Abstract
Burkholderia pseudomallei is a facultative intracellular pathogen capable of surviving and replicating within eukaryotic cells. Recent studies have shown that B. pseudomallei Bsa type III secretion system 3 (T3SS-3) mutants exhibit vacuolar escape and replication defects in J774.2 murine macrophages. In the present study, we characterized the interactions of a B. pseudomallei bsaZ mutant with RAW 264.7 murine macrophages. Following uptake, the mutant was found to survive and replicate within infected RAW 264.7 cells over an 18-h period. In addition, high levels of tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), granulocyte-macrophage colony-stimulating factor (GM-CSF), and RANTES, but not IL-1α and IL-1β, were detected in culture supernatants harvested from infected monolayers. The subcellular location of B. pseudomallei within infected RAW 264.7 cells was determined, and as expected, the bsaZ mutant demonstrated early-vacuolar-escape defects. Interestingly, however, experiments also indicated that this mutant was capable of delayed vacuolar escape. Consistent with this finding, evidence of actin-based motility and multinucleated giant cell formation were observed between 12 and 18 h postinfection. Further studies demonstrated that a triple mutant defective in all three B. pseudomallei T3SSs exhibited the same phenotype as the bsaZ mutant, indicating that functional T3SS-1 and T3SS-2 did not appear to be responsible for the delayed escape phenotype in RAW 264.7 cells. Based upon these findings, it appears that B. pseudomallei may not require T3SS-1, -2, and -3 to facilitate survival, delayed vacuolar escape, and actin-based motility in activated RAW 264.7 macrophages.
Burkholderia pseudomallei, the causative agent of melioidosis, is a gram-negative bacillus responsible for a variety of illnesses in both humans and animals. In regions where it is endemic, this environmental saprophyte can be routinely isolated from moist soils, stagnant waters, and rice paddies (12). The typical routes of infection include inhalation, aspiration, and cutaneous inoculation (10, 12). The clinical signs of melioidosis range from subacute and chronic suppurative infections to acute pneumonias and fulminating septicemias that can be rapidly fatal (6, 12, 25). B. pseudomallei strains are intrinsically resistant to a broad spectrum of antibiotics, a feature that can often complicate the treatment of melioidosis (17). Even with prolonged antibiotic treatment relapse is not uncommon (9). The mortality rate for septicemic melioidosis remains high (∼50%), and no licensed vaccine is currently available (45). Due to the severe course of infection and its potential for use as a biological weapon, B. pseudomallei is currently listed as a Centers for Disease Control category B select agent (32, 43).
Several studies have shown that the virulence of B. pseudomallei is multifactorial. In particular, capsular polysaccharide, lipopolysaccharide, flagella, and type III secretion have been shown to contribute to virulence in animal models of infection (11, 13, 14, 44). Additionally, B. pseudomallei is a facultative intracellular pathogen that can invade, survive, and replicate in epithelial and phagocytic cell lines (20, 21, 29). Several studies have demonstrated that this organism can escape from endocytic vacuoles and enter the cytoplasm of host cells, where it can polymerize actin and subsequently spread cell to cell (2, 3, 22, 36-38). It is possible that the ability of this organism to survive and replicate within both phagocytic and nonphagocytic host cells may provide an environment protected from host immune defenses, as well as from antibiotic treatment. At present, however, the specific contribution of intracellular survival and spread to disease pathogenesis is not fully understood.
Type III secretion systems (T3SSs) are molecular syringes that inject effector proteins into eukaryotic cells in order to subvert host cell processes to the benefit of bacteria. Three T3SS gene clusters have been identified in the B. pseudomallei genome (1, 30, 38). T3SS-1 and T3SS-2 are homologous to plant pathogen T3SSs, while T3SS-3 (bsa locus) is an animal pathogen-like T3SS similar to T3SSs of other facultative intracellular gram-negative pathogens (30). Previous studies have demonstrated that both the Bsa secretion apparatus and translocated effector proteins contribute to early vacuolar escape and replication within murine macrophage-like cells (38). More recently, B. pseudomallei bimA and the Burkholderia mallei bimBCADE locus have been identified and shown to be involved in actin-based motility in J774.2 cells (33, 36). Specifically, it has been determined that the autotransported protein BimA can induce actin polymerization within macrophages (36, 37). It is presumed that intra- and intercellular spread via actin-based motility facilitates cell-to-cell fusion and multinucleated giant cell (MNGC) formation. This phenomenon has been observed in a variety of B. pseudomallei-infected mouse and human cell lines, as well as in vivo (2, 22, 35, 46). Although the specific mechanism(s) involved in B. pseudomallei-induced MNGC formation remains unclear, both the T3SS-3 effector protein BipB and the global regulatory factor RpoS have been implicated in this process (40, 41).
In this study, we utilized insertional inactivation mutagenesis to construct a B. pseudomallei bsaZ mutant and examined the interactions of this strain with RAW 264.7 murine macrophages. Burkholderia bsaZ is homologous to Salmonella enterica serovar Typhimurium spaS and is predicted to encode a structural component of the type III secretion apparatus belonging to the YscU/HrpY protein family (38). Consistent with previous findings, the vacuolar escape of the bsaZ mutant was delayed compared to that of the parent strain following uptake by activated RAW 264.7 cells. Interestingly, however, results of light microscopy, laser scanning confocal microscopy, and transmission electron microscopy (TEM) experiments demonstrated that the bsaZ mutant was capable of delayed vacuolar escape, actin-based intra- and intercellular motility, and MNGC formation. Additional studies revealed that a triple mutant defective in T3SS clusters 1, 2 and 3 also exhibited the same phenotype as the bsaZ mutant.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
The bacterial strains used in this study are shown in Table 1. Escherichia coli and B. pseudomallei were grown at 37°C on LB-Lennox (LBL) (Difco) agar or in LBL broth. Brucella agar (Difco) supplemented with 4% glycerol (BB4G) was used for plate count and sterility analyses. When appropriate, zeocin (Invitrogen) was used at the following concentrations: 25 μg/ml for E. coli and 100 μg/ml for B. pseudomallei. For macrophage assays, bacteria were subcultured 1:100 into LBL broth from overnight cultures and grown at 37°C for ∼3 h. Bacterial stocks were maintained at −80°C as 20% glycerol suspensions. All studies utilizing viable B. pseudomallei were conducted under biosafety level 3 containment. Unless stated otherwise, all reagents were purchased from Sigma.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| TOP10 | High-efficiency transformation strain; Zeos | Invitrogen |
| SM10 | Mobilizing strain; Kmr Pms Zeos | 34 |
| B. pseudomallei strains | ||
| DD503 | 1026b derivative; Δ(amrR-oprA) rpsL; Pmr Zeos | 26 |
| ZP1220 | DD503 derivative; bsaZ::pZPbsaZ; Pmr Zeor Kms | This study |
| ΔsctUBp3 | DD503 derivative; ΔbsaZ mutant; Kms | 44 |
| ΔsctUBp1,2,3 | DD503 derivative; T3SS-1, -2, -3 mutant; Kms | 44 |
| Plasmids | ||
| pEM7/Zeo | Sh ble cassette vector; Zeor | Invitrogen |
| pGSV3 | Mobilizable suicide vector; Gmr | 14 |
| pZSV | Mobilizable suicide vector; pGSV3 derivative; Zeor | This study |
| pZPbsaZ | pZSV containing a 486-bp PCR fragment internal to bsaZ; Zeor | This study |
| Primers | ||
| SV1 | 5′-AAATAGCGCCGAGCTTTGTC-3′ | This study |
| BPbsaZ-F1 | 5′-GTACGCGAATTCCTCGTCGATCTGACGCGCATCGCG-3′a | This study |
| BPbsaZ-R1 | 5′-GTACGCGCTAGCGTGGTACAGGAAGTATTCGACGGC-3′a | This study |
| BPbsaZ-MF | 5′-CATCGTCGCGCTCATCGTGATCGC-3′ | This study |
| MB1ori-P1 | 5′-GAAGATCCTTTGATCTTTTCTACGG-3′ | 4 |
Underlining indicates the EcoRI and NheI restriction sites in the linker regions.
Recombinant DNA techniques.
DNA manipulations were performed using standard methods. Restriction enzymes and Klenow DNA polymerase were purchased from Promega and used according to manufacturer's instructions. PCR was performed using the Expand high-fidelity PCR system (Roche Applied Science). PCR and restriction enzyme-digested products were purified using a QIAquick gel extraction kit (QIAGEN). Ligation reactions were performed using a Fast-Link quick ligase kit (Epicenter Technologies). Plasmids were purified using a QIAprep spin miniprep kit (QIAGEN). Genomic DNA was purified using a Wizard genomic DNA purification kit (Promega). Chemically competent E. coli TOP10 cells were transformed according to the manufacturer's instructions (Invitrogen). DNA sequencing was performed by ACGT Inc. (Wheeling, IL).
The plasmids and primers used in this study are shown in Table 1. The suicide vector pZSV was constructed as follows. Briefly, pGSV3 was digested with SacI to remove the gentamicin resistance marker (aacC1 open reading frame [ORF]) and treated with Klenow DNA polymerase to produce blunt ends. pEM7/Zeo was digested with XhoI/EcoRI and treated with Klenow DNA polymerase, which resulted in a blunt-ended ∼450-bp DNA fragment harboring the zeocin resistance marker (Sh ble ORF). The Sh ble ORF was ligated into the pGSV3 backbone, creating pZSV. The SV1 primer was utilized for DNA sequence verification.
Mutant construction.
A bsaZ mutant, B. pseudomallei ZP1220, was constructed using a previously described insertional inactivation protocol (4, 7, 14). Briefly, a 486-bp fragment internal to the bsaZ allele was PCR amplified from purified B. pseudomallei DD503 chromosomal DNA using the BPbsaZ-F1 and BPbsaZ-R1 primers (Table 1). The amplified product was then cloned into pZSV. The resulting suicide plasmid, pZPbsaZ, was mobilized into DD503 via conjugative mating using E. coli SM10 for 8 h at 37°C. Site-specific recombination of the plasmid into the DD503 chromosome was verified via PCR using the BPbsaZ-MF and MB1ori-P1 primers (Table 1).
Cell culture.
Murine macrophage cell line RAW 264.7 (= ATCC TIB-71) was obtained from the American Type Culture Collection (Rockville, MD). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (Invitrogen) and a standard mixture of antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin, and 250 μg/ml amphotericin B) at 37°C under an atmosphere containing 5% CO2. For macrophage survival assays, RAW 264.7 cells were resuspended in DMEM supplemented with fetal bovine serum (DMEM-10), transferred into 24-well tissue culture plates at a density of ∼1 × 106 cells/well, and incubated overnight.
Macrophage survival assays.
Bacterial uptake and survival were quantitated utilizing modified kanamycin protection assays as previously described (4). In brief, bacterial suspensions were added in triplicate to RAW 264.7 cells at a multiplicity of infection (MOI) of 10. The monolayers were incubated with the bacteria for 1 h and then washed twice with Hanks' balanced salt solution (Invitrogen) to remove extracellular bacteria. Infected RAW 264.7 cells were then incubated in fresh DMEM-10 containing 250 μg/ml kanamycin to suppress the growth of residual extracellular bacteria. Monolayers were lysed at 3, 6, 12, and 18 h postinfection with 0.25% (vol/vol) Triton X-100, and serial dilutions of the lysates were plated onto BB4G and incubated at 37°C for 48 h. Plate counting was then used to enumerate bacterial loads. B. pseudomallei ZP1220 recovered from survival assays was confirmed to have maintained the Zeor marker, as assessed by growth in LBL containing 100 μg/ml of zeocin.
Cytokine and chemokine assays.
Culture supernatants were harvested from infected monolayers at 6, 12, and 18 h postinfection and sterilized using 0.45-μm Millex syringe-driven filter units (Millipore Corporation). Aliquots of each preparation were plated onto BB4G and incubated at 37°C for at least 48 h to confirm sterility. The sterilized supernatants were then assayed for the production of tumor necrosis factor alpha (TNF-α), interleukin-1α (IL-1α), IL-1β, IL-6, RANTES, and granulocyte-macrophage colony-stimulating factor (GM-CSF) using murine SearchLight custom multiplex arrays (Pierce).
Cytotoxicity assays.
Filter-sterilized B. pseudomallei-infected RAW 264.7 cell supernatants were assayed for lactate dehydrogenase (LDH) release using a CytoTox 96 nonradioactive cytotoxicity assay kit (Promega). Maximum release was achieved by lysis of monolayers with Triton X-100 at a final concentration of 1% (vol/vol). The LDH released by uninfected cells was designated spontaneous release. Cytotoxicity was calculated as follows: % cytotoxicity = (test LDH release − spontaneous release)/(maximal release − spontaneous release).
Light microscopy.
Light micrographs of RAW 264.7 monolayers infected with B. pseudomallei at an MOI of 10 were obtained at 18 h postinfection. Monolayers were visualized using a Nikon Eclipse TE300 inverted microscope with a ×40 objective.
TEM.
For TEM, RAW 264.7 cells were grown overnight on 13-mm-diameter Thermanox coverslips (Nunc, Inc., Naperville, IL) and infected at an MOI of 10 as described above for the macrophage survival assays. At 1, 3, 6, 12, and 18 h postinfection, monolayers were fixed overnight with 2.5% gluteraldehyde-4% paraformaldehyde in 100 mM sodium cacodylate buffer (pH 7.2). Cells were then washed three times with 100 mM cacodylate buffer and incubated in 0.5% osmium tetroxide-0.8% potassium ferricyanide in 100 mM cacodylate buffer for 1 h. Subsequently, cells were washed three times with 100 mM cacodylate buffer, followed by incubation for 1 h in 1% tannic acid. Samples were subsequently rinsed once with 100 mM cacodylate buffer, washed twice with distilled water, and stained with 4% uranyl acetate for 1 h. Cells were then washed with distilled water and dehydrated using an ethanol gradient (50 to 100% ethanol, 5 min for each step). The last step (100% ethanol) was repeated three times, after which cells were infiltrated with a 3:1 ethanol-Spurr resin mixture for 3 h, a 1:1 ethanol-Spurr resin mixture overnight, and finally a 1:3 ethanol-Spurr resin mixture for 4 h. Following a final infiltration in 100% Spurr resin under a vacuum for 3 h, samples were embedded in BEEM capsules and polymerized at 65°C overnight. Ultrathin sections were obtained using an MT-7000 ultramicrotome (Research and Manufacturing Company, Inc., Tucson, AZ) and collected on 200-mesh copper grids. Images were obtained using a Philips CM-10 TEM (Philips, Eindhoven, Netherlands) with a bottom-mounted AMT camera. Chemicals used for TEM sample preparation were obtained from either Electron Microscopy Services (Hatfield, PA) or Ted Pella, Inc. (Redding, CA).
Immunofluorescence staining and confocal microscopy.
RAW 264.7 cells were grown overnight on 12-mm glass coverslips (Fisher Scientific) and infected at an MOI of 40 as described above for the macrophage survival assays. Monolayers were immunostained at room temperature essentially as previously described (5, 23). Briefly, at 6, 12, and 18 h postinfection, monolayers were fixed in 2.5% paraformaldehyde for 15 min, followed by extensive washing in phosphate-buffered saline (PBS). Cells were then permeabilized in PBS containing 10% normal goat serum (Invitrogen) and 0.1% (wt/vol) saponin (SS-PBS) for 20 min. Cells were incubated with rabbit polyclonal antiserum raised against formalin-fixed Burkholderia thailandensis diluted 1:50 in SS-PBS for 45 min, washed several times in PBS-0.05% (wt/vol) saponin, and then incubated with Alexa Fluor 568 goat anti-rabbit immunoglobulin G (IgG) (1:800; Invitrogen), Alexa Fluor 488 phalloidin (1:200; Invitrogen), and DRAQ5 (1:1,000; Alexis Biochemicals) in SS-PBS for 45 min. Following extensive washing in PBS, coverslips were mounted on glass slides using Mowiol (Calbiochem). Laser scanning confocal microscopy was performed with a Zeiss 510 META confocal imaging system equipped with an Ar, HeNe laser on an inverted Axiovert 200 M microscope using a ×63 oil objective (Carl Zeiss MicroImaging Inc.). Images (1024 × 1024 pixels) were acquired using Zeiss 510 META software (Carl Zeiss MicroImaging Inc.).
Statistical analyses.
Data points were plotted using GraphPad Prism 4 (GraphPad Software Inc., San Diego, CA).
RESULTS
B. pseudomallei ZP1220 survives, replicates, and induces MNGC formation in RAW 264.7 murine macrophages.
Murine macrophage cell line RAW 264.7 is commonly utilized to examine the interactions of B. pseudomallei with phagocytic cells. In the present study, we assessed the importance of T3SS-3 for survival and replication of B. pseudomallei within RAW 264.7 cells. In order to facilitate these studies, bsaZ, encoding a putative structural component of the T3SS-3 apparatus (38), was mutated in B. pseudomallei DD503 by insertional inactivation with pZPbsaZ (Table 1). PCR was used to confirm that the resulting strain, ZP1220, harbored a stable insertion of the suicide plasmid at the correct location.
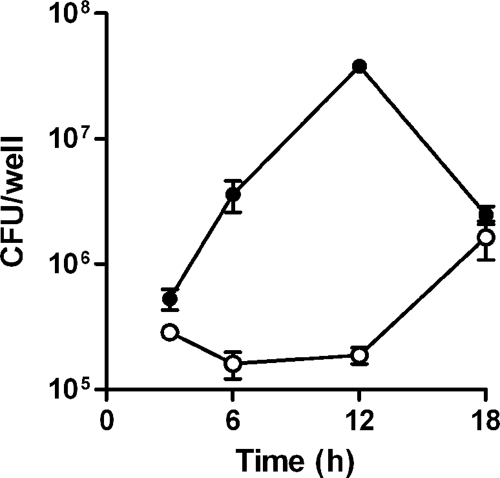
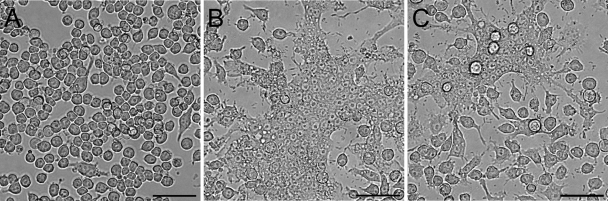
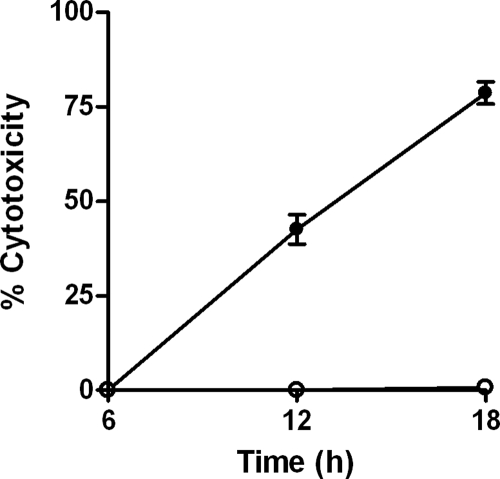
Survival of both DD503 and ZP1220 within RAW 264.7 cells was assessed at 3, 6, 12, and 18 h postinfection using modified kanamycin protection assays. At the initial time point (3 h), the two strains were taken up at comparable levels (Fig. 1). The results demonstrated that the numbers of intracellular DD503 bacteria increased up to 12 h but then decreased by 18 h postinfection (Fig. 1). In contrast, the intracellular loads of ZP1220 declined slightly over 12 h and then increased between 12 and 18 h postinfection (Fig. 1). These results correlated with the morphological appearance of the RAW 264.7 monolayers as determined by light microscopy. As shown in Fig. 2, by 18 h postinfection DD503-infected monolayers exhibited significant MNGC formation and monolayer sloughing compared to the control cells. In contrast, the ZP1220-infected monolayers appeared normal and intact, although MNGC formation was observed sporadically (Fig. 2C). To quantitate the integrity of the B. pseudomallei-infected RAW 264.7 cells, LDH release was measured. Consistent with the observed intracellular survival kinetics and light micrographs, cytotoxicity assays demonstrated that DD503 caused the RAW 264.7 cells to release significantly more LDH than ZP1220 at both 12 and 18 h postinfection (Fig. 3). These data indicated that B. pseudomallei ZP1220 was capable of survival and replication within RAW 264.7 cells. However, the kinetics of survival were different for the bsaZ mutant and the parent strain, as were the corresponding effects on the monolayer morphologies.
FIG. 1.
Survival characteristics of B. pseudomallei in RAW 264.7 cells. Monolayers were infected with B. pseudomallei DD503 (•) or ZP1220 (○) at an MOI of 10, and intracellular loads of bacteria were quantified at 3, 6, 12, and 18 h postinfection. The values are the means ± standard deviations of three independent experiments.
FIG. 2.
Light micrographs of RAW 264.7 cells infected with B. pseudomallei. The monolayers were visualized at 18 h postinfection and were infected as follows: (A) mock infected; (B) DD503 infected (MOI, 10); and (C) ZP1220 infected (MOI, 10). Bars = 50 μm. The micrographs are representative of at least three independent experiments.
FIG. 3.
Cellular integrity of RAW 264.7 cells infected with B. pseudomallei. Monolayers were infected with B. pseudomallei DD503 (•) or ZP1220 (○) at an MOI of 10. The percent cytotoxicity was determined by assaying for LDH release in culture supernatants at 6, 12, and 18 h postinfection. The values are the means ± standard deviations of three independent experiments performed in duplicate.
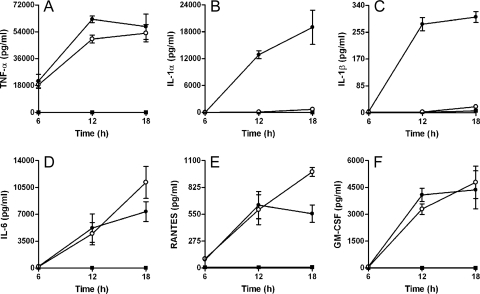
B. pseudomallei ZP1220 activates RAW 264.7 cells to produce proinflammatory cytokines and chemokines.
Activation of macrophages results in the production of a variety of cytokines and chemokines that are involved in the regulation of both innate and acquired host defense mechanisms. Previous studies have shown that B. pseudomallei can stimulate RAW 264.7 cells to produce high levels of TNF-α when they are infected at MOIs of ≥10 (42). To characterize the activation status of the DD503- and ZP1220-infected monolayers, culture supernatants harvested at 6, 12, and 18 h postinfection were assayed for the production of a variety of proinflammatory cytokines and chemokines. The results demonstrated that both DD503 and ZP1220 stimulated the RAW 264.7 cells to produce significant levels of TNF-α, IL-6, RANTES, and GM-CSF (Fig. 4A and D to F). Additionally, the kinetics of cytokine and chemokine production were similar for all time points assayed (Fig. 4A and D to F). In contrast, high levels of IL-1α and IL-1β were detected only for monolayers infected with DD503 (Fig. 4B and C). The presence of these cytokines correlated with the results of cytotoxicity assays demonstrating cell damage in DD503-infected monolayers but not in ZP1220-infected monolayers (Fig. 3). Based on these findings, it appears that the B. pseudomallei bsaZ mutant is capable of activating RAW 264.7 cells, but in a different way than parent strain DD503.
FIG. 4.
Cytokine and chemokine production by B. pseudomallei-infected RAW 264.7 cells. Culture supernatants from monolayers infected with B. pseudomallei DD503 (•) or ZP1220 (○) at an MOI of 10 or mock infected (▪) were harvested at 6, 12, and 18 h postinfection and assayed for the production of (A) TNF-α, (B) IL-1α, (C) IL-1β, (D) IL-6, (E) RANTES, and (F) GM-CSF. The values are the means ± standard deviations of three independent experiments performed in duplicate.
B. pseudomallei ZP1220 is capable of delayed vacuolar escape in RAW 264.7 cells.
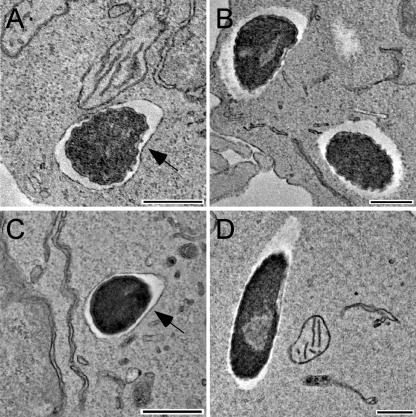
B. pseudomallei T3SS-3 mutants have previously been shown to exhibit vacuolar escape and replication defects in J774.2 murine macrophages (38). Interestingly, results obtained in this study demonstrate that by 18 h postinfection B. pseudomallei ZP1220 is capable of intracellular replication and MNGC formation in RAW 264.7 monolayers (Fig. 1 and 2), suggesting that there is vacuolar escape. To confirm the intracellular location of ZP1220 within RAW 264.7 cells at various time points postinfection, TEM was utilized. Consistent with previous studies, DD503 was observed within membrane-bound vesicles at 1 h postinfection but was able to escape by 3 h (Fig. 5A and B). TEM demonstrated that at 6 h postinfection ZP1220 remained within endocytic vesicles (Fig. 5C). At 12 h postinfection the majority of intracellular ZP1220 remained within membrane-bound compartments; however, individual bacteria that had escaped from endocytic vesicles were identified (Fig. 5D). By 18 h postinfection, most of the intracellular ZP1220 bacteria were observed to be in the cytoplasm of the RAW 264.7 cells (data not shown). Taken together, these results showed that the B. pseudomallei bsaZ mutant retains the ability to escape from endocytic vesicles but that this process is delayed by 6 to 12 h compared to the parent strain.
FIG. 5.
TEM micrographs of RAW 264.7 cells infected with B. pseudomallei. Fixed monolayers infected with B. pseudomallei DD503 (A and B) or ZP1220 (C and D) at an MOI of 10 were examined at 1 h (A), 3 h (B), 6 h (C), or 12 h (D) postinfection. The arrows indicate vacuolar membranes. Bars = 500 nm.
B. pseudomallei ZP1220 exhibits actin-based intra- and intercellular motility in RAW 264.7 cells.
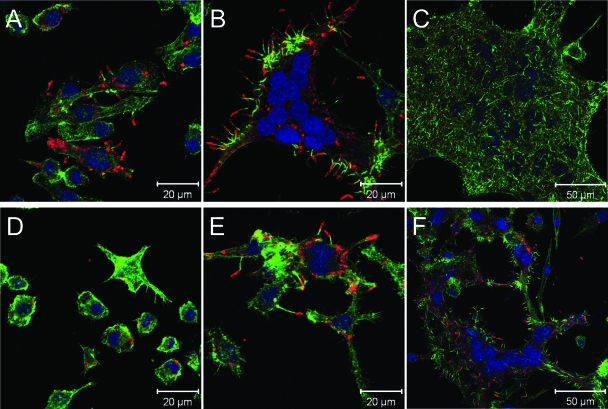
Similar to a variety of other intracellular pathogens, B. pseudomallei has been shown to stimulate actin polymerization within the cytoplasm of host cells (3, 22, 38). This process results in the formation of bacterium-containing host cell membrane protrusions that are thought to facilitate cell-to-cell spread (8). Previous studies have demonstrated that at 6 h postinfection, a B. pseudomallei bsaZ mutant is incapable of forming actin tails in J774.2 cells (38). This finding has been attributed to the inability of this mutant to escape from endocytic vesicles (38). To further support our finding that ZP1220 is capable of delayed vacuolar escape and entry into the cytoplasm, laser scanning confocal microscopy was utilized to assess whether this strain could polymerize actin within host cells. Both DD503 and ZP1220 were examined to determine their abilities to form actin tails in RAW 264.7 cells at 6, 12, and 18 h after inoculation. Consistent with previous reports, actin-based motility was evident in DD503-infected RAW 264.7 cells but not in ZP1220-infected monolayers at 6 h postinfection (Fig. 6A and D). MNGC formation was observed in DD503-infected monolayers at both 12 and 18 h postinfection (Fig. 6B and C). Interestingly, actin polymerization was apparent in ZP1220-infected RAW 264.7 cells at 12 h postinfection, and MNGC formation was observed by 18 h postinfection (Fig. 6E and F). These results were consistent with both light microscopy (Fig. 2) and TEM (Fig. 5) results and provided additional evidence that ZP1220 is capable of delayed escape from membrane-bound vacuoles and, upon entry into the cytoplasm, intra- and intercellular spread via actin-based motility.
FIG. 6.
Confocal micrographs of RAW 264.7 cells infected with B. pseudomallei. Fixed monolayers infected with B. pseudomallei DD503 (A to C) or ZP1220 (D to F) at an MOI of 40 were examined at 6 h (A and D), 12 h (B and E), and 18 h (C and F) postinfection. Bacteria were stained red with rabbit anti-B. thailandensis polyclonal sera and anti-rabbit IgG Alexa Fluor 568, actin was stained green with Alexa Fluor 488 phalloidin, and nuclei were stained blue with DRAQ5. The micrographs are representative of at least three independent experiments.
B. pseudomallei bsaZ deletion mutants ΔsctUBp3 and ΔsctUBp1,2,3 have the same delayed vacuolar escape phenotypes in RAW 264.7 cells as ZP1220.
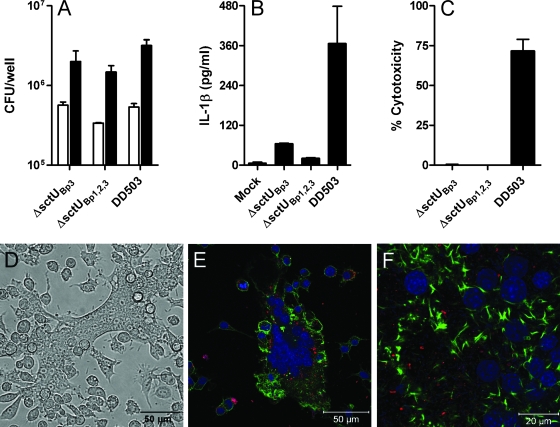
To confirm that the delayed escape phenotype exhibited by ZP1220 was not due to reversion, we assessed the interactions of B. pseudomallei ΔsctUBp3, a bsaZ deletion mutant constructed by allelic exchange (44), with RAW 264.7 cells. The results of kanamycin protection assays demonstrated that uptake (3 h) and survival (18 h) were similar for ZP1220 and ΔsctUBp3 (Fig. 1 and 7A). Additionally, like the findings for ZP1220, examination of ΔsctUBp3-infected monolayers by light microscopy revealed the presence of sporadic MNGC formation at 18 h postinfection (data not shown). Because vacuolar escape and actin-based motility are thought to be important for the formation of MNGCs, this observation was consistent with the ability of the organism to escape from membrane-bound compartments. Similar to the findings for ZP1220, low levels of IL-1β were detected is tissue culture supernatants of ΔsctUBp3-infected RAW 264.7 cells at 18 h postinfection (Fig. 7B). Likewise, ΔsctUBp3-infected monolayers exhibited very low levels of cytotoxicity compared to DD503-infected monolayers, as measured by LDH release at 18 h postinfection (Fig. 7C). These findings confirm that T3SS-3 may not be essential for escape into the cytoplasm of RAW 264.7 cells and suggest that other bacterial factors may be involved in late vacuolar escape.
FIG. 7.
Interactions of B. pseudomallei ΔsctUBp3, ΔsctUBp1,2,3, and DD503 with RAW 264.7 cells. Monolayers were infected at an MOI of 10 (A to D). (A) Bacterial uptake (open bars) and survival (filled bars) in RAW 264.7 cells were determined at 3 and 18 h postinfection, respectively. (B) Culture supernatants were assayed for IL-1β production at 18 h. (C) Percent cytotoxicity was determined by assaying for LDH release in culture supernatants at 18 h. The values are the means and standard deviations of three independent experiments performed in duplicate. (D) Light micrograph of ΔsctUBp1,2,3-infected RAW 264.7 cells at 18 h. Bar = 50 μm. (E and F) Confocal micrographs of RAW 264.7 cells infected with ΔsctUBp1,2,3 at an MOI of 40 at 18 h postinfection. Bacteria were stained red with rabbit anti-B. thailandensis polyclonal sera and anti-rabbit IgG Alexa Fluor 568, actin was stained green with Alexa Fluor 488 phalloidin, and nuclei were stained blue with DRAQ5. The micrographs are representative of at least two independent experiments.
In addition to the bsa locus, B. pseudomallei possesses two T3SSs similar to the plant-specific T3SS from Ralstonia solanacearum (1, 30). To determine if T3SS-1 or T3SS-2 could compensate for the loss of T3SS-3, a triple mutant with disruptions in all three T3SSs was assessed using RAW 264.7 cells. B. pseudomallei ΔsctUBp1,2,3 was constructed by allelic exchange and harbors stable deletions in the genes (sctU1, sctU2, and bsaZ) encoding the putative major inner membrane subunits of T3SS clusters 1, 2, and 3 and has previously been shown to be attenuated for virulence in the hamster model of melioidosis (44). The results of kanamycin protection assays demonstrated that both the uptake (3 h) and survival (18 h) of ΔsctUBp1,2,3 were similar to the uptake and survival observed for ZP1220, ΔsctUBp3, and DD503 (Fig. 1 and 7A). Analysis of cytokine levels revealed that similar to ZP1220, the ΔsctUBp1,2,3 mutant stimulated only low-level production of IL-1β at 18 h postinfection (Fig. 7B). Consistent with these findings, ΔsctUBp1,2,3-infected monolayers exhibited low levels of cytotoxicity compared to DD503-infected monolayers as quantitated by LDH release at 18 h postinfection (Fig. 7C). Similar to the findings for ZP1220, light microscopy revealed sporadic MNGC formation in ΔsctUBp1,2,3-infected monolayers (Fig. 7D). Likewise, confocal microscopy showed that ΔsctUBp1,2,3 retained the ability to induce actin polymerization and confirmed the presence of MNGCs. (Fig. 7E and F). Because vacuolar escape appears to be necessary for actin-based motility and subsequent formation of MNGCs, these observations were consistent with the ability of the organism to escape from membrane-bound compartments. Taken together, these results provide evidence that the components of T3SS-1 and T3SS-2 do not appear to be responsible for the late-escape phenotype observed in the bsaZ mutant and further support the notion that T3SS-3 may not be required for escape into the cytoplasm of host cells. Furthermore, these findings confirm that this phenotype observed with ZP1220 is not due to reversion and suggest that other bacterial factors likely facilitate delayed vacuolar escape in RAW 264.7 cells.
DISCUSSION
Survival and replication of B. pseudomallei within eukaryotic cells may be an important means by which this organism evades host innate and adaptive immune responses. Following uptake into phagocytic cells, B. pseudomallei is known to replicate intracellularly, escape endocytic vacuoles, and spread both intra- and intercellularly, resulting in the formation of MNGCs (20-22, 40). In addition, it has recently been shown that B. pseudomallei can induce pyroptosis (capase-1-dependent cell death) and, at high MOIs (≥50), oncosis (necrotic cell death) in macrophages (39). Previous reports have also demonstrated that T3SS-3 is important for early vacuolar escape in J774.2 macrophages, cell-to-cell spread, and virulence in the hamster model of melioidosis (38, 44). In the present study, we characterized the interactions of B. pseudomallei DD503 and derivatives of this strain deficient in T3SSs with RAW 264.7 macrophages. Our results showed that the kinetics of survival, replication, and bacterium-induced macrophage death were significantly different for DD503 and a bsaZ mutant. Modified kanamycin protection assays demonstrated that the number of intracellular DD503 bacteria initially increased but then decreased by 18 h postinfection. This decline in bacterial survival was likely due to bacterium-induced cell lysis of the monolayers and release of bacteria into the antibiotic-containing medium. In contrast to these findings, the number of intracellular ZP1220 bacteria initially declined slightly but then increased by 18 h postinfection. These findings are consistent with a report of Stevens et al., who showed that a bsaZ mutant did not replicate intracellularly in J774.2 cells within 12 h postinfection (38). However, by extending our studies to 18 h, we were able to observe intracellular replication and MNGC formation in ZP1220-infected RAW 264.7 monolayers, which suggested that there was delayed vacuolar escape. These results are consistent with a study that demonstrated survival of a Burkholderia cenocepacia T3SS mutant within RAW 264.7 cells (24, 31). In contrast, however, B. mallei T3SS-3 mutants have been shown to be rapidly cleared by J774.2 murine macrophages (31).
Previous studies have shown that the activation of RAW 264.7 cells by B. pseudomallei and B. mallei is significantly influenced by the MOI (5, 42). Results demonstrated that at an MOI of 10, both DD503 and the bsaZ mutant survived within activated RAW 264.7 cells. Additionally, the parent and mutant strains stimulated the production of similar levels of various proinflammatory cytokines and chemokines. The slight differences observed in cytokine levels (TNF-α, IL-6, RANTES, and GM-CSF) between DD503 and ZP1220 at 18 h were likely due to loss of monolayer integrity. Interestingly, however, only low levels of IL-1α and IL-1β were detected in the culture supernatants of ZP1220-infected RAW 264.7 cells. IL-1β is a secreted proinflammatory cytokine that is indicative of pyroptosis in infected macrophages (15, 16, 19, 27). IL-1α is a cell-associated cytokine that can be released from dying cells and is a key mediator of the inflammatory responses to necrotic cells (15, 18). The presence of these two cytokines in supernatants of DD503-infected RAW 264.7 cells but not in bsaZ mutant-infected RAW 264.7 cells directly correlated with the observed monolayer morphologies and quantitated cytotoxicities. These findings are consistent with a previous report of Sun et al. which suggested that T3SS-3 is necessary for IL-1β maturation and pyroptotic cell death of B. pseudomallei-infected THP-1 cells (39).
Microscopic studies have shown that B. pseudomallei can escape from endocytic vacuoles by 3 h following uptake by phagocytic cells (20, 21, 38). In addition, T3SS-3 mutants have been reported to be confined exclusively to endocytic vesicles associated with LAMP-1 in J774.2 cells at 6 h postinfection (38). Likewise, our results demonstrated that in RAW 264.7 cells, DD503 escaped from endocytic vacuoles by 3 h postinfection, while ZP1220 remained within membrane-bound vesicles at 6 h postinfection. However, results of TEM analyses at 12 and 18 h postinfection demonstrated that the bsaZ mutant retained the ability to escape from endocytic vacuoles. This result was unexpected based on previous reports but was consistent with the observed intracellular replication of ZP1220 in RAW 264.7 cells by 18 h postinfection. A recent study of Pilatz et al. demonstrated that a B. pseudomallei T3SS-3 mutant could multiply within HeLa cells, resulting in large vesicles packed with bacteria (28). It has been proposed that by 12 h postinfection these vacuoles become leaky, resulting in release of bacteria into the cytoplasm of host cells (28). This putative mechanism of escape is in contrast to the results obtained in this study, in which large vacuoles with multiple bacteria were rarely observed (data not shown). Most commonly, individual bacteria were observed free in the cytoplasm of RAW 264.7 cells at 12 and 18 h postinfection, suggesting a possible alternative mechanism for escape from endocytic vesicles.
Entry into the cytoplasm of host cells allows bacteria access to intracellular nutrients and actin pools. B. pseudomallei, like many other intracellular pathogens that replicate in the cytoplasm, displays actin-based motility in host cells. This strategy has been proposed to facilitate intra- and intercellular spread of bacteria without triggering extracellular immune responses within a host (8). Recent studies have demonstrated that B. pseudomallei BimA induces actin polymerization within murine macrophages (36, 37). Similarly, B. mallei bimA, bimB, bimC, bimE, and virAG mutants are deficient in actin tail formation in J774.2 cells (33). Previous studies have also shown that T3SS-3 mutants do not produce actin tails and membrane protrusions in J774.2 cells by 6 h postinfection (38). This is thought to be due to the fact that these mutants are confined to endocytic vesicles (38). More recently, it has been reported that a B. pseudomallei T3SS-3 mutant was observed in association with actin tails in the cytoplasm of HeLa cells at 12 h postinfection (28). Utilizing immunostaining in combination with confocal microscopy, we demonstrated that ZP1220 induced the formation of both actin tails and MNGCs in RAW 264.7 cells by 12 and 18 h postinfection. These findings were consistent with the observed escape from endocytic vesicles and intracellular replication by the B. pseudomallei T3SS-3 mutants. These data further suggest that a mechanism(s) other than the T3SS-3 facilitates the escape of B. pseudomallei from endocytic vesicles within RAW 264.7 cells.
B. pseudomallei possesses two additional T3SS gene clusters similar to the hrp-like T3SS gene clusters commonly associated with plant pathogens. Animal studies have shown that T3SS-1 and T3SS-2 do not significantly contribute to virulence in the hamster model of infection (44). To date, the ability of the various T3SSs in B. pseudomallei to cross-complement has not been investigated. Additionally, the involvement of T3SS-1 and T3SS-2 in late vacuolar escape has not been characterized. In the present study, RAW 264.7 macrophage survival assays utilizing a T3SS-1-T3SS-2-T3SS-3 triple mutant demonstrated that intracellular survival and replication, actin-based motility, and MNGC formation occurred by 18 h postinfection. These data suggested that under the conditions tested factors encoded by T3SS-1 or T3SS-2 could not account for the delayed vacuolar escape, actin-based motility, and MNGC formation observed in the ZP1220-infected RAW 264.7 monolayers. It is possible that T3SS-1 and T3SS-2 are involved in pathogenic or symbiotic interactions of B. pseudomallei with plants and the rhizosphere given that the environment is the principal reservoir for this organism. The roles, if any, of these T3SSs with respect to plants or other environmental hosts remain to be investigated.
In summary, we demonstrated that B. pseudomallei T3SS mutants displayed delayed vacuolar escape, actin-based motility, and MNGC formation kinetics in activated RAW 264.7 macrophages. In addition, it was shown that while RAW 264.7 monolayers infected with DD503 and ZP1220 produced similar levels of TNF-α, IL-6, RANTES, and GM-CSF, only the supernatants harvested from DD503-infected RAW 264.7 cells contained high levels of IL-1α and IL-1β. Further studies are necessary to determine the specific role of T3SS-3 with respect to cytokine production and macrophage death. Additional findings showed that the components of T3SS-1 and T3SS-2 did not appear to cross-complement for the delayed vacuolar escape phenotype of the bsaZ mutants in RAW 264.7 cell survival assays. While it is possible that the delayed vacuolar escape phenotype is related to residual activity associated with a defective T3SS-3, it is interesting to speculate that this phenotype may be due to a T3SS-independent mechanism(s). The delayed kinetics observed suggest that it is possible that the T3SSs encode functions that are redundant or that modulate virulence. The mechanism(s) utilized by B. pseudomallei to facilitate late vacuolar escape in RAW 264.7 cells remains to be defined.
Acknowledgments
We thank Joe Hinnebusch and Ted Hackstadt for critical reviews of the manuscript. We are grateful to Leigh Knodler for assistance with immunofluorescence staining and to Anita Mora for graphics support.
This research was supported by the Intramural Research Program of the NIH National Institute of Allergy and Infectious Diseases.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 28 April 2008.
REFERENCES
- 1.Attree, O., and I. Attree. 2001. A second type III secretion system in Burkholderia pseudomallei: who is the real culprit? Microbiology 1473197-3199. [DOI] [PubMed] [Google Scholar]
- 2.Boddey, J. A., C. J. Day, C. P. Flegg, R. L. Ulrich, S. R. Stephens, I. R. Beacham, N. A. Morrison, and I. R. Peak. 2007. The bacterial gene lfpA influences the potent induction of calcitonin receptor and osteoclast-related genes in Burkholderia pseudomallei-induced TRAP-positive multinucleated giant cells. Cell. Microbiol. 9514-531. [DOI] [PubMed] [Google Scholar]
- 3.Breitbach, K., K. Rottner, S. Klocke, M. Rohde, A. Jenzora, J. Wehland, and I. Steinmetz. 2003. Actin-based motility of Burkholderia pseudomallei involves the Arp 2/3 complex, but not N-WASP and Ena/VASP proteins. Cell. Microbiol. 5385-393. [DOI] [PubMed] [Google Scholar]
- 4.Brett, P. J., M. N. Burtnick, D. S. Snyder, J. G. Shannon, P. Azadi, and F. C. Gherardini. 2007. Burkholderia mallei expresses a unique lipopolysaccharide mixture that is a potent activator of human Toll-like receptor 4 complexes. Mol. Microbiol. 63379-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brett, P. J., M. N. Burtnick, H. Su, V. Nair, and F. C. Gherardini. 2008. iNOS activity is critical for the clearance of Burkholderia mallei from infected RAW 264.7 murine macrophages. Cell. Microbiol. 10487-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brett, P. J., and D. E. Woods. 2000. Pathogenesis of and immunity to melioidosis. Acta Trop. 74201-210. [DOI] [PubMed] [Google Scholar]
- 7.Burtnick, M. N., P. J. Brett, and D. E. Woods. 2002. Molecular and physical characterization of Burkholderia mallei O antigens. J. Bacteriol. 184849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlsson, F., and E. J. Brown. 2006. Actin-based motility of intracellular bacteria, and polarized surface distribution of the bacterial effector molecules. J. Cell Physiol. 209288-296. [DOI] [PubMed] [Google Scholar]
- 9.Chaowagul, W., Y. Suputtamongkol, D. A. Dance, A. Rajchanuvong, J. Pattara-arechachai, and N. J. White. 1993. Relapse in melioidosis: incidence and risk factors. J. Infect. Dis. 1681181-1185. [PubMed] [Google Scholar]
- 10.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chua, K. L., Y. Y. Chan, and Y. H. Gan. 2003. Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 711622-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Currie, B. J., D. A. Fisher, D. M. Howard, J. N. Burrow, D. Lo, S. Selva-Nayagam, N. M. Anstey, S. E. Huffam, P. L. Snelling, P. J. Marks, D. P. Stephens, G. D. Lum, S. P. Jacups, and V. L. Krause. 2000. Endemic melioidosis in tropical northern Australia: a 10-year prospective study and review of the literature. Clin. Infect. Dis. 31981-986. [DOI] [PubMed] [Google Scholar]
- 13.DeShazer, D., P. J. Brett, and D. E. Woods. 1998. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 301081-1100. [DOI] [PubMed] [Google Scholar]
- 14.DeShazer, D., D. M. Waag, D. L. Fritz, and D. E. Woods. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 30253-269. [DOI] [PubMed] [Google Scholar]
- 15.Dinarello, C. A. 1997. Interleukin-1. Cytokine Growth Factor Rev. 8253-265. [DOI] [PubMed] [Google Scholar]
- 16.Dinarello, C. A. 2005. Interleukin-1beta. Crit. Care Med. 33S460-462. [DOI] [PubMed] [Google Scholar]
- 17.Eickhoff, T. C., J. V. Bennett, P. S. Hayes, and J. Feeley. 1970. Pseudomonas pseudomallei: susceptibility to chemotherapeutic agents. J. Infect. Dis. 12195-102. [DOI] [PubMed] [Google Scholar]
- 18.Elkon, K. B. 2007. IL-1alpha responds to necrotic cell death. Nat. Med. 13778-780. [DOI] [PubMed] [Google Scholar]
- 19.Fink, S. L., and B. T. Cookson. 2007. Pyroptosis and host cell death responses during Salmonella infection. Cell. Microbiol. 92562-2570. [DOI] [PubMed] [Google Scholar]
- 20.Harley, V. S., D. A. Dance, B. S. Drasar, and G. Tovey. 1998. Effects of Burkholderia pseudomallei and other Burkholderia species on eukaryotic cells in tissue culture. Microbios 9671-93. [PubMed] [Google Scholar]
- 21.Jones, A. L., T. J. Beveridge, and D. E. Woods. 1996. Intracellular survival of Burkholderia pseudomallei. Infect. Immun. 64782-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kespichayawattana, W., S. Rattanachetkul, T. Wanun, P. Utaisincharoen, and S. Sirisinha. 2000. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect. Immun. 685377-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knodler, L. A., B. A. Vallance, M. Hensel, D. Jackel, B. B. Finlay, and O. Steele-Mortimer. 2003. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol. Microbiol. 49685-704. [DOI] [PubMed] [Google Scholar]
- 24.Lamothe, J., K. K. Huynh, S. Grinstein, and M. A. Valvano. 2007. Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacteria-containing vacuoles. Cell. Microbiol. 940-53. [DOI] [PubMed] [Google Scholar]
- 25.Leelarasamee, A., and S. Bovornkitti. 1989. Melioidosis: review and update. Rev. Infect. Dis. 11413-425. [DOI] [PubMed] [Google Scholar]
- 26.Moore, R. A., D. DeShazer, S. Reckseidler, A. Weissman, and D. E. Woods. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okemoto, K., K. Kawasaki, K. Hanada, M. Miura, and M. Nishijima. 2006. A potent adjuvant monophosphoryl lipid A triggers various immune responses, but not secretion of IL-1beta or activation of caspase-1. J. Immunol. 1761203-1208. [DOI] [PubMed] [Google Scholar]
- 28.Pilatz, S., K. Breitbach, N. Hein, B. Fehlhaber, J. Schulze, B. Brenneke, L. Eberl, and I. Steinmetz. 2006. Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect. Immun. 743576-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruksachartvuthi, S., N. Aswapokee, and K. Thankerngpol. 1990. Survival of Pseudomonas pseudomallei in human phagocytes. J. Med. Microbiol. 31109-114. [DOI] [PubMed] [Google Scholar]
- 30.Rainbow, L., C. A. Hart, and C. Winstanley. 2002. Distribution of type III secretion gene clusters in Burkholderia pseudomallei, B. thailandensis and B. mallei. J. Med. Microbiol. 51374-384. [DOI] [PubMed] [Google Scholar]
- 31.Ribot, W. J., and R. L. Ulrich. 2006. The animal pathogen-like type III secretion system is required for the intracellular survival of Burkholderia mallei within J774.2 macrophages. Infect. Immun. 744349-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schell, M. A., R. L. Ulrich, W. J. Ribot, E. E. Brueggemann, H. B. Hines, D. Chen, L. Lipscomb, H. S. Kim, J. Mrazek, W. C. Nierman, and D. Deshazer. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 641466-1485. [DOI] [PubMed] [Google Scholar]
- 34.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1784-791. [Google Scholar]
- 35.Sirikulchayanonta, V., and T. Subhadrabandhu. 1994. Melioidosis. Another etiology of granulomatous osteomyelitis. Report of 2 cases. Clin. Orthop. Relat. Res. 308183-186. [PubMed] [Google Scholar]
- 36.Stevens, J. M., R. L. Ulrich, L. A. Taylor, M. W. Wood, D. Deshazer, M. P. Stevens, and E. E. Galyov. 2005. Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J. Bacteriol. 1877857-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens, M. P., J. M. Stevens, R. L. Jeng, L. A. Taylor, M. W. Wood, P. Hawes, P. Monaghan, M. D. Welch, and E. E. Galyov. 2005. Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol. Microbiol. 5640-53. [DOI] [PubMed] [Google Scholar]
- 38.Stevens, M. P., M. W. Wood, L. A. Taylor, P. Monaghan, P. Hawes, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46649-659. [DOI] [PubMed] [Google Scholar]
- 39.Sun, G. W., J. Lu, S. Pervaiz, W. P. Cao, and Y. H. Gan. 2005. Caspase-1 dependent macrophage death induced by Burkholderia pseudomallei. Cell. Microbiol. 71447-1458. [DOI] [PubMed] [Google Scholar]
- 40.Suparak, S., W. Kespichayawattana, A. Haque, A. Easton, S. Damnin, G. Lertmemongkolchai, G. J. Bancroft, and S. Korbsrisate. 2005. Multinucleated giant cell formation and apoptosis in infected host cells is mediated by Burkholderia pseudomallei type III secretion protein BipB. J. Bacteriol. 1876556-6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Utaisincharoen, P., S. Arjcharoen, K. Limposuwan, S. Tungpradabkul, and S. Sirisinha. 2006. Burkholderia pseudomallei RpoS regulates multinucleated giant cell formation and inducible nitric oxide synthase expression in mouse macrophage cell line (RAW 264.7). Microb. Pathog. 40184-189. [DOI] [PubMed] [Google Scholar]
- 42.Utaisincharoen, P., N. Tangthawornchaikul, W. Kespichayawattana, P. Chaisuriya, and S. Sirisinha. 2001. Burkholderia pseudomallei interferes with inducible nitric oxide synthase (iNOS) production: a possible mechanism of evading macrophage killing. Microbiol. Immunol. 45307-313. [DOI] [PubMed] [Google Scholar]
- 43.Voskuhl, G. W., P. Cornea, M. S. Bronze, and R. A. Greenfield. 2003. Other bacterial diseases as a potential consequence of bioterrorism: Q fever, brucellosis, glanders, and melioidosis. J. Okla. State Med. Assoc. 96214-217. [PubMed] [Google Scholar]
- 44.Warawa, J., and D. E. Woods. 2005. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol. Lett. 242101-108. [DOI] [PubMed] [Google Scholar]
- 45.White, N. J. 2003. Melioidosis. Lancet 3611715-1722. [DOI] [PubMed] [Google Scholar]
- 46.Wong, K. T., S. D. Puthucheary, and J. Vadivelu. 1995. The histopathology of human melioidosis. Histopathology 2651-55. [DOI] [PubMed] [Google Scholar]