Abstract
Mrp antiporters catalyze secondary Na+(Li+)/H+ antiport and/or K+/H+ antiport that is physiologically important in diverse bacteria. An additional capacity for anion flux has been observed for a few systems. Mrp is unique among antiporters in that it requires all six or seven hydrophobic gene products (MrpA to MrpG) of the mrp operon for full antiporter activity, but MrpE has been reported to be dispensable. Here, the membrane complexes formed by Mrp proteins were examined using a cloned mrp operon from alkaliphilic Bacillus pseudofirmus OF4. The operon was engineered so that the seven Mrp proteins could be detected in single samples. Membrane extracts of an antiporter-deficient Escherichia coli strain expressing this construct were analyzed by blue native-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Mrp complexes of two sizes were identified containing all seven Mrp proteins. Studies of the single nonpolar mrp gene deletions in the construct showed that a subcomplex of MrpA, MrpB, MrpC, and MrpD was formed in the absence of MrpE, MrpF, or MrpG. By contrast, MrpE, MrpF, and MrpG were not observed in membranes lacking MrpA, MrpB, MrpC, or MrpD. Although MrpA and MrpD have been hypothesized to be the antiporter proteins, the MrpA-to-D complex was inactive. Every Mrp protein was required for an activity level near that of the wild-type Na+/H+ antiporter, but a very low activity level was observed in the absence of MrpE. The introduction of an MrpE(P114G) mutation into the full Mrp complex led to antiport activity with a greatly increased apparent Km value for Na+. The results suggested that interactions among the proteins of heterooligomeric Mrp complexes strongly impact antiporter properties.
Monovalent cation/proton antiporters of bacteria catalyze the efflux of ions such as Na+, Li+, K+, and NH4+ in exchange for extracellular H+ (7, 27, 29, 31). These antiporters prevent toxic levels of the cations from accumulating in the cytoplasm. They also support alkaline pH homeostasis and osmoregulation (27, 28, 45). The Mrp type antiporter studied here is among the cation/proton antiporter types most recently described (10). It is widespread in both gram-positive and gram-negative bacteria (39). Roles for Mrp antiporters have already been shown in alkaline pH homeostasis and Na+ resistance (10, 15, 39), sporulation (22), symbiotic nitrogen fixation (30), pathogenesis (21), arsenite resistance (19), and bile salt resistance (5, 15, 16). Mrp antiporters are classified in their own family, the cation/proton antiporter-3 family of the transporter classification system, because of their unique complexity (34, 39). This complexity is hypothesized to be structural as well as functional.
Mrp systems were hypothesized to form heterooligomeric complexes because Mrp antiporter activity depended upon the presence of all seven hydrophobic gene products of a typical mrp operon (13, 16). By contrast, the activity of other prokaryotic and eukaryotic monovalent cation/proton antiporters requires only a single hydrophobic gene product (7, 8, 32, 44). The idea of an Mrp complex was fostered by the sequence similarities of MrpA, MrpC, and MrpD to subunits that are found in membrane-embedded subcomplexes of ion-pumping NADH:quinone oxidoreductases and bacterial hydrogenases (2, 6, 10, 11, 20, 23, 24, 40). Recently, Kajiyama et al. (17) provided evidence for a physical complex of Mrp proteins. They used a panel of seven Bacillus subtilis mutants. Each mutant expressed a His-tagged version of a different mrp gene product. Partially purified membrane extracts from each mutant were fractionated by blue native-polyacrylamide gel electrophoresis (BN-PAGE). Immunoblot analyses that probed the His tag detected an ∼410-kDa band in each of the membranes, suggesting that an Mrp complex containing all seven Mrp proteins was formed in B. subtilis.
With respect to the functional complexity of Mrp systems, we hypothesized that Mrp was a consortium of cation and anion transporters (39). This was supported by evidence that the Mrp-dependent efflux of bile salts in B. subtilis was associated with MrpF (16), so the anion-transporting Mrp proteins are probably distinct from the MrpA and MrpD proteins that are proposed to carry out cation/proton antiporter activity (24). The individual Mrp transporter proteins might depend upon one another for stability and/or assembly into a catalytically active form. A consortium of transporters that forms a sizeable complex might also be advantageous for cation/proton antiport activity at the high pH values that are typical for Mrp systems (39, 41). The external surface of a large heterooligomeric complex could enhance proton gathering at the alkaline pH of the outer surface of the membrane. This would provide kinetic support for cation/proton antiport activity (40, 41).
The goals of the current study were to develop an experimental system in which each of the seven Mrp proteins could be monitored in single preparations. We could then characterize Mrp complexes and subcomplexes that have different subunit compositions and monitor membrane levels of each Mrp protein side by side with assays of the activity in mutant membranes. We constructed a plasmid-borne full mrp operon from alkaliphilic Bacillus pseudofirmus OF4, in which each of the seven gene products could be individually tracked via a combination of antibodies raised against synthetic peptides and commercial antibodies to epitope tags that were genetically introduced. The Mrp from an alkaliphilic Bacillus strain was of special comparative interest to the B. subtilis Mrp. The problem of proton gathering on the outer membrane surface in support of Na+/H+ antiport activity would be much greater at the optimal external pH of 10.5 for B. pseudofirmus OF4 growth than at the nearly neutral pH for B. subtilis growth (27). The construct was expressed in an antiporter-deficient strain of Escherichia coli so that properties of antiport could be assayed in a well-established vesicle system. The following specific goals were pursued. The first goal was to rigorously examine whether all seven Mrp proteins were present in an individual complex in single preparations, which was not demonstrated in the report of the B. subtilis Mrp complex (17). The second goal was to determine whether the Mrp complex(es) formed by the alkaliphile were comparable or different from that found for B. subtilis Mrp. The third goal was to determine whether each of the seven gene alkaliphile Mrp proteins was required for antiporter activity and/or complex formation, using individual mrp gene deletions. In addition, a panel of site-directed mrpE mutants was prepared to more fully assess the impact of MrpE on the antiporter activity of the full Mrp system. Although early studies indicated that full Mrp activity depends upon the presence of every protein, MrpE was suggested to be dispensable, since a B. subtilis mutant carrying a deletion of mrpE retained significant Na+ resistance and Na+ efflux capacity (46).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The medium used for routine growth of E. coli was LB medium (pH 7.0) (35) for strain DH5αMCR, and LBK medium (Luria broth with KCl instead of Nacl) plus 50 mM NaCl (pH 7.5) (9) was used for strain KNabc (26). Cells were grown with shaking at 37°C. Ampicillin at 100 μg/ml, Chloramphenicol at 5 μg/ml, and kanamycin at 25 μg/ml were added for growing plasmid-bearing cells and selecting transformants. Transformation of E. coli strains and all recombinant DNA manipulations was carried out by standard methods (35).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and description | Source and reference |
|---|---|---|
| E. coli strains | ||
| DH5αMCR | F−mcrAΔ1 (mrr-hsd RMS-mcrBC) Φ80dlacZ Δ(lacZYAargF)U169 deoR recA1 endA1 supE44 λthi-1 gyr-496 relA1 | Stratagene |
| KNabc | TG1 (ΔnhaA ΔnhaB ΔchaA) | 26 |
| B. pseudofirmus strains | ||
| OF4 | ||
| 811M | Wild type; Met− | 4 |
| Plasmids | ||
| pETDuet1 | ColE1 replicon plasmid; Apr | Novagen |
| pGEM-5zf(+) | Cloning vector; Apr | Promega |
| pGEM-7zf(+) | Cloning vector; Apr | Promega |
| pUC4K | E. coli plasmid vector; Kmr | Pharmacia |
| pMW119 | Cloning vector; Apr | Nippon Gene |
| pGEMKm | pGEM7Zf(+) + BamHI-digested Kmr fragment from pUC4K | This study |
| pMWKm | pMW119 + BamHI-digested Kmr fragment from pUC4K | This study |
| pMW-S | pMW119 + truncated mrp fragment (truncated MrpE, MrpF, and MrpG-S tag) | This study |
| pMWHis7 | pMWKm + truncated mrp fragment (truncated MrpD-His7 tag, MrpE, MrpF, and MrpG-S tag) | This study |
| pMWOFMrp | pMW118 + full mrp operon from B. pseudofirmus OF4 | 14 |
| pGEMOF4mrp16 | pGEM7Zf(+) + full mrp operon from B. pseudofirmus OF4 | This study |
| pDuetmrp5455 | pETDuet + full OF4 mrp operon (His-MrpA, MrpG-S tag) | This study |
| pDuetmrp5455ΔHis | pETDuet + full OF4 mrp operon (MrpG-S tag) | This study |
| pDuetmrpTS17 | pETDuet + full OF4 mrp operon (MrpA-T7 tag, MrpG-S tag) | This study |
| pDuetmrpTHS2 | pETDuet + full OF4 mrp operon (MrpA-T7 tag, MrpD-His10 tag with a mutation, MrpG-S tag) | This study |
| pDuetmrpTHS7 | pETDuet + full OF4 mrp operon (MrpA-T7 tag, MrpD-His7 tag, MrpG-S tag) | This study |
| pDuetmrpHS13 | pETDuet + full OF4 mrp operon (MrpD-His10 tag with a mutation, MrpG-S tag) | This study |
| pDuetmrpHS7 | pETDuet + full OF4 mrp operon (MrpD-His7 tag with a mutation, MrpG-S tag) | This study |
| pGEMmrpTH | pGEM-3Zf(+) + full OF4 mrp operon (MrpA-T7 tag, MrpD-His10 tag with a mutation)a | This study |
| pGEMmrpHS7 | pGEMOF4mrp16, MrpD-His7 tag, MrpG-S tag (mutation free)a | This study |
| pGEMmrpTCH | p pGEMmrpTH, MrpC-c-myc tag | This study |
| pGEMmrpTCHS | pGEMmrpTCH, MrpG-S tag | This study |
| pGEMmrpTFCHS | pGEMmrpTCHS, MrpB-FLAG tag | This study |
| pGEMmrpTFCHS7 | pGEMmrpTFCHS, MrpD-His7 tag (mutation free)a | This study |
| pMWΔA | pMW119 + ΔmrpA fragment | This study |
| pGEMΔB | pGEM5zf(+) + ΔmrpB fragment | This study |
| pGEMΔC | pGEM5zf(+) + ΔmrpC fragment | This study |
| pGEMmrpΔE | pGEM7zf(+) + ΔmrpE fragment | This study |
| pGEMmrpTFCHΔE | pGEMmrpTFCH + ΔmrpE fragment | This study |
| pGEMmrpΔF | pGEM7zf(+) + ΔmrpF fragment | This study |
| pGEMmrpΔG | pGEM7zf(+) + ΔmrpG fragment | This study |
| pGEMmrpTFCHSΔA | pGEMmrpTFCHS7, ΔmrpA | This study |
| pGEMmrpTFCHSΔB | pGEMmrpTFCHS7, ΔmrpB | This study |
| pGEMmrpTFCHSΔC | pGEMmrpTFCHS7, ΔmrpC | This study |
| pGEMmrpTFCHSΔD | pGEMmrpTFCHS7, ΔmrpD | This study |
| pGEMmrpTFCHSΔE | pGEMmrpTFCHS7, ΔmrpE | This study |
| pGEMmrpTFCHSΔF | pGEMmrpTFCHS7, ΔmrpF | This study |
| pGEMmrpTFCHSΔG | pGEMmrpTFCHS7, ΔmrpG | This study |
As detailed (see Fig. S1 in the supplemental material and the text), an intermediate in one of the constructions (pGEMmrpTH) had an error that was corrected to yield mutation-free pGEMmrpHS7 (another intermediate) and pGEMmrpTFCHS7 (final product).
Antibodies against MrpE and MrpF and the construction of MrpTFCHS7, a full Mrp system with five epitope tags on the other Mrp proteins.
Polyclonal antibodies were raised against synthetic peptides corresponding to regions of MrpE and MrpF and containing an N-terminal cysteine (C+ in the peptides shown below) for the conjugation of keyhole limpet hemocyanin. The peptide sequence for MrpE was C+AVPTKLKDWELS (corresponding to residues 92 to 104) and for MrpF was C+SIALSKFIERGVVFDRG (corresponding to residues 75 to 91). The conjugated peptides were injected into rabbits, and polyclonal antibodies were purified from serum using a Melon gel immunoglobulin G spin purification kit (Pierce). These antibodies were used to detect MrpE and MrpF in the immunoblotting analyses of this study.
A plasmid-borne full mrp operon was constructed such that the other five Mrp proteins, MrpA, MrpB, MrpC, MrpD, and MrpG, would be uniquely epitope tagged at their C-terminal ends with, respectively, a T7 tag, a FLAG tag, a c-myc tag, a hepta-histidine tag, and an S tag. Construction of the tagged full mrp construct, pGEMmrpTFCHS7, was carried out as shown in Fig. S1 in the supplemental material. All PCRs were carried out with AccuPrime Pfx DNA polymerase (Invitrogen) according to the manufacturer's instructions. The primers for the PCRs are listed in Table S1 in the supplemental material. The sequence of each plasmid was verified by analyses performed by Operon Biotechnologies (Tokyo, Japan), using an ABI-100 model 377 sequencer. The steps for the construction were as follows. For the construction of pDuetmrp5455 (with His6-MrpA and MrpG-S tags), PCR was performed with B. pseudofirmus OF4 genomic DNA with the set of primers OF4Mrp54NcoI and OF4Mrp55XhoI. OF4Mrp54NcoI has additional nucleotides encoding an NcoI site. OF4Mrp55XhoI has additional nucleotides encoding an XhoI site. The amplified PCR product was digested with NcoI and XhoI and then ligated into EcoRI-XhoI-digested pETDuet1 (Novagen), using Quick DNA ligase (New England Biolabs). This procedure resulted in mrpA with a His tag sequence fused at its N terminus and mrpG with an S tag sequence fused at its C terminus.
For the construction of pDuetmrp5455ΔHis (with the MrpG-S tag), two independent PCRs were performed withwith pDuetmrp5455 as the template with the sets of primers pETDuet-EcoNI and MrpA-del-His-R plus OF4Mrp28 and MrpA-del-His-F. The two purified PCR products were used as the template for a second PCR with primers pETDuet-EcoNI and OF4Mrp28. The purified PCR product of this reaction was digested with EcoNI and BglII and then cloned into EcoNI- and BglII-digested pDuetmrp5455. This procedure resulted in the deletion of the His tag region from the His6-mrpA fusion gene.
For construction of pDuetmrpTS17 (with the MrpA-T7 and MrpG-S tags), two independent PCRs were performed with B. pseudofirmus OF4 genomic DNA with the sets of primers OF4B-E1 and MrpA-T7-R plus OF4Mrp9 and MrpA-T7-F. The two purified PCR products were used as the template for a second PCR with primers OF4B-E1 and OF4Mrp9. The purified PCR product of this reaction was digested with NsiI and BamHI and then cloned into NsiI-BamHI-digested pDuetmrp5455ΔHis. This procedure resulted in the mrpA-T7 tag gene fusion in the operon.
For construction of pDuetmrpHS13 (with the MrpD-His10 and MrpG-S tags), two independent PCRs were performed with B. pseudofirmus OF4 genomic DNA and pDuetmrp5455 with the sets of primers OF4Mrp3 and MrpD-His10-R plus the T7 terminator primer and MrpD-His10-F, respectively. The two purified PCR products were used as the template for a second PCR with the primers OF4Mrp3 and T7 terminator. The purified PCR product of this reaction was digested with XhoI and BamHI and then cloned into XhoI-BamHI-digested pDuetmrp5455ΔHis. This procedure resulted in the mrpD-His10 tag gene fusion in the operon.
For the construction of pDuetmrpTHS2 (with the MrpA-T7, MrpD-His10, and MrpG-S tags), the truncated mrp fragment (2.2 kb, containing the mrpD-His10 region) of BamHII- and NsiI-digested pDuetmrpHS13 was ligated with the BamHI- and NsiI-digested large fragment (ca. 9 kb) of pDuetmrpTS17, yielding pDuetmrpTHS2.
For the construction of pGEMOF4mrp16 (tag-free plasmid), pMWOFMrp (14) was digested with EcoRI and XmaI, and then the purified product containing the mrp operon and its putative promoter region was ligated into EcoRI- and XmaI-digested pGEM-7Zf(+), yielding pGEMOF4mrp16.
For the construction of pGEMmrpTH (with MrpA-T7 and MrpD-His10 tags), the truncated mrp fragment (3.9 kb, containing parts of the mrpA-T7 tag, mrpB, mrpC, mrpD-His10, and part of the mrpE region) of SalI-digested pDuetmrpTHS2 was ligated with a SalI-digested large fragment (ca. 5.9 kb) of pGEMOF4mrp16 (tag free), yielding pGEMmrpTH.
For the construction of pGEMmrpTCH (with the MrpA-T7, MrpC-c-myc, and MrpD-His10 tags), two independent PCRs were performed with pGEMmrpTH DNA, with the sets of primers OF4Mrp37 and MrpC-c-myc-R plus OF4Mrp9 and MrpC-c-myc-F. The two purified PCR products (ca. 3.5 kb) were used as the template for a second PCR with primers OF4Mrp37 and OF4Mrp9. The purified PCR product of this reaction was digested with PstI and then cloned into PstI-digested pGEMmrpTH, yielding pGEMmrpTCH. This procedure produced a mrpC-c-myc tag gene fusion in the operon.
For the construction of pGEMmrpTCHS (with the MrpA-T7, MrpC-c-myc, MrpD-His10, and MrpG-S tags), PCR was performed with pDuetmrpTHS2 as the template with the sets of primers OF4Mrp3 and pDuet-KpnI. The amplified PCR product was digested with KpnI and BamHI and then ligated into KpnI- and BamHI-digested pMW119 (Nippon Gene), yielding pMW-S. The truncated mrp fragment (1.3 kb, containing parts of mrpE, mrpF, and the mrpG-S-tagged region) of KpnII- and NdeI-digested pMW-S was ligated with a KpnII- and NdeI-digested large fragment (ca. 8.0 kb) of pGEMmrpTCH, yielding pGEMmrpTCHS.
For the construction of pGEMmrpTFCHS (with the MrpA-T7, MrpB-FLAG, MrpC-c-myc, MrpD-His10, and MrpG-S tags), two independent PCRs were performed with pGEMmrpTCHS DNA, with the sets of primers OF4Mrp37 and MrpB-FLAG-R-2 plus OF4Mrp9 and MrpB-FLAG-F. The two purified PCR products were used as the template for a second PCR with primers OF4Mrp37 and OF4Mrp9. The purified PCR product (ca. 3.5 kb) of this reaction was digested with PstI, and then cloned into PstI-digested pGEMmrpTCHS, yielding pGEMmrpTFCHS. This procedure resulted in the addition of a FLAG-tagged gene at the end of mrpB.
During the construction of pGEMmrpTFCHS7, a point mutation was found just in front of the His10 tag in pGEMmrpTFCHS, prepared as described above. To produce a mutation-free version of pGEMmrpTFCHS7, PCR was performed with pDuetmrpTHS7 DNA, using primers OF4Mrp3 and pDuet-KpnI. pDuetmrpTHS7 contains the full mrp operon with an additional T7 tag at the end of mrpA, an additional His7 tag at end of mrpD, and an additional S tag at the end of mrpG. A BamHI-digested kanamycin resistance gene fragment from pUC4K (Pharmacia) was ligated with BamHI-digested pMW119, yielding pMWKm. The amplified PCR product was digested with KpnI and BamHI and then ligated into KpnI-BamHI-digested pMWKm, yielding pMWHis7. The truncated mrp fragment (2.0 kb, containing parts of the mrpD-His7 tag, mrpE, mrpF, and mrpG-S-tagged region) of NheI- and KpnI-digested pMWHis7 was ligated with an NheI- and KpnI-digested large fragment (ca. 8.0 kb) of pGEMOF4mrp16, yielding pGEMmrpHS7. The truncated mrp fragment (2.0 kb, containing parts of the mrpD-His7 tag, mrpE, mrpF, and the mrpG-S-tagged region) of BamHI-digested pGEMmrpHS7 was ligated with a BamHI-digested large fragment (ca. 8.0 kb) of pGEMmrpTFCHS, yielding pGEMmrpTFCHS7. The correction of the point mutation resulted in a final product in which a hepta-His tag was fused to mrpD.
Construction of a panel of individual mrp gene deletions in the pGEMmrpTFCHS7 plasmid.
The construction of each Mrp subunit gene deletion plasmid from pGEMmrpTFCHS7 created the deletions shown in Fig. S2. The constructions were carried out as follows. For the construction of pGEMmrpTFCHSΔA, two independent PCRs were performed with pGEMmrpTFCHS7 DNA and pGEMOF4mrp16 DNA with the sets of primers OF4Mrp22 and delta-A-F plus T7-F and delta-A-R (see Table S1 in the supplemental material). The two purified PCR products were used as the template for a second PCR with primers OF4Mrp22 and T7-F. The purified PCR product of this reaction (ca. 3.1 kb) was digested with XbaI and then cloned into XbaI-digested pMW119, yielding pMWΔA. The ΔmrpA fragment of XbaI-digested pMWΔA was ligated with an XbaI-digested small fragment (ca. 4.5 kb) of pGEMmrpTFCHS7, yielding pGEMmrpTFCHSΔA.
For the construction of pGEMmrpTFCHSΔB, two independent PCRs were performed with pGEMmrpTFCHS7 DNA and pGEMOF4mrp16 DNA with the sets of primers OF4Mrp43 and delta-B-F plus OF4Mrp37 and delta-B-R. The two purified PCR products were used as the template for a second PCR with primers OF4Mrp37 and OF4Mrp43. The purified PCR product of this reaction (ca. 2.5 kb) was digested with PstI and then cloned into PstI-digested pGEM-5zf(+), yielding pGEMΔB. The ΔmrpB fragment of PstI-digested pGEMΔB was ligated with a PstI-digested large fragment (ca. 7.6 kb) of pGEMmrpTFCHS7, yielding pGEMmrpTFCHSΔB.
For the construction of pGEMmrpTFCHSΔC, two independent PCRs were performed with pGEMmrpTFCHS7 DNA and pGEMOF4mrp16 DNA with the sets of primers OF4Mrp43 and delta-C-F plus OF4Mrp37 and delta-C-R. The two purified PCR products were used as the template for a second PCR with primers OF4Mrp37 and OF4Mrp43. The purified PCR product of this reaction (ca. 2.5 kb) was digested with PstI and then cloned into PstI-digested pGEM-5zf(+), yielding pGEMΔC. The ΔmrpC fragment of PstI-digested pGEMΔC was ligated with a PstI-digested large fragment (ca. 7.6 kb) of pGEMmrpTFCHS7, yielding pGEMmrpTFCHSΔC.
For the construction of pGEMmrpTFCHSΔD, two independent PCRs were performed with pGEMmrpTFCHS7 DNA and pGEMOF4mrp16 DNA with the sets of primers OF4Mrp24 and delta-D-F plus OF4Mrp37 and delta-D-R. The two purified PCR products were used as the template for a second PCR with primers OF4Mrp22 and T7-F. The purified PCR product of this reaction (ca. 3.0 kb) was digested with StuI and NdeI and then cloned into a StuI- and NdeI-digested large fragment (ca. 7.5kb) of pGEMmrpTFCHS7, yielding pGEMmrpTFCHSΔD.
For the construction of pGEMmrpTFCHSΔE, two independent PCRs were performed with OF4 genome DNA with the sets of primers OF4E-KpnI and delta-E-F plus OF4E-EcoRI and delta-E-R. The two purified PCR products were used as the template for a second PCR with primers OF4E-EcoRI and OF4E-KpnI. The purified PCR product of this reaction (ca. 1.7 kb) was digested with NheI and KpnI and then cloned into NheI- and KpnI-digested pGEMOF4mrp16, yielding pGEMmrpΔE. The truncated mrp fragment (3.9 kb, contains part of mrpA-T7 tag, mrpB, mrpC, mrpD-His10 and part of mrpE region) of SalI-digested pGEMmrpTFCHS7 was ligated with SalI-digested large fragment (ca. 5.7 kb) of pGEMmrpΔE, yielding pGEMmrpTFCHΔE. The truncated mrp fragment (6.3 kb, containing parts of the mrpA-T7 tag, mrpB, mrpC, mrpD-His10, ΔmrpE, mrpF, and part of the mrpG region) of EcoRI- and EagI-digested pGEMmrpTFCHΔE was ligated with a EcoRI- and EagI-digested small fragment (ca. 3.3 kb) of pGEMmrpTFCHS7, yielding pGEMmrpTFCHSΔE.
For the construction of pGEMmrpTFCHSΔF, two independent PCRs were performed with pGEMmrpTFCHS7 DNA and pGEMOF4mrp16 DNA with the sets of primers SP6-R and delta-F-F plus OF4Mrp71 and delta-F-R. The two purified PCR products were used as the template for a second PCR with primers OF4Mrp71 and SP6-R. The purified PCR product of this reaction (ca. 2.0 kb) was digested with BamHI and then cloned into BamHI-digested pGEM-7zf(+), yielding pGEMΔF. The ΔmrpF fragment of BamHI-digested pGEMΔF was ligated with a BamHI-digested large fragment (ca. 8.0 kb) of pGEMmrpTFCHS7, yielding pGEMmrpTFCHSΔF.
For the construction of pGEMmrpTFCHSΔG, two independent PCRs were performed with pGEMmrpTFCHS7 DNA and pGEMOF4mrp16 DNA with the sets of primers SP6-R and delta-G-F plus OF4Mrp71 and delta-G-R. The two purified PCR products were used as the template for a second PCR with primers OF4Mrp71 and SP6-R. The purified PCR product of this reaction (ca. 2.0 kb) was digested with BamHI and then cloned into BamHI-digested pGEM-7zf(+), yielding pGEMΔG. The ΔmrpF fragment of BamHI-digested pGEMΔG was ligated with a BamHI-digested large fragment (ca. 8.0 kb) of pGEMmrpTFCHS7, yielding pGEMmrpTFCHSΔG.
Introduction of the site-directed mutations in mrpE into the untagged full mrp construct pGEMOF4mrp16 and the tagged construct pGEMmrpTFCHS7.
A GeneTailor site-directed mutagenesis system (Invitrogen) was used for in vitro site-directed mutagenesis. The protocols used followed those of the instruction manual supplied by Invitrogen. For the construction of pGEMmrpNdeI-EagI, a truncated mrp fragment (ca. 0.7 kb, containing parts of mrpE, mrpF, and the mrpG region) of NdeI- and EagI-digested pGEMOF4mrp16 was ligated with the NdeII- and EagI-digested of pGEM7zf(+), yielding pGEMmrpNdeI-EagI. This plasmid was used as the template for the mutagenesis system. For the construction of pGEMmrpE-T113A, pGEMmrpE-P114G, pGEMmrpE-Q4A, pGEMmrpE-N8A, pGEMmrpE-K59A, pGEMmrpE-K66A, pGEMmrpE-E67A, and pGEMmrpE-H131A, PCR was performed with methylated pGEMmrpNdeI-EagI plasmid DNA, with the respective sets of primers that contained the target mutations MrpE-T113-R and MrpE-T113A-F, MrpE-P114-R and MrpE-P114G-F, MrpE-Q4-R and MrpE-Q4A-F, MrpE-N8-R and MrpE-N8A-F, MrpE-K59-R and MrpE-K59A-F, MrpE-K66-R and MrpE-K66A-F, MrpE-E67-R and MrpE-E67A-F, and MrpE-H131-R and MrpE-H131A-F. Each product was a linear, double-stranded DNA containing the mutation. Each mutagenesis mixture was transformed into E. coli DH5α-T1r. The host cell circularized the linear mutated DNA, and the McrBC endonucleases in the host cell digested the methylated template DNA, leaving only unmethylated, mutated product.
For the construction of pGEMOF4mrpE-T113A, pGEMOF4mrpE-P114G, pGEMOF4mrpE-Q4A, pGEMOF4mrpE-N8A, pGEMOF4mrpE-K59A, pGEMOF4mrpE-K66A, pGEMOF4mrpE-E67A, and pGEMOF4mrpE-H131A, each plasmid was digested with NdeI and EagI. The inserted fragment was then ligated with NdeI- and EagI-double-digested pGEMOF4mrp16, yielding pGEMOF4mrpE-T113A, pGEMOF4mrpE-P114G, pGEMOF4mrpE-Q4A, pGEMOF4mrpE-N8A, pGEMOF4mrpE-K59A, pGEMOF4mrpE-K66A, pGEMOF4mrpE-E67A, and pGEMOF4mrpE-H131A, respectively. For the construction of pGEMTFCHS-E-T113A and pGEMTFCHS-E-P114G, each plasmid was digested with NdeI and EagI. The inserted fragment was then ligated with NdeI- and EagI-double-digested pGEMTFCHS7, yielding pGEMTFCHS-E-T113A and pGEMTFCHS-E-P114G, respectively.
Preparation of membrane vesicles.
E. coli KNabc transformants expressing the empty vector pGEM-7zf(+) or the mrp operon with five epitope tags, pGEMmrpTFCHS7, or one of its mutant derivative were grown in LBK medium plus 50 mM NaCl at 37°C. Everted membrane vesicles were prepared by the method originally described by Rosen (33) and detailed recently (41), except that the buffer used was TCDG (10 mM Tris-HCl [pH 8.0], containing 140 mM choline chloride, 0.5 mM dithiothreitol, and 10% glycerol). The membrane vesicles were resuspended in TCDG buffer for antiporter assays and in ACA buffer {750 mM ɛ-aminocapronic acid, 50 mM BTP [bis-[tris(hydroxymethyl)-methylamino]-propane], 20% [wt/vol] glycerol [pH 7.0]} for BN-PAGE analyses. Vesicles were stored at −80°C. Protein content was determined by using the BCA assay (Pierce).
Antiport assays.
Fluorescence-based assays of the Na+/H+ antiport activities were conducted on everted membrane vesicles from E. coli KNabc transformants with a control vector, full Mrp constructs, or single-deletion mutant versions of the Mrp antiporter. Assays were conducted by the method described previously (9) at pH values indicated for particular experiments. Briefly, each vesicle preparation was assayed in 2 ml containing 50 mM BTP-sulfate buffer, 140 mM choline chloride, 5 mM MgCl2, 1 μM acridine orange (AO), and 66 μg of vesicle protein. Measurements were conducted using a Hitachi High-Technologies model F-4500 fluorescence spectrophotometer with excitation at 420 nm (using a 10-mm slit) and emission at 500 nm (with a 10-mm slit). Respiration was initiated by the addition of Tris-succinate to a final concentration of 2.5 mM. After steady-state fluorescence quenching was reached, NaCl was added to a final concentration of 2.5 mM. The Na+-dependent dequenching of fluorescence is the measure of Na+/H+ antiport activity. Assays were conducted in duplicate with three independent membrane preparations.
Immunoblotting analyses of each Mrp protein in transformant membrane fractions.
Five microliters of a membrane suspension (4 μg of membrane protein/μl) from each E. coli transformant was used for one-dimensional sodium dodecyl sulfate (SDS)-PAGE analyses on membrane samples. The same volume of SDS loading buffer was added to each sample, after which the proteins were separated on 12% polyacrylamide SDS gels (36). The gels were then transferred to nitrocellulose filters (Bio-Rad) electrophoretically by the application of 60 V for 3 h in Tris-glycine-methanol buffer (25 mM Tris, 192 mM glycine, 20% [vol/vol] methanol [pH 8.3]). The MrpA, MrpB, MrpC, MrpD, MrpE, MrpF, and MrpG proteins were detected by anti-T7 antibody (Sigma), anti-FLAG antibody (Abcam), anti-c-myc antibody (Abcam), anti-His antibody (Qiagen), anti-MrpE antibody (raised in this study), anti-MrpF antibody (raised in this study), and anti-S tag antibody (Abcam), respectively. Goat anti-rabbit horseradish peroxidase (Bio-Rad) was also used as the second antibody for detection of anti-MrpE and anti-MrpF antibodies. ECL solution (Amersham Biosciences) was the usual detection reagent. A set of ECL Plus solution (Amersham Biosciences) and Can Get Signal immunoreactions enhancer solution (Toyobo) was used in some experiments, as indicated. A quantitative imaging system, Pluor-S MAX (Bio-Rad), was used for detection and analysis of chemiluminescence images. For immunoblot protocols used for analyses of BN-SDS-PAGE analyses, conditions were optimized for each sample set and are provided in figure legends describing particular experiments.
BN-PAGE.
Protein complexes (10 mg/ml) were solubilized at 4°C for 20 min, in ACA buffer containing dodecyl maltoside (DDM) at concentrations from 0.5 to 2.0% (wt/vol). DDM (1.0% [wt/vol]) was determined to be the most effective, as evidenced by the number of complexes in the BN gel and their intensity and their molecular mass range. After samples were solubilized, they were cleared by centrifugation at 40,000 rpm for 90 min in a Beckman Ti70 rotor device at 4°C. The supernatant was divided into 20-μl aliquots which were stored at −80°C if they were not used immediately. BN-PAGE was performed in an XCell SureLock Mini-Cell, using native-PAGE, 4 to 16% gels, in a 15-well apparatus (Invitrogen). The details of the BN-PAGE protocols were those provided in the instruction manual from Invitrogen. High-molecular-mass markers, NativeMark unstained protein standards, were obtained from Invitrogen.
Two-dimensional SDS-PAGE.
Gel strips obtained from the one-dimensional BN-PAGE were soaked for 20 min in equilibration buffer (0.1% [wt/vol] SDS, 0.1% [vol/vol] 2-mercaptoethanol, 50 mM Tris-HCl [pH 6.8]). The SDS-PAGE and immunoblotting analyses were conducted as described above, with modifications for individual experiments described in the figure legends.
Nucleotide sequence accession numbers.
The sequence of the Bacillus pseudofirmus OF4 mrp operon with five epitope tags was deposited in GenBank under the accession number EF468713.
RESULTS
The full Mrp system with five epitope tags catalyzes Na+/H+ antiport activity, whereas all single-protein deletion forms are almost entirely inactive.
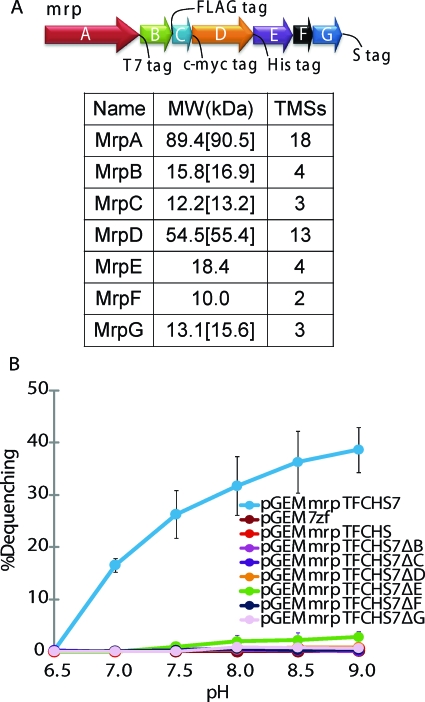
Polyclonal antibodies to synthetic peptides that could be used to monitor individual Mrp proteins were successfully raised against MrpE and MrpF but not for other Mrp proteins. Therefore, epitope tags were introduced genetically into the remaining five Mrp proteins, as described in Materials and Methods and shown in Fig. S1 in the supplemental material. The resulting construct, designated MrpTFCHS7, is shown schematically in Fig. 1A. MrpTFCHS7 conferred comparable Na+ resistance to a tag-free Mrp operon, OF4Mrp16, when they were expressed in pGEM7Zf(+) and tested in antiporter-deficient E. coli KNabc (ΔnhaA ΔnhaB ΔchaA) (data not shown). This E. coli mutant has been widely used for the characterization of heterologous antiporters (26, 27). MrpTFCHS7 also exhibited robust Na+/H+ antiport activity in the standard fluorescence-based assay of everted membrane vesicles of the E. coli KNabc transformant expressing pGEMmrpTFCHS7 (Fig. 1B). We compared the activity conferred by the tagged MrpTFCHS7 with that of the panel of deletions in which the individual, nonpolar mrp gene deletions were introduced. Except for membranes of the Mrp-ΔmrpE transformant, which showed a very low level of antiport activity, all the Mrp mutant membranes were negative for antiport activity (Fig. 1B).
FIG. 1.
A schematic diagram of the tagged, full MrpTFCHS7 construct and the Na+/H+ antiport activity conferred by this full Mrp system and the mutant forms with single gene deletions. (A) Diagram of the mrp locus in pGEMmrpTFCHS7. The tags that were introduced are indicated. The table beneath the diagram shows the predicted sizes of each untagged Mrp protein, the predicted sizes for each tagged protein (in brackets), and the predicted numbers of transmembrane segments (TMS) in each Mrp protein, as analyzed by SOUSUI software (http://bp.nuap.nagoya-u.ac.jp/sosui/). (B) The Na+/H+ antiport activities of MrpTFCHS7, of a negative control, and of the panel of single mrp gene deletions is shown as the percentage of dequenching. The fluorescence-based assay was conducted with everted membrane vesicles from E. coli KNabc transformants expressing the indicated plasmids. The results are averages of duplicate determinations from at least three independent preparations. The error bars indicate the standard deviations of the means.
The pattern of Mrp proteins found in membranes of mrp deletion mutants supports the interdependence of Mrp proteins for stable membrane incorporation.
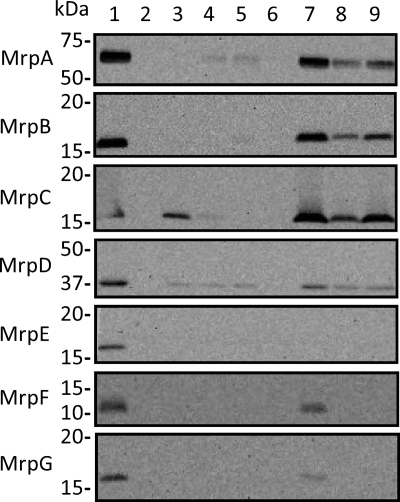
Earlier studies showed that each individual Mrp protein could be detected in E. coli membranes when it was overexpressed (38). The minimal antiport activity in membranes of single-deletion mutants of pGEMmrpTFCHS7 suggested that the Mrp proteins require one another for membrane incorporation in a stable and/or active form. SDS-PAGE analyses were conducted with membranes from the full MrpTFCHS7 and the mutant panel of E. coli KNabc transformants to determine how each of the single mrp gene deletions affected the level of the remaining Mrp proteins. All seven Mrp proteins were detected by immunoblotting analyses of membranes from the pGEMmrpTFCHS7 transformant (Fig. 2, lane 1). No cross-reacting material was detected in analyses of the empty vector control membranes (Fig. 2, lane 2). Only a single cross-reacting band was detected with each antibody in the MrpTFCHS7 membranes (data not shown). The positions of the largest Mrp proteins, MrpA and MrpD, corresponded to a smaller size than expected from the predicted molecular weight, e.g., the predicted versus the observed values were, respectively, 90.5 versus 65 for tagged MrpA and 55.4 versus 38 for tagged MrpD. These observations are consistent with findings with other large polytopic membrane proteins (25). The positions of the smaller Mrp proteins were closer to that expected from their calculated sizes.
FIG. 2.
Detection of the Mrp proteins in membranes from E. coli KNabc transformants expressing full MrpTFCHS7 or the single gene deletion mutants. Membrane protein (20 μg) from log-phase cells was used for SDS-PAGE. Immunoblotting was conducted as described in Materials and Methods. The sample preparations loaded were as follows: lanes 1, MrpTFCHS7; 2, plasmid control; 3, ΔA; 4, ΔB; 5, ΔC; 6, ΔD; 7, ΔE; 8, ΔF; 9, ΔG ΔA = ΔmrpA; all others are analogous. The Mrp proteins and molecular masses (kDa) being probed in each panel are indicated at the left of the figure.
The presence of each of the individual proteins was assessed for the panel of single mrp gene deletions. The results showed that neither MrpE, MrpF, nor MrpG was required for membrane incorporation of MrpA, MrpB, MrpC, or MrpD. These last four Mrp proteins were all found in membranes expressing Mrp-ΔmrpE, Mrp-ΔmrpF, or Mrp-ΔmrpG (Fig. 2, lanes 7, 8, and 9). The converse was not true. That is, MrpE, MrpF, and MrpG were all absent from membranes expressing Mrp systems with a deletion in either mrpA, mrpB, mrpC, or mrpD. These results suggested that MrpA to MrpD comprise a module that is required for MrpE to MrpG to incorporate stably into the membrane. Membranes from the Mrp-ΔmrpD transformant had no detectable levels of any of the seven Mrp proteins (Fig. 2, lane 6). This was the only membrane set from the panel of deletions that lacked all of the Mrp proteins. MrpD was the only protein that could be detected in the entire panel of single mrp mutants except for the mutant lacking mrpD itself. In addition to MrpD, MrpA and MrpC were both found in the absence of MrpB. Mrp-ΔmrpB membranes contained at least low levels of both MrpA and MrpC (Fig. 2, lane 4). MrpC was present in Mrp-ΔmrpA membranes (Fig. 2, lane 3), and low levels of MrpA were discernible in Mrp-ΔmrpC membranes (Fig. 2, lane 5). The relative amounts of a specific Mrp protein observed with different mutant strains may reflect the dependence of its stability on the particular Mrp proteins that are also present with it in the membrane. The MrpA-to-MrpD quartet of proteins appeared to comprise a stable combination. However, it was inactive with respect to antiport activity. No antiport activity was observed for membranes from the Mrp-ΔmrpF and ΔmrpG mutants, even though proteins MrpA to MrpD were all present in the membrane (Fig. 1B). Neither MrpF nor MrpG was observed in the membrane, unless the other was also present (Fig. 2, lanes 8 and 9). The presence of both MrpF and MrpG was required for observation of MrpE in the membrane (Fig. 2, lanes 8 and 9). The membranes from Mrp-ΔmrpE were the only ones in the deletion panel in which all the remaining proteins, MrpA to MrpD, MrpF, and MrpG were detected (Fig. 2, lane 7).
Two-dimensional BN-SDS-PAGE and immunoblotting analyses of MrpTFCHS7 identifies full MrpA-to-MrpG complexes of different sizes and substantial levels of an MrpA-to-MrpD subcomplex.
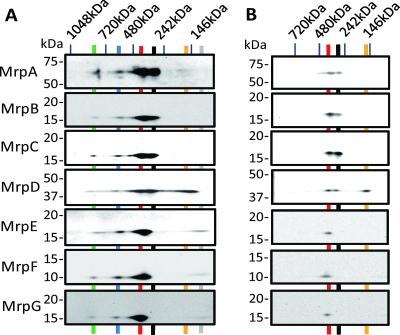
In order to probe complexes containing the full complement of alkaliphile Mrp proteins, we conducted BN-SDS-PAGE fractionation of membranes from E. coli KNabc expressing MrpTFCHS7. Immunoblotting analyses were conducted with SDS-PAGE-fractionated strips from BN gels, using the panel of antibodies that detected all seven Mrp proteins. No bands were observed for the analyses of the control membranes from a transformant with the empty vector (data not shown). The analyses of MrpTFCHS7 showed the consistent presence of two species at about 840 and 400 kDa. Both of those species contained all seven Mrp proteins (Fig. 3A). The smaller one was very close to the size of the Mrp complex estimated in B. subtilis (17). The estimates of molecular size from BN gels need to be corrected for detergent and dye binding, and a formula has been validated for this correction (12). The application of that formula to the 840- and 400-kDa Mrp complexes observed here resulted in estimated sizes of about 470 and 222 kDa, which are, respectively, close to the sizes expected for dimeric (440-kDa) and monomeric (220-kDa) MrpTFCHS7 complexes, in which all seven Mrp proteins are present in a single copy per monomer. Given their small sizes, the smallest Mrp proteins might be present in more than one copy. In some preparations, even larger species containing Mrp proteins were evident (e.g., see lane 1 of Fig. 4). In between the putative dimeric and monomeric Mrp complexes, lower levels of a species at about 570 kDa were usually detected (Fig. 3A). The corrected size of this species would be about 320 kDa. It could be a partial breakdown product of larger oligomeric Mrp complexes. Neither the larger complexes nor the 570 kDa species were visible under lower protein loading conditions (Fig. 3B).
FIG. 3.
Detection of Mrp complexes by immunoblotting analyses of BN-SDS-PAGE patterns of membranes from the E. coli KNabc expressing MrpTFCHS7. Protocols for loading and detection of proteins were optimized for specific immunoblotting analyses of membrane extracts that were fractionated by two-dimensional BN-SDS-PAGE. (A) Detection of complexes from E. coli KNabc membranes expressing full MrpTFCHS7. Gel strips obtained from the one-dimensional BN-PAGE analyses were soaked for 20 min in equilibration buffer. The proteins were then separated by 12% polyacrylamide SDS gels. For detection of the FLAG tag, the c-myc tag, the His tag, the MrpF, and the S tag, 20 μg of membrane protein was used. Enhanced chemiluminescence (ECL) was used for the detection. For the detection of the T7 tag and MrpE, 30 μg of membrane protein was used. Can Get Signal and ECL-Plus were used as the enhancers for the detection. The general SDS-PAGE and immunoblotting protocols are described in Materials and Methods. The green, blue, red, black, orange, and gray line, respectively, mark ca. 840-kDa, 570-kDa, 400-kDa, 320-kDa, 160-kDa, and 120-kDa bands. The Mrp proteins detected in each panel are listed on the left. (B) Detection of MrpTFCHS7 complexes in E. coli KNabc membranes in extracts, under conditions that resolve the full Mrp complex and MrpA-to-MrpD subcomplex. The experiment was conducted as described in the legend to panel A, except that 10 μg of membrane protein was used for each sample. The red, black, and orange lines, respectively, mark the ca. 400-kDa, 320-kDa, and 160-kDa bands.
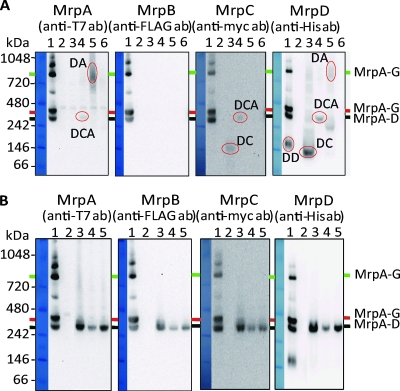
FIG. 4.
Detection of Mrp complexes by immunoblotting analyses of BN-PAGE patterns of membranes from the E. coli KNabc expressing mutant MrpTFHCS7 forms with single gene deletions. The methods are as described in the legend for Fig. 5A. The full MrpTFHCS7 is shown for comparison together with mutant membrane extracts. The locations corresponding to those of the two full Mrp heterooligomeric species (MrpA to MrpG, marked by green and red lines) and the MrpA-to-MrpD subcomplex (marked by the black line) are shown at the right side of the two sets of panels. (A) Detection of complexes in immunoblots of BN-PAGE-fractionated samples from MrpTFCHS7 and mrpA, mrpB, mrpC, and mrpD deletion mutants of the tagged Mrp construct. Each panel was probed with the antibody indicated at the top of the panel. The lanes contain full Mrp (lane 1), negative control (lane 2), followed by samples from four single-deletion mutant forms of Mrp (ΔA, lane 3; ΔB, lane 4; ΔC, lane 5; ΔD, lane 6, where ΔA = ΔmrpA [all others are analogous]). For detection of the T7 tag, the FLAG tag, and the His tag, 60 μg of membrane protein were used from the samples of the mrpA, mrpB, mrpC, and mrpD deletion plasmids and the empty vector transformants, and 1 μg were used from the samples of the full mrp plasmid. Can Get Signal and ECL-Plus were used as the enhancers for these detections. For detection of the c-myc tag, 30 μg of each membrane protein was used. Enhanced chemiluminescence was used as the detection method. Individual bands that were detected in at least two gels were circled in red, except for the MrpDD band in lane 1, since this band was observed whenever a full Mrp complex was found. The apparent composition of each circled band is indicated near the band. DA, MrpD plus MrpA; DCA, MrpD plus MrpC plus MrpA; DC, MrpD plus MrpC; DD, putative dimeric MrpD. (B) Detection of complexes in immunoblots of BN-PAGE-fractionated samples from Mrp-ΔmrpE, Mrp-ΔmrpF, and Mrp-ΔmrpG deletion mutants. Each panel was probed with the antibody indicated at the top of the panel. The lanes contain full Mrp (lane 1), negative control (lane 2), followed by samples from three single-deletion mutant forms of Mrp (ΔE, lane 3; ΔF, lane 4; ΔG, lane 5). For detection of the T7 tag, the FLAG tag, and the His tag, 60 μg of membrane protein was used. Can Get Signal and ECL-Plus were used as the enhancers for detection. For detection of the c-myc tag, 30 μg of membrane protein was used. ECL was used for detection.
The BN-SDS-PAGE analyses of the complete alkaliphile Mrp system also identified three additional species that were smaller than any of those carrying the full Mrp complexes. The major one was a species that contained a MrpA-to-MrpD subcomplex (Fig. 3A), and the least intense and smallest one was a MrpE-MrpG subcomplex (Fig. 3A). The MrpA-to-MrpD subcomplex was a major band in every experiment, often overlapping with the 400-kDa full Mrp species, unless the loading conditions were modified (Fig. 3B). On the BN gels the molecular size of the MrpA-to-MrpD subcomplex was ∼320 kDa, correcting to 178 kDa. This correlates well with the predicted 176 kDa of a species containing one of each of the four proteins. The MrpE-to-MrpG subcomplex was less intense and barely detectable in some repeats of the experiments shown in Fig. 3A. The MrpE-to-MrpG subcomplex was not detected under the lower loading conditions of the analyses shown in Fig. 3B. The remaining oligomeric Mrp species, detected at about 160 kDa, contained only MrpD. It was readily observed in every experiment in which full Mrp complexes were also found (Fig. 3A and B). Its size, when corrected by using the same factor used for the larger species, is closer to a dimeric than a monomeric MrpD species. We tentatively designated it MrpDD. MrpDD was found only when full Mrp complexes were present. Thus, MrpDD could be a partial breakdown product of larger species as noted above as a possibility for the 570 kDa species (Fig. 3A and B).
The panel of single mrp deletion mutants in MrpTFCHS7 exhibits a spectrum of subcomplex patterns in BN-PAGE and immunoblotting analyses.
Full Mrp complexes were found only, as expected, in membranes in which the full Mrp system was expressed (Fig. 4A and B, lanes 1). BN-PAGE analyses of the mrpA, mrpB, mrpC or mrpD deletion mutant indicated that the proteins that were observed in the one-dimensional SDS-PAGE analyses of membranes expressing these mutant forms (Fig. 2) associated with each other in the membrane. Apparent MrpDC and MrpDA species were observed, respectively, in membranes from mrpA and mrpC mutants (Fig. 4A, lanes 3 and 5), and a MrpDCA species was observed in membranes from the mrpB mutant (Fig. 4A, lane 4). Analyses of the MrpE-to-MrpG content were not conducted with these samples, since those proteins were not observed in the one-dimensional analyses (Fig. 2). No subcomplexes were evident in membranes expressing the mrpD deletion mutant (Fig. 4A, lane 6). This was consistent with the absence of any Mrp proteins in the one-dimensional analysis (Fig. 2). The BN-PAGE patterns of the membranes from the mrpE, mrpF, and mrpG mutants were also consistent with those of the association among the Mrp proteins found in the membranes of deletion mutants. There was a strong band corresponding to a MrpA-to-MrpD subcomplex in the membranes of all three of these mutants (Fig. 4B, lanes 3, 4, and 5). No MrpE, MrpF, or MrpG was detected in association with the MrpA-to-MrpD species (data not shown). We noted, however, that in the BN-PAGE analyses, there was a weak, fuzzy MrpA-to-MrpD-containing band that ran above the usual discrete MrpA-to-MrpD band in the Mrp-ΔmrpE membranes. It was not observed with the full Mrp or any other mutant membranes (Fig. 4B, compare lane 3 versus lanes 4 and 5).
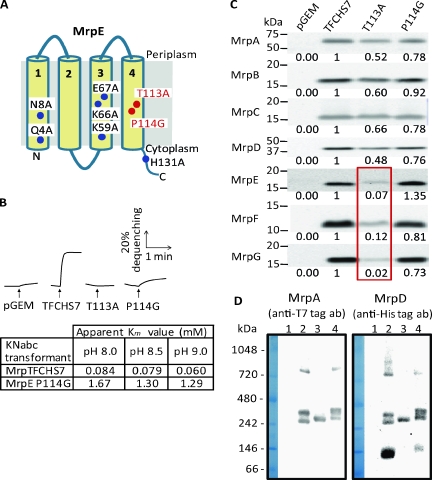
Two mrpE point mutations, T113A and P114G, respectively, abolish antiport activity and raise the apparent Km value of Mrp-dependent Na+/H+ antiporter activity.
Deletion of mrpE from the full tagged mrpTFCHS7 construct resulted in the loss of almost all antiport activity (Fig. 1B). Nonetheless, the retention of some residual activity distinguished this deletion mutant from others in the panel. We sought to further probe the impact of changes in MrpE on the Na+/H+ antiport activity of Mrp by introducing eight site-directed mutations in conserved residues (Fig. 5A). Only two of the mutations, T113A and P114G, had an effect on the ability of the mutant operon to complement the Na+-sensitive phenotype of E. coli KNabc. Both of these mutant Mrp systems failed to complement at all (data now shown). In assays of Na+/H+ antiport activity with everted vesicles, the T113A mutant of MrpTFCHS7 exhibited no activity, whereas the P114G mutant exhibited greatly reduced antiporter activity relative to that of MrpTFCHS7 (Fig. 5B). More detailed studies of the activity of the P114G mutant form showed that its apparent Km value for Na+ was 16.5 to 21.5 times higher than that of its tagged Mrp parent, depending upon the assay pH (table in Fig. 5B). The apparent Km value for the untagged full Mrp (OF4Mrp16) was also assayed and found to be comparable to that of the tagged form (data not shown). The P114G mutant Mrp retained responsiveness to increasing pH, i.e., the apparent Km value for Na+ decreased for both the parent and the mutant as the assay pH was raised from pH 8.0 to 9.0 (Fig. 5B). Immunoblotting analyses indicated that the T113A mutation in MrpE led to significantly lower levels of MrpE, MrpF, and MrpG in the membranes of the E. coli KNabc host, whereas the T114G mutation did not have a significant effect on the membrane levels of any of the Mrp proteins (Fig. 5C). The MrpA-to-MrpD subcomplex was the only species observed in BN-PAGE analyses of the T113A mutant (Fig. 5D, lane 3, data shown only for MrpA and MrpD). By contrast, membranes from the P114G mutant contained the two full Mrp forms that were observed consistently in preparations of the wild type. That is, the P114G mutant membranes contained the species observed in gels at positions corresponding to 840 kDa and 400 kDa and, respectively, are calculated to be ∼470 kDa and ∼222 kDa. The P114G mutant membranes also contained proteins MrpA to MrpD and MrpDD, as had preparations in which the full wild type, MrpTFCHS7, was expressed (Fig. 5D, lanes 2 and 4).
FIG. 5.
Properties of MrpTFCHS7 with point mutations in mrpE. (A) The residues that were mutated in the putative transmembrane segments of MrpE molecule (as predicted by SOSUI software, as described in the legend to Fig. 1). These conserved residues were changed to alanines, except for Pro-114, which was changed to glycine. The mutants that lost the capacity to complement the Na+ sensitivity of E. coli KNabc are indicated in red. (B) Antiport activity is shown as the percentage of dequenching of AO fluorescence when 10 mM Na+ was added (at the arrow) to energized everted vesicles that had achieved a steady-state pH, acid in, by respiration. Na+/H+ antiport activity was assayed at pH 8.5 in vesicles from transformants with the empty vector pGEM7zf(+) (pGEM) or the vector expressing full Mrp (pGEMmrpTFCHS7) or mutant forms with T113A or P114G point mutations in mrpE. The table below the tracings shows the apparent Km values of MrpTFCHS7 and the P114G mutant for Na+ at several pH values. (C) Western analyses of SDS-PAGE-fractionated membranes from the wild type and the mutant transformants using antibodies against each Mrp subunit. The plasmid present in the membranes is indicated above each lane, i.e., E. coli KNabc transformed by pGEM7zf(+) (pGEM), pGEMTFCHS7 (TFCHS7), pGEMTFCHS-E-T113A (T113A), and pGEMTFCHS-E-P114G (P114G). The Mrp protein being probed is shown at the left. The MrpE, MrpF, and MrpG proteins detected in the T113A mutant preparation are outlined with a red square. A quantitative imaging system, Pluor-S MAX (Bio-Rad), was used for the detection and analysis of a chemiluminescence image. (D) Detection of Mrp complexes from the membrane fraction of E. coli KNabc transformed with the empty vector (lane 1), pGEMmrpTFCHS7 (lane 2), and pGEMmrpTFCHS7 with one of two point mutations in mrpE, T113A (lane 3) or P114G (lane 4). BN-PAGE analyses were conducted as in the experiments shown in Fig. 3. All of the Mrp proteins were probed. The bands in these gels were found to include the same protein complements as the bands of the same size shown in Fig. 3. Only the data for analyses of MrpA and MrpD are shown here. For detection of both the T7 tag of MrpA and the His tag of MrpD, 30 μg of membrane protein was used.
DISCUSSION
The studies of Mrp from alkaliphilic B. pseudofirmus OF4 reported here have identified full Mrp complexes of estimated sizes, ∼470 kDa (840 kDa species on the gel) and ∼222 kDa (400 kDa species on the gel). These sizes were consistent with those expected for dimers and monomers containing copies of all seven Mrp proteins. The two species of complete Mrp heterooligomers were consistently observed (Fig. 3 and 4), unless the conditions of the SDS-PAGE analyses were deliberately chosen to produce smaller bands of the major species. In many preparations, species larger than the putative dimer were evident, but they might not have contained all seven Mrp proteins. For example, MrpD appears depleted in the species seen above the putative dimeric Mrp (840-kDa band) at about 950 kDa in lane 1 of Fig. 4. It is possible that such larger oligomers are remnants of complete heterooligomeric complexes that are larger than dimers. Such larger Mrp complexes may be fragile and prone to the loss of fragments. That could account for the observation of the putative breakdown products, i.e., the 570-kDa species (without correction) that was observed between the putative dimeric and monomeric complexes and the MrpDD species. Only one other demonstration of a full Mrp complex has been reported. Those analyses of the B. subtilis Mrp complex revealed a single complete heterooligomeric form whose 410-kDa mass on the gel (uncorrected) corresponded well to the putative 400-kDa (uncorrected, ∼222 kDa corrected) species found here for B. pseudofirmus OF4 Mrp. Larger complete Mrp complexes were not noted in the B. subtilis study (17). Perhaps differences between the specific conditions of the other study and those of ours, such as the bacterial host or conditions of the analyses, underpin the absence versus presence of larger heterooligomers. Alternatively, it may prove to be a real difference between Mrp from neutrophilic B. subtilis and alkaliphilic B. pseudofirmus OF4 when comparisons are made under the same conditions. Larger Mrp heterooligomers could be an adaptation that best supports proton gathering by the alkaliphile Mrp system.
A MrpA-to-MrpD subcomplex and much smaller amounts of a MrpE-to-MrpG complex were observed with BN-PAGE analyses of membranes expressing MrpTFCHS7 (Fig. 3A). The MrpA-to-MrpD subcomplex is apparently a stable subcomplex. It contains all three of the proteins that have homology with membrane subunits of respiratory complexes that also form a subcomplex domain, i.e., MrpA, MrpD, and MrpC. However, it has no antiport activity, in spite of the presence of the two proteins thought to possess that activity, MrpA and MrpD (23, 24). The finding of a MrpE-to-MrpG subcomplex, albeit at lower levels than the MrpA-to-MrpD subcomplex, is consistent with earlier observations of this subcomplex in fractionations of an extract from cells expressing a full Mrp construct with a single tag (His6 tag) on MrpG (38). MrpE, MrpF, and MrpG may constitute a functional module, since many archaeal operons that contain multiple mrp gene homologues begin with mrpEFG instead of the normal eubacterial order, mrpA-G (39).
In the absence of either MrpE, MrpF, or MrpG, the MrpA-to-MrpD subcomplex was the only species produced in appreciable amounts (Fig. 4B). MrpF and MrpG are also critical for antiport activity, and MrpE is required for substantial antiport activity (Fig. 1B). We hypothesize that the modest activity found for the MrpE deletion mutant corresponds to a small amount of a species that contains all the Mrp proteins except MrpE. Support for this idea comes from two observations. First, in one-dimensional SDS-PAGE analyses of the panel of mutant membranes, the membranes from the MrpE deletion were the only ones in the panel in which the remaining six Mrp proteins could be detected (Fig. 2). Second, in the BN-PAGE analyses of that mutant, a fuzzy region was noted above the MrpA-to-MrpD subcomplex that was not observed with analyses of other mutants (Fig. 4B, lane 3 versus lanes 4 and 5). Although we did not detect MrpF or MrpG in that region, we hypothesize that the “fuzzy” material is indeed a small amount of full Mrp minus MrpE. This species could be a labile six-protein species that has low antiporter activity that accounts for the low activity of membranes from the ΔmrpE transformant. In B. subtilis, an analogous pattern could account for the residual Na+ resistance and Na+ efflux capacity in a ΔmrpE mutant (46).
Overall, the studies of both the ΔmrpE deletion and the two site-directed mutants in the alkaliphile mrpE support a major role for MrpE in the formation of a stable Mrp complex that is fully active. The T113A mutant strongly affected the membrane levels of MrpE. Even the low residual activity observed with the ΔmrpE mutant (Fig. 1B) was absent from membranes expressing Mrp(T113A) (Fig. 5B). This suggests that details of the MrpE interaction with other Mrp proteins have a pronounced effect on activity. The findings with the P114G mutant further support this idea. The levels of Mrp proteins were not greatly affected by the P114G mutation, yet the kinetic properties of antiport activity were strongly affected (Fig. 5B and C). We cannot rule out the possibility that MrpE itself is an antiport protein and that the effect of the P114G mutation of MrpE, therefore, has a direct effect on the properties of the antiporter activity catalyzed by this protein. However, this seems unlikely since MrpE is the only Mrp protein whose deletion is compatible with at least some residual antiport activity. It seems more likely that the P114G mutation prevents the MrpE-to-MrpG module from exerting some crucial effect on the MrpA-to-MrpD subcomplex that results in an antiport-active conformation by the proteins that actually catalyze the antiport activity. The assignment of direct catalytic roles in Na+/H+ antiport activity and anion flux to specific Mrp proteins is a major task remaining in studies of this important and complicated antiporter system.
Another important area for future work with Mrp is prompted by the different effects of individual mrp gene deletions on the levels of the six remaining Mrp proteins that are stably incorporated into the membranes. Studies with individual His-tagged Mrp proteins indicated that each of them can insert into the membrane without the presence of the other Mrp proteins (38). Kinetic experiments will be required to assess how the different effects of single deletions on levels of the other Mrp proteins relate to specific interdependencies among the Mrp proteins with respect to assembly and/or stability within the membrane. The results of the current work establish that those Mrp proteins found in the membranes of deletion strains are associated with one another in subcomplexes. The results also indicate that MrpD has special importance in making it possible for all the other proteins to successfully assemble in the membrane in a stable form. We note recent evidence that a mutation in a human homologue of MrpD that encodes ND2 of human mitochondrial respiratory complex I results in the neurodegenerative Leigh syndrome, in which an assembly intermediate specifically lacking the Mrp-like proteins is observed (42, 43). Further studies of both the Mrp proteins and the related proteins of a membrane-embedded domain of complex I should clarify the evolutionary paths and their divergence in service of different ion-translocating complexes (1, 3, 18, 37).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health research grant GM28454 (to T.A.K.), a Grant-in-Aid for Young Scientists (B), and a grant from the 21st Century Center of Excellence program of the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M.I.).
We thank Blanca Barquera, Arthur Guffanti, and Joel Morgan for reading draft versions of the manuscript.
Footnotes
Published ahead of print on 11 April 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baranova, E. A., P. J. Holt, and L. A. Sazanov. 2007. Projection structure of the membrane domain of Escherichia coli respiratory complex I at 8 A resolution. J. Mol. Biol. 366140-154. [DOI] [PubMed] [Google Scholar]
- 2.Blanco-Rivero, A., F. Leganes, E. Fernandez-Valiente, P. Calle, and F. Fernandez-Pinas. 2005. mrpA, a gene with roles in resistance to Na+ and adaptation to alkaline pH in the cyanobacterium Anabaena sp. PCC7120. Microbiology 1511671-1682. [DOI] [PubMed] [Google Scholar]
- 3.Brandt, U. 2006. Energy converting NADH:quinone oxidoreductase (Complex I). Annu. Rev. Biochem. 7569-92. [DOI] [PubMed] [Google Scholar]
- 4.Clejan, S., A. A. Guffanti, M. A. Cohen, and T. A. Krulwich. 1989. Mutation of Bacillus firmus OF4 to duramycin resistance results in substantial replacement of membrane lipid phosphatidylethanolamine by its plasmalogen form. J. Bacteriol. 1711744-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dzioba-Winogrodzki, J., O. Winogrodzki, T. A. Krulwich, M. A. Boin, C. C. Hase, and P. Dibrov. 2008. The Vibrio cholerae Mrp system: cation/proton antiport properties and enhancement of bile salt-resistance in a heterologous host. J. Mol. Microbiol. Biotechnol., in press. [DOI] [PMC free article] [PubMed]
- 6.Friedrich, T., and H. Weiss. 1997. Modular evolution of the respiratory NADH:ubiquinone oxidoreductase and the origin of its modules. J. Theor. Biol. 187529-540. [DOI] [PubMed] [Google Scholar]
- 7.Fujisawa, M., M. Ito, and T. A. Krulwich. 2007. Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity. Proc. Natl. Acad. Sci. USA 10413289-13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerchman, Y., A. Rimon, M. Venturi, and E. Padan. 2001. Oligomerization of NhaA, the Na+/H+ antiporter of Escherichia coli in the membrane and its functional and structural consequences. Biochemistry 403403-3412. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg, E. B., T. Arbel, J. Chen, R. Karpel, G. A. Mackie, S. Schuldiner, and E. Padan. 1987. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 842615-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamamoto, T., M. Hashimoto, M. Hino, M. Kitada, Y. Seto, T. Kudo, and K. Horikoshi. 1994. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol. Microbiol. 14939-946. [DOI] [PubMed] [Google Scholar]
- 11.Hedderich, R., and L. Forzi. 2005. Energy-converting [NiFe] hydrogenases: more than just H2 activation. J. Mol. Microbiol. Biotechnol. 1092-104. [DOI] [PubMed] [Google Scholar]
- 12.Heuberger, E. H., L. M. Veenhoff, R. H. Duurkens, R. H. Friesen, and B. Poolman. 2002. Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J. Mol. Biol. 317591-600. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu, T., K. Kodama, T. Kuroda, T. Mizushima, and T. Tsuchiya. 1998. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J. Bacteriol. 1806642-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, M., A. A. Guffanti, and T. A. Krulwich. 2001. Mrp-dependent Na+/H+ antiporters of Bacillus exhibit characteristics that are unanticipated for completely secondary active transporters. FEBS Lett. 496117-120. [DOI] [PubMed] [Google Scholar]
- 15.Ito, M., A. A. Guffanti, B. Oudega, and T. A. Krulwich. 1999. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 1812394-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, M., A. A. Guffanti, W. Wang, and T. A. Krulwich. 2000. Effects of nonpolar mutations in each of the seven Bacillus subtilis mrp genes suggest complex interactions among the gene products in support of Na+ and alkali but not cholate resistance. J. Bacteriol. 1825663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajiyama, Y., M. Otagiri, J. Sekiguchi, S. Kosono, and T. Kudo. 2007. Complex formation of the mrp (sha)ABCDEFG gene products that constitute the principal Na+/H+ antiporter in Bacillus subtilis. J. Bacteriol. 1897511-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao, M. C., E. Nakamaru-Ogiso, A. Matsuno-Yagi, and T. Yagi. 2005. Characterization of the membrane domain subunit NuoK (ND4L) of the NADH-quinone oxidoreductase from Escherichia coli. Biochemistry 449545-9554. [DOI] [PubMed] [Google Scholar]
- 19.Kashyap, D. R., L. M. Botero, C. Lehr, D. J. Hassett, and T. R. McDermott. 2006. A Na+:H+ antiporter and a molybdate transporter are essential for arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 1881577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kikuno, R., and T. Miyata. 1985. Sequence homologies among mitochondrial DNA-coded URF2, URF4 and URF5. FEBS Lett. 18985-88. [DOI] [PubMed] [Google Scholar]
- 21.Kosono, S., K. Haga, R. Tomizawa, Y. Kajiyama, K. Hatano, S. Takeda, Y. Wakai, M. Hino, and T. Kudo. 2005. Characterization of a multigene-encoded sodium/hydrogen antiporter (Sha) from Pseudomonas aeruginosa: its involvement in pathogenesis. J. Bacteriol. 1875242-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosono, S., Y. Ohashi, F. Kawamura, M. Kitada, and T. Kudo. 2000. Function of a principal Na+/H+ antiporter, ShaA, is required for initiation of sporulation in Bacillus subtilis. J. Bacteriol. 182898-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathiesen, C., and C. Hagerhall. 2003. The ‘antiporter module’ of respiratory chain Complex I includes the MrpC/NuoK subunit ′a revision of the modular evolution scheme. FEBS Lett. 54597-13. [DOI] [PubMed] [Google Scholar]
- 24.Mathiesen, C., and C. Hagerhall. 2002. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta 1556121-132. [DOI] [PubMed] [Google Scholar]
- 25.Newman, M. J., D. L. Foster, T. H. Wilson, and H. R. Kaback. 1981. Purification and reconstitution of functional lactose carrier from Escherichia coli. J. Biol. Chem. 25611804-11808. [PubMed] [Google Scholar]
- 26.Nozaki, K., T. Kuroda, T. Mizushima, and T. Tsuchiya. 1998. A new Na+/H+ antiporter, NhaD, of Vibrio parahaemolyticus. Biochim. Biophys. Acta 1369213-220. [DOI] [PubMed] [Google Scholar]
- 27.Padan, E., E. Bibi, M. Ito, and T. A. Krulwich. 2005. Alkaline pH homeostasis in bacteria: New insights. Biochim. Biophys. Acta 171767-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padan, E., and S. Schuldiner. 1996. Bacterial Na+/H+ antiporters - molecular biology, biochemistry and physiology., p. 501-531. In W. N. Konings, H. R. Kaback, and J. Lolkema (ed.), The Handbook of Biological Physics, Vol. II. Transport processes in membranes. Elsevier Science, Amsterdam. [Google Scholar]
- 29.Padan, E., M. Venturi, Y. Gerchman, and N. Dover. 2001. Na+/H+ antiporters. Biochim. Biophys. Acta 1505144-157. [DOI] [PubMed] [Google Scholar]
- 30.Putnoky, P., A. Kereszt, T. Nakamura, G. Endre, E. Grosskopf, P. Kiss, and A. Kondorosi. 1998. The pha gene cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol. Microbiol. 281091-1101. [DOI] [PubMed] [Google Scholar]
- 31.Radchenko, M. V., R. Waditee, S. Oshimi, M. Fukuhara, T. Takabe, and T. Nakamura. 2006. Cloning, functional expression and primary characterization of Vibrio parahaemolyticus K+/H+ antiporter genes in Escherichia coli. Mol. Microbiol. 59651-663. [DOI] [PubMed] [Google Scholar]
- 32.Rimon, A., T. Tzubery, and E. Padan. 2007. Monomers of the NhaA Na+/H+ antiporter of Escherichia coli are fully functional yet dimers are beneficial under extreme stress conditions at alkaline pH in the presence of Na+ or Li+. J. Biol. Chem. 28226810-26821. [DOI] [PubMed] [Google Scholar]
- 33.Rosen, B. P. 1986. Ion extrusion systems in E. coli. Methods Enzymol. 125328-386. [DOI] [PubMed] [Google Scholar]
- 34.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Frisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
- 36.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 37.Steuber, J. 2003. The C-terminally truncated NuoL subunit (ND5 homologue) of the Na+-dependent complex I from Escherichia coli transports Na+. J. Biol. Chem. 27826817-26822. [DOI] [PubMed] [Google Scholar]
- 38.Swartz, T. H. 2006. Mrp Systems of Gram-Positive Bacteria: Properties of the Monovalent Cation/Proton Antiport. Ph.D. Dissertation, Mount Sinai School of Medicine of New York University, New York.
- 39.Swartz, T. H., S. Ikewada, O. Ishikawa, M. Ito, and T. A. Krulwich. 2005. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles. 9345-354. [DOI] [PubMed] [Google Scholar]
- 40.Swartz, T. H., M. Ito, D. B. Hicks, M. Nuqui, A. A. Guffanti, and T. A. Krulwich. 2005. The Mrp Na+/H+ antiporter increases the activity of the malate:quinone oxidoreductase of an Escherichia coli respiratory mutant. J. Bacteriol. 187388-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swartz, T. H., M. Ito, T. Ohira, S. Natsui, D. B. Hicks, and T. A. Krulwich. 2007. Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3 (Mrp) family are revealed by an optimized assay in an Escherichia coli host. J. Bacteriol. 1893081-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ugalde, C., R. Hinttala, S. Timal, R. Smeets, R. J. Rodenburg, J. Uusimaa, L. P. van Heuvel, L. G. Nijtmans, K. Majamaa, and J. A. Smeitink. 2007. Mutated ND2 impairs mitochondrial complex I assembly and leads to Leigh Syndrome. Mol. Genet. Metab. 9010-14. [DOI] [PubMed] [Google Scholar]
- 43.Ugalde, C., R. Vogel, R. Huijbens, B. Van Den Heuvel, J. Smeitink, and L. Nijtmans. 2004. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum. Mol. Genet. 132461-2472. [DOI] [PubMed] [Google Scholar]
- 44.Veenhoff, L. M., E. H. Heuberger, and B. Poolman. 2002. Quaternary structure and function of transport proteins. Trends Biochem. Sci. 27242-249. [DOI] [PubMed] [Google Scholar]
- 45.Verkhovskaya, M. L., B. Barquera, and M. Wikstrom. 2001. Deletion of one of two Escherichia coli genes encoding putative Na+/H+ exchangers (ycgO) perturbs cytoplasmic alkali cation balance at low osmolarity. Microbiology 1473005-3013. [DOI] [PubMed] [Google Scholar]
- 46.Yoshinaka, T., H. Takasu, R. Tomizawa, S. Kosona, and T. Kudo. 2003. A shaE deletion mutant showed lower Na+ sensitivity compared to other deletion mutants in the Bacillus subtilis sodium/hydrogen antiporter (Sha) system. J. Biosci. Bioeng. 95306-309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.