Abstract
Streptococcus mutans, a major oral pathogen responsible for dental caries formation, possesses a variety of mechanisms for survival in the human oral cavity, where the conditions of the external environment are diverse and in a constant state of flux. The formation of biofilms, survival under conditions of acidic pH, and production of mutacins are considered to be important virulence determinants displayed by this organism. Biofilm formation is facilitated by the production of GbpC, an important cell surface-associated protein that binds to glucan, an adhesive polysaccharide produced by the organism itself. To better understand the nature of the environmental cues that induce GbpC production, we examined the roles of 14 sensor kinases in the expression of gbpC in S. mutans strain UA159. We found that only the LiaS sensor kinase regulates gbpC expression, while the other sensor kinases had little or no effect on gbpC expression. We also found that while LiaS negatively regulates gbpC expression, the inactivation of its cognate response regulator, LiaR, does not appear to affect the expression of gbpC. Since both gbpC expression and mutacin IV production are regulated by a common regulatory network, we also tested the effect of the liaS mutation on mutacin production and found that LiaS positively regulates mutacin IV production. Furthermore, reverse transcription-PCR analysis suggests that LiaS does so by regulating the expression of nlmA, which encodes a peptide component of mutacin IV, and nlmT, which encodes an ABC transporter. As with the expression of gbpC, LiaR did not have any apparent effect on mutacin IV production. Based on the results of our study, we speculate that LiaS is engaged in cross talk with one or more response regulators belonging to the same family as LiaR, enabling LiaS to regulate the expression of several genes coding for virulence factors.
Two-component signal transduction systems (TCS) are one of the primary mechanisms by which bacteria sense and respond to external stimuli. Various cellular processes are regulated by TCS, including biofilm formation, bacteriocin production, chemotaxis, competence development, sporulation, and stress tolerance responses (for recent reviews, see references 11, 48, and 52). Some of the TCS are also essential for the survival of cells (14, 29). In pathogenic bacteria, TCS are one of the key signaling systems that regulate the expression of genes that are necessary for pathogenesis (6, 11). TCS are typically composed of a membrane-anchored sensor histidine kinase and its cognate effector, a response regulator protein that resides in the cytoplasm. The recognition of specific environmental cues leads to the autophosphorylation of the senor kinase at a conserved histidine residue (48). The phosphate group is then transferred from the sensor kinase to a conserved aspartic acid residue present on the cognate response regulator. Response regulators typically function as transcriptional regulators with DNA binding activity and, upon phosphorylation, modulate the expression of certain target genes. The number of TCS varies greatly among bacteria, ranging from a few TCS to hundreds (2, 45).
Streptococcus mutans, which resides in the human oral cavity, is the primary causative organism of dental caries, an important and economically costly human disease. This pathogenic bacterium exhibits a biofilm lifestyle when residing in dental plaque, a highly dense and diverse bacterial community that forms on the tooth surface (28). S. mutans displays several unique mechanisms that enable it to survive and persist in dental plaque (3, 24). For example, the ability to produce and bind to extracellular polysaccharides such as glucan allows S. mutans to colonize and to maintain a dominant presence in the oral plaque (5). Glucan is a polymer of glucose that is synthesized by S. mutans from the disaccharide sucrose ingested by its host through the activity of various glucosyltransferases (Gtfs) (5, 54). A significant constituent of the plaque biofilm is water-insoluble glucans, which are formed primarily through the activity of GtfB/GtfC (54). S. mutans also expresses at least four glucan binding proteins (Gbps) on the cell surface, which directly binds to the glucans produced by the bacterium (4, 5, 41, 44, 47). The adhesive nature of glucans, along with the surface-associated Gbps, facilitates the adherence and accumulation of this bacterium as a stable biofilm on the tooth surface.
Among all the Gbps, GbpC appears to be the most important, since it is directly involved in biofilm formation and cariogenicity of S. mutans (41). GbpC has also been shown to be involved in the rapid dextran-dependent aggregation of bacteria (41). The dextran-dependent aggregation phenotype is growth phase independent and was not observed under standard growth conditions; however, this phenotype was induced in cells grown under various stress conditions (41, 43). The results of a recent expression study suggest that the presence of sucrose in growth medium induces low levels of repression of gbpC expression, while the presence of other carbohydrates had little to no effect on gbpC expression (7). Maximal expression of gbpC is observed at the mid-exponential growth phase, but the gbpC transcript produced is unstable, with a short half-life of less than 2 min (7). Expression of the gbpC gene is also dependent on the growth temperature as well as the pH of the growth medium (7). Moreover, the expression of gbpC is also under the transcriptional control of CovR (7, 42), a global response regulator that regulates virulence factors in many streptococci. CovR represses the expression of gbpC by directly binding to the promoter region and preventing binding by RNA polymerase.
While we are beginning to gain further insight into the regulation of gbpC expression, the environmental signals that induce the expression of gbpC and the sensor kinase(s) that receives the signal are currently unknown. S. mutans encodes a total of 14 TCS systems (8), some of which are involved in the synthesis of glucans and the expression of Gbps (7, 10, 21). In this communication, we studied the roles of all 14 sensor kinases in the regulation of gbpC expression and found that LiaS represses gbpC transcription. Furthermore, our studies revealed that LiaS also activates the production of mutacin IV, which is also under the control of a common regulatory network along with gbpC.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strain DH5α was grown in Luria-Bertani medium supplemented, when necessary, with ampicillin (100 μg/ml), erythromycin (300 μg/ml), kanamycin (100 μg/ml), and/or spectinomycin (100 μg/ml). S. mutans UA159 and its derivatives were used for all genetic experiments. The various S. mutans strains were routinely grown in Todd-Hewitt medium (BBL; Becton Dickson) supplemented with 0.2% yeast extract (THY). When necessary, erythromycin (10 μg/ml), chloramphenicol (10 μg/ml), kanamycin (300 μg/ml), and/or spectinomycin (300 μg/ml) was added to the sterile growth medium.
Construction of sensor kinase deletion strains.
The wild-type transcriptional reporter strain IBS131, which contains the PgbpC-gusA fusion construct inserted at the SMu1405 locus of its genome (7), was used for the in vivo analysis of the inactivated sensor kinase genes. Internal fragments (approximately 500 bp) corresponding to the open reading frames of 14 putative sensor kinase genes were cloned into pBSKEry (9) to generate 14 mutagenic plasmids; these plasmids were previously described in detail by Biswas et al. (8) and were used to systematically inactivate the corresponding sensor kinase genes in IBS131 via natural transformation. The resulting mutant strains were confirmed by PCR analysis using flanking primers as described previously (8). The names of the various strains generated, along with the corresponding mutated sensor kinases, are listed in Table 1.
TABLE 1.
S. mutans strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| UA159 | Wild type, serotype c | R.A. Burne, University of Florida |
| IBS131 | UA159 with Pgbpc-gusA reporter at SMu1405 locus | 7 |
| IBS148 | UA159 SMu486 (liaS)::aad9 | This study |
| IBS149 | UA159 SMu487 (liaR)::aad9 | This study |
| IBS183 | UA159 SMu486-SMu487 (liaSR)::aad9 | This study |
| IBS151 | IBS148 with Pgbpc-gusA reporter at SMu1405 locus | This study |
| IBS152 | IBS149 Pgbpc-gusA reporter at SMu1405 locus | This study |
| IBS155 | IBS183 Pgbpc-gusA reporter at SMu1405 locus | This study |
| IBS326 | IBS131 with SMu45 inactivated by pIB12 | This study |
| IBS339 | IBS131 with SMu486 inactivated by pIB14 | This study |
| IBS328 | IBS131 with SMu577 inactivated by pIB15 | This study |
| IBS329 | IBS131 with SMu660 inactivated by pIB16 | This study |
| IBS330 | IBS131 with SMu928 inactivated by pIB17 | This study |
| IBS331 | IBS131 with SMu1009 inactivated by pIB18 | This study |
| IBS332 | IBS131 with SMu1037 inactivated by pIB19 | This study |
| IBS333 | IBS131 with SMu1128 inactivated by pIB20 | This study |
| IBS334 | IBS131 with SMu1145 inactivated by pIB21 | This study |
| IBS335 | IBS131 with SMu1516 inactivated by pIB22 | This study |
| IBS336 | IBS131 with SMu1548 inactivated by pIB23 | This study |
| IBS337 | IBS131 with SMu1814 inactivated by pIB24 | This study |
| IBS338 | IBS131 with SMu1916 inactivated by pIB25 | This study |
| IBS327 | IBS131 with SMu1965 inactivated by pIB13 | This study |
To delete the SMu486 (liaS) locus, a 1.54-kb fragment spanning the entire SMu486 region was PCR amplified from UA159 genomic DNA using primers Bam-Smu486-F1 and Bam-Smu487-R1. This fragment was cloned into the pGemT-Easy vector (Promega, WI) to create pIB38. A 0.87-kb spectinomycin resistance gene (aad9) was amplified from pUCSpec (20) using primers Spec-P-For and Spec-Rev and cloned into XhoI-XbaI-digested/T4 polymerase-blunted pIB38 to generate pIB72. The orientation of the aad9 insert was verified by PCR and found to be in the same direction as the SMu486 locus. Plasmid pIB72 was then linearized by NotI and used for the transformation of UA159. Spectinomycin-resistant transformants were selected, and the deletion of the SMu486 (liaS) locus was verified by PCR; a successful transformant was selected and named IBS148 (see Fig. 2). The transcriptional fusion construct PgbpC-gusA was then inserted into the chromosome of IBS148 via insertion at the SMu1405 locus after transformation with plasmid pIB121 (7), yielding strain IBS151.
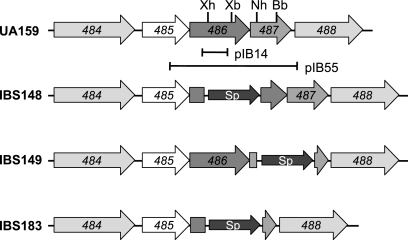
FIG. 2.
Schematic diagram illustrating the deletions performed on the loci coding for the SMu486/SMu487 TCS in S. mutans UA159. Open reading frames and orientations of transcription are indicated with block arrows. The locations of the DNA sequence fragments that were used for insertion mutagenesis (pIB14) and construction of the complementing plasmid pIB55 are indicated by horizontal lines below SMu486/SMu487. The SMu486, SMu487, and SMu486/SMu487 genes were deleted and replaced with the spectinomycin resistance gene aad (Sp).
For the deletion of the SMu487 (liaR) locus, a 1.48-kb fragment containing the entire SMu487 region was PCR amplified from UA159 genomic DNA using primers Bam-Smu486-F2 and Bam-Smu487-R2. This fragment was then cloned into the pGemT-Easy vector to create pIB39. The aad9 gene, as described above, was cloned into NheI-BbsI-digested/T4 polymerase-blunted pIB39 to generate pIB74. Plasmid pIB74 was then linearized with NotI and transformed into UA159. Transformants were selected on the basis of spectinomycin resistance, and the deletion of the SMu487 (liaR) locus was confirmed by PCR. One of the selected clones was named IBS149 (see Fig. 2) and used for subsequent analysis and for transformation with pIB121 to generate strain IBS152, which contains a PgbpC-gusA reporter construct in its genomic DNA at SMu1405.
For the simultaneous deletion of both SMu486 and SMu487 (ΔliaSR), a 2.43-kb fragment spanning both open reading frames was PCR amplified from UA159 genomic DNA using primers Bam-Smu486-F1 and Bam-Smu487-R2 and cloned into the pGemT-Easy vector to generate pIB66. The aad9 gene was then cloned into XhoI-BbsI-digested/T4 polymerase-blunted pIB66 to generate pIB68. The SMu486/SMu487 (liaSR) locus was inactivated after the transformation of strains UA159 and IBS131 with NotI-linearized pIB68, generating strains IBS183 and IBS155, respectively (see Fig. 2).
To determine whether the expression of SMu487 leads to the regulation of gbpC expression, wild-type strain IBS131 was transformed with pIB181 or pIB169, pJRS1315 derivatives with or without SMu487, respectively. Briefly, SMu487 was amplified from genomic DNA from wild-type UA159 using EcoR-Smu487-F1 (5′-GCGGAATTCATGCTGATGAGTAAAACAAAAGTTATACTGG-3′) and Bam-Smu487-R2 (CGCGGATCCTTTAGCACCTGCTTCAATGACAGG) and cloned into EcoRI-BamHI-digested pIB169, which contains the Pveg promoter, the vegetative promoter of Bacillus. The resulting plasmid contained a Pveg-SMu487 fusion and was designated pIB181.
Semiquantitative RT-PCR.
Total RNA samples were isolated from liquid cultures at different growth phases for analyses of gbpC transcript levels. The isolation of RNA from liquid culture was performed as described previously (7). Since the production of mutacin IV was analyzed by monitoring the growth of the various strains on solid medium, total RNA samples were isolated from cultures grown on THY agar to measure nlmA or nlmT transcript levels. For the isolation of RNA from solid medium, THY agar plates containing appropriate antibiotics, if required, were inoculated with UA159 or its derivatives and grown overnight at 37°C at 5% CO2. Samples were collected by scraping the cultures from the agar plates and resuspending the bacterial biomass in RNAProtect solution (Qiagen); RNA was purified according to the supplier's suggested protocol. The RNA concentrations were determined by UV spectrophotometry, in addition to Bioanalyzer analysis. Semiquantitative analyses of transcript levels of various genes, as well as gyrA (as control), were performed by use of a one-step reverse transcription (RT)-PCR assay using the Titan One Tube RT-PCR system (Roche) as described previously (7). Fifty nanograms of RNA was used for each RT-PCR, analyzed in 1% agarose gel, and quantitated using Doc-It-LS software (UVP).
The primers specific for the gbpC transcript (GbpC-F5 and GbpC-R2) (7) produce a 308-bp PCR product, the primers specific for the nlmA transcript (NlmAF1 [5′-ATGGATACACAGGCATTTGAACAATTTGATGTA-3′] and NlmAR1 [5′-TGAGATCGAATGAGTCCCCAAGTGCCTA-3′]) produce a 200-bp PCR product, and the primers specific for the nlmT transcript (NlmTF [5′-TTAATCGTCATAGCCGTTAACATTCTCTTAGAGA-3′] and NlmTR [5′-CCTTACTCATCCTAGTCACCTTAACTGAAGGAT-3′]) produce a 481-bp PCR product. The primers specific for the gyrA transcript (GyrA For and Gyr Rev) (7) generate a 470-bp PCR product. Expression of the gyrA gene served as an internal control to ensure that equal amounts of RNA were used in all RT-PCRs.
GusA assays.
β-Glucuronidase (Gus) assays were performed after the S. mutans cultures reached the mid-exponential growth phase (70 Klett units) (9) as described previously by Biswas and Biswas (10). The activities of the culture lysates were quantified by comparison with the activity of glucuronidase standards (Sigma-Aldrich). The protein concentration of the lysate was determined using a Micro BCA protein assay kit (Pierce) standardized with bovine serum albumin (Sigma).
Deferred antagonism assay.
Deferred antagonism assays were used to evaluate the production of mutacin IV by the S. mutans cultures (8). Mutacin IV-producing tester strains were stabbed into THY agar plates and grown overnight at 37°C under anaerobic conditions. Streptococcus gordonii DL-1 and Streptococcus cristatus 5100 were used as indicator strains. These strains were grown in THY broth; 0.4 ml of each culture was mixed with 4 ml of soft agar (0.4%) and poured over the surface of the THY agar plate stabbed with the tester strains. The inoculated plates were incubated overnight at 37°C under anaerobic conditions, and the diameter of the zone of inhibition around the mutacin-producing strains was measured the following morning.
RESULTS
Identification of a putative sensor kinase that regulates gbpC expression.
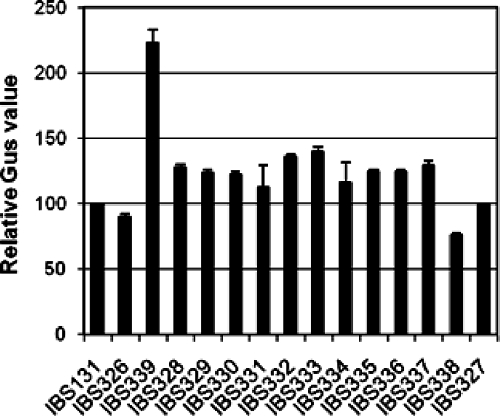
Since GbpC is essential for biofilm formation and the cariogenesis of S. mutans, we wanted to gain more information about the nature of the signal that is involved in the regulation of expression of gbpC. S. mutans encodes at least 14 complete TCS (1, 8); thus, any sensor kinase belonging to one or more of these 14 TCS may potentially modulate the expression of gbpC. In order to study whether any of these putative sensor kinases are involved in the regulation of gbpC expression, we used strain IBS131, the wild-type strain that contains the promoter region of gbpC fused to a gusA reporter gene, at the SMu1405 locus (7). Small PCR fragments (∼500 bp) derived from each of the sensor kinase genes were cloned into suicide vector pBSKEry (9) to generate 14 inactivating plasmids (8) for the transformation of IBS131. Sensor kinase mutants were systematically generated through single-crossover chromosomal integration of the inactivating plasmid at the specific sensor kinase locus. Transcription from PgbpC was then measured in the wild type and 14 sensor kinase mutant strains by assaying the GusA activity produced from PgbpC-gusA. Only one sensor kinase mutant, SMu486 (IBS339), exhibited increased activity (∼2.2-fold) compared to that of wild-type strain IBS131 (Fig. 1). The Gus activity for the other sensor kinase mutants was similar to that of the wild-type strain. Taken together, the results suggest that SMu486 represses expression from PgbpC, either directly or through an interaction with another system; no other sensor kinase appears to be associated with the regulation of expression from PgbpC.
FIG. 1.
Expression from PgbpC-gusA in the sensor kinase mutants. Fourteen different sensor kinases were inactivated in IBS131 in order to measure their effect on PgbpC-gusA expression. Strains were grown in THY broth at 37°C and harvested at the mid-exponential phase, and gusA activity was measured. The values were normalized with Gus activity obtained with IBS131. Experiments were repeated at least twice, and the mean values are shown.
Analysis of the region surrounding the SMu486 locus suggests that it is the second gene of a multicistronic operon (Fig. 2). SMu486 encodes a putative protein that exhibits a high degree of homology to the recently characterized LiaS from Bacillus subtilis (33). The immediate downstream gene, SMu487, encodes a putative response regulator with a high degree of homology to LiaR of B. subtilis, while the immediate upstream gene, SMu485, shows extensive homology with LiaF of B. subtilis. Therefore, this locus was renamed LiaFSR to reflect the sequence conservation with the B. subtilis operon.
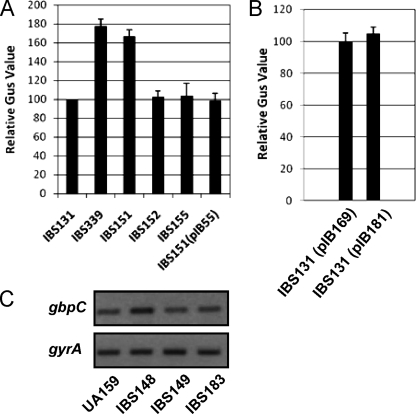
The inactivation of liaS via single-crossover chromosomal integration with plasmid pIB14 (Table 1) may have a polar effect on the downstream liaR gene, which encodes the cognate response regulator. To eliminate the possibility of polar effects, nonpolar mutants of liaS and liaR were generated by direct gene replacement. Expression of liaR in the single- and double-crossover liaS mutants was confirmed by RT-PCR analysis; the levels of liaR transcript did not differ in either mutant (data not shown). Both liaS and liaR were simultaneously inactivated to study whether this TCS is involved in the expression of gbpC. Gus activity from the wild-type, ΔliaS, ΔliaR, and ΔliaSR strains was measured. We observed that nonpolar mutant strain IBS151 (liaS) had a 1.7-fold-increased Gus activity compared to that of wild-type strain IBS131 (Fig. 3A). This increase in Gus activity is comparable with that of the single-crossover mutant IBS339, which resulted in an approximately twofold increase in Gus activity. On the other hand, no differences in activity were observed between the ΔliaR (IBS152) or ΔliaSR double mutant strain (IBS155) and wild-type strain IBS131.
FIG. 3.
Regulation of gbpC expression by SMu486/SMu487. (A) Expression from PgbpC-gusA was measured for the wild type, (IBS131); the ΔSMu486 (IBS151), ΔSMu487 (IBS152), and ΔSMu486/SMu487 (IBS155) strains; the ΔSMu486 strain complemented with pIB55 (IBS151/pIB55); and the single-crossover ΔSMu486 strain (IBS339). Experiments were performed in triplicate, and the mean values with standard deviations are shown. (B) Expression from PgbpC-gusA was measured with the wild-type strain (IBS131) containing either pIB169 or pIB181, as described in the text. Experiments were performed in triplicate, and the mean values with standard deviations are shown. (C) Semiquantitative RT-PCR results showing the expression of gbpC from the wild type (UA159), the ΔSMu486 mutant (IBS148), the ΔSMu487 mutant (IBS149), and the ΔSMu486 mutant complemented with pIB55 (IBS148/pIB55). The data are representative of RT-PCR analyses resulting from at least two different RNA isolations.
To verify the results of the transcriptional fusion reporter analysis described above, the transcription of gbpC was directly measured using semiquantitative RT-PCR analysis. RNA was extracted from strains UA159, IBS148 (ΔliaS), IBS149 (ΔliaR), and IBS183 (ΔliaSR) after growth reached mid-exponential phase. Semiquantitative RT-PCR was performed using gbpC-specific primers to measure the level of gbpC expression; the level of gyrA transcript produced was also measured to ensure that equal amounts of RNA were being used in the RT-PCR assay. As expected, the gbpC transcript level of IBS148 was about twofold higher than that of wild-type strain UA159 (Fig. 3C). This increased level of gbpC expression in the ΔliaS mutant strain was also observed for cultures grown to early stationary phase (data not shown), indicating that LiaS-mediated gbpC repression is growth phase independent. The levels of gbpC transcript produced from the other mutant strains were equivalent to those of wild-type strain UA159, consistent with the results of the PgbpC-gusA fusion reporter assays.
To confirm that the observed increase in the expression of gbpC was due to the inactivation of liaS, the level of gbpC transcript produced was also measured in IBS148 and IBS151 transformed with pIB55, a plasmid containing the full-length liaSR operon (8). The level of gbpC transcript produced by the complemented strain was comparable to that of the wild-type strain (Fig. 3A and data not shown). Taken together, these results suggest that LiaS is involved in the regulation of gbpC expression. However, the role of the cognate response regulator LiaR in the regulation of gbpC is unclear. The overexpression of liaR from a heterologous promoter, Pveg, in IBS131, a wild-type strain that contains the PgbpC-gusA fusion reporter, did not lead to an increase in expression from PgbpC (Fig. 3B), suggesting that under the conditions tested, LiaR does not regulate the expression of gbpC; similar results were observed during semiquantitative RT-PCR analysis, as an increase in gbpC expression was not observed between the wild-type and LiaR-overexpressing strains (data not shown).
Mutacin IV production is down-regulated in the liaS mutant.
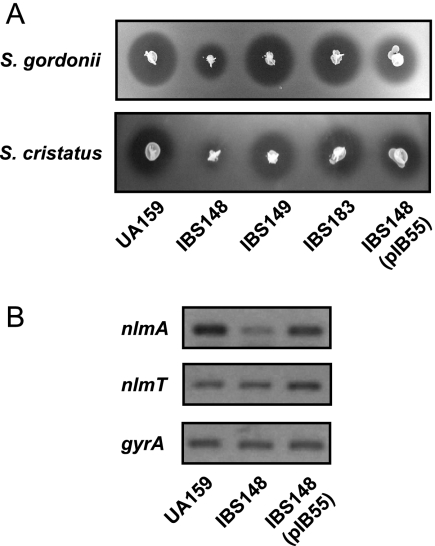
S. mutans has the capacity to produce mutacin, a bacteriocin, to suppress the growth of other competitor bacteria present in the dental plaque community (12, 19, 36, 38). In particular, S. mutans strain UA159 produces a mutacin that is active against sanguinis group and mitis group streptococci, the main competitors of S. mutans in the oral cavity (17, 18, 50). It was previously shown that both gbpC expression and mutacin production are modulated by the same regulatory network controlled by LuxS (35). Moreover, it was also shown that a regulatory gene for mutacin expression is present immediately upstream of gbpC (35). Therefore, it was of great interest to determine whether LiaS also regulates mutacin production. For these experiments, IBS148 (ΔliaS), IBS149 (ΔliaR), IBS183 (ΔliaSR), and the isogenic parental strain UA159 were subjected to deferred antagonism assays using S. cristatus (5100), and S. gordonii (DL-1) as indicator strains. As shown in Fig. 4A, the production of mutacin IV was drastically reduced in IBS148 but not in the other mutant strains. To verify that this effect was due to the inactivation of liaS, mutacin production was also measured in IBS148 transformed with pIB55. The level of production of mutacin was fully restored to the wild-type level in the complemented strain (Fig. 4A). Thus, in addition to gbpC, LiaS also appears to regulate mutacin production in S. mutans.
FIG. 4.
Regulation of mutacin IV production by SMu486 in S. mutans. (A) Deferred antagonism assay for mutacin IV production. S. mutans cultures were stabbed into THY agar and incubated overnight at 37°C under microaerophilic conditions. The plates were overlaid with soft agar containing the indicator strain. The zone of inhibition of the indicator strains was evaluated after overnight incubation. (B) Semiquantitative RT-PCR results showing the expression of nlmA and nlmT by the wild type (UA159), the ΔSMu486 mutant (IBS148), and the ΔSMu486 mutant complemented with pIB55 (IBS148/pIB55). The data are representative of RT-PCR analyses resulting from at least two different RNA isolations.
On the basis of genome sequence analysis, five groups of mutacins (groups I to V) have been identified in S. mutans. While several different kinds of mutacin are encoded by S. mutans, most of the clinical isolates of S. mutans produce at least one kind of mutacin (13, 16). The strain used in this study, UA159, produces only one type, mutacin IV, which is a nonlantibiotic (18, 50). Mutacin IV is a two-component bacteriocin that is composed of the NlmA and NlmB peptides, which are encoded by two transcriptionally linked genes (50). The dramatic decrease in the level of mutacin IV produced by the liaS mutant could be due to several factors including a reduction in the transcription of nlmA and nlmB or a deficiency in the export of the NlmA and NlmB peptides by the ABC transporter encoded by nlmT (17); therefore, the levels of transcription of nlmA and nlmT were measured in strains UA159, IBS148, and IBS148/pIB55. As shown in Fig. 4B, the level of nlmA expression was drastically reduced in IBS148, while the level of nlmA in the complemented strain was not significantly different from that in the wild-type strain. This suggests that LiaS either directly or indirectly activates the transcription of nlmA. The level of expression of nlmT also appeared to increase approximately twofold in strain IBS148/pIB55 relative to those of the wild-type strain and ΔliaS mutant strain IBS148, suggesting that LiaS may also have a role in the regulation of nlmT expression.
DISCUSSION
To gain a better understanding of the signal(s) that modulates gbpC expression, we systematically inactivated each of the 14 sensor kinases encoded in the genome of S. mutans. While most of the mutations of each of the sensor kinases had no significant effect on the expression of gbpC, a mutation in liaS led to an increase in gbpC expression. LiaSR belongs to the family of “bacitracin-responsive” TCS (31), which have been described in detail for Bacillus subtilis and Staphylococcus aureus. In B. subtilis, this family also includes BceSR and YvcQP (22), while the LiaSR homolog of S. aureus is known as VraSR (25). The histidine kinases in this family belong to a family of intramembrane sensing kinases that contain an unusually short sensing domain that is completely buried in the cytoplasmic membrane (31). The LiaS-like sensor kinases are induced under cell wall stress signals generated by cell wall antibiotics such as bacitracin and vancomycin (15, 32). In addition, it was previously reported that LiaS (known as HK11) of S. mutans is responsible for acid stress tolerance (27). It is possible that acid stress alters the cell wall integrity in S. mutans, activating the LiaSR system and the expression of genes involved in the acid stress response. Indeed, cell wall stress in B. subtilis induces the expression of general stress response genes, including various acid stress response genes (32). Similarly, the VraSR regulon of S. aureus also includes many genes involved in acid stress tolerance (15).
Although we found that LiaS controls gbpC expression, the cognate response regulator LiaR did not have any apparent effect on gbpC expression under the conditions tested. This phenomenon was also observed for mutacin IV production as well. The inactivation of liaS, whether through single- or double-crossover chromosomal integration, yielded similar results in our studies, suggesting that polar effects downstream of liaS were not a factor during single-crossover recombination. Moreover, semiquantitative RT-PCR analysis indicated that equivalent levels of LiaR were produced using either recombination technique (data not shown). It is surprising to find that only a mutation of the sensor kinase affected gbpC expression, while a mutation of the cognate response regulator did not have any noticeable effect. Although the mechanism is unclear, there are many precedents. For example, the inactivation of the sensor kinase of the TCS08 TCS of Streptococcus pneumoniae leads to the down-regulation of the genes involved with cellobiose utilization, but the inactivation of the cognate response regulator has no effect (34). Similarly, the inactivation of the CiaR response regulator of S. mutans has little effect on mutacin I production, whereas the inactivation of CiaH, the cognate sensor kinase, abolishes mutacin I production (39). Finally, in S. mutans, it was also reported previously that the inactivation of LiaS results in decreased acid tolerance, while the inactivation of LiaR does not affect acid tolerance (27).
While our results indicate that LiaS is involved in the regulation of gbpC expression, the role of LiaR is not immediately clear. A mutant strain containing inactivated liaR did not show significant differences in the level of expression of gbpC compared to that of the wild-type strain (Fig. 3), while the overexpression of liaR in a wild-type strain did not lead to significant increases in expression from PgbpC (Fig. 3B); this was also confirmed via semiquantitative RT-PCR analysis (data not shown). One possible explanation for our findings is that the phosphorylated form of LiaR is inactive and unable to activate the expression of gbpC; however, when the cognate sensor kinase LiaS is inactivated, LiaR is not phosphorylated, such that it is able to stimulate the transcription of gbpC. In the absence of LiaR (ΔliaR and ΔliaSR), the expression of gbpC is not observed. Further experimental studies will be required to confirm our speculation.
Another possible explanation could be that LiaS is involved in cross talk with a noncognate response regulator. In fact, recent reports in the literature support signal transduction from a sensor kinase to a noncognate response regulator in bacteria (40, 46, 51), and cross talk has also been suggested to be involved in the regulation of the S. mutans acid tolerance response by a TCS (27). A recently reported in vitro study suggested that approximately 50% of sensor kinases can cross talk with other response regulators in E. coli (53). Although the potential cross talk partners for LiaS were not identified in this study, there are several prospective candidates, such as SMu1008. The SMu1008/SMu1009 system, also known as MbrC/MbrD, belongs to the family of “bacitracin-responsive” TCS and is required for bacitracin resistance (49). The sensor kinase SMu1009 shows a high degree of similarity with LiaS and, like LiaS, contains a very short membrane-spanning domain (30). Thus, SMu1008 may cross talk with LiaS in addition to its role as the cognate response regulator for the sensor kinase SMu1009.
In contrast to the expression of gbpC, which is up-regulated in the ΔliaS mutant, the expression of nlmA is drastically reduced in the liaS mutant but was restored upon complementation with full-length liaS; as before, the inactivation of liaR did not have any noticeable effect on the expression of nlmA. This suggests that LiaS may act as a sensor kinase that stimulates either two distinct regulators with opposite functions or a single response regulator, such as LiaR, with different activities. There are several examples in the literature in which a single sensor kinase activates two different response regulators. For example, the CheA sensor kinase of E. coli has been shown to interact with CheB and CheY to regulate chemotaxis (26). On the other hand, response regulators with two disparate functions have also been identified. As an example, OmpR of E. coli functions both as an activator and as a repressor to differentially regulate the expression of the ompC and ompF genes (23, 37). Therefore, whether LiaS interacts with a single response regulator with opposing functions or with two different regulators remains to be studied. The expression of nlmT also appeared to be stimulated upon complementation with wild-type liaS, although the degree of expression was much lower than that with nlmA. Overall, the results suggest that LiaS is involved in the synthesis and export of mutacin IV in S. mutans.
In summary, we have shown that a putative regulatory locus, liaSR, may be important for the regulation of several virulence factors in S. mutans UA159. It was previously reported that the sensor kinase LiaS regulates the expression of genes involved in acid stress tolerance (8); our results indicated that LiaS also has a role in the regulation of expression of gbpC, which is essential for biofilm formation, and nlmA, which encodes one of the peptide components of mutacin IV. LiaS might also regulate other genes necessary for biofilm formation and cariogenesis, such that further studies are required to identify these genes. Furthermore, the function of LiaR is unclear and requires further experimental analysis to determine its physiological role in virulence and biofilm formation. A complete understanding of the LiaSR TCS and its regulon is essential in order to elucidate the regulation of virulence factors encoded by S. mutans and to gain a better understanding of the pathogenesis of disease induced by this organism.
Acknowledgments
This publication was made possible in part by funding by NIDCR grant DE016686 to I.B.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 5 May 2008.
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashby, M. K. 2004. Survey of the number of two-component response regulator genes in the complete and annotated genome sequences of prokaryotes. FEMS Microbiol. Lett. 231277-281. [DOI] [PubMed] [Google Scholar]
- 3.Banas, J. A. 2004. Virulence properties of Streptococcus mutans. Front. Biosci. 91267-1277. [DOI] [PubMed] [Google Scholar]
- 4.Banas, J. A., R. R. Russell, and J. J. Ferretti. 1990. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutans Ingbritt. Infect. Immun. 58667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banas, J. A., and M. M. Vickerman. 2003. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 1489-99. [DOI] [PubMed] [Google Scholar]
- 6.Beier, D., and R. Gross. 2006. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9143-152. [DOI] [PubMed] [Google Scholar]
- 7.Biswas, I., L. Drake, and S. Biswas. 2007. Regulation of gbpC expression in Streptococcus mutans. J. Bacteriol. 1896521-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas, I., L. Drake, D. Erkina, and S. Biswas. 2007. Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. J. Bacteriol. 19068-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biswas, I., and J. R. Scott. 2003. Identification of rocA, a positive regulator of covR expression in the group A streptococcus. J. Bacteriol. 1853081-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas, S., and I. Biswas. 2006. Regulation of the glucosyltransferase (gtfBC) operon by CovR in Streptococcus mutans. J. Bacteriol. 188988-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calva, E., and R. Oropeza. 2006. Two-component signal transduction systems, environmental signals, and virulence. Microb. Ecol. 51166-176. [DOI] [PubMed] [Google Scholar]
- 12.Chen, P., F. Qi, J. Novak, and P. W. Caufield. 1999. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl. Environ. Microbiol. 651356-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabio, U., M. Bondi, G. Manicardi, P. Messi, and R. Neglia. 1987. Production of bacteriocin-like substances by human oral streptococci. Microbiologica 10363-370. [PubMed] [Google Scholar]
- 14.Fabret, C., and J. A. Hoch. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 1806375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardete, S., S. W. Wu, S. Gill, and A. Tomasz. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 503424-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gronroos, L., M. Saarela, J. Matto, U. Tanner-Salo, A. Vuorela, and S. Alaluusua. 1998. Mutacin production by Streptococcus mutans may promote transmission of bacteria from mother to child. Infect. Immun. 662595-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale, J. D., N. C. Heng, R. W. Jack, and J. R. Tagg. 2005. Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans. J. Bacteriol. 1875036-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale, J. D., Y. T. Ting, R. W. Jack, J. R. Tagg, and N. C. Heng. 2005. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection. Appl. Environ. Microbiol. 717613-7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillman, J. D., J. Novak, E. Sagura, J. A. Gutierrez, T. A. Brooks, P. J. Crowley, M. Hess, A. Azizi, K. Leung, D. Cvitkovitch, and A. S. Bleiweis. 1998. Genetic and biochemical analysis of mutacin 1140, a lantibiotic from Streptococcus mutans. Infect. Immun. 662743-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Husmann, L. K., D. L. Yung, S. K. Hollingshead, and J. R. Scott. 1997. Role of putative virulence factors of Streptococcus pyogenes in mouse models of long-term throat colonization and pneumonia. Infect. Immun. 651422-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Idone, V., S. Brendtro, R. Gillespie, S. Kocaj, E. Peterson, M. Rendi, W. Warren, S. Michalek, K. Krastel, D. Cvitkovitch, and G. Spatafora. 2003. Effect of an orphan response regulator on Streptococcus mutans sucrose-dependent adherence and cariogenesis. Infect. Immun. 714351-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan, S., A. Junker, J. D. Helmann, and T. Mascher. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 1885153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenney, L. J. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5135-141. [DOI] [PubMed] [Google Scholar]
- 24.Kuramitsu, H. 1993. Virulence factors of mutans streptococci: role of molecular genetics. Crit. Rev. Oral Biol. Med. 4159-176. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49807-821. [DOI] [PubMed] [Google Scholar]
- 26.Li, J., R. V. Swanson, M. I. Simon, and R. M. Weis. 1995. The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry 3414626-14636. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y. H., P. C. Lau, N. Tang, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 1846333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 1813666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mascher, T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264133-144. [DOI] [PubMed] [Google Scholar]
- 31.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 501591-1604. [DOI] [PubMed] [Google Scholar]
- 33.Mascher, T., S. L. Zimmer, T. A. Smith, and J. D. Helmann. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 482888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKessar, S. J., and R. Hakenbeck. 2007. The two-component regulatory system TCS08 is involved in cellobiose metabolism of Streptococcus pneumoniae R6. J. Bacteriol. 1891342-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merritt, J., J. Kreth, W. Shi, and F. Qi. 2005. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol. Microbiol. 57960-969. [DOI] [PubMed] [Google Scholar]
- 36.Paul, D., and H. D. Slade. 1975. Production and properties of an extracellular bacteriocin from Streptococcus mutans bacteriocidal for group A and other streptococci. Infect. Immun. 121375-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20911-917. [DOI] [PubMed] [Google Scholar]
- 38.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 6715-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi, F., J. Merritt, R. Lux, and W. Shi. 2004. Inactivation of the ciaH gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect. Immun. 724895-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saini, D. K., V. Malhotra, and J. S. Tyagi. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 56575-80. [DOI] [PubMed] [Google Scholar]
- 41.Sato, Y., Y. Yamamoto, and H. Kizaki. 1997. Cloning and sequence analysis of the gbpC gene encoding a novel glucan-binding protein of Streptococcus mutans. Infect. Immun. 65668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato, Y., Y. Yamamoto, and H. Kizaki. 2000. Construction of region-specific partial duplication mutants (merodiploid mutants) to identify the regulatory gene for the glucan-binding protein C gene in vivo in Streptococcus mutans. FEMS Microbiol. Lett. 186187-191. [DOI] [PubMed] [Google Scholar]
- 43.Sato, Y., Y. Yamamoto, and H. Kizaki. 2000. Xylitol-induced elevated expression of the gbpC gene in a population of Streptococcus mutans cells. Eur. J. Oral Sci. 108538-545. [DOI] [PubMed] [Google Scholar]
- 44.Shah, D. S., G. Joucla, M. Remaud-Simeon, and R. R. Russell. 2004. Conserved repeat motifs and glucan binding by glucansucrases of oral streptococci and Leuconostoc mesenteroides. J. Bacteriol. 1868301-8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi, X., S. Wegener-Feldbrügge, S. Huntley, N. Hamann, R. Hedderich, and L. Søgaard-Andersen. 2008. A bioinformatics and experimental analysis of proteins of two-component systems in Myxococcus xanthus. J. Bacteriol. 190613-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silva, J. C., A. Haldimann, M. K. Prahalad, C. T. Walsh, and B. L. Wanner. 1998. In vivo characterization of the type A and B vancomycin-resistant enterococci (VRE) VanRS two-component systems in Escherichia coli: a nonpathogenic model for studying the VRE signal transduction pathways. Proc. Natl. Acad. Sci. USA 9511951-11956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, D. J., H. Akita, W. F. King, and M. A. Taubman. 1994. Purification and antigenicity of a novel glucan-binding protein of Streptococcus mutans. Infect. Immun. 622545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 49.Tsuda, H., Y. Yamashita, Y. Shibata, Y. Nakano, and T. Koga. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob. Agents Chemother. 463756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Ploeg, J. R. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 1873980-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhamme, D. T., J. C. Arents, P. W. Postma, W. Crielaard, and K. J. Hellingwerf. 2002. Investigation of in vivo cross-talk between key two-component systems of Escherichia coli. Microbiology 14869-78. [DOI] [PubMed] [Google Scholar]
- 52.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26369-376. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto, K., K. Hirao, T. Oshima, H. Aiba, R. Utsumi, and A. Ishihama. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 2801448-1456. [DOI] [PubMed] [Google Scholar]
- 54.Yamashita, Y., W. H. Bowen, R. A. Burne, and H. K. Kuramitsu. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 613811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]