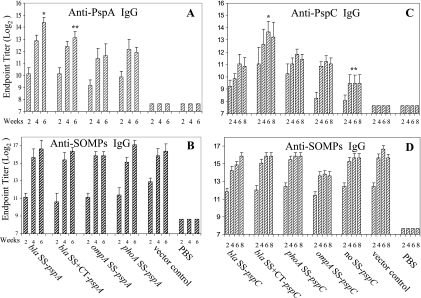
Serum IgG responses to PspA and SOMPs from immunized mice were measured by ELISA (Fig. 5). IgG responses to PspA were observed after 2 weeks postimmunization and increased over time. Maximal anti-SOMP IgG and anti-PspA IgG levels were detected at 6 weeks postimmunization, similar to previous results (39). χ9241 (pYA4088; bla SS-pspA) reached the highest anti-PspA IgG endpoint titer of 214.4 at week 6, compared to the other three signal sequence groups (P < 0.03). No anti-PspA IgG was detected in sera obtained from mice immunized with the vector control or PBS. The anti-SOMP IgG responses in all groups, including the vector control, were similar in both kinetics and titer and were not significantly different (P > 0.05). These results indicate that the bla SS-pspA fusion protein was the most effective immunogen when delivered by our RASV.
FIG. 5.
Kinetic analysis of anti-PspA, anti-PspC, and anti-SOMP serum IgG responses in mice. Mice were orally immunized with 1 × 109 CFU of serovar Typhimurium χ9241 harboring the indicated SS-pspA or SS-pspC fusions. The IgG titers were determined biweekly by ELISA. The numbers listed below the x axis indicate weeks after immunization. (A) Statistical differences in IgG titer were evaluated at 6 weeks after immunization. *, P < 0.03 for comparison with all other vaccine groups; **, P < 0.05 for comparison with ompA SS-pspA and phoA SS-pspA groups. All vaccine groups were significantly different from the vector and PBS controls (P < 0.05). (B) There were no significant differences in anti-SOMP serum IgG titers at week 6 between groups (P > 0.05). (C) Statistical significances were determined at week 6. *, P < 0.001 for comparison with all other vaccine groups; **, P < 0.01 for comparison with all other vaccine groups. All vaccine groups were significantly different from the vector and PBS controls (P < 0.05). (D) There were no significant differences in anti-SOMPs serum IgG titers between vaccine groups and vector controls at week 6 (P > 0.05). Titers are the log2 geometric mean titers ± standard deviations for eight mice.
Immune responses to PspC were investigated (Fig. 5) in groups of mice immunized with a single dose of one of the PspC fusion strains. IgG responses were determined for 8 weeks postimmunization. All groups developed serum responses against PspC and SOMPs. Maximal anti-PspC IgG and anti-SOMP IgG levels were detected at 6 weeks postimmunization for all signal sequence groups. The anti-PspC IgG serum response elicited by χ9241 (pYA4269; bla SS+CT-pspC) peaked at 6 weeks, with a titer of 213.6, which was significantly higher than the titers observed in the other groups (P < 0.001) (Fig. 5).
When PspC was delivered without a signal sequence, anti-PspC titers were significantly lower than those for any of the signal sequence groups at 6 weeks postimmunization (P < 0.01). No anti-PspC IgG was detected in sera obtained from mice immunized with the vector control. The anti-SOMP IgG responses in all strains, including vector controls, were similar in both kinetics and levels, and there were no significant differences between groups (P > 0.05). As seen above with PspA, these results show that the magnitude of serum IgG response is influenced by the particular signal sequence to which an antigen is fused.
IgG isotype analyses and IFN-γ and IL-4 T-cell cytokine assay.
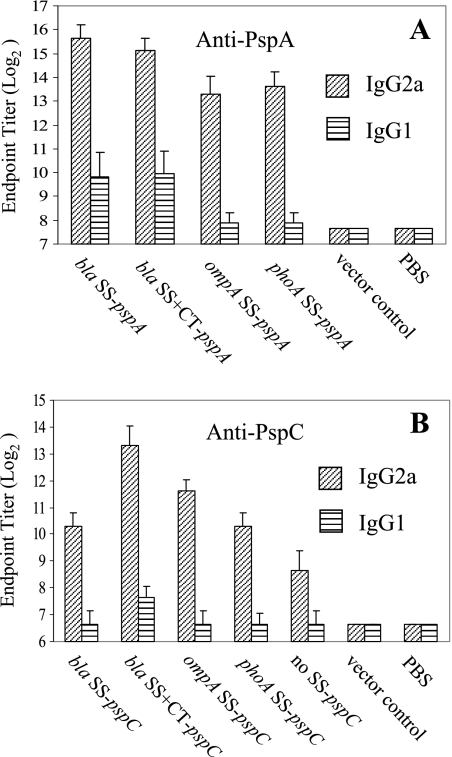
The immune responses to PspA and PspC were further examined by measuring the levels of IgG isotype subclasses IgG2a and IgG1 at 6 weeks after immunization. Th1 helper cells direct cell-mediated immunity and promote IgG class switching to IgG2a, and Th2 cells provide potent help for B-cell antibody production and promote IgG class switching to IgG1 (49, 57). Dominant Th1-type immune responses were observed for PspA and PspC in all vaccine strains (Fig. 6). The IgG2a titers for PspA and PspC were much higher than the IgG1 titers at 6 weeks, indicating that the Salmonella vaccines induced a strong cellular immune response against PspA and PspC. Dominant Th1 responses were also observed for the SOMP antigens in all strains, including vector controls (data not shown).
FIG. 6.
Serum IgG2a and IgG1 responses to PspA (A) and PspC (B) in mice. The IgG2a and IgG1 titers were determined for BALB/c mice orally immunized with 1 × 109 CFU of serovar Typhimurium χ9241 harboring different SS-pspA and SS-pspC fusions at 6 weeks after immunization. Values are the geometric mean titers ± standard deviations for eight mice.
T-lymphocyte function was evaluated by examining production of antigen-specific T-cell cytokines at 6 weeks after immunization. Th1 immune responses are associated with the production of cytokines, such as IFN-γ and tumor necrosis factor alpha, by T cells. Th2 immune responses produce more cytokines, such as IL-4 (49, 58). CFCs were assessed by a cytokine ELISPOT assay of spleen cells derived from Salmonella-immunized BALB/c mice. Lymphocytes were stimulated for 1 day with 5 μg/ml of recombinant PspA or PspC. ConA was used as a positive control for IFN-γ and IL-4 stimulation, and RPMI 1640 medium was used as a negative control.
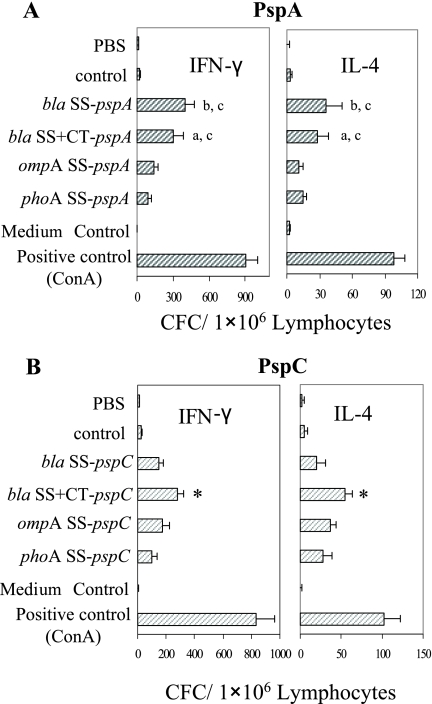
Strong antigen-specific T-lymphocyte activity was detected in all mice immunized with the PspA- and PspC-expressing RASV strains (Fig. 7). All the vaccine strains induced significantly higher antigen-specific IFN-γ and IL-4 cytokine-forming T cells than the vector and PBS controls (P < 0.01). The numbers of IFN-γ-specific CFCs from mice immunized with Salmonella PspA and Salmonella PspC strains were higher than the numbers of IL-4-specific CFCs.
FIG. 7.
T-lymphocyte function in mice immunized with serovar Typhimurium χ9241 harboring different SS-pspA (A) or SS-pspC (B) fusions. Antigen-specific IFN-γ and IL-4 cytokine-forming T cells were determined by an ELISPOT assay of spleen cells derived from Salmonella PspA or PspC-immunized BALB/c mice at 6 weeks after immunization. (A) All vaccine groups were significantly different from the pYA3493 and PBS controls (P < 0.05). Different letters (a, P < 0.01; b, P < 0.05) indicate significant differences in numbers of CFCs between vaccine groups at week 6. Groups that share letters are not significantly different (c, P > 0.05). (B) All vaccine groups were significantly different from the vector and PBS controls (P < 0.01). The asterisk indicates a significant difference in numbers of CFCs between the bla SS+CT-pspC vaccine group and other groups at week 6 (P < 0.01). Depicted are the mean numbers of CFCs/1 × 106 lymphocytes ± standard errors of the means. ConA and RPMI 1640 media were used as positive and negative controls, respectively, for IFN-γ and IL-4 stimulation.
RASV expressing bla SS-pspA induced the highest numbers of IFN-γ-specific CFCs (398 CFCs/106 cells) and IL-4-producing CFCs (36 CFCs/106 cells) at week 6. There was no significant difference in IFN-γ- and IL-4-specific CFC between mice immunized with bla SS-pspA- and bla SS+CT-pspA-expressing strains (P > 0.05), but both of these strains were more effective at eliciting CFCs than the RASV strains expressing ompA SS-pspA and phoA SS-pspA (P < 0.05).
S. enterica expressing bla SS+CT-pspC induced significantly more IFN-γ CFCs (398 CFCs/106 cells) and IL-4 CFCs (36 CFCs/106 cells) than the other Salmonella PspC strains (P < 0.05). But there were no significant differences between the remaining strains (P > 0.05). Overall, all of the immunized mice produced fewer IL-4-producing cells than IFN-γ-producing cells, indicating that a Th1 immune response was dominant for all the vaccine strains, consistent with our results from the serum IgG analysis.
Mucosal immune responses.
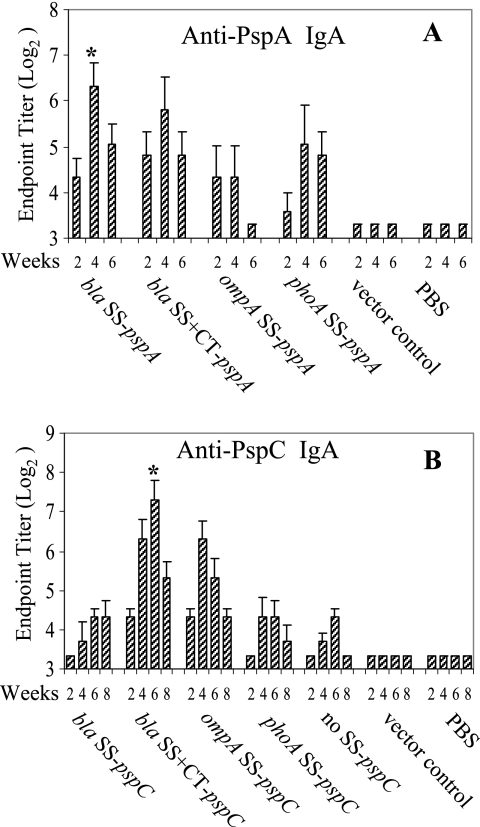
Mucosal immunity acts as the primary immune defense against natural infection by S. pneumoniae (66, 70). IgA responses to PspA and PspC in the vaginal fluids of immunized mice were detected by ELISA. All the Salmonella SS-pspA vaccines elicited anti-PspA IgA in vaginal washes (Fig. 8). Both of the bla fusions (bla SS+CT-pspA and bla SS-pspA) elicited similar IgA responses at week 4, and their titers were significantly higher than those of the phoA SS-pspA and ompA SS-pspA vaccinates (P < 0.05). All the Salmonella SS-pspC vaccines elicited anti-PspC IgA in vaginal washes (Fig. 8). However, bla SS+CT-pspC induced the highest IgA responses at week 6 (P < 0.05). These data indicate that the specific leader sequence can dramatically influence the mucosal immune response elicited against Salmonella-delivered antigens.
FIG. 8.
Mucosal IgA responses to PspA or PspC in vaginal secretions. BALB/c mice were orally immunized with S. enterica expressing PspA or PspC signal sequence fusion proteins. IgA endpoint titers in vaginal washes were measured by ELISA at the indicated time after immunization. The numbers listed below the x axis indicate weeks after immunization. Statistical significance was determined at week 4 (A) or week 6 (B). (A) Anti-PspA IgA responses. The asterisk indicates a significant difference from ompA SS and phoA SS groups (P < 0.05). (B) Anti-PspC IgA responses. The asterisk indicates that the bla SS+CT-pspC group was significantly different from other vaccine groups (P < 0.05). Titers are the geometric mean titers ± standard deviations for eight mice.
Evaluation of protective immunity.
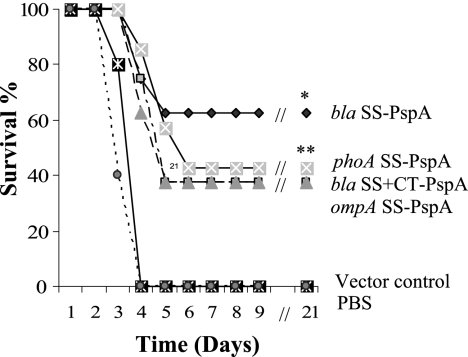
To ascertain whether the secretion signal sequences affected protective efficacy, we evaluated the abilities of PspA- and PspC-expressing RASVs to protect mice against pneumococcal infection in a sepsis model. BALB/c mice orally immunized with the PspA vaccine strains were challenged i.p. with 100 LD50 of S. pneumoniae WU2. All of the PspA vaccines provided significant protection against pneumococcal challenge compared with vector and PBS controls (Fig. 9). χ9241 (pYA4088; bla SS-pspA) provided the greatest efficacy compared with the control groups (P ≤ 0.002), with 62.5% survival, while the remaining strains all provided about the same level of protection as the controls (P ≤ 0.01), with 37.5 to 42.8% survival. The differences between vaccinate groups were not statistically significant (P > 0.05). Vaccination also increased the mean survival time until death compared to the levels for the controls. None of the mice immunized with the vector control strain were protected. Our results showed that the bla SS-pspA fusion provided the greatest efficacy, consistent with the immune response data.
FIG. 9.
Oral immunization of BALB/c mice with Salmonella PspA vaccines confers protection against S. pneumoniae challenge. BALB/c mice were orally immunized with 1 × 109 CFU of serovar Typhimurium strain χ9241 harboring different SS-pspA fusions. Mice were challenged i.p. with approximately 2 × 104 CFU of virulent S. pneumoniae WU2 (100 LD50) at week 6 after immunization. Mortality was monitored for 3 weeks after pneumococcal challenge. All vaccine groups were significantly different from the pYA3493 and PBS controls (*, P ≤ 0.002; **, P ≤ 0.01). There were no significant differences between vaccine groups (P > 0.05).
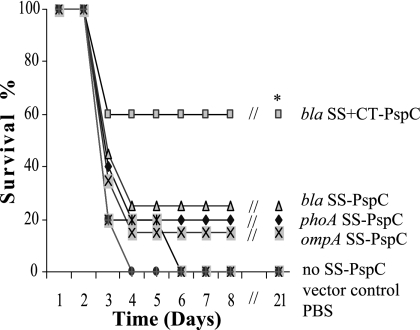
Because the pspC gene of S. pneumoniae WU2 is unrelated to the pspC expressed in our vaccine strains, vaccinated mice were challenged with 10 LD50 of S. pneumoniae strain D39 (6, 46). All vaccinates were partially protected against pneumococcal challenge (Fig. 10). Strain χ9241 (pYA4269) expressing the Bla SS+CT-PspC fusion protein showed significantly greater protection against challenge than the other vaccine strains and the control groups (P < 0.05). Mice vaccinated with the other vaccine strains were partially protected, but these results were not significantly different from those for the control group (P > 0.05). None of the mice immunized with the vector control strains survived after challenge. Taken together, these results showed that the RASV expressing the Bla SS+CT-PspC fusion was superior to the other constructs, as judged by induction of serum and mucosal antibody responses, stimulation of CFCs, and protection against virulent pneumococcal challenge. As was the case with the PspA vaccines, protection results are correlated with the immune response data; the vaccine that induced the highest antigen-specific titers provided the best protection against challenge.
FIG. 10.
Salmonella PspC vaccines confer protection against S. pneumoniae challenge to orally immunized BALB/c mice. BALB/c mice were orally immunized with 1 × 109 CFU of Salmonella strain χ9241 harboring different SS-pspC fusions. Mice were challenged i.p. with approximately 4 × 103 CFU of virulent S. pneumoniae D39 (10 LD50) at week 8 after immunization. Mortality was monitored for 3 weeks after pneumococcal challenge. Mice vaccinated with strain χ9241(pYA4029; bla SS+CT-pspC) showed significant protection against challenge compared to those vaccinated with other vaccine strains and the pYA3493 and PBS controls (*, P < 0.05). Other vaccine strains were not significantly different from controls (P > 0.05).
DISCUSSION
Successful delivery of protein antigens for vaccination purposes requires presentation in an optimal form and to suitable compartments of the host immune system. Some observations indicate that enveloped and secreted proteins are highly immunogenic and more readily interact with antigen-presenting cells because of their subcellular locations (58). In the development of attenuated Salmonella-based multivalent vaccines, a preferable system would have a recombinant antigen secreted from the cytoplasm (26, 43, 58).
A number of secretion systems have been utilized to secrete heterologous antigens in Salmonella vaccines. Gram-negative bacteria have evolved five different secretion systems, types I, II, III, IV, and V, to secrete proteins to the external environment, with the type II system being the most prevalent (2, 10). The E. coli α-hemolysin type I secretion system has been used in several Salmonella-based live vaccines to secrete antigens of bacterial, viral, and parasitic origins and has shown promising results in different animal models (25, 26). Type III secretion systems have been used for delivery into the major histocompatibility complex class I (MHC-I)-restricted antigen-processing pathway by attenuated Salmonella vaccines to elicit CD8+ T-cell responses (8, 44). Pneumococcal surface protein A (PspA) fusion with the type II secretion signal sequence Bla SS elicited higher PspA-specific immune responses than synthesis of the protein without the signal sequence and protected mice against virulent S. pneumoniae challenge (38). Interestingly, the greatest degree of protection was observed after vaccination with recombinant vaccine carriers when the heterologous antigens were secreted; it was either low or absent when the corresponding antigens remained in the cytoplasmic compartment (30, 31).
In this work, we performed a direct comparison of four T2SS signal sequences with two different protein antigens, PspA and PspC, evaluating the protein expression, immunogenicity, and efficacy observed when these antigens were expressed in RASV. The strain carrying pYA4088 (bla SS-pspA) produced more PspA than the other fusion plasmids and secreted more PspA into the periplasmic fraction than strains expressing the ompA SS and phoA SS fusions, indicating that bla SS is more efficient at directing PspA secretion than other signal sequences. The addition of the C-terminal end of bla (bla SS+CT) did not have much effect on PspA secretion (Fig. 2). Although there was less PspA in the periplasm than with the bla SS fusion, this was compensated for by the additional fraction of protein in the supernatant. In the case of PspC, however, the inclusion of the C-terminal end of bla did have an effect. The Salmonella strain carrying the bla SS+CT-pspC fusion showed the highest levels of protein expression and secretion, with approximately 60% of the protein being secreted to either the periplasm or the supernatant. PspC expression was drastically reduced when the protein was cloned without a signal sequence.
There are many advantages to directing protein antigens to the periplasm. It is generally believed that secreted proteins, particularly those that are surface exposed in their native states, will be correctly folded in periplasm space (35, 61), so the immune responses directed against these proteins are more likely to include relevant conformational epitopes. In addition, it has been shown that the stability of proteins can be affected by the cellular compartment in which they are located (61). Heterologous N-terminal signal sequences are often used in recombinant gene expression for the purpose of achieving translocation of the protein of interest, typically leading to increases in the expression levels of these proteins in E. coli (52, 56). The reason for this may be that the N-terminal sequence is important for stabilizing mRNA secondary structure or for enhancing translation initiation (56). Moreover, it has been reported that the codon immediately following the translation initiation codon (ATG) can have strong effects on translation initiation efficiency in E. coli, but this positive effect was highly sensitive to sequence alterations in the upstream ribosome binding site region (56, 59). The presence of an AAA (Lys) codon at position +2 has been shown to enhance protein expression. However, despite the facts that ompA SS and phoA SS in our constructions carry the AAA triplet (Lys) and the bla SS does not (AGT at position +2), all constructs appear to make comparable amounts of their respective fusion proteins (Fig. 2), indicating that other factors are influencing protein expression. Nevertheless, it may be possible to increase expression of the bla SS constructs by modifying the second codon to AAA. In any case, these results support our hypothesis that in the absence of additional information, finding the optimal signal sequence for a given antigen is an empirical process requiring a trial-and-error approach.
In our constructs, most of the secreted protein was directed to the periplasm, with only a fraction directed to the supernatant (Fig. 4). This result was not surprising for bla SS and phoA SS, since these signal sequences were derived from periplasmic proteins. These results showed that type II signal sequences are efficient in the translocation step from the cytoplasm to the periplasm. It is not clear how these antigens are transported from the periplasm to the outside medium. However, it has been shown that β-lactamase can be packaged in membrane vesicles and exported into the extracellular medium by Pseudomonas aeruginosa (11). Therefore, this seems to be a likely explanation for our results. Some studies have indicated that antigens secreted into the extracellular milieu, such as HlyA and the Bacillus anthracis protective antigen-ClyA fusion protein, are highly immunogenic. So it is possible that improving the secretion of protein from the periplasm to outside the cell could be of benefit in inducing the immune response. This possibility will also be addressed in future studies.
All four PspA-expressing RASV strains were able to induce strong serum IgG responses in immunized mice after a single oral dose (Fig. 5). pYA4088 (bla SS-pspA) induced a slightly higher response at all times, but the difference was not statistically significant until week 6 (P < 0.05). Although the amounts of secreted Bla SS-PspA and Bla SS+CT-PspA fusion proteins were similar, there was a significant difference in their induced antibody titers and a small difference in protective efficacy. The reason for these differences is not clear. However, it is possible that the addition of the bla C-terminal sequence resulted in the misfolding of one or more conformational epitopes in PspA.
In the development of pneumococcal vaccines, IgG isotype switching to a mixed or Th2-type humoral immune response, along with a mucosal IgA response, is preferred to prevent colonization of S. pneumoniae in the respiratory tract (pneumonia) or ear mucosa (otitis media) (66, 70). Previous studies have found a mixed Th1/Th2-type immune response for PspA fused with bla SS (39). In this study, a strong Th1-type immune response was observed based on the antibody subtype IgG2a/IgG1 ratios and antigen-specific T-cell cytokines IFN-γ and IL-4. One reason for this difference in results may be that the strain that we used, χ9241, has a different mode of attenuation than the strain used in the previous study, χ8501 (hisG Δcrp-28 Δasd16) (39), and different modes of attenuation can influence the immune response (22). An additional factor may be related to the timing of antigen expression. Salmonella bacteria normally survive within the phagosomal compartments of antigen-presenting cells; secretion of antigens mainly targets them into the MHC-II antigen-processing pathway. PspA or PspC expression in χ9241 was initially low because the inoculum was grown in the presence of arabinose, whereas expression in strain χ8501 was constitutive during growth. Therefore, there may not have been adequate levels of PspA or PspC secreted early during infection to trigger an MHC-II response. Antigen produced inside host cells would be primarily processed by the MHC-I pathway. But PspA or PspC could be presented by both MHC-II and MHC-I molecules in the χ8501 vaccine because PspA or PspC secretion would occur at every step during infection.
Mucosal immunity acts as a primary defense against natural infection by S. pneumoniae (66, 70). One clear advantage of antigen delivery by S. enterica is the stimulation of mucosal immunity (16, 21, 33, 54, 55). In keeping with our expectations, we found that our strains were able to induce strong mucosal responses against PspA and PspC (Fig. 8). Despite the fact that the mice were challenged i.p., there was a correlation between the level of mucosal IgA and the degree of protection. More importantly, we found that the induction of mucosal IgA was influenced by the signal sequence, with the bla SS and bla SS+CT fusions inducing the strongest responses. These mucosal responses should play an even bigger role in immunity against natural pneumococcal challenge, which occurs at the mucosal surface.
There were slight variations in expression results between the two antigens with regard to the signal sequences. The bla SS-pspA fusion was expressed at a higher level than the bla SS+CT-pspA fusion, while the opposite results were observed with PspC (Fig. 2). Although the differences in expression between fusions were less that twofold in both cases, the impacts on expression were different. For PspA, both fusions provided significant protection (Fig. 9), while in the case of PspC, only the bla SS+CT-pspC fusion was efficacious (Fig. 10). The differences in protection were also correlated with the anti-PspA and anti-PspC titers. Taken together, our data show that in both cases, the construct that produced the highest level of antigen expression induced the highest antigen-specific titer and the greatest protective efficacy. In addition, the optimal fusion sequence was different for each antigen, indicating that individual antigens should be tested empirically to determine the signal sequence that will yield the optimal results. The amount of antigen expression could serve as a reasonable indicator of which fusion partner is best, although it is likely that other parameters, such as growth rate and mode of attenuation (22), could also influence the immunogenicity.
Acknowledgments
We thank Kenneth Roland for critically reading and revising the manuscript, Melha Mellata and Shamaila Ashraf for editorial help, Susan Hollingshead (University of Alabama at Birmingham) for providing the S. pneumoniae strain WU2, Wei Kong for suggestions on the animal tests, Vidya Ananthnarayan for protein purifications, Bonnie Gunn and Javier Santander for animal test help, and Xiangmin Zhang and Ascencion Torres-Escobar for suggestions on plasmid construction.
This research project was supported by grants from the National Institutes of Health (R01 AI056289) and the Bill and Melinda Gates Foundation (no. 37863).
Editor: F. C. Fang
Footnotes
Published ahead of print on 5 May 2008.
REFERENCES
- 1.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochkareva, E. S., M. E. Solovieva, and A. S. Girshovich. 1998. Targeting of GroEL to SecA on the cytoplasmic membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 95478-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briles, D. E., S. K. Hollingshead, J. King, A. Swift, P. A. Braun, M. K. Park, L. M. Ferguson, M. H. Nahm, and G. S. Nabors. 2000. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J. Infect. Dis. 1821694-1701. [DOI] [PubMed] [Google Scholar]
- 4.Briles, D. E., S. K. Hollingshead, G. S. Nabors, J. C. Paton, and A. Brooks-Walter. 2000. The potential for using protein vaccines to protect against otitis media caused by Streptococcus pneumoniae. Vaccine 19(Suppl. 1)S87-S95. [DOI] [PubMed] [Google Scholar]
- 5.Briles, D. E., J. D. King, M. A. Gray, L. S. McDaniel, E. Swiatlo, and K. A. Benton. 1996. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine 14858-867. [DOI] [PubMed] [Google Scholar]
- 6.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 676533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatfield, S. N., I. G. Charles, A. J. Makoff, M. D. Oxer, G. Dougan, D. Pickard, D. Slater, and N. F. Fairweather. 1992. Use of the nirB promoter to direct the stable expression of heterologous antigens in Salmonella oral vaccine strains: development of a single-dose oral tetanus vaccine. Bio/Technology 10888-892. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L. M., G. Briones, R. O. Donis, and J. E. Galán. 2006. Optimization of the delivery of heterologous proteins by the Salmonella enterica serovar Typhimurium type III secretion system for vaccine development. Infect. Immun. 745826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, J. H., and S. Y. Lee. 2004. Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl. Microbiol. Biotechnol. 64625-635. [DOI] [PubMed] [Google Scholar]
- 10.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13581-588. [DOI] [PubMed] [Google Scholar]
- 11.Ciofu, O., T. J. Beveridge, J. Kadurugamuwa, J. Walther-Rasmussen, and N. Hoiby. 2000. Chromosomal β-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 459-13. [DOI] [PubMed] [Google Scholar]
- 12.Curtiss, R., III. 1965. Chromosomal aberrations associated with mutations to bacteriophage resistance in Escherichia coli. J. Bacteriol. 8928-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtiss, R., III, L. J. Charamella, C. M. Berg, and P. E. Harris. 1965. Kinetic and genetic analyses of d-cycloserine inhibition and resistance in Escherichia coli. J. Bacteriol. 901238-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtiss, R., III, X. Zhang, S. Y. Wanda, H. Y. Kang, V. Konjufca, Y. Li, B. Gunn, S. Wang, G. Scarpellini, and I. S. Lee. 2007. Induction of host immune responses using Salmonella-vectored vaccines, p. 297-313. In K. A. Brogden, N. Cornick, T. B. Stanton, Q. Zhang, L. K. Nolan, and M. J. Wannemuehler (ed.), Virulence mechanisms of bacterial pathogens, 4th ed. ASM Press, Washington, DC.
- 15.Curtiss, R., III, S. M. Kelly, and J. O. Hassan. 1993. Live oral avirulent Salmonella vaccines. Vet. Microbiol. 37397-405. [DOI] [PubMed] [Google Scholar]
- 16.Curtiss, R., III, S. M. Kelly, S. A. Tinge, C. O. Tacket, M. M. Levine, J. Srinivasan, and M. Koopman. 1994. Recombinant Salmonella vectors in vaccine development. Dev. Biol. Stand. 8223-33. [PubMed] [Google Scholar]
- 17.Curtiss, R., III, K. Nakayama, and S. M. Kelly. 1989. Recombinant avirulent Salmonella vaccine strains with stable maintenance and high level expression of cloned genes in vivo. Immunol. Investig. 18583-596. [DOI] [PubMed] [Google Scholar]
- 18.Curtiss, R., III, S.-Y. Wanda, X. Zhang, and B. Gunn. 2007. Salmonella vaccine vectors displaying regulated delayed in vivo attenuation to enhance immunogenicity, abstr. E-061, p 278. Abstr. 107th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 19.Curtiss, R., III. 2005. Antigen delivery systems: development of live recombinant attenuated bacterial antigen and DNA vaccine delivery vector vaccines, p. 1009-1037. In J. Mestecky, W. Strober, J. Bienenstock, J. R. McGhee, and L. Mayer (ed.), Mucosal immunology, 3rd ed., vol. 1. Academic Press, San Diego, CA. [Google Scholar]
- 20.Dodt, J., T. Schmitz, T. Schafer, and C. Bergmann. 1986. Expression, secretion and processing of hirudin in E. coli using the alkaline phosphatase signal sequence. FEBS Lett. 202373-377. [DOI] [PubMed] [Google Scholar]
- 21.Doggett, T. A., and R. Curtiss III. 1992. Delivery of antigens by recombinant avirulent Salmonella strains. Adv. Exp. Med. Biol. 327165-173. [DOI] [PubMed] [Google Scholar]
- 22.Dunstan, S. J., C. P. Simmons, and R. A. Strugnell. 1998. Comparison of the abilities of different attenuated Salmonella typhimurium strains to elicit humoral immune responses against a heterologous antigen. Infect. Immun. 66732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dvorak, H. F., B. K. Wetzel, and L. A. Heppel. 1970. Biochemical and cytochemical evidence for the polar concentration of periplasmic enzymes in a “minicell” strain of Escherichia coli. J. Bacteriol. 104543-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galan, J. E., K. Nakayama, and R. Curtiss III. 1990. Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 9429-35. [DOI] [PubMed] [Google Scholar]
- 25.Gentschev, I., I. Glaser, W. Goebel, D. J. McKeever, A. Musoke, and V. T. Heussler. 1998. Delivery of the p67 sporozoite antigen of Theileria parva by using recombinant Salmonella dublin: secretion of the product enhances specific antibody responses in cattle. Infect. Immun. 662060-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentschev, I., H. Mollenkopf, Z. Sokolovic, J. Hess, S. H. Kaufmann, and W. Goebel. 1996. Development of antigen-delivery systems, based on the Escherichia coli hemolysin secretion pathway. Gene 179133-140. [DOI] [PubMed] [Google Scholar]
- 27.Ghrayeb, J., H. Kimura, M. Takahara, H. Hsiung, Y. Masui, and M. Inouye. 1984. Secretion cloning vectors in Escherichia coli. EMBO J. 32437-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenwood, B. 1999. The epidemiology of pneumococcal infection in children in the developing world. Philos. Trans. R. Soc. Lond. B 354777-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hennequin, C., F. Porcheray, A. Waligora-Dupriet, A. Collignon, M. Barc, P. Bourlioux, and T. Karjalainen. 2001. GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology 14787-96. [DOI] [PubMed] [Google Scholar]
- 30.Hess, J., I. Gentschev, D. Miko, M. Welzel, C. Ladel, W. Goebel, and S. H. Kaufmann. 1996. Superior efficacy of secreted over somatic antigen display in recombinant Salmonella vaccine induced protection against listeriosis. Proc. Natl. Acad. Sci. USA 931458-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hess, J., L. Grode, I. Gentschev, J. Fensterle, G. Dietrich, W. Goebel, and S. H. Kaufmann. 2000. Secretion of different listeriolysin cognates by recombinant attenuated Salmonella typhimurium: superior efficacy of haemolytic over non-haemolytic constructs after oral vaccination. Microbes Infect. 21799-1806. [DOI] [PubMed] [Google Scholar]
- 32.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 685889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hopkins, S., J. P. Kraehenbuhl, F. Schödel, A. Potts, D. Peterson, P. de Grandi, and D. Nardelli-Haefliger. 1995. A recombinant Salmonella typhimurium vaccine induces local immunity by four different routes of immunization. Infect. Immun. 633279-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iannelli, F., M. R. Oggioni, and G. Pozzi. 2002. Allelic variation in the highly polymorphic locus pspC of Streptococcus pneumoniae. Gene 28463-71. [DOI] [PubMed] [Google Scholar]
- 35.Jonda, S., M. Huber-Wunderlich, R. Glockshuber, and E. Mossner. 1999. Complementation of DsbA deficiency with secreted thioredoxin variants reveals the crucial role of an efficient dithiol oxidant for catalyzed protein folding in the bacterial periplasm. EMBO J. 183271-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaderbhai, M. A., C. C. Ugochukwu, D. C. Lamb, and S. L. Kelly. 2000. Targeting of active human cytochrome P4501A1 (CYP1A1) to the periplasmic space of Escherichia coli. Biochem. Biophys. Res. Commun. 279803-807. [DOI] [PubMed] [Google Scholar]
- 37.Kadonaga, J. T., A. Pluckthun, and J. R. Knowles. 1985. Signal sequence mutants of β-lactamase. J. Biol. Chem. 26016192-16199. [PubMed] [Google Scholar]
- 38.Kang, H. Y., and R. Curtiss III. 2003. Immune responses dependent on antigen location in recombinant attenuated Salmonella typhimurium vaccines following oral immunization. FEMS Immunol. Med. Microbiol. 3799-104. [DOI] [PubMed] [Google Scholar]
- 39.Kang, H. Y., J. Srinivasan, and R. Curtiss III. 2002. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar Typhimurium vaccine. Infect. Immun. 701739-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karamyshev, A. L., Z. N. Karamysheva, A. V. Kajava, V. N. Ksenzenko, and M. A. Nesmeyanova. 1998. Processing of Escherichia coli alkaline phosphatase: role of the primary structure of the signal peptide cleavage region. J. Mol. Biol. 277859-870. [DOI] [PubMed] [Google Scholar]
- 41.Kaufmann, S. H., and J. Hess. 1999. Impact of intracellular location of and antigen display by intracellular bacteria: implications for vaccine development. Immunol. Lett. 6581-84. [DOI] [PubMed] [Google Scholar]
- 42.Khokhlova, O. V., and M. A. Nesmeianova. 2003. Interaction of SecB and SecA with the N-terminal region of mature alkaline phosphatase on its secretion in Escherichia coli. Mol. Biol. (Moscow) 37712-718. [PubMed] [Google Scholar]
- 43.Kochi, S. K., K. P. Killeen, and U. S. Ryan. 2003. Advances in the development of bacterial vector technology. Expert Rev. Vaccines 231-43. [DOI] [PubMed] [Google Scholar]
- 44.Konjufca, V., S. Y. Wanda, M. C. Jenkins, and R. Curtiss III. 2006. A recombinant attenuated Salmonella enterica serovar Typhimurium vaccine encoding Eimeria acervulina antigen offers protection against E. acervulina challenge. Infect. Immun. 746785-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koshland, D., and D. Botstein. 1980. Secretion of β-lactamase requires the carboxy end of the protein. Cell 20749-760. [DOI] [PubMed] [Google Scholar]
- 46.Kwon, H. Y., A. D. Ogunniyi, M. H. Choi, S. N. Pyo, D. K. Rhee, and J. C. Paton. 2004. The ClpP protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge. Infect. Immun. 725646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehnhardt, S., N. S. Pollitt, J. Goldstein, and M. Inouye. 1988. Modulation of the effects of mutations in the basic region of the OmpA signal peptide by the mature portion of the protein. J. Biol. Chem. 26310300-10303. [PubMed] [Google Scholar]
- 48.Nayak, A. R., S. A. Tinge, R. C. Tart, L. S. McDaniel, D. E. Briles, and R. Curtiss III. 1998. A live recombinant avirulent oral Salmonella vaccine expressing pneumococcal surface protein A induces protective responses against Streptococcus pneumoniae. Infect. Immun. 663744-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Garra, A., and N. Arai. 2000. The molecular basis of T helper 1 and T helper 2 cell differentiation. Trends Cell Biol. 10542-550. [DOI] [PubMed] [Google Scholar]
- 50.Rentier-Delrue, F., D. Swennen, and J. Martial. 1988. pIN-III-ompA secretion vectors: modification of the ompA signal peptide sequence for easier insert cloning. Nucleic Acids Res. 168726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts, M., J. Li, A. Bacon, and S. Chatfield. 1998. Oral vaccination against tetanus: comparison of the immunogenicities of Salmonella strains expressing fragment C from the nirB and htrA promoters. Infect. Immun. 663080-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saini, M., and S. Vrati. 2003. High-level synthesis of Johnson grass mosaic virus coat protein in Escherichia coli and its auto-assembly to form virus-like particles. Protein Expr. Purif. 2886-92. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Schödel, F., S. Kelly, S. Tinge, S. Hopkins, D. Peterson, D. Milich, and R. Curtiss III. 1996. Hybrid hepatitis B virus core antigen as a vaccine carrier moiety. II. Expression in avirulent Salmonella spp. for mucosal immunization. Adv. Exp. Med. Biol. 39715-21. [PubMed] [Google Scholar]
- 55.Sirard, J. C., F. Niedergang, and J. P. Kraehenbuhl. 1999. Live attenuated Salmonella: a paradigm of mucosal vaccines. Immunol. Rev. 1715-26. [DOI] [PubMed] [Google Scholar]
- 56.Sletta, H., A. Tondervik, S. Hakvag, T. E. Aune, A. Nedal, R. Aune, G. Evensen, S. Valla, T. E. Ellingsen, and T. Brautaset. 2007. The presence of N-terminal secretion signal sequences leads to strong stimulation of the total expression levels of three tested medically important proteins during high-cell-density cultivations of Escherichia coli. Appl. Environ. Microbiol. 73906-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spellberg, B., and J. E. Edwards, Jr. 2001. Type 1/type 2 immunity in infectious diseases. Clin. Infect. Dis. 3276-102. [DOI] [PubMed] [Google Scholar]
- 58.Spreng, S., G. Dietrich, and G. Weidinger. 2006. Rational design of Salmonella-based vaccination strategies. Methods 38133-143. [DOI] [PubMed] [Google Scholar]
- 59.Stenstrom, C. M., E. Holmgren, and L. A. Isaksson. 2001. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene 273259-265. [DOI] [PubMed] [Google Scholar]
- 60.Summers, R. G., and J. R. Knowles. 1989. Illicit secretion of a cytoplasmic protein into the periplasm of Escherichia coli requires a signal peptide plus a portion of the cognate secreted protein. Demarcation of the critical region of the mature protein. J. Biol. Chem. 26420074-20081. [PubMed] [Google Scholar]
- 61.Talmadge, K., and W. Gilbert. 1982. Cellular location affects protein stability in Escherichia coli. Proc. Natl. Acad. Sci. USA 791830-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanji, Y., J. Gennity, S. Pollitt, and M. Inouye. 1991. Effect of OmpA signal peptide mutations on OmpA secretion, synthesis, and assembly. J. Bacteriol. 1731997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tinge, S. A., and R. Curtiss III. 1990. Isolation of the replication and partitioning regions of the Salmonella typhimurium virulence plasmid and stabilization of heterologous replicons. J. Bacteriol. 1725266-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, S., Y. Li, G. Scarpellini, W. Kong, and R. Curtiss III. 2007. Salmonella vaccine vectors displaying regulated delayed antigen expression in vivo to enhance immunogenicity, abstr. E-064, p. 278. Abstr. 107th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 65.Witholt, B., M. Boekhout, M. Brock, J. Kingma, H. V. Heerikhuizen, and L. D. Leij. 1976. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal. Biochem. 74160-170. [DOI] [PubMed] [Google Scholar]
- 66.Wu, H. Y., M. H. Nahm, Y. Guo, M. W. Russell, and D. E. Briles. 1997. Intranasal immunization of mice with PspA (pneumococcal surface protein A) can prevent intranasal carriage, pulmonary infection, and sepsis with Streptococcus pneumoniae. J. Infect. Dis. 175839-846. [DOI] [PubMed] [Google Scholar]
- 67.Xu, R., P. Du, J. J. Fan, Q. Zhang, T. P. Li, and R. B. Gan. 2002. High-level expression and secretion of recombinant mouse endostatin by Escherichia coli. Protein Expr. Purif. 24453-459. [DOI] [PubMed] [Google Scholar]
- 68.Yang, X., B. J. Hinnebusch, T. Trunkle, C. M. Bosio, Z. Suo, M. Tighe, A. Harmsen, T. Becker, K. Crist, N. Walters, R. Avci, and D. W. Pascual. 2007. Oral vaccination with Salmonella simultaneously expressing Yersinia pestis F1 and V antigens protects against bubonic and pneumonic plague. J. Immunol. 1781059-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zekarias, B., H. Mo, and R. Curtiss III. 2008. Recombinant attenuated Salmonella enterica serovar Typhimurium expressing the carboxy-terminal domain of alpha toxin from Clostridium perfringens induces protective responses against necrotic enteritis in chickens. Clin. Vaccine Immunol. 15805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, Q., J. Bernatoniene, L. Bagrade, A. J. Pollard, T. J. Mitchell, J. C. Paton, and A. Finn. 2006. Serum and mucosal antibody responses to pneumococcal protein antigens in children: relationships with carriage status. Eur. J. Immunol. 3646-57. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, X., S. M. Kelly, W. S. Bollen, and R. Curtiss III. 1997. Characterization and immunogenicity of Salmonella typhimurium SL1344 and UK-1 Δcrp and Δcdt deletion mutants. Infect. Immun. 655381-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]