Abstract
Endotoxins are amphipathic lipopolysaccharides (LPSs), major constituents of the outer membrane of gram-negative bacteria. They consist of a lipid region, covalently linked to a core oligosaccharide, to which may be linked a repetitive glycosidic chain carrying antigenic determinants. Most of the biological activities of endotoxins have been associated with the lipid moiety of the molecule: unique to gram-negative bacteria, LPS is a ligand of the mammalian TLR4-MD2-CD14 pathogen recognition receptor complex. Lipid A preparations are often heterogeneous with respect to both the numbers and the lengths of fatty acids and the natures of substituents on the phosphate groups when present. The variants can significantly affect host immune responses. Nine species in the Bordetella genus have been described, and the fine LPS structures of seven of them have been published. In this report, lipids A from Bordetella pertussis Tohama I and B. bronchiseptica strain 4650 were further characterized and revealed to have a glucosamine substituting both lipid A phosphate groups of the diglucosamine backbone. These substitutions have not been previously described for bordetellae. Moreover, a B. pertussis transposon mutation that maps within a gene encoding a Bordetella ArnT (formerly PmrK) glycosyl transferase ortholog does not carry this substitution, thus providing a genetic basis for the modification. Reverse transcriptase PCR of this locus showed that it is Bvg regulated, suggesting that the ability of Bordetella to modify lipid A via this glucosamine modification is a potential virulence trait.
Endotoxins are lipopolysaccharides (LPSs), the major components of the external membrane of gram-negative bacteria. LPSs may cause several pathophysiological symptoms: some are beneficial at small doses by activating the host defense system, but at higher doses, LPSs may lead to septic shock and death (16, 60).
The LPS molecular architecture has three regions: a hydrophobic moiety called lipid A, a core oligosaccharide containing 2-keto-3-deoxyoctonoic acid (Kdo), and a serospecific O polysaccharide composed of repeating oligosaccharide units. Lipid A is the LPS anchor in the outer leaflet of the external bacterial membrane. LPS is a so-called pathogen-associated molecular pattern. Unique to bacteria, fungi, and viruses, pathogen-associated molecular patterns are ligands of a family of mammalian transmembrane Toll-like receptors (TLRs), which play a major role in pathogen recognition by the host (34). LPS is a ligand of the TLR4-MD2-CD14 complex (51), and stimulation of this receptor complex leads to activation of signaling pathways, resulting in induction of antimicrobial genes and release of cytokines, thereby initiating inflammatory and immune defense responses. Although most of the biological activities have been associated with the LPS lipid moiety, the role of the polysaccharide moiety is not negligible (42). Indeed, LPS consisting of lipid A carrying only two Kdo residues induces stronger biological activities than isolated or synthetic lipid A (16, 52). The conformation of at least part of lipid A is modified by the Kdos and their charges (8). Furthermore, lipid A itself can be modified via mechanisms such as acylation, deacylation, hydroxylation, and phosphate group substitution with aminoarabinose, galactosamine (GalN), or phosphoethanolamine (51). These modifications can play a significant role in modulating host responses to infection (23).
The Bordetella genus currently contains nine species, most of which are respiratory tract pathogens: Bordetella pertussis causes whooping cough in humans; B. bronchiseptica is commonly found associated with atrophic rhinitis in pigs, snuffles in rabbits, and kennel cough in dogs; and B. avium causes bordetellosis in birds (49, 64).
The LPS structures of six of the Bordetella species (B. pertussis [12, 14, 40, 41, 44, 46], B. parapertussis [22, 45, 58], B. bronchiseptica [22, 45, 58], B. hinzii [5], B. avium [39], and B. trematum [73]) have been reported. As we have shown, Bordetella lipid A structures have a common bisphosphorylated β-1,6 glucosamine (GlcN) disaccharide backbone with two amide-linked C14-OH substituents (4, 17, 24, 79). The nature and distribution of ester-linked fatty acids have so far proved to be species or strain specific. One of the unusual features of Bordetella lipid A compared to those of most other lipids A is the absence of symmetry at the C-3 and C-3′ positions. Hypoacylation and the presence of short-chain fatty acids (C10-OH) observed only in the two human Bordetella pathogens further add to the less common structural properties of Bordetella lipid A (4, 11, 79). It is likely that this hypoacylation in the genus, which is known to reduce cytokine induction (38), is an adaptation reducing TLR4 activation and resulting responses.
On the other hand, B. bronchiseptica lipid A is palmitoylated (79), an acylation attributed to a late biosynthetic enzymatic step mediated by PagP. This modification is required for persistent colonization of the mouse respiratory tract (57) and for resistance to antibody-mediated complement lysis during B. bronchiseptica respiratory infection (56).
It has long been shown that lipid A structures differ not only between different genera but often also between species of the same genus as well as among strains of the same species (6, 11, 36, 43, 67). Moreover, a given strain may express different LPS species simultaneously with varying abundance (26, 59). All these variations may affect both the susceptibility to penetration by antibiotics and the immunostimulatory activities of the outer leaflets of gram-negative bacteria. The increasing number of LPS modifications that are being unveiled is partly due to the availability of more-sensitive methods for LPS extraction and analysis and also to the larger screening of different batches of strains.
ArnT (formerly PmrK) is a glycosyl transferase that has been shown to modify lipids A of Salmonella and Pseudomonas with aminoarabinose and those of Francisella with GalN (59). All the sequenced Bordetella strains have a gene encoding an ortholog of ArnT, but it was not known whether these orthologs are functional or whether they produce similarly modified lipids A. The lipid A structures of three B. bronchiseptica strains published earlier (79) were highly heterogeneous and variable among strains; the variability was mostly in the positioning of the fatty acids, but the phosphate groups were not substituted. However, using a recently developed microanalysis technique that is capable of rapid analysis of lipid A under mild conditions, we have shown that the matrix-assisted laser desorption ionization (MALDI) mass spectrometry (MS) spectrum from one batch of B. bronchiseptica strain 4650 lipid A displays additional peaks corresponding to a compound of 161 atomic mass units (amu), which we suggested is likely due to a hexosamine modification of the distal phosphate groups (68). In this report, we provide further characterization of this modification. We show that lipid A from B. bronchiseptica 4650 has a GlcN residue as a substituent on both distal phosphate groups, and we demonstrate that B. pertussis Tohama I lipid A is similarly modified. In addition, a transposon insertion within the B. pertussis arnT locus abolishes this modification. Furthermore, this locus is regulated by the Bordetella BvgAS two-component system, a master virulence regulatory system, suggesting that this ability to modify Bordetella lipid A is a virulence trait. The ability to modify the structure of its lipid A components may allow Bordetella to escape or alter TLR4-dependent host defense mechanisms (23, 51).
(Part of this work was presented at the 8th International Symposium Saga of the Genus Bordetella at the Pasteur Institute, Paris, France, 7 to 10 November 2006. It was also published, in part, in Alina Tirsoaga's doctoral thesis, Paris-Sud University, Orsay, France, 19 January 2007, and in part in Nico Marr's doctoral thesis, University of Würzburg, Würzburg, Germany, 20 June 2007.)
MATERIALS AND METHODS
Bacterial strains and cultures.
A Tohama I derivative of B. pertussis, BP338 (76), and its isogenic Tn5lac mutant BPM2859 (77) or Tn5 mutant BP347 (76) were obtained from Alison Weiss (University of Cincinnati). B. bronchiseptica strain 4650 batches 1 and 2 were obtained from the National Research Council Collection (NRCC). The B. pertussis strains were grown on Bordet-Gengou (BG) agar (BD Biosciences) supplemented with 15% sheep blood (Dalynn) or in Stainer-Scholte (SS) broth (65) containing 0.06% bovine serum albumin (Sigma) to mid- to late-log phase in the presence of 30 μg/ml of nalidixic acid for BP338 and its derivatives and 50 μg/ml kanamycin for the transposon mutants. For LPS or lipid A studies, SS broth-grown bacteria of B. pertussis were heat inactivated for 40 min at 56°C and the heat-killed bacteria were lyophilized. B. bronchiseptica was grown as reported earlier (79), and the cells were killed in 2% phenol before harvesting.
LPS and lipid A extraction, purification, and structural analysis.
LPSs from B. bronchiseptica strain 4650 (batches 1 and 2) were extracted by a modified enzyme-phenol-water method (35). The preparations were obtained as precipitated gels by ultracentrifugation (105,000 × g, 4°C, 12 h) and then purified by extraction with solvents to remove phospholipids and free fatty acids. B. pertussis LPSs were extracted by an ammonium hydroxide isobutyrate method (24) and further purified. All preparations were treated with proteases and nucleases until thin-layer chromatography (TLC) and UV spectra showed no detectable contaminants (69).
B. pertussis lipids A were obtained by an ammonium hydroxide isobutyrate hydrolysis method (24) and further purified by solvent extractions.
Lipid A dephosphorylation.
Lipid A samples (1 mg) isolated from BP338 and BPM2859 were suspended in aqueous hydrofluoric acid (0.4 ml) and kept under stirring in a Teflon tube at 4°C for 48 h as previously described (4, 5). After solvent removal under vacuum in a polypropylene desiccator with a NaOH trap, the residue was suspended in water (0.5 ml) and ultracentrifuged at 4°C and 200,000 × g for 45 min in a Beckman TL100 apparatus. The supernatants containing the free soluble compounds were lyophilized and subjected to hexosamine analysis. The pellet containing the dephosphorylated lipids A was recovered, hydrolyzed with 6 M HCl for 6 h at 95°C, and subjected to hexosamine analysis as well.
Hexosamine analysis.
The hexosamine contents of samples were analyzed with a Hitachi L-8800 amino acid analyzer equipped with a 2620MSC-PS column (ScienceTec, Les Ulis, France). The elution protocol recommended by the manufacturer for the separation of amino acids and hexosamines was used. Under these conditions, GlcN, GalN, and mannosamine (ManN) were eluted at 58.8, 60.2, and 60.0 min, respectively, between phenylalanine and lysine (56.4 and 67.0 min, respectively).
TLC.
TLC was done on aluminum-backed silica gel plates (Merck). Products were visualized by charring them (in an oven at 150°C for 5 min) after spraying them with 10% sulfuric acid in ethanol or by spraying them with ninhydrin solution. Ten micrograms of lipid A was deposited on the origin of a TLC plate and chromatographed in a solvent mixture of chloroform-methanol-water-triethylamine (12:6:1:0.04) (17).
Sodium dodecyl sulfate (SDS)-polyacrylamide gel of LPS.
Fifteen-percent polyacrylamide gels were prepared and loaded with samples of 0.2 to 0.5 μg of the starting LPS preparation and its silica gel fractions, electrophoresed as previously described, and then stained (72).
Reduction.
Samples were reduced at room temperature with excess sodium tetrahydroborate. The reagent was destroyed with acetic acid and the mixture taken to dryness under reduced pressure.
Peracetylation.
Samples were peracetylated with acetic anhydride-dry sodium acetate in sealed tubes at 100°C for 1 h. After drying (50°C, reduced pressure), the product was extracted three times with ethyl acetate.
N acetylation.
Samples (1 mg) were suspended in 0.5 ml water mixed with 0.1 ml methanol. One drop of acetic anhydride was added and the mixture left under stirring at room temperature for 1/2 h. After evaporation under vacuum, the process was repeated twice.
Fatty acids.
Fatty acids were analyzed after hydrolysis of LPS or lipids A with 4 M HCl for 2 h at 100°C, neutralization, treatment with 2 M NaOH for 2 h at 100°C (4), extraction with ethyl acetate, methylation of the extract by diazomethane, and identification by gas chromatographic (GC) retention time on an HP5 column (30 m by 0.32 mm; Hewlett Packard), using a temperature gradient of 150°C to 300°C at 6°C/min. GC-MS analyses were performed as previously described (67), using an internal standard of octadecanoic acid for quantification. Synthetic acyloxyacyl fatty acids provided by D. Charon were used as a reference.
Nitrous deamination.
Nitrous deamination of O-deacylated lipid A (5 mg/ml) was performed as described previously (15). The reaction mixture was centrifuged at 200,000 × g for 1 h to separate these two fractions. After neutralization of the supernatant with sodium hydroxide, reduction with sodium tetrahydroborate was performed as described above. The pellet was analyzed by MALDI-MS, and the reduced and peracetylated supernatant was analyzed by GC-MS.
Identification of glycose absolute configurations.
Lipids A (2 mg) were hydrolyzed with 0.5 ml of 4 M HCl at 100°C for 2 h. After cooling and extraction of fatty acids with ethyl acetate, the mixture was concentrated to dryness under reduced pressure, and water was added and evaporated from the residue repeatedly until it became neutral. After N acetylation, the GlcN residue was treated with trifluoroacetic acid-R-(-)-2-butanol (3) and analyzed by gas-liquid chromatography on a BP10 (30 m by 0.32 mm; Scientific Glass Engineering) GC column, using a program of 160°C (1 min) to 220°C at 5°C min−1 and 0.6 kPa.
GC.
Alditol acetates were analyzed by GC with an HP5 column (30 m by 0.32 mm), using a program of 180°C (2 min) to 240°C at 2°C min−1.
Fatty acid analysis.
To determine the total lipid composition, fatty acids were released after hydrolysis of the LPS or lipids A with 4 M HCl for 2 h at 100°C, neutralization, treatment with 2 M NaOH for 2 h at 100°C, extraction with hexane, and methylation of the extract by diazomethane. Identification was performed by GC analysis on an HP5 column (30 m by 0.32 mm) with a program of 150°C to 300°C at 6°C/min. GC-MS was performed with a DB5ms capillary column (30 m) coupled to a Finnigan MAT 95.S MS.
SDS-promoted hydrolysis.
Lipid A was prepared by hydrolyzing LPS in 20 mM sodium acetate (pH 4.5)-1% SDS at 100°C for 1 h, followed by lyophilization. Detergent was removed by repeated extraction with acidified ethanol, the LPS/lipid A was recovered by centrifugation and dried under a stream of N2, and the lipid A in the dried pellet was extracted with chloroform-methanol-water (12:6:1) (17).
Lipid A isolation from whole cells.
As described by El Hamidi et al. (24), 10 mg of lyophilized, heat-killed bacteria of B. pertussis wild-type and mutant strains BP338 and BPM2859, respectively, were suspended in 400 μl of a mixture of isobutyric acid and 1 M ammonium hydroxide (5:3, vol/vol) and were kept for 2 h at 100°C in a screw-cap test tube under magnetic stirring. The mixture was cooled in ice water and centrifuged (2,000 × g for 15 min). The supernatant was diluted with water (1:1, vol/vol) and lyophilized. The sample was then washed twice with 400 μl of methanol and centrifuged (2,000 × g for 15 min). Finally, the insoluble lipid A was solubilized and extracted once in 100 to 200 μl of a mixture of chloroform-methanol-water (3:1.5:0.25, vol/vol/vol). For 1-mg samples, 100 μl of the different solvent mixtures was used at each step.
Sequential liberation of ester-linked fatty acids by mild-alkali treatment.
As described previously (68), the following conditions were applied for the first-step liberation of primary ester-linked fatty acids. Lipid A (200 μg) was suspended at 1 mg/ml in 35% ammonium hydroxide and kept under stirring for 5 h at 50°C. For liberation of the secondary ester-linked fatty acids, lipid A was suspended in 41% methylamine and kept under stirring for 5 h at 37°C. The solutions were dried with a stream of nitrogen, and the residue was taken up in a mixture of chloroform-methanol-water (3:1.5:0.25, vol/vol), followed by TLC and analysis by MALDI-MS.
MALDI-MS.
MALDI-MS was carried out in the linear mode under delayed-extraction conditions with a Perseptive Voyager STR (PE Biosystem, France) time-of-flight MS (IBBMC, Orsay, France). Gentisic acid (2,5-dihydroxybenzoic acid) was used as a matrix; a suspension of lipid A in chloroform-methanol-water (12:6:1; 1 mg/ml) was desalted with a few grains of Dowex 50W-X8 (H+), and 1 μl was deposited on the target, mixed with 1 μl of the matrix, suspended at 10 mg/ml in water or in 0.1 M aqueous citric acid (66), and dried. Analyte ions were desorbed from the matrix with pulses from a 337-nm nitrogen laser. Spectra were obtained in the negative-ion mode at 20 kV with an average of 128 pulses.
MALDI PSD.
MALDI postsource decay (PSD) time-of-flight MS experiments were performed to localize the HexN residues in O-deacylated molecular species. Samples were prepared as described above. The reflectron configuration with delayed-ion extraction was used to obtain the fragment-ion spectrum by metastable decomposition of a preselected ion. The laser power used was the minimum necessary to obtain adequate fragmentation, and the reflectron voltage was stepped down from 20 kV in five steps.
Sequencing of the Tn5lac insertion site.
To amplify the flanking regions of the Tn5lac insertion site in B. pertussis mutant BPM2859, inverse PCR (53) was carried out. Genomic DNA was isolated using a DNeasy tissue kit (Qiagen) and digested with DdeI (New England Biolabs). The restriction enzymes were heat inactivated, and the digested DNA fragments were circularized using T4 DNA ligase (New England Biolabs).
A 2.5-μl volume of the ligation reactions was used as a template for a standard PCR with primers Tn5in (CTGGGCTAAATCTGTGTTCTCTTCG) and Tn5out (TCAGATCCTGGAAAACGGGAAAGG). The following cycles were used for the inverse PCR: an initial denaturation step of 3 min at 94°C; 5 cycles of 30 s at 94°C, 1 min at 65°C, and 3 min at 72°C; followed by 30 cycles of 30 s at 94°C, 1 min at 60°C, and 3 min at 72°C; and ending with a last delay of 5 min at 72°C. The PCR products were subjected to 1% agarose gel electrophoresis, excised from a Sybr Safe (Invitrogen)-stained agarose gel, purified by use of a QIAquick gel extraction kit (Qiagen), and sequenced by use of the primer Tn5out at the Nucleic Acid Protein Services (NAPS), University of British Columbia. The resulting sequences were used to query the B. pertussis genome sequence by using BLAST (78).
Semiquantitative reverse transcriptase PCR.
B. pertussis strains were grown in SS broth to mid-logarithmic phase or on BG agar and then harvested in SS broth to give optical densities at 600 nm of ∼0.3 to 0.6. Total RNA was isolated using an RNAqueous kit (Ambion), and contaminating genomic DNA was removed using a DNA-free kit (Ambion). Absence of contaminating genomic DNA was verified by PCR using >0.1 μg of total RNA and exactly the same conditions as for PCR amplification of cDNA. Reverse transcriptase (RT) PCR analysis of 1 to 2 μg total RNA was carried out by use of SuperScript II reverse transcriptase (Invitrogen) and random primers (Invitrogen). The cDNA generated from exactly 0.1 μg of total RNA was PCR amplified with the gene-specific primers BP0398fw1 (TTCTTCGTCCACCAGCATTTCG), BP0398rev1 (AGCTGCAGGTCGAACGGATAGG), BP0399fw1 (ATCTGTCTGGACGCCGACATGC), BP0399rev1 (CAGGTAACCGCCGTAGGACAGC), Vag8fw1 (CCCCAAGCTTCGTCCGAGCACGGTATCAACG), and Vag8rev1 (CGCTCTAGACACATAGATCCCGGCGACTTCC) and GC melt reagent (Clontech) at a 20% final concentration. Simultaneously, control PCRs were carried out by replacing the cDNA template with either 0.1 to 0.2 μg of total RNA, 0.1 μg of genomic DNA of B. pertussis strain BP338, or distilled H2O. The following cycles were used: an initial denaturation step of 2 min (for reactions with cDNA or total RNA or without a template) or 5 min (for reactions with genomic DNA) at 94°C; followed by 30 cycles of 45 s at 94°C, 45 s at 63°C, and 1 min at 72°C; and a last delay of 10 min at 72°C. PCR products were subjected to 1% agarose gel electrophoresis and visualized by Sybr Safe staining (Invitrogen).
RESULTS
Structural analysis of the lipid A isolated from B. bronchiseptica strain 4650.
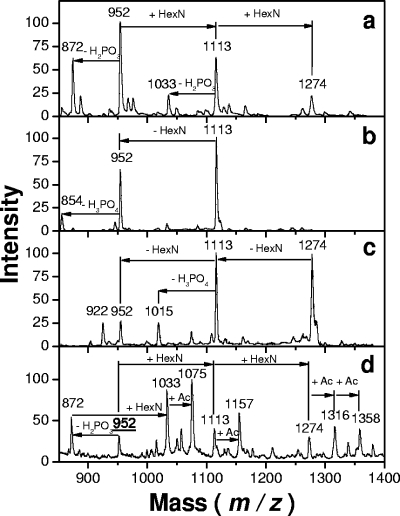
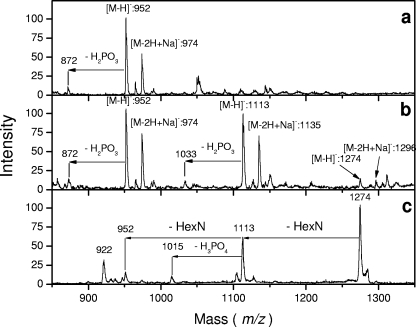
Previous work performed as a screening test on laboratory Bordetella samples revealed the presence of additional peaks in the lipid A backbone of B. bronchiseptica strain 4650 batch 2 (68). The present work was done in order to better characterize this component and focus on such substitutions in different Bordetella strains and species suspected to carry such modification (48). We first reexamined the lipid A acylation patterns of B. bronchiseptica strain 4650 and found them to be identical to those previously reported (79). We next used methylamine mild-alkali treatment to liberate the ester-linked fatty acids, as shown by Tirsoaga et al. (68). The residual lipid A obtained after this treatment was analyzed by MALDI in the negative-ion mode (Fig. 1a). A major peak at an m/z of 952 was attributed to two GlcNs, two phosphates, and two C14-OH groups in an amide linkage. Another major peak was observed at an m/z of 1,113, 161 amu heavier, and a third, smaller one was observed with the same increase over the second, at an m/z of 1,274. The peaks at m/z values of 952 and 1,113 were doubled by the monophosphorylated forms (−80 amu). The absence of such a doublet for the peak at an m/z of 1,274 suggested the presence of a hexosamine addition on both phosphate groups, as the corresponding molecular species could not release phosphate alone (80 amu) without its linked hexosamine. PSD experiments performed on both peaks at m/z values of 1,113 and 1,274 confirmed the release of substituents at −161 amu, generating daughter peaks at m/z values of 952 and 1,113 (Fig. 1b and c). To verify the distribution of the hexosamine substituents on both phosphate groups, mild-acid hydrolysis (used for selective liberation of the glycosidic phosphate group) was applied and led to the release of this substituent, as demonstrated by MALDI-MS (not shown).
FIG. 1.
MALDI negative-ion mass spectra of O-deacylated B. bronchiseptica strain 4650 batch 2 lipid A: direct time-of-flight spectrum (a), PSD spectrum of the 1,113-m/z precursor ions (b), and PSD spectrum of the 1,274-m/z precursor ions (c). (d) Negative-ion mass spectrum of N-acetylated (+Ac), O-deacylated B. bronchiseptica lipid A.
Detection of free amino groups in the lipid A structure by MS after N acetylation.
In order to demonstrate the presence of two free amino groups whose presence would be generated by the addition of two hexosamines in the lipid A backbone, a ninhydrin color test was performed, followed by a search for added N-acetyl compounds by MS after N acetylation. Lipid A samples were deposited on silica gel plates and sprayed with ninhydrin. Spots corresponding to molecular species in which the free amino groups were present were stained, and the corresponding products isolated on a preparative scale were scraped off. MS analysis confirmed that the peaks at +161 amu appeared in the spots positive for ninhydrin.
The deesterified B. bronchiseptica strain 4650 lipid A was N acetylated and reanalyzed by MALDI-MS. Additional peaks were observed at +42 amu and +84 amu, indicating N acetylation of the two free amino groups present in this lipid A sample (Fig. 1d).
Identification of a B. pertussis arnT locus.
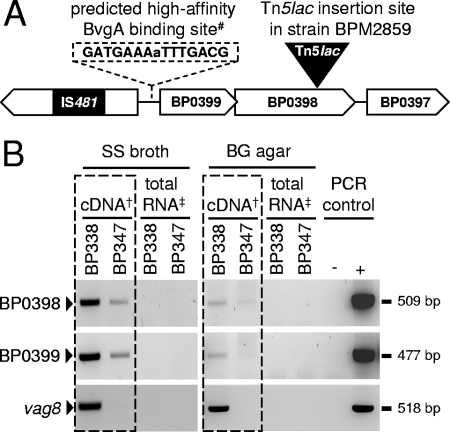
Weiss et al. (77) identified mutants of B. pertussis deficient in BvgAS-regulated genes by use of the promoter fusion transposon Tn5lac. A later study (48) demonstrated that in one of these transposon mutants, BPM2859, Tn5lac maps to a previously uncharacterized genomic region of B. pertussis (locus tag BP0398), encoding a putative glycosyl transferase with significant homology to ArnT proteins of gram-negative bacteria, including those of Salmonella, Pseudomonas, and Francisella species (51, 59). The annotated genome sequence of B. pertussis Tohama I (54) revealed BP0398 to be part of an as-yet-uncharacterized putative glycosylation locus that comprises another predicted glycosyl transferase gene (locus tag BP0399) just upstream of BP0398 (Fig. 2A). The putative gene product of BP0399 is homologous to ArnC (formerly PmrF) as well as to GtrB proteins, which are also glycosyl transferases found in gram-negative bacteria (2, 10). These loci appear to be present and intact in all other Bordetella strains sequenced so far, including B. bronchiseptica RB50 (locus tags BB4269 and BB4268), B. parapertussis 12822 (locus tags BPP3825 and BPP3824), and B. avium 197N (locus tags BAV2928 and BAV2927) (54, 63).
FIG. 2.
(A) Schematic representation of the genomic organization of the glycosylation loci BP0399 and BP0398 in B. pertussis strain Tohama I. Orthologous genes are found in other bordetellae, including B. bronchiseptica RB50 (locus tags BB4269 and BB4268), B. parapertussis hu 12822 (locus tags BPP3825 and BPP3824), and B. avium 197N (locus tags BAV2928 and BAV2927) (54, 63). BP0399 and BP0398 encode putative glycosyl transferases belonging to CAZy families 2 and 83, respectively (1). BP0397 and BP0396 encode putative proteins of as-yet-unknown function. #, characters in capital indicate imperfect heptads identified by the BvgA binding site motif, and characters in lowercase indicate a base between the heptads (19). (B) Results of semiquantitative RT-PCR analysis (boxed) and PCR controls of BP0399, BP0398, and vag8 with B. pertussis strain BP338 (wild type) and its isogenic mutant BP347 (bvgS::Tn5) after growth in SS broth and on BG agar. †, RT-PCRs using cDNA from 100 ng total RNA as a template; ‡, control PCRs using 100 to 200 ng total RNA as a template; +, control PCRs using 100 ng genomic DNA as a template; −, control PCRs without a template.
Regulation of the B. pertussis arnT locus.
The intergenic region upstream of the Bordetella arnT locus harbors a putative BvgA binding site, and interestingly, the predicted start site of BP0398 overlaps with the stop codon of BP0399, suggesting that they are cotranscribed (Fig. 2A) (19, 54). The promoter trap used to identify locus BP0398 was designed to find Bvg-regulated genes (77). To test whether locus BP0399 was also Bvg regulated and to verify that this was indeed true for locus BP0398, semiquantitative RT-PCR was performed on total RNA extracted from B. pertussis wild-type strain BP338 and its isogenic BvgS mutant BP347 upon growth in SS broth and on BG agar. RT-PCRs of vag8, known to be positively regulated by the BvgAS two-component system (27), were performed as controls. As shown in Fig. 2B, both glycosyl transferase genes, carried by loci BP0398 and BP0399, were found to be transcribed in wild-type strain BP338 and showed reduced levels of transcription in BvgS mutant BP347. Interestingly, in contrast to that of vag8, transcription of both glycosyl transferase genes was not abolished in strain BP347, as low levels of mRNA were detectable, suggesting the existence of an additional Bvg-independent promoter. In addition, expression levels of the two putative glycosyl transferase genes seemed to be higher upon growth of strains BP338 and BP347 in SS broth than upon cultivation on BG agar (Fig. 2B). PCRs using genomic DNA of strain BP338 or no DNA template provided positive or negative controls, respectively, and no PCR products were detectable when total RNA preparations prior to reverse transcription were used as a template, confirming the absence of genomic DNA contamination.
Comparison of LPS and lipid A structures isolated from B. pertussis wild-type and mutant strains.
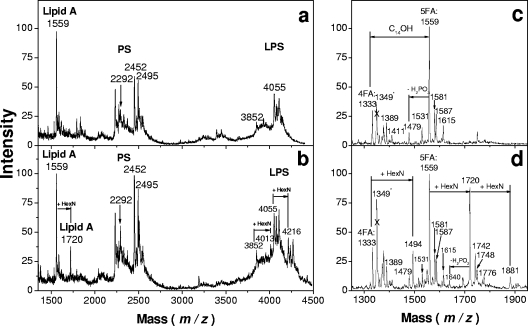
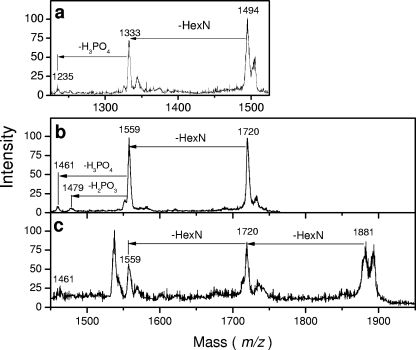
Given that bordetellae possess an ArnT ortholog, we hypothesized that lipid A from wild-type B. pertussis had a hexosamine modification, whereas the transposon mutant BPM2859, encoding a disrupted BP0398, did not. LPSs from B. pertussis wild-type strain BP338 and its isogenic transposon mutant BPM2859 were examined. LPS was extracted from lyophilized, heat-killed lysates of SS broth-grown bacteria by a mixture of isobutyric acid and 1 M ammonium hydroxide (5:3, vol/vol) (24). They were compared by TLC and SDS-polyacrylamide gel electrophoresis, and no major migration difference was observed under these conditions (not shown). However, comparison of LPS MALDI mass spectra showed two additional peaks at +161 amu at m/z values of 1,720 and 4,216 in the wild-type strain BP338 (Fig. 3a) that were absent from mutant strain BPM2859 (Fig. 3b). The peak at an m/z of 1,720 (1,559 + 161) corresponded to the lipid fragment containing hexosamine substituents, whereas the second peak, at an m/z of 4,216 (4,055 + 161), was clearly identified in the region corresponding to B. pertussis LPS molecular species (Fig. 3b).
FIG. 3.
Comparison of negative-ion MALDI mass spectra of B. pertussis LPS and lipid A. Results are shown for LPS isolated from the mutant BPM2859 (a) and the wild-type BP338 (b) strains and lipid A isolated from the mutant BPM2859 (c) and the wild-type BP338 (d) strains. The crossed peaks at m/z values of 1,349 in panels c and d correspond to a contaminant.
Lipids A were obtained by microhydrolysis of whole bacterial cells and analyzed by MALDI-MS (24). A quick comparison showed that the spectrum obtained in the negative-ion mode from the wild-type strain exhibited peaks corresponding to the main molecular species (m/z values of 1,333 and 1,559) plus the mass of one HexN (m/z values of 1,494 and 1,720) or two HexNs (m/z of 1,881) (Fig. 3c and d). Smaller peaks corresponded to residual sodiated molecular ions (m/z values of 1,581 and 1,742).
PSD MALDI analysis of the lipid A peaks at m/z values of 1,494, 1,720, and 1,881.
All the extra peaks observed in B. pertussis wild-type lipid A were tested by PSD fragmentation (Fig. 4a, b, and c). The peak at an m/z of 1,881 generated the two other peaks, while peaks at m/z values of 1,720 and 1,494 gave peaks at m/z values of 1,559 and 1,333, respectively. This confirmed that all extra peaks were related to the free phosphate lipid A molecular species and corresponded to the newly described lipid A molecules.
FIG. 4.
PSD MALDI negative-ion mass spectra of the extra-HexN-containing molecular species of lipid A isolated from the B. pertussis wild-type strain BP338: 1,494-m/z (a), 1,720-m/z (b), and 1,881-m/z (c) precursor ions.
After complete O deacylation of the B. pertussis lipids A tested in this study under the conditions described, a major peak was observed at an m/z of 952, corresponding to two GlcNs, two phosphate groups, and two C14-OH amide-linked fatty acids. Another major peak appeared at an m/z of 1,113, corresponding to the molecular species at an m/z of 952 plus the mass of one HexN unit and a smaller peak, at an m/z of 1,274, plus a second HexN unit. These data indicated that lipid A of the B. pertussis wild-type strain BP338 (Fig. 5b and c) is modified by the presence of two extra HexNs, whereas its isogenic mutant BPM2859 is not (Fig. 5a).
FIG. 5.
MALDI negative-ion mass spectra of O-deacylated lipid A from BPM2859 (a) and BP338 (b) strains. (c) PSD MALDI spectrum of the precursor ion from spectrum b at an m/z of 1,274.
Characterization of the hexosamine substituents.
Purified samples of the different lipids A from the B. pertussis and B. bronchiseptica strains used in this study were hydrolyzed with strong acid, and after removal of the fatty acids by extraction, the reduced sugars were peracetylated and the alditol acetates tested by GC as described above (not shown). A single peak corresponding to peracetylated glucosaminol was observed, indicating that the substituents were not different from the two GlcNs constituting the lipid A backbone, and these substituents were assumed to be GlcNs.
To confirm these data, the lipid A samples were subjected to nitrous deamination as previously described (12), and the hexosamines with free amino groups present in the lipid A were converted after reduction to 2,5-anhydro mannitol, easily detected and differentiated by GC. The presence of other sugars would have generated different compounds under these conditions, like glucitol or 2,5-anhydro-talitol from ManN or GalN residues, respectively (33). The deaminated lipid A residue was tested by MALDI-MS, and it was confirmed that both GlcNs had been removed from the phosphate groups by the process, a single peak at an m/z of 952 being observed (not shown).
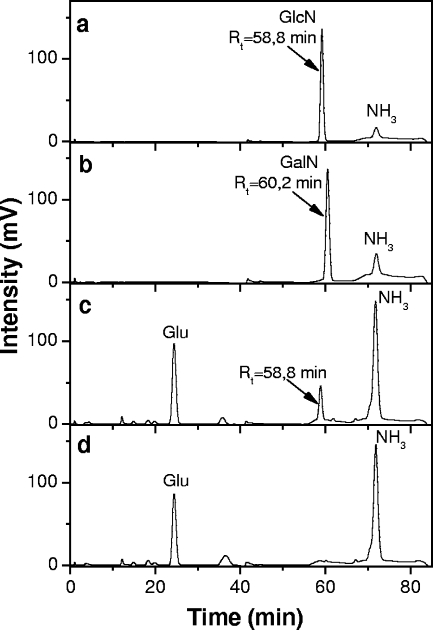
To further substantiate that the HexN was indeed GlcN, lipids A isolated from the B. pertussis wild-type strain BP338 and mutant BPM2859 were treated with hydrofluoric acid and the soluble released substituents were injected into an amino acid analyzer (Fig. 6) (3). A peak retention time of 58.8 min, corresponding to GlcN (Fig. 6a), was seen in the wild-type supernatant sample (Fig. 6c), whereas no HexN peak was detected in the supernatant sample of the mutant strain (Fig. 6d). Both dephosphorylated lipid A residues present in the pellet were hydrolyzed with hydrochloric acid and tested under the same conditions. This revealed peaks corresponding to the standard GlcN retention time (not shown), as expected for the composition of the disaccharide lipid A backbone. Taken together, these data indicate that lipid A of Bordetella is modified by a GlcN at both phosphate groups.
FIG. 6.
Amino acid analyzer elution profiles of hydrofluoric acid-released lipid A substituents. (a) Standard GlcN; (b) standard GalN; (c) derivatives released from wild-type strain BP338 lipid A; (d) derivatives released from mutant strain BPM2859. Rt, retention time.
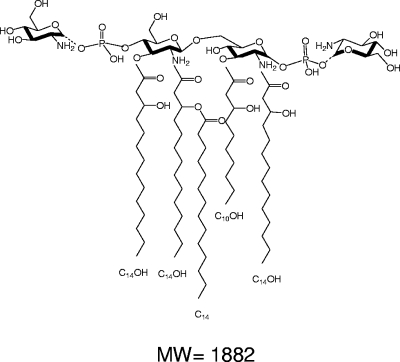
We next characterized the anomeries of the new phosphate-GlcN bonds. Since the limited amount and heterogeneous nature of the sample precluded analysis by nuclear magnetic resonance, we used a chemical method that was previously developed using synthetic, phosphorylated, N-acetylated GlcN to examine the anomery of GlcN I of lipid A. In that study, it was concluded that the glycoside had an α linkage (17). The stability of the phosphate anomers on the B. pertussis lipid A sample was thus defined under mild-acidic conditions. The two amino groups in the newly isolated lipid A were acetylated (Fig. 1d), and the preparation was exposed to the acidic conditions that break β bonds. MALDI-MS analysis after the hydrolysis showed that the sample was unmodified, allowing us to conclude that both additional GlcNs present in the lipid A were α linked and to propose the structure presented in Fig. 7.
FIG. 7.
Proposed structure for strain BP338 B. pertussis lipid A penta-acyl molecular species representing both phosphate groups substituted with GlcNs. Dotted-line bonds indicate incompleteness of the substitutions, leading to structures with one or no substituting GlcNs (m/z = 1,720 or 1,559, respectively). MW, molecular weight.
DISCUSSION
Structural analysis of lipids A from B. pertussis and B. bronchiseptica revealed the presence, in strains examined here, of HexNs substituting both lipid A phosphate groups that form the di-GlcN backbone. Analyses of GC and amino acid analyzer results identified these substituents as GlcNs. To our knowledge, this is the first description of a GlcN modification of lipid A in these positions in any bacterial species. The importance of phosphate groups in LPS biological activity interactions has been well documented (13, 18, 32, 42). Phosphate groups and an occasional pyrophosphate are usually present at positions C-1 and C-4′ in the lipid A moiety. However, exceptions with no or only one phosphate group or with a negatively charged sugar exist (75); few LPS examples have neutral sugars at this position (61). The negative charges of the phosphate groups can be neutralized by substitution with phosphorylethanolamine, GalN, or 4-amino-l-arabinose (l-Ara4N) (37). The presence of anionic groups in the lipid core region constitutes a strongly stabilized area, and positively charged substituents on the phosphate groups are known to increase the resistance of the bacteria to cationic antibiotics. The three-dimensional supramolecular structures of lipid A (conical, cylindrical, or lamellar) have been described in connection with the presence or absence of the substituents mentioned here. It has been postulated that these different shapes are deciding factors in endotoxic activity (9). Of note, lipid A with one phosphate group is much less toxic, while keeping its adjuvant properties (7, 13), whereas lipid A lacking both phosphate groups is inactive (47, 55, 62). In addition to l-Ara4N transfer to lipid A, S. enterica serovar Typhimurium can alter the number of its lipid A acyl chains by the action of the palmitoyl transferase PagP (31) and the deacylase PagL (70) or modify lipid A acyl chains by addition of a hydroxyl group catalyzed by the dioxygenase LpxO (29). Homologous genes of all these lipid A-modifying enzymes are found in other gram-negative bacteria as well (51), yet in most cases it is not known whether these genes are expressed and/or how they are regulated. In bordetellae, pagP is expressed in B. bronchiseptica in a Bvg-dependent manner, which is required for persistent colonization of the mouse respiratory tract (57) and for resistance to antibody-mediated complement lysis during B. bronchiseptica respiratory infection (56). However, pagP of B. pertussis strain Tohama I (locus tag BP3006) seems to be transcriptionally silent due to an IS481 insertion within its promoter region (54). Similarly, although pagL is intact in B. bronchiseptica and the functionality of the B. bronchiseptica pagL gene product in E. coli has been demonstrated (28), pagL (locus tag BP3592) is a pseudogene in B. pertussis strain Tohama I (54). Thus, these two mechanisms for modifying lipid A are unlikely to occur in B. pertussis, at least with respect to the Tohama I strain. The low endotoxicity of the Bordetella human strains has been noted previously and has been attributed to their low fatty acid substitution as well as to the shortness of their fatty acid chains and substituted Kdo (11). It remains to be determined whether the presence of positively charged substituents on the phosphate groups of Bordetella lipid A further impacts its endotoxic activity, perhaps by decreasing its propensity to act as a ligand of the TLR4-MD2-CD14 pathogen recognition receptor complex. We are currently testing this hypothesis.
The GlcN modification of Bordetella lipid A described in this report is reminiscent of the l-Ara4N lipid A modifications seen in Salmonella and Pseudomonas and the GalN lipid A modification seen in Francisella (51, 59). The glycosyl transferase ultimately responsible for catalyzing the addition of a pentosamine/hexosamine to lipid A in these bacteria is ArnT (formerly PmrK) (71).
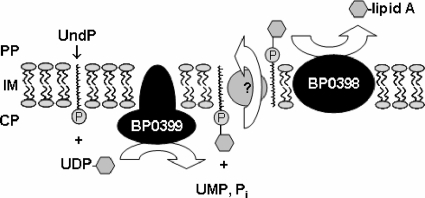
The genetic basis for a hexosamine modification on the phosphate moiety of lipid A has been determined for Salmonella, Pseudomonas, and Francisella. The seven- or eight-gene pmrF operon, which is regulated by PmrA/B, functions to synthesize and transfer l-Ara4N to Salmonella and Pseudomonas lipids A, thereby conferring resistance to polymyxin B (25). In vitro data suggest that the glycosyl transferase ArnC (formerly PmrF) catalyzes the formation of undecaprenyl phosphate (UndP)-β-sugar by use of activated UDP-sugar and the lipid carrier UndP as substrates (10), whereas the glycosyl transferase ArnT (formerly PmrK) transfers the sugar moiety (in the case of Salmonella, l-Ara4N) from bactoprenol (UndP-sugar) to lipid A, which is believed to proceed in the periplasm (71). The modified lipid A then transits to the outer membrane, where it is incorporated into the outer leaflet. More recently, Francisella tularensis subspecies novicida lipid A was shown to be modified by GalN, a process mediated by an ArnT ortholog (74). Here, we showed that the presence of positively charged substituents on the phosphate groups of lipid A of B. pertussis requires the expression of a Bvg-regulated gene with the locus tag BP0398, encoding a putative protein orthologous to ArnT of other gram-negative bacteria. Moreover, transcription analysis indicates that B. pertussis locus BP0398 is cotranscribed with an adjacent gene, locus BP0399, encoding a putative ArnC-like protein. This is in accordance with results of a previous study by Cummings et al. (19), who identified these genes as Bvg activated in B. pertussis and B. bronchiseptica by the use of whole-genome microarray analysis. We propose that the gene products of B. pertussis loci BP0398 and BP0399 as well as their counterparts in other bordetellae likely serve a function similar to that of ArnT and ArnC proteins of other gram-negative bacteria (Fig. 8).
FIG. 8.
Model for the catalytic reaction mediated by the gene products of BP0399 and BP0398 of B. pertussis Tohama I derivatives, representative of the orthologous proteins of other bordetellae. Hypothesized reactions are based on amino acid sequence similarities of the gene products of BP0399 and BP0398 to ArnC/GtrB and ArnT proteins, respectively. GlcN is symbolized by a hexagon. PP, periplasm; IM, inner membrane; CP, cytoplasm; Pi, phosphate (inorganic).
Analogous to the Bvg-dependent transcription of the BP0399/BP0398 loci in B. pertussis Tohama I (Fig. 2) and other bordetellae (19), lipid A modifications in S. enterica serovar Typhimurium, Pseudomonas aeruginosa, and other gram-negative bacteria are also regulated by two-component systems and therefore depend on environmental stimuli (25, 50). Interestingly, we observed that transcription of both glycosyl transferase genes (loci BP0398 and BP0399) was not abolished in B. pertussis BvgS mutant BP347, as low levels of mRNA were detectable, and expression levels seemed to be higher upon growth of strains BP338 and BP347 in SS broth than upon cultivation on BG agar. This phenomenon was not assessed in a whole-genome analysis performed by Cummings et al. (19) and suggests that the expression of loci BP0398 and BP0399 of B. pertussis wild-type strain BP338 is part of an additional, Bvg-independent regulatory mechanism.
None of the four B. pertussis strains analyzed earlier in this laboratory or samples received from other collaborators carried substituents on the lipid A phosphate groups, suggesting strain-specific variations in expression of the lipid A modification locus studied here. This is not unusual in Bordetella, as recent studies have demonstrated substantial transcriptional and genetic diversity among different isolates of the same Bordetella species (19, 20, 21). Little is known of how this affects virulence, but neutralization of phosphate groups is known to strengthen bacterial resistance to antibacterial peptides (30). It remains to be seen whether this is true for B. pertussis.
In the present work, in order to preserve the phosphate groups and their substituents during lipid A separation from the oligosaccharide, especially the one on GlcN I on account of its lability, we used SDS-promoted mild-acid hydrolysis or, alternatively, β elimination. The latter procedure also offers mild conditions when the glycosidic phosphate group and possible substituent have to be preserved. It requires more LPS and confirmation that the adjacent Kdo molecule has free OH groups at C-7 and C-8. When only small amounts of bacteria were available, LPS extraction or lipid A cleavage was performed directly on bacterial cells; this is why we applied these methods to the B. pertussis wild-type and mutant strains. This method was shown to be as mild as, if not milder than, the SDS-promoted hydrolysis. We do not believe that hydrolyses could be responsible for a total loss of GlcN substituents when present and be the reason for the absence of these elements in previous reports. GlcN at C-4 is acid resistant, and the one at C-1 could be only partially lost together with the phosphate group under classical mild-acid conditions.
The structural modification capacities of Bordetella lipids A now attain seven positions of a single molecule: (i) the characteristic fatty acid asymmetry and variability at C-3 and C-3′, shown to be specific in B. pertussis and B. parapertussis but variable with B. bronchiseptica (79); (ii) the 2-hydroxylation of C14 in the acyloxyacyl position at C-2′ with B. avium, B. trematum, and B. hinzii (4, 11) (unpublished data); and (iii) the additional substitution of a C16 in the secondary position at C-3′, reported to occur in B. bronchiseptica and B. hinzii (57), and this newly described GlcN substitution of the phosphate groups in B. bronchiseptica and B. pertussis, also detected in other bordetellae (M. Caroff, unpublished data).
Acknowledgments
This work was funded in part by grants from the Natural Sciences and Engineering Research Council of Canada (R.C.F.).
The participation of Alexey Novikov (IBBMC, Orsay, France) for MS experiments and work on figures was greatly appreciated. We thank Lando Robillo for help with the B. pertussis sample preparation.
Footnotes
Published ahead of print on 18 April 2008.
REFERENCES
- 1.AFMB-CNRS. 9 February 2007, posting date. CAZy—carbohydrate-active enzymes. AFMB-CNRS, Universités Aix-Marseille I and II, Marseille, France. http://afmb.cnrs-mrs.fr/CAZY/.
- 2.Allison, G. E., and N. K. Verma. 2000. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 817-23. [DOI] [PubMed] [Google Scholar]
- 3.Auger, G., J. van Heijenoort, D. Mengin-Lecreulx, and D. Blanot. 2003. A MurG assay which utilises a synthetic analogue of lipid I. FEMS Microbiol. Lett. 219115-119. [DOI] [PubMed] [Google Scholar]
- 4.Aussel, L., J. R. Brisson, M. B. Perry, and M. Caroff. 2000. Structure of the lipid A of Bordetella hinzii ATCC 51730. Rapid Commun. Mass Spectrom. 14595-599. [DOI] [PubMed] [Google Scholar]
- 5.Aussel, L., R. Chaby, K. Le Blay, J. Kelly, P. Thibault, M. B. Perry, and M. Caroff. 2000. Chemical and serological characterization of the Bordetella hinzii lipopolysaccharides. FEBS Lett. 48540-46. [DOI] [PubMed] [Google Scholar]
- 6.Aussel, L., H. Therisod, D. Karibian, M. B. Perry, M. Bruneteau, and M. Caroff. 2000. Novel variation of lipid A structures in strains of different Yersinia species. FEBS Lett. 46587-92. [DOI] [PubMed] [Google Scholar]
- 7.Ayme, G., M. Caroff, R. Chaby, N. Haeffner-Cavaillon, A. Le Dur, M. Moreau, M. Muset, M. C. Mynard, M. Roumiantzeff, D. Schulz, and L. Szabo. 1980. Biological activities of fragments derived from Bordetella pertussis endotoxin: isolation of a nontoxic, Shwartzman-negative lipid A possessing high adjuvant properties. Infect. Immun. 27739-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brade, L., R. Engel, W. J. Christ, and E. T. Rietschel. 1997. A nonsubstituted primary hydroxyl group in position 6′ of free lipid A is required for binding of lipid A monoclonal antibodies. Infect. Immun. 653961-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandenburg, K., H. Mayer, M. H. Koch, J. Weckesser, E. T. Rietschel, and U. Seydel. 1993. Influence of the supramolecular structure of free lipid A on its biological activity. Eur. J. Biochem. 218555-563. [DOI] [PubMed] [Google Scholar]
- 10.Breazeale, S. D., A. A. Ribeiro, A. L. McClerren, and C. R. Raetz. 2005. A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-L-arabinose. Identification and function oF UDP-4-deoxy-4-formamido-L-arabinose. J. Biol. Chem. 28014154-14167. [DOI] [PubMed] [Google Scholar]
- 11.Caroff, M., L. Aussel, H. Zarrouk, A. Martin, J. C. Richards, H. Therisod, M. B. Perry, and D. Karibian. 2001. Structural variability and originality of the Bordetella endotoxins. J. Endotoxin Res. 763-68. [PubMed] [Google Scholar]
- 12.Caroff, M., J. Brisson, A. Martin, and D. Karibian. 2000. Structure of the Bordetella pertussis 1414 endotoxin. FEBS Lett. 4778-14. [DOI] [PubMed] [Google Scholar]
- 13.Caroff, M., J. M. Cavaillon, C. Fitting, and N. Haeffner-Cavaillon. 1986. Inability of pyrogenic, purified Bordetella pertussis lipid A to induce interleukin-1 release by human monocytes. Infect. Immun. 54465-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caroff, M., R. Chaby, D. Karibian, J. Perry, C. Deprun, and L. Szabo. 1990. Variations in the carbohydrate regions of Bordetella pertussis lipopolysaccharides: electrophoretic, serological, and structural features. J. Bacteriol. 1721121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caroff, M., C. Deprun, and D. Karibian. 1993. 252Cf plasma desorption mass spectrometry applied to the analysis of underivatized rough-type endotoxin preparations. J. Biol. Chem. 26812321-12324. [PubMed] [Google Scholar]
- 16.Caroff, M., D. Karibian, J. M. Cavaillon, and N. Haeffner-Cavaillon. 2002. Structural and functional analyses of bacterial lipopolysaccharides. Microbes Infect. 4915-926. [DOI] [PubMed] [Google Scholar]
- 17.Caroff, M., A. Tacken, and L. Szabo. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175273-282. [DOI] [PubMed] [Google Scholar]
- 18.Cavaillon, J. M., C. Fitting, M. Caroff, and N. Haeffner-Cavaillon. 1989. Dissociation of cell-associated interleukin-1 (IL-1) and IL-1 release induced by lipopolysaccharide and lipid A. Infect. Immun. 57791-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cummings, C. A., H. J. Bootsma, D. A. Relman, and J. F. Miller. 2006. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 1881775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummings, C. A., M. M. Brinig, P. W. Lepp, S. van de Pas, and D. A. Relman. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 1861484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diavatopoulos, D. A., C. A. Cummings, L. M. Schouls, M. M. Brinig, D. A. Relman, and F. R. Mooi. 2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Fabio, J. L., M. Caroff, D. Karibian, J. C. Richards, and M. B. Perry. 1992. Characterization of the common antigenic lipopolysaccharide O-chains produced by Bordetella bronchiseptica and Bordetella parapertussis. FEMS Microbiol. Lett. 76275-281. [DOI] [PubMed] [Google Scholar]
- 23.Dixon, D. R., and R. P. Darveau. 2005. Lipopolysaccharide heterogeneity: innate host responses to bacterial modification of lipid a structure. J. Dent. Res. 84584-595. [DOI] [PubMed] [Google Scholar]
- 24.El Hamidi, A., A. Tirsoaga, A. Novikov, A. Hussein, and M. Caroff. 2005. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J. Lipid Res. 461773-1778. [DOI] [PubMed] [Google Scholar]
- 25.Ernst, R. K., T. Guina, and S. I. Miller. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 31327-1334. [DOI] [PubMed] [Google Scholar]
- 26.Erridge, C., E. Bennett-Guerrero, and I. R. Poxton. 2002. Structure and function of lipopolysaccharides. Microbes Infect. 4837-851. [DOI] [PubMed] [Google Scholar]
- 27.Finn, T. M., and D. F. Amsbaugh. 1998. Vag8, a Bordetella pertussis bvg-regulated protein. Infect. Immun. 663985-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geurtsen, J., L. Steeghs, J. T. Hove, P. van der Ley, and J. Tommassen. 2005. Dissemination of lipid A deacylases (pagL) among gram-negative bacteria: identification of active-site histidine and serine residues. J. Biol. Chem. 2808248-8259. [DOI] [PubMed] [Google Scholar]
- 29.Gibbons, H. S., S. Lin, R. J. Cotter, and C. R. Raetz. 2000. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J. Biol. Chem. 27532940-32949. [DOI] [PubMed] [Google Scholar]
- 30.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 271171-1182. [DOI] [PubMed] [Google Scholar]
- 31.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95189-198. [DOI] [PubMed] [Google Scholar]
- 32.Haeffner-Cavaillon, N., M. Caroff, and J. M. Cavaillon. 1989. Interleukin-1 induction by lipopolysaccharides: structural requirements of the 3-deoxy-D-manno-2-octulosonic acid (KDO). Mol. Immunol. 26485-494. [DOI] [PubMed] [Google Scholar]
- 33.Hase, S., and Y. Matsushima. 1969. Amino sugar analysis by gas-liquid chromatography. J. Biochem. (Tokyo) 6657-62. [DOI] [PubMed] [Google Scholar]
- 34.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20197-216. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, K. G., and M. B. Perry. 1976. Improved techniques for the preparation of bacterial lipopolysaccharides. Can. J. Microbiol. 2229-34. [DOI] [PubMed] [Google Scholar]
- 36.Karibian, D., C. Deprun, and M. Caroff. 1993. Comparison of lipids A of several Salmonella and Escherichia strains by 252Cf plasma desorption mass spectrometry. J. Bacteriol. 1752988-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly, J., H. Masoud, M. B. Perry, J. C. Richards, and P. Thibault. 1996. Separation and characterization of O-deacylated lipooligosaccharides and glycans derived from Moraxella catarrhalis using capillary electrophoresis-electrospray mass spectrometry and tandem mass spectrometry. Anal. Biochem. 23315-30. [DOI] [PubMed] [Google Scholar]
- 38.Kirikae, T., F. U. Schade, U. Zahringer, F. Kirikae, H. Brade, S. Kusumoto, T. Kusama, and E. T. Rietschel. 1994. The significance of the hydrophilic backbone and the hydrophobic fatty acid regions of lipid A for macrophage binding and cytokine induction. FEMS Immunol. Med. Microbiol. 813-26. [DOI] [PubMed] [Google Scholar]
- 39.Larocque, S., J. R. Brisson, H. Therisod, M. B. Perry, and M. Caroff. 2003. Structural characterization of the O-chain polysaccharide isolated from Bordetella avium ATCC 5086: variation on a theme. FEBS Lett. 53511-16. [DOI] [PubMed] [Google Scholar]
- 40.Lasfargues, A., M. Caroff, and R. Chaby. 1993. Structural features involved in the mitogenic activity of Bordetella pertussis lipopolysaccharides for spleen cells of C3H/HeJ mice. FEMS Immunol. Med. Microbiol. 7119-129. [DOI] [PubMed] [Google Scholar]
- 41.Lebbar, S., M. Caroff, L. Szabo, C. Merienne, and L. Szilogyi. 1994. Structure of a hexasaccharide proximal to the hydrophobic region of lipopolysaccharides present in Bordetella pertussis endotoxin preparations. Carbohydr. Res. 259257-275. [DOI] [PubMed] [Google Scholar]
- 42.Lebbar, S., J. M. Cavaillon, M. Caroff, A. Ledur, H. Brade, R. Sarfati, and N. Haeffner-Cavaillon. 1986. Molecular requirement for interleukin 1 induction by lipopolysaccharide-stimulated human monocytes: involvement of the heptosyl-2-keto-3-deoxyoctulosonate region. Eur. J. Immunol. 1687-91. [DOI] [PubMed] [Google Scholar]
- 43.Lebbar, S., D. Karibian, C. Deprun, and M. Caroff. 1994. Distribution of lipid A species between long and short chain lipopolysaccharides isolated from Salmonella, Yersinia, and Escherichia as seen by 252Cf plasma desorption mass spectrometry. J. Biol. Chem. 26931881-31884. [PubMed] [Google Scholar]
- 44.Le Blay, K., M. Caroff, F. Blanchard, M. B. Perry, and R. Chaby. 1996. Epitopes of Bordetella pertussis lipopolysaccharides as potential markers for typing of isolates with monoclonal antibodies. Microbiology 142971-978. [DOI] [PubMed] [Google Scholar]
- 45.Le Blay, K., M. Caroff, J. C. Richards, M. B. Perry, and R. Chaby. 1994. Specific and cross-reacting monoclonal antibodies to Bordetella parapertussis and Bordetella bronchiseptica lipopolysaccharides. Microbiology 1402459-2465. [DOI] [PubMed] [Google Scholar]
- 46.Le Dur, A., M. Caroff, R. Chaby, and L. Szabo. 1978. A novel type of endotoxin structure present in Bordetella pertussis. Isolation of two different polysaccharides bound to lipid A. Eur. J. Biochem. 84579-589. [DOI] [PubMed] [Google Scholar]
- 47.Loppnow, H., H. Brade, I. Durrbaum, C. A. Dinarello, S. Kusumoto, E. T. Rietschel, and H. D. Flad. 1989. IL-1 induction-capacity of defined lipopolysaccharide partial structures. J. Immunol. 1423229-3238. [PubMed] [Google Scholar]
- 48.Marr, N. 2007. Evasion of complement-mediated host defenses by Bordetella pertussis via recruitment of human C1-esterase inhibitor. Doctoral Thesis. University of Wuerzburg, Wuerzburg, Germany.
- 49.Mattoo, S., and J. D. Cherry. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18326-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McPhee, J. B., S. Lewenza, and R. E. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50205-217. [DOI] [PubMed] [Google Scholar]
- 51.Miller, S. I., R. K. Ernst, and M. W. Bader. 2005. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 336-46. [DOI] [PubMed] [Google Scholar]
- 52.Muroi, M., and K. Tanamoto. 2002. The polysaccharide portion plays an indispensable role in Salmonella lipopolysaccharide-induced activation of NF-κB through human toll-like receptor 4. Infect. Immun. 706043-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 3532-40. [DOI] [PubMed] [Google Scholar]
- 55.Persing, D. H., R. N. Coler, M. J. Lacy, D. A. Johnson, J. R. Baldridge, R. M. Hershberg, and S. G. Reed. 2002. Taking toll: lipid A mimetics as adjuvants and immunomodulators. Trends Microbiol. 10S32-S37. [DOI] [PubMed] [Google Scholar]
- 56.Pilione, M. R., E. J. Pishko, A. Preston, D. J. Maskell, and E. T. Harvill. 2004. pagP is required for resistance to antibody-mediated complement lysis during Bordetella bronchiseptica respiratory infection. Infect. Immun. 722837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Preston, A., E. Maxim, E. Toland, E. J. Pishko, E. T. Harvill, M. Caroff, and D. J. Maskell. 2003. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol. Microbiol. 48725-736. [DOI] [PubMed] [Google Scholar]
- 58.Preston, A., B. O. Petersen, J. O. Duus, J. Kubler-Kielb, G. Ben-Menachem, J. Li, and E. Vinogradov. 2006. Complete structures of Bordetella bronchiseptica and Bordetella parapertussis lipopolysaccharides. J. Biol. Chem. 28118135-18144. [DOI] [PubMed] [Google Scholar]
- 59.Raetz, C. R., C. M. Reynolds, M. S. Trent, and R. E. Bishop. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76295-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rau, H., U. Seydel, M. Freudenberg, J. Weckesser, and H. Mayer. 1995. Lipopolysaccharide of Rhodospirillum salinarum 40: structural studies on the core and lipid A region. Arch. Microbiol. 164280-289. [DOI] [PubMed] [Google Scholar]
- 62.Rietschel, E. T., T. Kirikae, F. U. Schade, U. Mamat, G. Schmidt, H. Loppnow, A. J. Ulmer, U. Zahringer, U. Seydel, F. Di Padova, et al. 1994. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 8217-225. [DOI] [PubMed] [Google Scholar]
- 63.Sebaihia, M., A. Preston, D. J. Maskell, H. Kuzmiak, T. D. Connell, N. D. King, P. E. Orndorff, D. M. Miyamoto, N. R. Thomson, D. Harris, A. Goble, A. Lord, L. Murphy, M. A. Quail, S. Rutter, R. Squares, S. Squares, J. Woodward, J. Parkhill, and L. M. Temple. 2006. Comparison of the genome sequence of the poultry pathogen Bordetella avium with those of B. bronchiseptica, B. pertussis, and B. parapertussis reveals extensive diversity in surface structures associated with host interaction. J. Bacteriol. 1886002-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skeeles, J. K., and L. H. Arp. 1997. Bordetellosis (turkey coryza), p. 275-288. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. R. McDougal, and Y. M. Saif (ed.), Diseases of poultry. Iowa State University Press, Ames, IA.
- 65.Stainer, D. W., and M. J. Scholte. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63211-220. [DOI] [PubMed] [Google Scholar]
- 66.Therisod, H., V. Labas, and M. Caroff. 2001. Direct microextraction and analysis of rough-type lipopolysaccharides by combined thin-layer chromatography and MALDI mass spectrometry. Anal. Chem. 733804-3807. [DOI] [PubMed] [Google Scholar]
- 67.Therisod, H., M. A. Monteiro, M. B. Perry, and M. Caroff. 2001. Helicobacter mustelae lipid A structure differs from that of Helicobacter pylori. FEBS Lett. 4991-5. [DOI] [PubMed] [Google Scholar]
- 68.Tirsoaga, A., A. El Hamidi, M. B. Perry, M. Caroff, and A. Novikov. 2007. A rapid, small-scale procedure for the structural characterization of lipid A applied to Citrobacter and Bordetella strains: discovery of a new structural element. J. Lipid Res. 482419-2427. [DOI] [PubMed] [Google Scholar]
- 69.Tirsoaga, A., A. Novikov, M. Adib-Conquy, C. Werts, C. Fitting, J. M. Cavaillon, and M. Caroff. 2007. Simple method for repurification of endotoxins for biological use. Appl. Environ. Microbiol. 731803-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trent, M. S., W. Pabich, C. R. Raetz, and S. I. Miller. 2001. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 2769083-9092. [DOI] [PubMed] [Google Scholar]
- 71.Trent, M. S., A. A. Ribeiro, S. Lin, R. J. Cotter, and C. R. Raetz. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 27643122-43131. [DOI] [PubMed] [Google Scholar]
- 72.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119115-119. [DOI] [PubMed] [Google Scholar]
- 73.Vinogradov, E., and M. Caroff. 2005. Structure of the Bordetella trematum LPS O-chain subunit. FEBS Lett. 57918-24. [DOI] [PubMed] [Google Scholar]
- 74.Wang, X., A. A. Ribeiro, Z. Guan, S. C. McGrath, R. J. Cotter, and C. R. Raetz. 2006. Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida. Biochemistry 4514427-14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weintraub, A., U. Zahringer, H. W. Wollenweber, U. Seydel, and E. T. Rietschel. 1989. Structural characterization of the lipid A component of Bacteroides fragilis strain NCTC 9343 lipopolysaccharide. Eur. J. Biochem. 183425-431. [DOI] [PubMed] [Google Scholar]
- 76.Weiss, A. A., E. L. Hewlett, G. A. Myers, and S. Falkow. 1983. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect. Immun. 4233-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weiss, A. A., A. R. Melton, K. E. Walker, C. Andraos-Selim, and J. J. Meidl. 1989. Use of the promoter fusion transposon Tn5 lac to identify mutations in Bordetella pertussis vir-regulated genes. Infect. Immun. 572674-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wellcome Trust Sanger Institute. 31 October 2005, posting date. Bordetella pertussis, B. parapertussis and B. bronchiseptica. Genome project. Wellcome Trust Sanger Institute Hinxton, Cambridge, United Kingdom. http://www.sanger.ac.uk/Projects/B_pertussis/.
- 79.Zarrouk, H., D. Karibian, S. Bodie, M. B. Perry, J. C. Richards, and M. Caroff. 1997. Structural characterization of the lipids A of three Bordetella bronchiseptica strains: variability of fatty acid substitution. J. Bacteriol. 1793756-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]