Abstract
Ers has been qualified as the PrfA-like transcriptional regulator of Enterococcus faecalis. In a previous study we reported that Ers is important for the survival within macrophages of this opportunist pathogenic bacterium. In the present work we have used proteomic and microarray expression profiling of E. faecalis JH2-2 and an ers-deleted mutant (Δers mutant) strains to define the Ers regulon. In addition to EF_0082 (encoding a putative facilitator family transporter), already known to be under Ers regulation, three genes or operons displayed a significant decrease (confirmed by reverse transcription quantitative PCR) in expression in the Δers mutant. The first locus corresponds to three genes: arcA, arcB, and arcC1 (arcABC). These genes are members of the ADI operon, encoding enzymes of the arginine deiminase system. The second is the EF_1459 gene, which encodes a hypothetical protein and is located within a putative phage genetic element. Lastly, Ef_3319 is annotated as the alpha subunit of the citrate lyase encoded by citF. citF is a member of a putative 12-gene operon involved in citrate catabolism. Moreover, the promoter sequence, similar to the “PrfA box” and found in the promoter regions of ers and EF_0082, has been shown to be included in the DNA segment recognized by Ers. Phenotypic analysis of the Δers mutant strain revealed a growth defect when cultured with arginine or citrate as the energy source; this was not seen for the wild type. As expected, similar results were obtained with mutants in which arcA and citF were inactivated. In addition, in the mouse peritonitis model of virulence, the Δers mutant appeared significantly less lethal than the JH2-2 wild-type strain. Taken together, these results indicate that the regulator Ers has a pleiotropic effect, especially in the cellular metabolism and virulence of E. faecalis.
Enterococcus faecalis is a human commensal of the lactic acid bacterium group sometimes used in the dairy industry and even as a probiotic (9) but also associated with nosocomial infections. Indeed, E. faecalis is now one of the leading causes of surgical site, urinary tract, and bloodstream acquired infections (32). Because of this “Janus-faced” status of E. faecalis, the use of enterococci in the food process has been increasingly criticized (8, 9). Thus, it appears important to understand how this “safe” bacterium can become a dangerous pathogen. Shankar and collaborators have found that some virulence determinants were clustered on a large pathogenicity island (PAI) (36). This 150-kb PAI (with 129 open reading frames [ORFs]) is not systematically present in E. faecalis strains. Moreover, some strains also harbor a 17-kb transposon-like sequence within the PAI which contains the esp gene (encoding a surface protein known to be related to virulence in E. faecalis), the cyl operon (encoding the cytolysine), and one paralog of the gls24 operon (encoding a general stress protein and qualified as a virulence factor) (11, 35, 38). Other virulence factors in E. faecalis have been characterized. However, despite these data and the increasing number of infections of this bacterium, virulence mechanisms remain poorly understood.
From the 3,337 predicted protein-encoding ORFs in E. faecalis V583, 217 have or may have regulatory functions (28). In addition, no general stress sigma factors of the RNA polymerase have been found (28). So, in order to cope with hostile host and nonhost environments, E. faecalis has to develop mechanisms of adaptation involving specific transcriptional regulators. Some of them have already been shown to be correlated with virulence, such as Fsr, EtaRS, CylR, HypR, and PerR (13, 29, 39, 41, 42). In a previous study, we have identified a new transcriptional regulator of E. faecalis, named Ers (for enterococcal regulator of survival) (12). Ers is a member of the Crp/Fnr family and showed 69% amino acid similarity to Srv, a PrfA-like regulator of Streptococcus pyogenes implicated in virulence (30). Moreover, Ers appears as the most homologous protein of PrfA, the major positive regulator of virulence genes in Listeria monocytogenes (19). Phenotypical analysis of the ers mutant revealed that the Ers protein is important for survival within macrophages and in relation to lethal oxidative challenge in E. faecalis. Moreover, transcriptional analysis using reverse transcription quantitative PCR (RT-qPCR) showed that ers itself and EF_0082 (located near ers) are regulated by Ers. Interestingly, one sequence similar to the “PrfA box” of Listeria was observed in both promoter regions (12). Nevertheless, mutation of EF_0082 (encoding a putative transporter of unknown function) did not result in changes in survival within murine macrophages or in relation to H2O2 stress (12).
A few groups have used global approaches (microarray and proteomic) to investigate regulatory networks by taking snapshots during growth or stress or by comparing mutant and wild-type strains of enterococci. In 2005, DNA microarrays were used in order to identify genes whose expressions are modified by the presence of erythromycin in E. faecalis (1). More recently, microarrays were carried out to compare transcriptional profiles of the OG1RF wild type versus the regulator fsrB-deleted mutant (4).
In the present study, in order to gain insight into the role of Ers and the possible mechanisms by which it is required for virulence, we compared the global transcription and protein profiles of the wild type and an ers-deleted mutant strain of E. faecalis. In cultures harvested in the middle of exponential growth phase, four loci have been confirmed to be downregulated in the Δers mutant strain. The first is, as expected, EF_0082; the second locus corresponds to the arcABC member of the ADI operon encoding enzymes of the arginine deiminase system (3). The third is the EF_1459 gene, encoding a hypothetical protein, and the last, citF, produces the alpha subunit of the citrate lyase. Based on these data, we then demonstrated that Ers is involved in arginine and citrate catabolism. Moreover, the deletion of ers affected the virulence of E. faecalis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The E. faecalis strain JH2-2 (16, 43) and its derivatives were grown at 37°C in M17 medium (40) supplemented with 0.5% glucose (GM17) or in an M17 medium lacking of any sources of carbon (ccM17MOPS) (17) supplemented with 0.3% glycerol, 0.5% citrate or 1% arginine, and 0.05% glucose. When required, erythromycin (50 or 150 μg ml−1) was added. Escherichia coli strains were cultured under vigorous shaking at 37°C in LB medium (33) with ampicillin (100 μg ml−1), kanamycin (25 μg ml−1), or erythromycin (150 μg ml−1) when required.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| E. faecalis strains | ||
| JH2-2 | Fusr Rifr; plasmid-free wild-type strain | 16 |
| JH2-2 pVE 6007 | Strain harboring pVE 6007 plasmid [CmrrepA(Ts)] | 43 |
| Δers mutant | JH2-2 isogenic Δers mutant | This study |
| Complemented Δers | ers insertion in the JH2-2 Δers mutant | This study |
| arcA mutant | JH2-2 arcA::pORI19-1; Emr | This study |
| citF mutant | JH2-2 citF::pORI19-1; Emr | This study |
| EF_1459 mutant | JH2-2 EF_1459::pORI19-1; Emr | This study |
| EF_0082 mutant | JH2-2 EF_0082::pORI19-1; Emr | 12 |
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15] Tn10 (Tetr) | Stratagene |
| M15(pREP4) | pQE30 host strain | Qiagen |
| Ec101 | pORI19-1 host strain | 18 |
| Plasmids | ||
| pORI19-1 | Emr, lacZ′; cloning vector | 18 |
| pMAD | oripE194(Ts); Emr AmprbgaB | 2 |
| pQE30 | Ampr; expression vector | Qiagen |
| pGEM-T easy | f1 ori lacZ; Ampr | Promega |
| pMad-Δers | Construction for the deleted ers mutant | This study |
| pMad-ers | Construction for the ers complemented strain | This study |
| pQE-ers | ers cloned into the expression vector pQE30 | This study |
| pORI-arcA | Internal fragment of arcA cloned into pORI19-1 | This study |
| pORI-citF | Internal fragment of ciF cloned into pORI19-1 | This study |
| pORI-1459 | Internal fragment of EF_1459 cloned into pORI19-1 | This study |
| pGEM-T-Pers | Promoter of ers cloned into pGEM-T | This study |
| pGEM-T-P82 | Promoter of EF_0082 cloned into pGEM-T | This study |
| pGEM-T-ParcA | Promoter of arcA cloned into pGEM-T | This study |
| pGEM-T-P1459 | Promoter of EF_1459 cloned into pGEM-T | This study |
Fusr, fusidic acid resistance; Rifr, rifampin resistance; Cmr, chloramphenicol resistance; Emr, erythromycin resistance; Ampr, ampicillin resistance; Tetr, tetracycline resistance.
General molecular methods.
PCR was carried out in a reaction volume of 25 μl with 5 μg of chromosomal DNA of E. faecalis JH2-2 by use of PCR “master mix” (Eppendorf, Hamburg, Germany). The annealing temperature was 5°C below the melting temperature of the primers, 30 cycles were performed, and PCR products were purified using a NucleoSpin extract II kit (Macherey-Nagel, Düren, Germany). Primers used for this study are listed in Table 2. Plasmids were purified using a NucleoSpin plasmid kit (Macherey-Nagel). Restriction endonucleases, alkaline phosphatase, and T4 DNA ligase were obtained from Amersham Biosciences (Amersham Biosciences, Piscataway, NJ), Promega (Promega, Madison, WI), and Roche Applied Science (Roche, Indianapolis, IN) and used according to the manufacturers' instructions. E. coli and E. faecalis were transformed using a Gene Pulser apparatus (Bio-Rad Laboratories, Richmond, CA). Genomic DNA extraction and other standard techniques were carried out as described by Sambrook et al. (33).
TABLE 2.
Primers used in this study
| Locusa or primer name | Sequence (5′-3′) of indicated primera:
|
Use | |
|---|---|---|---|
| Forward | Reverse | ||
| EF_0082 | TTGACGTCAGCACCTTCTTC | CGTAGCGTTCACCTTTGACA | RT-qPCR |
| arcA | CGGTGAACACCGTAAATTCAT | AAACAACCAAACCACCTTCG | RT-qPCR |
| arcB | ACTGTTTGCCAGCCTTTCAT | GCGGAAGACTTCATCCGTAA | RT-qPCR |
| arcC-1 | TTGTCCAATGCGCTTAATCA | ATGCCTCATCTGCTGGATCT | RT-qPCR |
| EF_1459 | TTTGCTAACAGTCAATTGGATTACA | TGCTTTATTATTTTGCAGGGTTT | RT-qPCR |
| citF | ACTTGTTCGTGGACGGATTC | TGCAATGCCGACTTCTGTTA | RT-qPCR |
| Prom-ers | TAAGTTTGGTTCTGTCATTA | CGACTCGAAAGGAATGTTCA | Cloning in pGEM-T |
| Prom-ef0082 | CTTTTTCTATGTATGAGGAAG | CACCGCAATAATAACCATTA | Cloning in pGEM-T |
| Prom-arcAb | CTGTCGCTGGCTTCGACACTGTTA | CACTGTTTTCAATTTCCGATTTCAG | Cloning in pGEM-T |
| Prom-ef1459b | CAGTGATGACCTAGTGAGGTTTGC | CTGTGTAATCCAATTGACTGTTAGC | Cloning in pGEM-T |
| Prom-citfb | ATTGCGTTAGCATTGGCGGC | CTTCCTAAAAGTTTTGTCTCCCGAG | Cloning in pGEM-T |
| Prom-ef3326b | GTAAGCGTTAACAACCGGTGTTAA | CCTGTGTGTTGCTCTTTCTTG | Cloning in pGEM-T |
| ersDC-Uc | CCGGAATTCCCATGATCTAACTGTAACGGTGTCC | CGCGGATCCTAGTCTTGTAAAACATTTCTCATTGGC | Cloning in pMAD |
| ersDC-Dc | CGCGGATCCTTAACATCAGCACCCCCCGCAAGT | GAAGATCTGGGGGATATTATGGATGTGCTAGG | Cloning in pMAD |
| arcA-intd | TTGACGACAACGAAGAACTGATTCAA | CGGTGTTCACCGATATCAAAAGCC | Cloning in pORI19-1 |
| citF-intd | TCTTGGATATGCGATGGTCGATG | GCGGTGAGGCATACATATTGGC | Cloning in pORI19-1 |
| ef1459-intd | AAAGCATGTAGTTGATTTTGCTAAC | TTCTCCACTTTTTATAAGCATCG | Cloning in pORI19-1 |
| pQE30-Ers | GCGGATCCATGAGAAATGTTTTACAAGACTAC | CGCGCGCTGCAGTTAAACAATAATGTTATCTCTAATC | Cloning in pQE30 |
| mad1F | TCTAGCTAATGTTACGTTACAC | Cloning verification | |
| mad2R | TCATAATGGGGAAGGCCATC | Cloning verification | |
| Pu | GTTGTAAAACGACGGCCAGT | Cloning verification | |
| Pr | CACAGGAAACAGATATGACC | Cloning verification | |
From the annotated sequence available at http://www.tigr.org.
Primers also used for RT-PCR experiments.
Primers used to clone upstream (U) and downstream (D) sequences of ers in order to construct a deleted mutant by a double crossing-over (DC) event.
Primers used to clone internal fragments of arcA, citF, and EF_1459 in order to construct insertional mutants.
Construction of Δers and complemented Δers mutants.
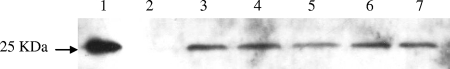
For the deletion and complementation assays, a DNA fragment containing ligated upstream (1,012-bp) and downstream (1,100-bp) sequences of the ers gene or including the entire ers gene obtained by PCR using the Pfu ultra-high-fidelity DNA polymerase (Stratagene, La Jolla, CA) was cloned into plasmid pMAD (2) (Table 1). One microgram of recombinant plasmid was finally used to transform competent cells. After electroporation, 300 μl of cell suspension was plated onto GM17 agar containing 50 μg ml−1 of erythromycin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (100 μg ml−1). Plates were incubated for 48 h at 30°C or 37°C. In both cases, a few dark blue colonies were obtained and analyzed for presence of the plasmid by PCR using primers mad1F and mad2R (Table 2). Some blue colonies were then cultured twice in GM17 liquid medium with erythromycin (50 μg ml−1) at 45°C overnight. In the next step, the cultures were used to inoculate (0.05% [vol/vol]) GM17 liquid medium without antibiotic. The tubes were incubated for 6 h at 30°C followed by incubation at 45°C overnight. This step was repeated two or three times. Serial dilutions of the culture were plated on GM17 agar containing 100 μg ml−1 of X-Gal and incubated for 48 h at 45°C. Among a vast majority of still-dark and -light blue clones, 0.1 to 0.3 percent of white colonies was present and represented candidate clones resulting from a double crossover event. These white colonies were isolated on GM17 agar with or without erythromycin. Antibiotic-sensitive clones were analyzed by PCR in the presence of a deleted or intact ers gene. Western blotting was also used to confirm the absence of Ers from the Δers mutant strain (Fig. 1, lane 2). Ers is a 217-amino-acid-residue protein, and the mutant has a deletion of 209 residues.
FIG. 1.
Western blot analysis of E. faecalis JH2-2 (lanes 3 to 7) and Δers mutant (lane 2) protein extracts (40 μg) with polyclonal antibody against His6-Ers. Purified His6-Ers protein (0.1 μg) was loaded on lane 1. Proteins were obtained from cells harvested at OD600s of 0.25 (lane 3), 0.5 (lanes 2 and 4), 0.75 (lane 5), 1 (lane 6), and 1.5 (lane 7).
Microarray analysis.
Three samples of total RNA from E. faecalis wild-type and Δers mutant strains harvested at mid-exponential growth phase in M17 were isolated using an RNeasy Midi kit (Qiagen, Valencia, CA). Ten micrograms of RNA was mixed with 750 ng of random hexamers (Invitrogen, Carlsbad, CA) and cDNA generated by reverse transcription (RT) using SuperScript II reverse transcriptase (1,500 U; Invitrogen). The reaction mixtures were incubated at 25°C for 10 min, 37°C for 60 min, 42°C for 60 min, and 70° for 10 min. Following RNase H (Invitrogen) and RNase A (Ambion, Austin, TX) digestion for 1 h, cDNA was purified with a QIAquick PCR purification kit (Qiagen) according to the recommendation of the manufacturer. The three cDNA samples coming from the same strain (wild type or Δers mutant) were then mixed together. The pool of cDNA was sheared enzymatically by the protocol of NimbleGen Systems, Inc. (Madison, WI) and biotinylated using biotin-N6-ddATP and terminal transferase. Labeled cDNA samples were individually hybridized to the E. faecalis V583-specific microarray according to the NimbleGen standard operating procedure. Following washes and labeling with a streptavidin-Cy3 complex according to the NimbleGen procedure, microarrays were scanned at a 5-μm resolution and data were extracted using NimbleScan (NimbleGen). Data were normalized and converted to estimates of transcript abundance, using the total signal intensity, to allow comparison of individual microarrays. The E. faecalis V583-specific DNA microarray was designed and produced by NimbleGen Systems, Inc. The array includes 3,265 ORFs (or targets) loaded in triplicate. For each target, 19 probe pairs of 60-mer in situ-synthesized oligonucleotides are present on the slide. A pair consists of a sequence perfectly matched to the ORF and another that differed from the original sequence at the two center positions.
Two-dimensional protein gel electrophoresis and protein identification.
Protein preparation from cells harvested in mid-exponential growth phase in M17 and two-dimensional electrophoresis were performed as described by Giard et al. (10). First, spots of interest were visually identified, and then the intensity was determined using the OptiQuant image analysis software (Packard Instrument Company, Downers Grove, IL). We considered positive match a difference between the mutant and the wild type of up to 3. These spots were excised from the gel, and peptides were digested by trypsin as described by Budin-Verneuil et al. (5). An electrospray ion trap spectrometer (LCQ DecaXP; ThermoFinnigan, San Jose, CA) coupled online with high-performance liquid chromatography was used for peptide analysis. Mass spectrometry (MS) spectra were acquired in a mode that alternated a full MS scan (mass range, 400 to 1,600) and a collision-induced dissociation tandem mass MS of the most abundant ion. Data were analyzed using the SEQUEST algorithm incorporated with the ThermoFinnigan BioWorks software.
RT-qPCR.
Specific primers designed to produce amplicons of equivalent lengths (100 bp) were designed using Primer3 software (available at the website http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and are listed in Table 2. RT-qPCR was carried out as previously described (41). Quantification of 23S rRNA levels was used as an internal control. Amplification, detection (with automatic calculation of the threshold value), and data analysis were performed twice and in duplicate with three different RNA samples by use of the Bio-Rad iCycler iQ detection system (Bio-Rad Laboratories).
The value used for the comparison of gene expression in various strains was the number of PCR cycles required to reach the threshold cycle (CT) (between 17 and 26). To relate the CT value back to the abundance of an mRNA species, CT was converted to “n-fold difference” by comparing mRNA abundance in the JH2-2 wild-type strain to that obtained with the Δers mutant strain. The n-fold difference was calculated by the formula n = 2−x when the CT for the mutant was less than the CT for JH2-2 and by the formula n = −2x when the CT for the mutant was more than the CT for JH2-2; in these formulas, x equals the CT for the mutant minus the CT for JH2-2. Then, values greater than 1 reflect a relative increase in mRNA abundance compared to the wild type, and negative values reflect a relative decrease. Statistical comparison of means was performed using Student's t test with values for ΔCT (CT for gene/CT for 23S) obtained for the wild type and the Δers mutant. Relative changes of at least 2 and a P value less than or equal to 0.05 were considered significant.
Construction of arcA, citF, and EF_1459 mutants by homologous recombination.
Internal fragments of the arcA, citF, and EF_1459 genes (466 bp, 503 bp, and 212 bp, respectively) were first amplified by PCR using chromosomal DNA of E. faecalis JH2-2 as the template (primers are listed in Table 2) and cloned into the conditional suicide vector pORI19-1 (18) (Table 1). The resulting plasmid, obtained after transformation of E. coli Ec101, was introduced into E. faecalis JH2-2 in which plasmid pG+host3 (pVE6007) (21), encoding a thermosensitive RepA protein and allowing the replication of pORI19-1, had previously been introduced (Table 1). After electroporation, both plasmids were maintained together at the permissive temperature of 30°C by plating cells on GM17 agar medium containing erythromycin (150 μg ml−1). Several clones were grown for 2 h at 30°C in GM17 broth without antibiotic and then incubated for 3 h at 42°C before being plated on GM17 agar medium containing erythromycin (150 μg ml−1). Integrations by single-crossover recombination within arcA, citF, and EF_1459 in erythromycin-resistant colonies were verified by PCR and by Southern blotting.
RT-PCR analysis and mapping of the transcriptional start site.
Reverse transcription was realized with RNA previously obtained and using Omniscript enzyme (Qiagen) at 37°C for 1 h. The cDNA was denatured and amplified by PCR using primers listed in Table 2. Positive controls were carried out using genomic DNA instead of cDNA. If cDNA amplifications yielded no PCR products, this may indicate that a putative promoter region is located between the two primers used. The transcriptional start points of EF_0082 and citF were determined by using a rapid amplification of cDNA ends (RACE) 5′/3′ kit (Roche, Mannheim, Germany) according to the manufacturer's instructions.
Overproduction and purification of Ers.
E. faecalis Ers was overproduced and purified as a hybrid protein with a His6 tag fused to its N terminus. First, the ers gene was amplified by PCR using primers listed in Table 2 and cloned into the pQE30 vector (Qiagen) (Table 1). Overproduction of the protein was realized in the E. coli M15(pREP4) strain (Table 2) carrying pQE30::ers. E. coli was grown in 200 ml of LB medium supplemented with ampicillin (100 μg ml−1) and kanamycin (25 μg ml−1) to an optical density at 600 nm (OD600) of 0.5. The production of His6-Ers was induced by the addition of 1 mM of isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 37°C. Cells were harvested by centrifugation and resuspended in 1 ml of Lew buffer (Protino kit; Macherey-Nagel) supplemented with lysozyme (1 mg ml−1). Cells were then disrupted by two passages through a “one-shot cell disrupter” system (ConstantSystem, Northants, United Kingdom) at 215 × 106 Pa. The lysate was centrifuged at 13,000 × g for 30 min at 4°C and the soluble fraction of proteins was recovered. His-tagged Ers was purified using Ni-nitrilotriacetic acid columns from the Protino kit (Macherey-Nagel) according to the manufacturer's recommendations. Then, samples were desalted on PD-10 columns (Amersham Biosciences). Protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad Laboratories).
The His-tagged Ers protein was purified under native conditions and was sent to GIP Plate-forme Technologique d'Evreux (Evreux, France), where immune rabbit serum was generated by intravenous immunization with the protein in phosphate-buffered saline.
Western blotting.
Ten milliliters of bacterial culture was harvested at an OD600 of 0.5 by centrifugation. Cells resuspended in 125 μl of Tris buffer at 0.25 M (pH 7.5) were broken by the addition of glass beads (0.1- to 0.25-mm diameter) and by vortexing for 4 min. Unbroken cells were removed by centrifugation, the supernatant was transferred to another tube, and protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories). Sodium dodecyl sulfate Laemmli buffer was added before loading. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane and blocked for 1 h with Tris-buffered saline-Tween (TBS-T) buffer (20 mM Tris, 137 mM NaCl, 0.1% Tween 20, pH 7.5) containing 5% skim milk. Then, the membrane was washed twice 10 min with TBS-T and incubated for 1 h with antisera against His-tagged Ers in TBS-T. After three 5-min washes with TBS-T, the blot was incubated for 1 h with a rabbit immunoglobulin conjugated to peroxidase (ECL detection kit; GE Healthcare, Little Chalfont, United Kingdom). Finally, the membrane was washed three times with TBS-T and developed using the ECL procedure according to the manufacturer's instructions.
Footprinting experiments.
DNase I footprinting assays were performed using a method based on that previously described by Yindeeyoungyeon and Schell (45). The binding reactions were carried out at room temperature for 5 min in 70-μl reaction volumes containing 33 mM Tris-HCl (pH 7.5), 2 mM CaCl2, 2 mM dithiothreitol, 20 μg ml−1 poly(dI-dC), 0.02% bovine serum albumin, 40 μg His6-Ers, and 250 ng of D4-labeled DNA fragment. The DNase treatment was then performed by the addition of 70 μl of a solution containing 80 mM Tris-HCl (pH 7.5), 12 mM MgCl2, and 250 U of DNase I (Amersham Biosciences) and incubation for 1 min at room temperature. The reaction was stopped by the addition of 35 μl of 25 mM EDTA and incubation for 5 min at 94°C. DNA was then purified using the NucleoSpin extract II kit (Macherey-Nagel); precipitated by the addition of 10% (vol/vol) 100 mM EDTA, 10% (vol/vol) 3 M sodium acetate (pH 3.2), 5% (vol/vol) glycogen, and 3 volumes of ethanol (100%); and resuspended in 40 μl of SLS buffer (Beckman Coulter, Fullerton, CA). Subsequently, capillary electrophoresis was done using a CEQ8000 sequencing apparatus (Beckman Coulter). The determination of the DNA sequence of the protected region was performed after comigration of the footprinting assay and the corresponding sequence reaction.
Mouse peritonitis model.
Testing of the JH2-2 and mutant and complemented strains was performed as described by Teng et al. (39). Briefly, the strains were incubated in brain heart infusion broth overnight at 37°C under constant agitation. The cells were harvested by centrifugation, twice washed with ice-cold 0.85% saline solution, and resuspended in the same solution to reach a density of ∼1.5 × 1010 CFU/ml. The inoculum size was confirmed by plating onto brain heart infusion agar. Dilutions (2- to 10-fold) of the bacterial suspension, prepared in chilled 0.85% saline solution, were used as inocula, after 10-fold diluting them in 25% sterile rat fecal extract (from a single batch) (27). Outbred ICR female mice of 4 to 6 weeks of age (Harlan Italy S.r.l., San Pietro al Natisone, Udine, Italy) were used. Mice were injected intraperitoneally with 1 ml of each bacterial inoculum made in 25% sterile rat fecal extract and then housed five per cage and fed ad libitum. Mice were monitored every 3 h, and the number of surviving mice was recorded. Survival curves were obtained by the Kaplan-Meier method and compared by log rank test using the GraphPad Prism software (GraphPad Software Inc., San Diego, CA). Comparisons with P values of <0.05 were considered to be significant. All strains were tested more than once.
RESULTS
Characterization of the Ers regulon of E. faecalis by use of DNA microarray, proteomic analysis, and RT-PCR.
Ers is the PrfA-like transcriptional regulator of E. faecalis and is involved in the survival of oxidative stress and within macrophages (12). In order to determine which genes are regulated by Ers, global transcriptional and proteomic approaches have been carried out. Gene expression in a wild-type E. faecalis JH2-2 strain was compared with that in an isogenic Δers mutant strain by analysis on E. faecalis V583 genome microarrays. Total RNA used for these experiments was isolated from wild-type and Δers mutant strain cultures that had been grown to mid-exponential phase. The choice of this time point was driven by our Western blot results, in which the protein Ers is clearly present during the exponential growth phase (Fig. 1). Figure 1 also shows that Ers appears constitutively produced during the growth phase. From this transcriptomic analysis, we selected the genes whose expression significantly differed by more than twofold. This difference was then confirmed using RT-qPCR. Our results derive from the combination of DNA chip hybridization (using a pool of three different samples of cDNA) and RT-PCR carried out with three other samples. This study revealed that only five genes are differentially regulated (with P values of <0.05) between the wild type and the Δers mutant (Table 3). All of these genes were expressed at lower levels in the Δers mutant (2.2- to 14.1-fold), indicating an activation role of Ers. Among these, EF_0082, annotated as encoding a putative transporter, has been shown in a previous study to be a member of the Ers regulon (12). The other genes include arcA, arcB, and arcC1, encoding the arginine deiminase, the ornithine carbamoyltransferase, and the carbamate kinase, respectively. Barcelona-Andrés et al. describe that these loci are included in a five-gene transcriptional unit, called the ADI operon, involved in arginine catabolism in E. faecalis (3). Lastly, the EF_1459 gene encodes a hypothetical protein.
TABLE 3.
List of gene members of the Ers regulon identified by DNA microarray assay or proteomic and confirmed by RT-qPCR
| E. faecalis locus | Gene symbol | Known or putative functiona | JH2-2/ers mutantb | RT-qPCRc | Student's t test |
|---|---|---|---|---|---|
| EF_0074 | ers | Transcriptional regulator, Crp/Fnr family | —e | — | — |
| EF_0082 | Major facilitator family transporter | 3.22 | −6.2 | 0.000 | |
| EF_0104 | arcA | Arginine deiminase | 4.62 | −14.1 | 0.001 |
| EF_0105 | arcB | Ornithine carbamoyltransferase | 5.19 | −6.7 | 0.002 |
| EF_0106 | arcC-1 | Carbamate kinase | 5.29 | −10.3 | 0.007 |
| EF_1459 | Hypothetical protein | 5.7 | −2.2 | 0.031 | |
| EF_3319d | citF | Citrate lyase alpha subunit | — | −13.4 | 0.002 |
| EF_1002d | divIVA | Cell division protein DivIVA | — | +1.7f | |
| EF_1045d | pfk | 6-Phosphofructokinase | — | +1.1f | |
| EF_1353d | pdhA | Pyruvate dehydrogenase complex E1 component, alpha subunit | — | +1.3f | |
| EF_3293d | guaB | Inosine-5-monophosphate dehydrogenase | — | −1.9f |
From reference 28.
Average ratios of normalized data of fluorescence determined by DNA microarray essay. Ratios correspond to the fluorescence measured for the wild-type strain compared to that of the Δers mutant.
Factor of repression determined by RT-qPCR.
Identified by proteomic experiments.
—, Transcription of ers is regulated by Ers (reference 12 and this study) but because of its absence in the mutant strain, no values are given.
No significant difference.
The proteomic approach provides another way of defining gene products influenced by Ers during exponential growth phase. Two-dimensional gel electrophoresis of proteins from growing E. faecalis JH2-2 and Δers mutant strains have been carried out. By MS, after extraction of proteins from the gel, we identified five other putative members of the Ers regulon (Table 3). EF_1002, EF_1045, EF_3319, and EF_3293 correspond to a cell division protein, the 6-phosphofructokinase, the alpha subunit of the citrate lyase, and the inosine-5-monophosphate dehydrogenase, respectively. These proteins are absent from the Δers mutant. On the other hand, EF_1353, corresponding to the alpha subunit of the pyruvate dehydrogenase, is present in high amounts in the mutant strain compared to what is seen for the wild type. In order to determine whether the role of Ers occurs at the transcriptional or posttranscriptional level, RT-qPCRs were carried out. Only citF transcription appears highly (13.4-fold) and significantly (P = 0.002) repressed in the Δers mutant (Table 3). No transcriptional modifications were observed with EF_1002, EF_1045, EF_1353, or EF_3293 (Table 3). The genome regions of E. faecalis strain V583 found around each member gene of the Ers regulon are shown in the maps illustrated in Fig. 2A.
FIG. 2.
(A) Genetic organization of ers, EF_0082, EF_1459, and ADI and citrate operon chromosomal regions of E. faecalis V583. Large arrows indicate the different ORFs and their orientations show the transcriptional direction. Gene members of the Ers regulon are in black. The known or possible locations of the promoter regions (P) of those loci are indicated with bent arrows. Arrowheads indicate mutation sites by insertion of the pORI19-1 plasmid (see Materials and Methods for details). (B) Identification of the putative promoter region by RT-PCR experiments. PCRs using primers symbolized by black arrowheads in panel A were realized with cDNA (odd-numbered lanes) or chromosomal DNA (even-numbered lanes) matrices of E. faecalis JH2-2. The absence of amplification using the cDNA (lanes 1, 3, and 7) suggested that a putative promoter is located between the two primers used.
Role of Ers in metabolic pathways.
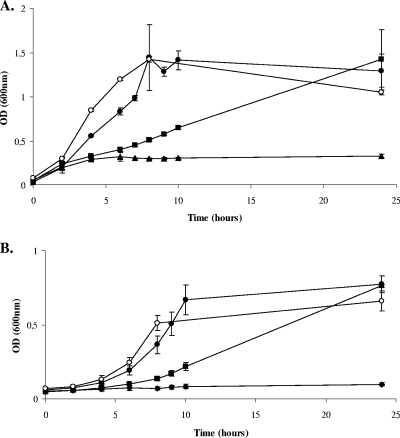
Since Ers regulates citF and arcABC in E. faecalis, we decided to investigate the role of these loci and ers in metabolic pathways. arcA and citF mutant strains were constructed via the insertion of a suicide vector into the corresponding ORFs (Fig. 2A). The growth rates of the arcA, citF, and Δers mutant strains and of the JH2-2 wild type were indistinguishable in rich media at 37°C (data not shown). Figure 3 shows that E. faecalis JH2-2 is able to utilize arginine or citrate as an energy source. arcA and citF mutants are unable to grow when arginine and citrate, respectively, are the sole sources of carbohydrate (Fig. 3). The weak increase in OD600 observed during the first growth hours of the arcA mutant is probably due to the presence of the small quantity of glucose added to the media (Fig. 3A). Indeed, Deibel describes that the growth of E. faecalis in a medium containing arginine still requires a small concentration of fermentable carbohydrate, most likely for the biosynthesis of carbon-containing molecules (7). Moreover, under these conditions, the growth rates of Δers mutant cells are greatly impaired compared to those of the wild type (Fig. 3). This is in strong agreement with the ability of Ers to regulate the expression of arcABC and citF. The transcription of arcABC or citF is downregulated but not totally suppressed in this strain. Hence, it is not surprising that culture of the Δers mutant manages to reach the same cell density as the wild type after 24 h of growth (Fig. 3). In order to verify that observed Δers mutant phenotypes were solely due to the lack of ers expression, we complemented the mutant with an insertion of an intact copy of ers (see Materials and Methods for details). In both cases, this complementation allows for the recovery of growth in the presence of arginine or citrate (Fig. 3).
FIG. 3.
Growth curves of different E. faecalis strains in ccM17MOPS medium (17) complemented with 1% arginine and 0.05% glucose (A) or 0.5% citrate (B). OD600 was measured during the growth of E. faecalis strains JH2-2 (•), the Δers mutant (▪), the complemented Δers mutant (○), and the arcA mutant (▴) (A) or the citF mutant (⧫) (B).
Interactions of Ers with promoter regions and Ers DNA-binding sites.
In a previous study, we characterized the promoter region of ers by RACE-PCR experiments (12). Barcelona-Andrés and collaborators (3) have demonstrated that the ADI operon expression is controlled by a promoter located upstream from arcA in E. faecalis, a finding confirmed by our RT-PCR experiments (Fig. 2B, lanes 1 and 2). RT-PCR experiments were carried out in order to determine the putative locations of the other gene promoters. A putative promoter was identified upstream of EF_1459 (Fig. 2B, lanes 3 and 4). In addition, we observed that the putative promoter region of citF is located upstream from the EF_3326 gene (Fig. 2B, lanes 5 to 8). This corresponds well to the spacing between ORFs. Transcriptional start sites of EF_0082 and EF_1459 have been mapped using 5′RACE-PCR. These “+1” sites are located 116 and 34 bp downstream of the ATG codon, respectively (see Fig. 5 below).
FIG. 5.
Sequence comparison of promoters of ers, EF_0082, EF_0104 (arcA), and EF_1459 from E. faecalis JH2-2. Ers binding sequences identified by footprinting are indicated in large boldface letters. Transcriptional start sites are in boldface and underlined. Those of ers and EF_0104 are from references 12 and 3, respectively. ATG start codons and putative ribosomal binding sites are underlined. “−10” and “−35” boxes are underlined and in italic. The putative consensus sequence was obtained using Gibbs software, available at the website http://melina2.hgc.jp/public/index.html. The corresponding sequences in the promoter are boxed.
In order to study the interactions of Ers with DNA, we overproduced and purified Ers harboring an N-terminal six-histidine tag (His6-Ers). We then used this purified protein for DNase I footprinting experiments. DNA targets consisted of ∼450-bp DNA fragments corresponding to the putative promoter regions of ers, EF_0082, arcABC, EF_1459, and citF loci. DNase I footprinting experiments were then performed in order to determine the sequence of the Ers binding sites in these putative DNA targets. No protected sequences have been determined for the citF promoter. Thus, this gene might be indirectly regulated by Ers.
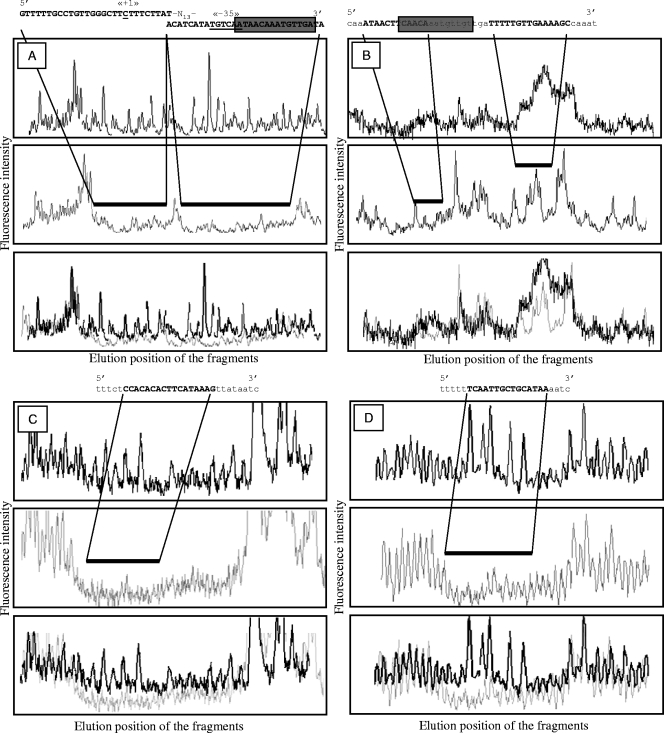
As shown in Fig. 4A, His6-Ers protects two regions separated by 13 bp in the ers promoter extending from position −50 to −21 and from −8 to +19 relative to the transcription start site. These regions thus overlap the putative RNA polymerase binding sites as well as the “+1” of transcription. One interesting feature of ers is the presence in its promoter region of a sequence closely related to the PrfA box of Listeria (12). As expected, this sequence (TCAACATTTGTTAT) is included in the protected region (Fig. 4A). Secondly, we tested the binding of His6-Ers on the EF_0082 promoter (Fig. 4B). Here, the Ers binding site is composed of two sequences spaced by 11 bp (Fig. 4B). These sequences run from −106 to −90 and from −79 to −67 relative to the transcription start site. Interestingly, one sequence, partially included in the protected fragment, displays strong similarity to the “PrfA-like box” identified above (ACAACATTTGTTGA; divergent bases are underlined) (Fig. 4B). The DNA target of Ers in the promoter of the ADI operon (CTTTATGAAGTGTGTGG) corresponds to a sequence located 186 bp upstream from the transcriptional start site (Fig. 4C). In this space, three Arg boxes and one cis-acting replication element site are also present (3). Ers is also able to interact with the promoter of EF_1459 (Fig. 4D). The protected sequence, consisting of 15 bp (TTATGCAGCAATTGA), is found 50 bp from the transcriptional start site. For EF_0082, arcA, and EF_1459, the Ers binding site is located upstream of the promoters (−67, −186, and −50 bp, respectively), which is in agreement with the activator role of Ers. Concerning the ers promoter, the binding region overlaps both “−10” and “−35” regions, which also confirms its own repression.
FIG. 4.
DNase I footprinting assay of His6-Ers with a D4-labeled DNA fragment containing the ers (A) or EF_0082 (B), arcA (C), and EF_1459 (D) promoter regions. Amplification was performed with PU and D4-PR (Table 2). Electrophoregrams of reactions performed without (in black at the top) or with (in gray in the middle) His6-Ers give fluorescence intensity (fragment abundance) along the y axis and elution position of the fragments (size) along the x axis. At the bottom is a composite picture mixing the two electrophoregrams. The solid line indicates the DNA region protected from DNase I by His6-Ers. The corresponding DNA sequence with PR primer (complementary sequence) is indicated. (A and B) The sequences close to the PrfA box are boxed.
An alignment of the promoter regions recognized by Ers allowed a characterization of a putative consensus sequence (AACATTTGTTG) (Fig. 5). Except for the promoter of arcA, this sequence is retrieved at least partially in the region that interacts with the regulator Ers. Moreover, for the ers and EF_0082 promoters, it is included in the sequence similar to the “PrfA box” (Fig. 5). No perfect match with the consensus sequence has been observed in the promoter region from the genome sequence of E. faecalis V583. Work is in progress to analyze putative promoters harboring sequences with one or more divergences with the “Ers box”.
These results, in addition to those showing Ers dependency in the transcription of these genes, provide support for the ideas of both a direct involvement of the Ers regulator in the activation of EF_0082, arcABC1, and EF_1459 and an indirect one for citF.
Ers mutant strains decreased the virulence of E. faecalis.
Ers has been shown to be involved in the ability of E. faecalis to survive within macrophages (12). This finding correlates with the high sensitivity of the ers mutant toward H2O2 challenge (12) compared to that of the wild type. In an effort to determine whether or not the absence of Ers also affects the virulence of E. faecalis, we compared the capabilities of the wild-type and Δers mutant strains to cause mouse mortality with intraperitoneal inoculation. As shown in Fig. 6, the rate of mortality is slightly but significantly reduced in mice infected with the Δers mutant strain (log rank test, P = 0.0121). After 50 hours of infection, fewer than 10% of wild-type-infected mice were still alive, whereas 60% of mice infected with the mutant had survived. Observing that JH2-2 will take all animal life 70 h after inoculation versus 100 h for the ers mutant (Fig. 6), we conclude that Ers is required for the full virulence of E. faecalis in the mouse peritonitis model. Experiments conducted with the ers-complemented strain restored the virulence to the wild-type level (data not shown). Likewise, mouse mortalities were the same for the EF_0082, arcA, EF_1459, and citF mutant strains as for the JH2-2 wild type (data not shown).
FIG. 6.
Kaplan-Meier survival curves of peritoneally infected mice after injection of E. faecalis JH2-2 (6.2 × 107 CFU) (•) and Δers mutant (6.9 × 107 CFU) (▪) strains.
DISCUSSION
In this study, we have examined the influence of Ers (annotated as EF_0074 in the genome of E. faecalis V583) on the transcriptome and proteome of E. faecalis. The transcriptional regulation of four loci appears controlled by Ers. Ers has already been qualified as the PrfA-like regulator of E. faecalis (12). This qualification is based not only upon the fact that Ers belongs to the same Crp/Fnr regulator family but also upon the fact that several amino acid residues critical for the full function of PrfA in L. monocytogenes are present in the Ers sequence (12). In the present study, we provide further evidence of the parallels between Ers and PrfA. First, as with PrfA and Srv (the PrfA-like regulator of S. pyogenes), Ers clearly appears to be involved in the virulence of E. faecalis (19, 30). The lesser ability of the Δers mutant to kill mice may be linked to its dramatic sensitivity to macrophage aggression (12). It does not seem to be strain dependent, since ers mutation in another wild-type background (OG1RF) also affects survival within macrophages (unpublished results). The second similarity is the presence of sequences similar to the “PrfA box” in the promoter regions of ers and EF_0082, genes known to be under Ers regulation (12). Crp-Fnr-like regulators are site-specific DNA-binding proteins. In PrfA-dependent promoters, PrfA binds to a palindromic recognition sequence (PrfA box) (TTAACANNTGTTAA) located at position −41 from the transcription start site (25, 37). Nevertheless, the position of putative PrfA boxes in some PrfA box-containing genes appears to be variable (−30 to −206 bp from the start codon) (26). ers gene expression has previously been shown to be autoregulated, and footprint experiments reveal that sequences similar to the “PrfA box” in ers and EF_0082 promoters are included in the DNA fragment protected by Ers. Except for EF_0082, such a sequence has not been found in the promoter regions of the other identified members of the Ers regulon. However, our results show that Ers is obviously able to directly regulate the expression of these genes, probably in addition to other transcriptional regulators. Analysis of the PrfA regulon of L. monocytogenes has allowed the identification of 73 member genes, only 15 of which harbored a PrfA box (26). A final parallel between Ers and PrfA, interesting for our study, is the arginine deiminase system belonging to both regulons (reference 26 and this study).
In our previous study, transcriptional analyses were carried out with genes showing homology with PrfA-regulated loci from Listeria. One of these, namely, EF_0082, is clearly under Ers positive control during the exponential growth phase. We confirm this result here using microarrays (12). Ef_0082 displays weak similarity (36%) with the hexose phosphate transporter UhpT (encoded by the hpt gene) of L. monocytogenes, which is involved in intracellular proliferation and virulence in mice as well as in resistance to fosfomycin (6, 34). As opposed to what is seen for the Listeria hpt product, the lack of Ef_0082 in E. faecalis does not alter bacterial survival within macrophages or susceptibility to fosfomycin (12; our unpublished results). It is interesting that the EF_0082 homologous genes of S. pyogenes and Lactococcus lactis correspond to the genes directly adjacent to srv and rcfB, respectively (20, 30). SPy1856 of S. pyogenes is 56% identical to Ef_0082 and is annotated as a putative antibiotic resistance protein, NorA. This type of transporter is involved in resistance against quinolones such as norfloxacin in Staphylococcus aureus (44). The MICs of norfloxacin for ers and for EF_0082 mutant strains are not different from those for the wild type (unpublished results). Since seven EF_0082 paralogs are present in the E. faecalis chromosome, it is possible that other transporters from this family may compensate for the EF_0082 deficiency.
The arginine deiminase system catalyzes the conversion of arginine to ornithine, ammonia, and carbon dioxide, with 1 mol of ATP generated per mol of arginine (15). For E. faecalis as well as for S. suis it has been suspected that the ADI operon is under the control of multiple factors (3, 14). Barcelona-Andrés and colleagues have identified three putative binding sequences for ArgR and one for CcpA but no “ArcR box” upstream of arcA of E. faecalis (3). Recently, Bourgogne and collaborators showed that the arcABC operon is regulated by the Fsr system of the E. faecalis OG1RF strain (4). Nevertheless, this cannot be the case for our JH2-2 wild-type strain, since it is a natural fsrB mutant. In this study, we have shown that Ers is also directly involved in the transcriptional regulation of this operon, pointing to a very complex regulation of this metabolic pathway. Using a Northern blot analysis, Barcelona-Andrés et al. have shown that cells produce an mRNA in the presence of arginine, corresponding to the transcript of the arcABCD-arcR cluster (3). For unknown reasons, only arcA, arcB, and arcC, and not arcR and arcD, were deregulated in our study. Similarly, for E. faecalis OG1RF, arcR was not deregulated in the fsr mutant strain (4).
The proteomic approach has allowed us to identify a new member of the Ers regulon, namely, EF_3319 (citF), which encodes the alpha subunit of the citrate lyase. The other spots differentially expressed in the Δers mutant seem indirectly and not transcriptionally regulated by Ers, while two of them are also implicated in metabolic pathways. Citrate metabolism is carried out by only a few strains of lactic acid bacteria. E. faecalis growth on citrate requires uptake followed by the breakdown of citrate to acetate and oxaloacetate by citrate lyase, with the latter decarboxylated to pyruvate (15). For Weissella paramesenteroides (previously named Leuconostoc paramesenteroides), the transcription of the operon containing genes involved in citrate fermentation is induced by the presence of citrate in the medium (22). The expression of the cit operon depends on posttranscriptional regulation and on the transcriptional activator CitI (24). This regulator also seems to be responsible for the acid-inducible transcription of the operon encoding the citrate lyase complex of L. lactis (23). From the results of our study, we know that the regulation of such an operon is different for E. faecalis, in which Ers plays a role (even if it is an indirect one) and in which a gene homologous to citI is undiscoverable in the genome sequence. The use of several different substrates (such as arginine or citrate) contributes significantly to the growth advantage for cells harboring these metabolic pathways. Hence, in addition to virulence features, it is important for opportunistic species like E. faecalis to be able to colonize a wide range of environments. Our footprint experiments revealed that Ers can be active in vitro without cofactor, but we cannot exclude the possibility that Ers, as other Crp/Fnr family regulators, may require a cofactor or posttranslational activation for full activity. So, some Ers-dependent genes may have been missed in our analyses.
The survival within macrophages of the E. faecalis ers mutant and JH2-2 wild-type strains was monitored using an in vivo-in vitro infection model (12). A spectacular decrease of the survival of the ers mutant compared to that of the wild-type strain was observed. Furthermore, as we have seen, mice infected with the Δers mutant survive 30 h longer than those infected with JH2-2. Based on these results, Ers may be considered as a virulence factor in E. faecalis. These observations may be correlated with the ability to survive under oxidative stress conditions, since effectors of the immune response have a bactericidal activity through the production of reactive oxygen species (for a review, see reference 31). Indeed, Ers is clearly involved in the resistance to oxidative stress due to H2O2 (12). Nevertheless, in our model, genes from the Ers regulon identified in the present study do not seem to be individually involved in virulence. Moreover, assessments of survival of the EF_0082, arcA, and citF mutants within macrophages were performed but no difference between the mutants and the wild type has been observed (12; data not shown). The hope is that the identification of other genes directly or indirectly regulated by Ers will explain the phenotype observed with the ers mutant of E. faecalis. For this we have we constructed a wild-type strain harboring a recombined plasmid where the expression of ers can be induced by the addition of nisin. For such conditions, preliminary data confirm that the overproduction of Ers also induces the expression of EF_0082. This tool will be used to identify new Ers-dependent genes.
In sum, Ers, a PrfA-like regulator of E. faecalis, appears to be involved in fitness and in virulence. Its implication in metabolic pathways should provide a growth advantage favorable for dissemination into the environment and host colonization. Moreover, Ers plays a role in survival within macrophages and in the rate of mouse death. The availability of global methods allowing us to verify deregulation at the whole-chromosome level will lead to investigation of the impact of ers deletion in other growth or stress conditions. In such investigations, we expect to find new members of the regulon. Moreover, this will allow us to understand the cross talk between different metabolic pathways and the establishment of the pathogenicity.
Acknowledgments
The expert technical assistance of Annick Blandin, Béatrice Gillot, Patricia Marquet, Johann Peltier, and Marie-Jeanne Pigny was greatly appreciated. We thank A. Benachour, J.-M. Laplace, V. Pichereau, A. Rincé, and N. Sauvageot for helpful discussions.
Editor: F. C. Fang
Footnotes
Published ahead of print on 21 April 2008.
REFERENCES
- 1.Aakra, A., H. Vebo, L. Snipen, H. Hirt, A. Aastveit, V. Kapur, G. Dunny, B. E. Murray, and I. F. Nes. 2005. Transcriptional response of Enterococcus faecalis V583 to erythromycin. Antimicrob. Agents Chemother. 492246-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaud, M., A. Chastanet, and M. Débarbouillé. 2004. A new vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 706887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcelona-Andrés, B., A. Marina, and V. Rubio. 2002. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J. Bacteriol. 1846289-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgogne, A., S. G. Hilsenbeck, G. M. Dunny, and B. E. Murray. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 1882875-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budin-Verneuil, A., V. Pichereau, Y. Auffray, D. S. Ehrlich, and E. Maguin. 2005. Proteomic characterization of the acid tolerance response in Lactococcus lactis MG1363. Proteomics 54794-4807. [DOI] [PubMed] [Google Scholar]
- 6.Chico-Calero, I., M. Suarez, B. Gonzalez-Zorn, M. Scortti, J. Slaghuis, W. Goebel, and J. A. Vazquez-Boland. 2002. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. USA 99431-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deibel, R. H. 1964. Utilization of arginine as an energy source for growth of Streptococcus faecalis. J. Bacteriol. 87988-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton, T. J., and M. J. Gasson. 2001. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 671628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foulquié Moreno, M. R., P. Sarantinopoulos, E. Tsakalidou, and L. De Vuyst. 2006. The role and application of enterococci in food and health. Int. J. Food Microbiol. 1063-29. [DOI] [PubMed] [Google Scholar]
- 10.Giard, J.-C., J.-M. Laplace, A. Rincé, V. Pichereau, A. Benachour, C. Leboeuf, S. Flahaut, Y. Auffray, and A. Hartke. 2001. The stress proteome of Enterococcus faecalis. Electrophoresis 142947-2954. [DOI] [PubMed] [Google Scholar]
- 11.Giard, J.-C., A. Rincé, H. Capiaux, Y. Auffray, and A. Hartke. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotropic effect on cell morphology, stress sensitivity and gene expression in Enterococcus faecalis. J. Bacteriol. 1824512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giard, J.-C., E. Riboulet, N. Verneuil, M. Sanguinetti, Y. Auffray, and A. Hartke. 2006. Characterization of Ers, a PrfA-like regulator of Enterococcus faecalis. FEMS Immunol. Med. Microbiol. 46410-418. [DOI] [PubMed] [Google Scholar]
- 13.Gilmore, M. S., P. S. Coburn, S. R. Nallapareddy, and B. E. Murray. 2002. Enterococcal virulence, p. 301-354. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. American Society for Microbiology, Washington, DC.
- 14.Gruening, P., M. Fulde, P. Valentin-Weigand, and R. Goethe. 2006. Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J. Bacteriol. 188361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huycke, M. M. 2002. Physiology of enterococci, p. 133-175. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. American Society for Microbiology, Washington, DC.
- 16.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Carbona, S., N. Sauvageot, J.-C. Giard, A. Benachour, B. Posteraro, Y. Auffray, M. Sanguinetti, and A. Hartke. 2007. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, alkylhydroperoxide reductase and thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol. Microbiol. 661148-1163. [DOI] [PubMed] [Google Scholar]
- 18.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 1777011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leimeister-Wachter, M., C. Haffner, E. Domann, W. Goebel, and T. Chakraborty. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 878336-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madsen, S. M., T. Hindré, J.-P. Le Pennec, H. Israelsen, and A. Dufour. 2005. Two acid inducible promoters from Lactococcus lactis require the cis-acting ACiD-box and the transcription regulator RcfB. Mol. Microbiol. 56735-746. [DOI] [PubMed] [Google Scholar]
- 21.Maguin, E., P. Duwat, T. Hege, and D. Ehrlich. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 1745633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin, M., C. Magni, P. Lopez, and D. de Mendoza. 2000. Transcriptional control of the citrate-inducible citMCDEFGRP operon, encoding genes involved in citrate fermentation in Leuconostoc paramesenteroides. J. Bacteriol. 1823904-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, M. G., P. D. Sender, S. Peiru, D. de Mendoza, and C. Magni. 2004. Acid-inducible transcription of the operon encoding the citrate lyase complex of Lactococcus lactis biovar diacetylactis CRL264. J. Bacteriol. 1865649-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, M. G., C. Magni, D. de Mendoza, and P. Lopez. 2005. CitI, a transcription factor involved in regulation of citrate metabolism in lactic acid bacteria. J. Bacteriol. 1875146-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mengaud, J., M. F. Vicente, and P. Cossart. 1989. Transcriptional mapping and nucleotide sequence of the Listeria monocytogenes hlyA region reveal structural features that may be involved in regulation. Infect. Immun. 573695-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 471613-1625. [DOI] [PubMed] [Google Scholar]
- 27.Pai, S. R., K. V. Singh, and B. E. Murray. 2003. In vivo efficacy of the ketolide ABT-773 (cethromycin) against enterococci in a mouse peritonitis model. Antimicrob. Agents Chemother. 472706-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2992071-2074. [DOI] [PubMed] [Google Scholar]
- 29.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 1833372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid, S. D., A. G. Montgomery, and J. M. Musser. 2004. Identification of srv, a PrfA-like regulator of group A Streptococcus that influences virulence. Infect. Immun. 721799-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riboulet, E., N. Verneuil, S. La Carbona, N. Sauvageot, Y. Auffray, A. Hartke, and J.-C. Giard. 2007. Relationships between oxidative stress and virulence in Enterococcus faecalis. J. Mol. Microbiol. Biotechnol. 13140-146. [DOI] [PubMed] [Google Scholar]
- 32.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21510-515. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Scortti, M., L. Lacharme-Lora, M. Wagner, I. Chico-Calero, P. Losito, and J. A. Vàzquez-Boland. 2006. Coexpression of virulence and fosfomycin susceptibility in Listeria: molecular basis of an antimicrobial in vitro-in vivo paradox. Nat. Med. 12515-517. [DOI] [PubMed] [Google Scholar]
- 35.Shankar, N., C. V. Lockatell, A. S. Baghdayan, C. Drachenberg, M. S. Gilmore, and D. E. Johnson. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 694366-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417746-750. [DOI] [PubMed] [Google Scholar]
- 37.Sheehan, B., A. Klarsfeld, R. Ebright, and P. Cossart. 1996. A single substitution in the putative helix-turn-helix motif of the pleiotropic activator PrfA attenuates Listeria monocytogenes virulence. Mol. Microbiol. 20785-797. [DOI] [PubMed] [Google Scholar]
- 38.Teng, F., E. C. Nannini, and B. E. Murray. 2005. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the Gls24 protein as a possible immunotherapy target. J. Infect. Dis. 191472-480. [DOI] [PubMed] [Google Scholar]
- 39.Teng, F., L. Wang, K. V. Singh, B. E. Murray, and G. M. Weinstock. 2002. Involvement of PhoP-PhoS homologs in Enterococcus faecalis virulence. Infect. Immun. 701991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terzaghi, B. E., and W. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 2807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verneuil, N., A. Rincé, M. Sanguinetti, B. Posteraro, G. Fadda, Y. Auffray, A. Hartke, and J.-C. Giard. 2005. Contribution of PerR-like regulator in oxidative stress response and virulence of Enterococcus faecalis. Microbiology 1513997-4004. [DOI] [PubMed] [Google Scholar]
- 42.Verneuil, N., M. Sanguinetti, Y. Le Breton, B. Posteraro, G. Fadda, Y. Auffray, A. Hartke, and J.-C. Giard. 2004. Effects of Enterococcus faecalis hypR gene encoding a new transcriptional regulator on oxidative stress response and intracellular survival within macrophages. Infect. Immun. 724424-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada, H., S. Kurose-Hamada, Y. Fukuda, J. Mitsuyama, M. Takahata, S. Minami, Y. Watanabe, and H. Narita. 1997. Quinolone susceptibility of norA-disrupted Staphylococcus aureus. Antimicrob. Agents Chemother. 412308-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yindeeyoungyeon, W., and M. A. Schell. 2000. Footprinting with an automated capillary DNA sequencer. BioTechniques 291034-1040. [DOI] [PubMed] [Google Scholar]