Abstract
Orthologs of RecG and RuvABC are highly conserved among prokaryotes; in Escherichia coli, they participate in independent pathways that branch migrate Holliday junctions during recombinational DNA repair. RecG also has been shown to directly convert stalled replication forks into Holliday junctions. The bacterium Helicobacter pylori, with remarkably high levels of recombination, possesses RecG and RuvABC homologs, but in contrast to E. coli, H. pylori RecG limits recombinational repair. We now show that the RuvABC pathway plays the prominent, if not exclusive, repair role. By introducing an E. coli resolvase (RusA) into H. pylori, the repair and recombination phenotypes of the ruvB mutant but not the recG mutant were improved. Our results indicate that RecG and RuvB compete for Holliday junction structures in recombinational repair, but since a classic RecG resolvase is absent from H. pylori, deployment of the RecG pathway is lethal. We propose that evolutionary loss of the H. pylori RecG resolvase provides an “antirepair” pathway allowing for selection of varied strains. Such competition between repair and antirepair provides a novel mechanism to maximize fitness at a bacterial population level.
Prokaryotic DNA replication is rapid and highly faithful, but replication fork halts, arising from direct DNA damage, secondary structure formation, or DNA-bound proteins, are common (22, 31, 32). Failure to accurately resume replication after such lesions causes cell death or, due to strand breakage, results in genomic instability, including DNA rearrangement and mutation (35). Although homologous recombination is critical for restoring damaged replication forks (33), conversion into Holliday junctions is required for recombination to occur. In Escherichia coli, one pathway involves RecG, a DNA helicase that converts stalled replication forks into Holliday junctions (7, 12, 45).
RecG also is involved in the branch migration of Holliday junctions (34), whereas other E. coli proteins resolve the Holliday junctions to active replication forks. An alternative pathway involving RuvABC also branch migrates Holliday junctions, with RuvC providing the analogous Holliday junction resolution (51). E. coli strains with null mutations in ruvAB or recG have moderately increased sensitivity to UV (26, 28, 37) and defective Hfr recombination and transduction (27, 28), consistent with impaired recombinational repair. Mutations in both recG and ruvAB lead to severely deficient recombinational repair and extreme UV sensitivity (8, 14, 25, 28); this synergistic effect implies RecG and RuvABC involvement in independent recombinational repair pathways.
RecG and RuvABC orthologs are highly conserved in prokaryotes (39, 41). Helicobacter pylori, a gram-negative bacterium that colonizes the human gastric mucosa, possesses homologs of RecG and RuvABC (1, 19, 36, 48). Although H. pylori RecG can complement the UV sensitivity phenotype of an E. coli recG mutant, it is not involved in H. pylori repair of UV damage (19). In contrast to E. coli, H. pylori RecG mutants have improved recombinational repair, as measured by assays of ciprofloxacin resistance, transformation frequencies, and frequency of deletions between direct repeats (19). That each of these processes requires branch migration and resolution of Holliday junctions (30) and that recombinational repair is improved in RecG-null mutants suggest that in H. pylori, RecG initiates a “dead-end” pathway that competes with RuvABC for Holliday junctions. A prior study done in H. pylori found ruvC mutants to have increased susceptibility to oxidative stress, with decreased survival in a mouse host, suggesting the importance of the RuvABC pathway in recombinational repair (29).
Based on these observations, we hypothesized that RuvABC is most critical to H. pylori recombinational repair. We now demonstrate that Ruv mutants are similar in this phenotype to RecA strains. We further hypothesized that the lack of an appropriate RecG resolvase may explain the dichotomy between H. pylori and E. coli RecG mutant phenotypes. To test this hypothesis, we introduced an E. coli Holliday junction resolvase, RusA, into H. pylori ruvB mutants to attempt to complement the incomplete H. pylori RecG recombinational repair pathway. There is no known homolog of RusA in H. pylori or in other Epsilonproteobacteria, including Campylobacter jejuni (1, 36). Our results show that E. coli RusA can resolve RecG helicase-mediated repair in H. pylori, thus demonstrating the necessary role of an appropriate Holliday junction resolvase in H. pylori RecG-mediated recombinational repair pathway. The presence of an incomplete recombinational repair pathway involving RecG, leading to cell death, implies an intrinsic tension between the Ruv- and RecG-mediated Holliday junction helicase activities and indicates alternative selection for DNA repair pathways among prokaryotes.
MATERIALS AND METHODS
Statistical analyses.
Student's t test, unpaired with equal variance, was used to determine significance in all cases. A P value of <0.05 was defined as significant.
Bacterial strains and plasmids.
The H. pylori and E. coli strains and plasmids used in this study are listed in Table S1 in the supplemental material. E. coli strains AB1157 and HRS2301 (ruvB::cat), kindly provided by Hideo Shinagawa, were routinely grown on Luria-Bertani (LB) agar plates at 37°C supplemented with appropriate antibiotics. H. pylori strains were routinely grown at 37°C in 5% CO2 on plates with trypticase soy agar (TSA) or brucella agar (BA) with 10% newborn calf serum (NCS) supplemented with streptomycin (20 μg/ml), chloramphenicol (20 μg/ml), and/or kanamycin (25 μg/ml).
Construction of H. pylori mutants used to assess susceptibility to DNA damage and intergenomic recombination frequencies.
A fragment of the Hp1059 (ruvB homolog) open reading frame (ORF) was amplified by PCR using primers RuvBF and RuvBR, based on sequenced strain 26695 (see Table S2 in the supplemental material), and cloned into pGEMT-Easy (Promega, Madison, WI) to create pRuvB. The construction of pRecAKm and pRecG has been described previously (19). A unique BamHI site was created in pRuvB via inverse PCR using primers RuvBinvF and RuvBinvR (see Table S2 in the supplemental material). The aphA cassette, conferring kanamycin resistance (Kanr), was used to interrupt ruvB within pRuvB to create pRuvBKm. The cat cassette, conferring chloramphenicol resistance (Camr) was used to interrupt the recG region of pRecG, creating pRecGCat. H. pylori strain JP26 was transformed to Kanr or Catr with pRuvBKm, pRecAKm, or pRecGCat to create JP26 ruvB::aphA, JP26 recA::aphA, and JP26 recG::cat, respectively, as described previously (50). The RecG RuvB double mutant, JP26 recG ruvB, was constructed by transforming JP26 recG::cat with pRuvBKm.
Chromosomal DNA was isolated from transformants, and the correct insertion of the aphA and/or cat cassette into the expected ORFs was confirmed by PCR in each case; the double mutant JP26 recG ruvB was further confirmed with Southern hybridizations (see Fig. S3 in the supplemental material).
Construction of H. pylori mutants used to assess deletion frequencies.
A unique BamHI site was created in pRuvB and pVacA by inverse PCR using primers RBinvR and RBinvF, based on sequenced strain 26695 (see Table S2 in the supplemental material), and each ORF was subsequently interrupted with a deletion cassette containing either 0-, 50-, or 100-bp repeats to create pRuvB-0, -50, and -100 and pVacA-0, -50, and -100. The construction and use of the deletion cassettes have been described previously (3, 13). H. pylori strain JP26 was subsequently transformed to chloramphenicol resistance with these plasmids to create JP26 ruvB::0, JP26 ruvB::50, and JP26 ruvB::100 and JP26 vacA::0, JP26 vacA::50, and JP26 vacA::100. Chromosomal DNA was isolated from transformants, and correct insertion of the deletion cassette into the expected ORF was confirmed by PCR.
RusA expression in H. pylori.
A plasmid with E. coli rusA placed downstream of the ureAB promoter (pADC-RusA) was constructed using primers RusAF and RusAR (see Table S2 in the supplemental material) and used to introduce E. coli rusA in trans into the genome of H. pylori strain JP26 via natural transformation, as described previously (2), to create JP26 WT+RusA. JP26 WT+RusA was transformed with pRuvBKm or pRecGKm (19) to create JP26 RB+rusA and JP26 RG+rusA, respectively. Correct insertions were confirmed by PCR of chromosomal DNA.
Complementation of the JP26 ruvB::aphA mutant.
Plasmids pADC-HPrB and pADC-ECrB, with ORF Hp1059 or E. coli ruvB placed downstream of the ureAB promoter, were constructed using primers HPruvBcombF and HPruvBcombR or ECruvBcompF and ECruvBcompR, respectively (see Table S2 in the supplemental material), and used to introduce Hp1059 or E. coli ruvB in trans into the genome of JP26 via natural transformation, as described previously (2, 50), to create JP26 HPruvBcomb and JP26 ECruvBcomp, respectively. Transformants were selected based on Catr, and the correct insertion was confirmed by PCR of the chromosomal DNA. Mutants were subsequently transformed with pRuvBKm and confirmed by PCR.
Complementation of E. coli rusA mutant.
E. coli strain HRS2301 (ruvB::cat) was transformed with either pADC-HPrB, pADC-ECrB, or pAD1 (no insert) to create strains 2301-HPuD, 2301-ECuD, and 2301-AD1, respectively.
Recovery from DNA damage.
E. coli or H. pylori cells to be tested were grown on TSA plates for 24 h or 48 h, respectively, and suspended in 1 ml phosphate buffered solution (PBS). Wild-type H. pylori cells are more sensitive to UV irradiation than are E. coli cells (17, 19). Equal amounts of suspension were inoculated on TSA plates at dilutions that would produce 100 to 500 CFU per plate postexposure. Cells then were exposed to UV at 312 nm (Stratagene Transluminator; Stratagene, La Jolla, CA) for 0 to 1,400 kJ/m2 and incubated at 37°C, colonies were counted, and percent survival was calculated.
Ciprofloxacin Etest strips (AB Biodisk, Solna, Sweden) were used to determine MICs for both wild-type, mutant, and complemented strains of H. pylori, according to the manufacturer's instructions. Plates were incubated for 48 h, and MIC determinations were repeated at least six times for each sample.
Streptomycin resistance frequency assay to assess intergenomic recombination.
H. pylori strains were grown on TSA plates for 48 h and harvested into 1 ml of PBS, and 25 μl was spotted onto a fresh TSA plate combined with 50 ng of donor DNA and incubated for 18 h at 37°C in 5% CO2. Donor DNA was an 800-bp PCR product of H. pylori rpsL from streptomycin-resistant (Str) strain JP26 with A128G (Torii et al., unpublished observation). The transformation mixture then was harvested into 1 ml PBS, and 100 μl of the appropriate serial dilutions was plated onto either TSA or BA plates containing 10% NCS and 25 μg/μl streptomycin. Plates were incubated for 4 days at 37 C in 5% CO2, and total recombination frequency was determined by dividing the number of Str colonies by the total number of CFU. As a negative control, H. pylori strains with no DNA added were tested in parallel in each experiment; no colonies were seen in any case.
Deletion frequency assays in H. pylori.
To assess recombination frequencies in the H. pylori strains containing the deletion or control cassettes, the cells were grown on TSA plates for 48 h at 37°C (5% CO2), allowing for deletions to occur, and then harvested into PBS. Total CFU and numbers of (Kanr) deletion mutants were determined by plating serial dilutions onto TSA plates or BA plates with kanamycin (25 μg/ml), respectively. As a control, 200 μl from each suspension was inoculated onto BA plates containing NCS, kanamycin (25 μg/ml), and chloramphenicol (20 μg/ml); as expected, in no experiments were strains with double resistance identified, confirming the specificity of the deletion process (3). Plates were incubated at 37°C in a 5% CO2 environment for 96 h, colonies were counted, and deletion frequencies were calculated.
RESULTS
Analyses of RuvB family homologs.
We first sought to establish the relationship of the annotated H. pylori RuvB homolog (HP1059 and JHP0366 in sequenced strains 26695 and J99, respectively) to known bacterial RuvB orthologs. The product of HP1059 has 97.3% sequence identity to JHP0366 and 52.0% sequence identity to E. coli RuvB. Alignment of the amino acid sequences of H. pylori and E. coli RuvB (see Fig. S1A in the supplemental material) showed extensive conservation, particularly within the helicase domains (11), and the H. pylori RuvB pylogenetic relationships are consistent with orthology (see Fig. S1B in the supplemental material). In E. coli, ruvA and ruvB are located on the same operon, with ruvC nearly adjacent (see Fig. S2A in the supplemental material), but in H. pylori, ruvB is not located near either ruvA (HP0883 in strain 26695 and JHP0815 in strain J99) or RuvC (HP0877 and JHP0811) (see Fig. S2B in the supplemental material); these genomic differences predict possible functional differences.
Role of H. pylori RuvB in DNA repair.
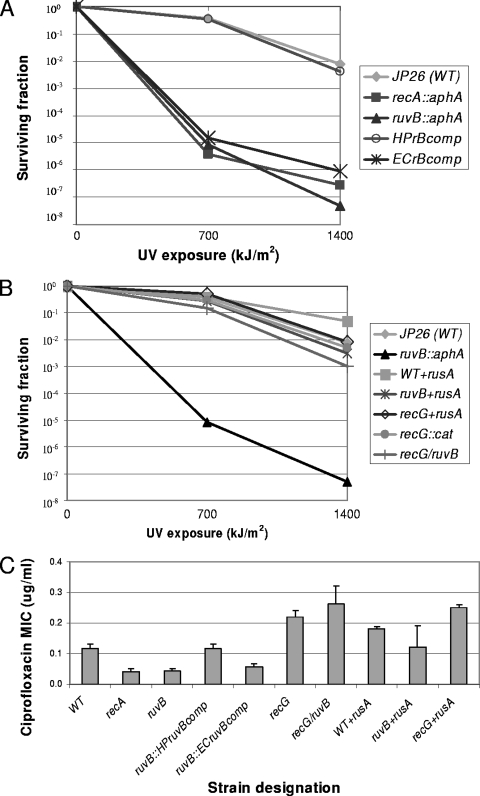
Confirming prior results (19), wild-type H. pylori cells are more susceptible to UV irradiation (Fig. 1A) than are E. coli cells (see Fig. S4 in the supplemental material). To determine the role of the RuvABC pathway in DNA repair, we constructed an H. pylori ruvB mutant and examined survival after exposure to UV irradiation. In contrast to E. coli ruvB mutants, which have moderate susceptibility to DNA damage (26), the H. pylori ruvB mutant has intense susceptibility to UV, similar to that of a recA mutant (Fig. 1A). That the phenotype is due to ruvB mutation was confirmed using the in trans-complemented strain, JP26 HPruvBcomb, in which wild-type ruvB was introduced into the ureA locus; in this strain, there was restoration of the wild-type phenotype. These results are consistent with the hypothesis that the RuvABC pathway is predominant, since RecG cannot complete recombinational repair (19). Complementation of the ruvB mutant with the E. coli ruvB product did not change the ruvB phenotype; the converse cross-species studies show that H. pylori ruvB does not complement the E. coli ruvB mutation (see Fig. S4 in the supplemental material). Thus, despite their orthology, the ruvB products from E. coli and H. pylori have different phenotypes, consistent with their divergent genomic relationships.
FIG. 1.
Susceptibility of H. pylori wild-type, mutant and complemented strains to genotoxic stress. (A) UV exposure. H. pylori cells were assayed for survival after exposure to UV (312 nm) for 0 to 1,400 kJ/m2. The strains were H. pylori JP26 (wild type [WT]), mutants JP26 ruvB::aphA (ruvB) and JP recA::aphA (recA), and complemented strains JP26 HPruvBcomb and JP26 ECruvBcomp. (B) Effect of RusA on UV exposure. The strains were H. pylori JP26 (WT), JP26 ruvB::aphA (ruvB), JP26 WT+RusA, JP26 ruvB+rusA, JP26 recG+rusA, JP26 recG::cat (recG), and double mutant JP26 recG ruvB. The results shown are representative of ≥3 experiments. (C) Ciprofloxacin exposure. For the above H. pylori strains, MICs were determined using Etest strips (AB Biodisc, Solna, Sweden) in ≥6 separate trials. JP26 recA::aphA, JP26 ruvB::aphA, and JP26 ECruvBcomp all show significant (P < 0.05) decreases in MICs compared to wild-type JP26. JP26 recG::cat, JP26 recG+rusA, JP26 recG ruvB, and JP26 WT+RusA all show significant (P < 0.05) MIC increases compared to wild-type JP26.
Next, we asked whether introduction of a resolvase (RusA from E. coli) would be able to complete the RecG repair pathway and thereby rescue the UV susceptibility of H. pylori ruvB mutants. To address this, rusA was introduced into the genome of H. pylori wild-type, recG, and ruvB strains and cells were assayed for survival after exposure to UV (Fig. 1B). As expected from prior studies (19), the H. pylori recG mutant shows no difference in UV susceptibility from the wild type. Expression of rusA had no effect on UV survival in the wild-type or recG background. However, rusA expression in the ruvB mutant essentially restored UV survival. When we assessed whether mutation of both recG and ruvB would have synergistic effects on UV susceptibility, as occurs in E. coli, we found that the H. pylori ruvB recG double mutant shows no significant difference from the wild type (Fig. 1B), demonstrating that recG mutation suppresses the ruvB phenotype. These studies indicate that a ruvB-dependent pathway is sufficient for DNA repair in H. pylori, but in its absence, rusA may complete an alternative pathway. The results of these experiments involving mutants suggest that recG and ruvB participate in opposing pathways.
As another test of control of DNA damage, susceptibility to ciprofloxacin (Fig. 1C) was determined for H. pylori wild-type and mutant strains. Ciprofloxacin, similar to other fluoroquinolones, creates a ternary structure with double-stranded DNA and either topoisomerase or gyrase, blocking replication fork progression (9, 20). Susceptibility to ciprofloxacin provides a measure of recombinational repair, a primary mechanism for overcoming replication fork blocks; E. coli ruv mutants have decreased quinolone MICs (42). The H. pylori ruvB mutant has a significantly diminished ciprofloxacin MIC, to the same extent as the recA mutant (Fig. 1C), findings that parallel the UV susceptibility results. The H. pylori ruvB-complemented strain (JP26 HPruvBcomb) has restoration of the wild-type phenotype, confirming the importance of ruvB in recombinational repair, whereas introduction of the E. coli ruvB (JP26 ECruvBcomp) does not complement, similar to findings for UV-induced injury (Fig. 1C). As now expected (19), the recG mutant has a significantly increased ciprofloxacin MIC. When rusA was introduced into the wild-type H. pylori strain (JP26 WT+RusA), there was no significant difference from the wild type alone. Introduction of rusA into the ruvB mutant restores ciprofloxacin MICs to the level of wild-type H. pylori, demonstrating in another assay that RusA is able to restore an incomplete H. pylori repair pathway. Introduction of rusA into a recG mutant had no effect on phenotype, indicating that RusA does not interact with any other recombinational repair pathway; it appears to complete only the RecG pathway. The double mutant JP26 recG ruvB also displays a significant (P < 0.01) increase in ciprofloxacin MIC compared to the wild type, providing further evidence that the “antirepair” RecG phenotype suppresses the “repair” RuvB phenotype.
Effect of RuvB on intergenomic recombination.
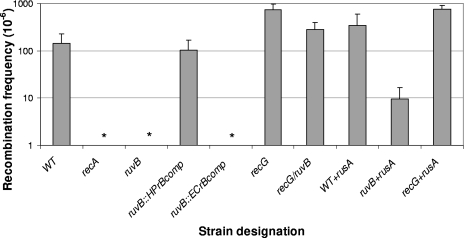
To determine whether ruvB mutants were impaired in recombination, we examined their ability to be naturally transformed by exogenous DNA (Fig. 2). No transformants were detected in the ruvB mutant (frequency of <10−8), similar to those obtained for the recA mutant (19, 40). Complementation with H. pylori ruvB (JP26 HPruvBcomp) restores the wild-type phenotype, whereas complementation with E. coli ruvB (JP26 ECruvBcomp) again had no effect. The double mutant (JP26 recG ruvB) showed a substantially higher level of recombination than ruvB, approximating the wild-type level, as for DNA repair (Fig. 1). These experiments provide evidence that RecG and RuvB have opposing functions with regard to a transformation phenotype and that recG mutation suppresses the ruvB phenotype.
FIG. 2.
Intergenomic recombination frequencies of H. pylori wild-type, mutant, and complemented strains. H. pylori strains were transformed to streptomycin resistance with an 800-bp A128G rpsL PCR product (23); bars represent means ± standard deviations of transformation frequency. The strains transformed were H. pylori JP26 (wild type [WT]) and isogenic recA, recG, ruvB, and recG ruvB mutants, an ruvB mutant complemented with either H. pylori ruvB (JP26 HPruvBcomb), or E. coli ruvB (JP26 ECruvBcomp) downstream of a strong (ureA) promoter, and H. pylori wild-type and recG and ruvB mutant strains expressing RusA (JP26 WT+RusA, JP26 recG+rusA, and JP26 ruvB+rusA, respectively). As expected, no transformants were observed in the recA mutant control. The recG::cat strain exhibits a significantly higher transformation frequency than the wild type (P < 0.05). Asterisks indicate that no transformants were detected for the recA, ruvB, and ECruvBcomp mutants (frequency, <10−8). Complementation with H. pylori but not E. coli ruvB restored transformation. Addition of RusA to either the wild type or ruvB mutants enhanced resistance but had no effect in the recG mutant.
Introduction of rusA into wild-type H. pylori (JP26 WT+RusA) results in a small but not significant (P = 0.09) difference in recombination frequency compared to the wild type. Introduction of rusA into the H. pylori ruvB mutant (with intact RecG) partially restored recombination; since intergenomic recombination requires branch migration and resolution of Holliday junctions, the higher recombination rates may reflect a now completed pathway. As now expected, introduction of rusA into the the recG strain had no effect on the recG phenotype.
Effect of RuvB on deletions between direct repeats.
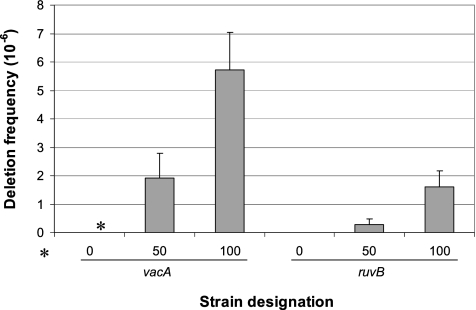
Since in H. pylori, the RecG helicase limits deletions between direct repeats (19), we next examined the phenotype of the RuvB helicase by using a previously characterized deletion cassette, with identical repeats of either 0 bp (control), 50 bp, or 100 bp (3, 16, 18, 19). Since deletion produces a phenotypic change from Camr to Kanr, it is simple to score (3). A strain that is wild type for ruvB but has the deletion cassette inserted into a control gene (vacA) showed the expected (3) wild-type deletion frequencies (Fig. 3). However, the ruvB mutants displayed significant (P < 0.05), four- to sevenfold-lower deletion frequencies than the control (vacA) strain with the identical cassettes (Fig. 3). These data indicate that the RuvB helicase facilitates deletions between direct repeats, in contrast to RecG, which lowers the frequency (19). Thus, in a fourth phenotype, the helicases show opposing functions.
FIG. 3.
Deletion frequency of H. pylori wild-type and ruvB mutant strains. The chloramphenicol/kanamycin resistance cassette flanked by identical repeats of 0 (control), 50, or 100 bp, as described by Aras et al. (3), was inserted into vacA (control) or ruvB, and deletion frequencies calculated, as described previously (3). As expected, no deletions were detected (*, frequency of <10−8) for the strains with the 0-bp cassette, and strains with the cassette in vacA or ruvB showed progressively higher deletion frequencies with increasing size of the identical repeats. Strains with the 50-bp or 100-bp deletion cassette in ruvB display significant (P < 0.05) seven- and fourfold decreases, respectively, in deletion frequency compared to the control (vacA) with comparable cassettes. Bars represent means ± standard deviations from ≥6 replicate experiments.
DISCUSSION
In E. coli, the ruvAB operon is induced by DNA damage as part of the SOS response (43, 44), whereas there is no SOS response in H. pylori (49). Unlike in E. coli, ruvA, ruvB, and ruvC are located in separate regions of the H. pylori chromosome. These differences predict that the levels of regulation of ruvABC expression in E. coli and in H. pylori are substantially different and that the functions of the genes in their respective backgrounds also differ.
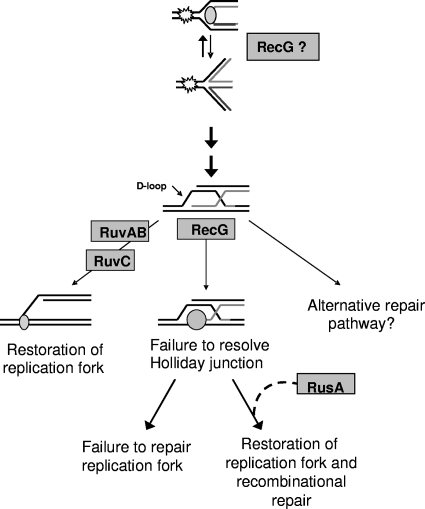
Both RuvB and RecG are helicases that can branch migrate Holliday junctions, which are common recombination intermediates (24). In the RuvABC pathway, the RuvC endonuclease nicks DNA, catalyzing Holliday junction resolution into double-stranded DNA (Fig. 4). Although the resolvase in the RecG pathway has not been completely elucidated, it has been hypothesized that RusA may serve this function in E. coli (6, 41). Since E. coli possesses complementary RuvABC- and RecG-mediated pathways of Holliday junction branch-migration and resolution, separate ruv or recG mutants display moderate defects in DNA repair, whereas ruv recG double mutants have severe defects (8, 14, 25, 28).
FIG. 4.
Proposed mechanism of RuvB/RecG Holliday junction resolution in H. pylori. Our results suggest that two H. pylori DNA processing pathways may compete for the same intermediate, a Holliday junction, which can be branch migrated (left pathway) by RuvAB and resolved by RuvC, leading to restoration of an intact replication fork, enabling loading of the replisome (small oval). Alternatively, (middle pathway) RecG (gray circle) can branch migrate Holliday junctions, but due to lack of resolution, there is no restoration of template leading to cell death. Addition of RusA restores the RecG pathway, leading to recombinational repair. RecG may act upstream of RuvABC to directly convert stalled forks into Holliday junctions in H. pylori, as it does in E. coli. Although this has not yet been shown to occur in H. pylori, this is depicted as a possibility.
In marked contrast to E. coli, H. pylori recG mutants do not have defective DNA repair, as measured by UV and ciprofloxacin susceptibility, which demonstrates the lack of involvement of recG in H. pylori recombinational repair (19). Results from a prior study (19) predicted a more prominent role for RuvABC in recombinational repair in H. pylori. Consistent with this hypothesis, we now show that RuvABC is the predominant pathway for H. pylori recombinational repair, with ruvB strains showing phenotypes similar to those of recA strains. These findings also indicate the absence of other functional pathways that can compensate for ruvB mutation under the conditions tested. However, with mutation of both ruvB and recG, both resistance to DNA damage and intergenomic recombination are restored to near-wild-type levels, indicating that recG mutation suppresses the ruvB phenotype; again, this is in marked contrast to E. coli (25, 28). One hypothesis is that RecG interferes with recombinational repair, but because of a strong Ruv phenotype, this cannot be detected. Thus, the double mutant has loss of a repair enzyme (ruvB) and an antirepair enzyme (recG), which compete for the same substrate DNA ends.
Support for this hypothesis comes from examination of other phenotypes. H. pylori recG mutants have diminished susceptibility to ciprofloxacin and increased frequencies of intergenomic recombination and deletion (19), processes that are facilitated by branch migration and Holliday junction resolution. Such observations suggest that branch migration and Holliday junction resolution are more efficient in the absence of RecG function; however, with ruvB mutation, the predominant H. pylori pathway for Holliday junction branch migration and resolution is clearly lost. The lower deletion frequencies in ruvB mutants indicate the role of branch migration/Holliday junction resolution in their processing; that ruvB mutants have a far-less-extreme phenotype than for recombinational repair implies that H. pylori can use alternate pathways prior to the RecA-dependent step. In total, the results are consistent with the hypothesis that RecG competes with RuvABC for DNA substrates but initiates an incomplete repair pathway in H. pylori, interfering with repair (Fig. 1). What is the missing step? In E. coli, a RecG-specific resolvase, rusA, resolves RecG branch migration products (6, 41). By introducing E. coli rusA into H. pylori ruvB mutants, the observed restoration of wild-type phenotypes for DNA repair and recombination and the lack of effect in the recG mutant (Fig. 1 and 2) indicate that RecG pathway resolution may be missing in H. pylori.
In conclusion, we have shown that RuvABC is involved in the major recombinational repair pathway in H. pylori critical for DNA damage repair and facilitates intergenomic (and intragenomic) recombination events to a far greater extent than in E. coli. Although RecG has a role in branch migration, RecG interferes with recombinational repair. Why has a “dead-end” RecG pathway been preserved in H. pylori? H. pylori is an organism with remarkable genomic plasticity (46), arising through natural transformation (23) and recombination (10, 47), spontaneous point mutations, and deletions/duplications involving repetitive DNA sequences (15). Our results suggest that the presence of a helicase such as RecG, which limits deletions and recombination, preserves genomic integrity (19), but that a (antirepair) RecG-mediated pathway (without a resolvase) may permit selection against H. pylori cells with DNA injuries.
We hypothesize that the enhanced genomic integrity afforded by RecG balances the diminished recombinational repair its deployment yields. In the stomach, which resembles a “chemostat” for the H. pylori cells (5, 21) that dominate their ecological niche (4), fluctuations in mutation rate may be detrimental to individual cells but profit the overall H. pylori population by providing more varied phenotypic substrates in the face of changing selection pressures (13). The tension between RecG and RuvB in H. pylori may be the molecular solution that evolved in this mismatch repair-deficient organism (17, 38), to solve the problem of generating diversity in a way that maximizes preservation of essential genomic features.
Supplementary Material
Footnotes
Published ahead of print on 28 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397176-180. [DOI] [PubMed] [Google Scholar]
- 2.Ando, T., D. A. Israel, K. Kusugami, and M. J. Blaser. 1999. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 1815572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aras, R. A., J. Kang, A. I. Tschumi, Y. Harasaki, and M. J. Blaser. 2003. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc. Natl. Acad. Sci. USA 10013579-13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 103732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser, M. J., and D. Kirschner. 1999. Dynamics of Helicobacter pylori colonization in relation to the host response. Proc. Natl. Acad. Sci. USA 968359-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolt, E. L., and R. G. Lloyd. 2002. Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol. Cell 10187-198. [DOI] [PubMed] [Google Scholar]
- 7.Dillingham, M. S., and S. C. Kowalczykowski. 2001. A step backward in advancing DNA replication: rescue of stalled replication forks by RecG. Mol. Cell 8734-736. [DOI] [PubMed] [Google Scholar]
- 8.Donaldson, J. R., C. T. Courcelle, and J. Courcelle. 2004. RuvAB and RecG are not essential for the recovery of DNA synthesis following UV-induced DNA damage in Escherichia coli. Genetics 1661631-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falush, D., C. Kraft, N. S. Taylor, P. Correa, J. G. Fox, M. Achtman, and S. Suerbaum. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. USA 9815056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3419-429. [Google Scholar]
- 12.Gregg, A. V., P. McGlynn, R. P. Jaktaji, and R. G. Lloyd. 2002. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell 9241-251. [DOI] [PubMed] [Google Scholar]
- 13.Huang, S., J. Kang, and M. J. Blaser. 2006. Antimutator role of the DNA glycosylase mutY gene in Helicobacter pylori. J. Bacteriol. 1886224-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishioka, K., H. Iwasaki, and H. Shinagawa. 1997. Roles of the recG gene product of Escherichia coli in recombination repair: effects of the delta recG mutation on cell division and chromosome partition. Genes Genet. Syst. 7291-99. [DOI] [PubMed] [Google Scholar]
- 15.Kang, J., and M. J. Blaser. 2006. Bacterial populations as perfect gases: genomic integrity and diversification tensions in Helicobacter pylori. Nat. Rev. Microbiol. 4826-836. [DOI] [PubMed] [Google Scholar]
- 16.Kang, J., and M. J. Blaser. 2006. UvrD helicase suppresses recombination and DNA damage-induced deletions. J. Bacteriol. 1885450-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, J., S. Huang, and M. J. Blaser. 2005. Structural and functional divergence of MutS2 from bacterial MutS1 and eukaryotic MSH4-MSH5 homologs. J. Bacteriol. 1873528-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang, J., N. M. Iovine, and M. J. Blaser. 2006. A paradigm for direct stress-induced mutagenesis in prokaryotes. FASEB J. 202476-2485. [DOI] [PubMed] [Google Scholar]
- 19.Kang, J., D. Tavakoli, A. Tschumi, R. A. Aras, and M. J. Blaser. 2004. Effect of host species on RecG phenotypes in Helicobacter pylori and Escherichia coli. J. Bacteriol. 1867704-7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khodursky, A. B., and N. R. Cozzarelli. 1998. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J. Biol. Chem. 27327668-27677. [DOI] [PubMed] [Google Scholar]
- 21.Kirschner, D. E., and M. J. Blaser. 1995. The dynamics of Helicobacter pylori infection of the human stomach. J. Theor. Biol. 176281-290. [DOI] [PubMed] [Google Scholar]
- 22.Kuzminov, A. 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16373-384. [DOI] [PubMed] [Google Scholar]
- 23.Levine, S. M., E. A. Lin, W. Emara, J. Kang, M. DiBenedetto, T. Ando, D. Falush, and M. J. Blaser. 2007. Plastic cells and populations: DNA substrate characteristics in Helicobacter pylori transformation define a flexible but conservative system for genomic variation. FASEB J. 213458-3467. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Y., and S. C. West. 2004. Happy Hollidays: 40th anniversary of the Holliday junction. Nat. Rev. Mol. Cell Biol. 5937-944. [DOI] [PubMed] [Google Scholar]
- 25.Lloyd, R. G. 1991. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J. Bacteriol. 1735414-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd, R. G., F. E. Benson, and C. E. Shurvinton. 1984. Effect of ruv mutations on recombination and DNA repair in Escherichia coli K12. Mol. Gen. Genet. 194303-309. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd, R. G., and C. Buckman. 1995. Conjugational recombination in Escherichia coli: genetic analysis of recombinant formation in Hfr × F− crosses. Genetics 1391123-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd, R. G., and C. Buckman. 1991. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J. Bacteriol. 1731004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loughlin, M. F., F. M. Barnard, D. Jenkins, G. J. Sharples, and P. J. Jenks. 2003. Helicobacter pylori mutants defective in RuvC Holliday junction resolvase display reduced macrophage survival and spontaneous clearance from the murine gastric mucosa. Infect. Immun. 712022-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovett, S. T. 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 521243-1253. [DOI] [PubMed] [Google Scholar]
- 31.Maisnier-Patin, S., K. Nordstrom, and S. Dasgupta. 2001. Replication arrests during a single round of replication of the Escherichia coli chromosome in the absence of DnaC activity. Mol. Microbiol. 421371-1382. [DOI] [PubMed] [Google Scholar]
- 32.McCool, J. D., and S. J. Sandler. 2001. Effects of mutations involving cell division, recombination, and chromosome dimer resolution on a priA2::kan mutant. Proc. Natl. Acad. Sci. USA 988203-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGlynn, P., and R. G. Lloyd. 2002. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 18413-419. [DOI] [PubMed] [Google Scholar]
- 34.McGlynn, P., and R. G. Lloyd. 1999. RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 273049-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh, J. D., H. Kling-Backhed, M. Giannakis, J. Xu, R. S. Fulton, L. A. Fulton, H. S. Cordum, C. Wang, G. Elliott, J. Edwards, E. R. Mardis, L. G. Engstrand, and J. I. Gordon. 2006. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. USA 1039999-10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otsuji, N., H. Iyehara, and Y. Hideshima. 1974. Isolation and characterization of an Escherichia coli ruv mutant which forms nonseptate filaments after low doses of ultraviolet light irradiation. J. Bacteriol. 117337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto, A. V., A. Mathieu, S. Marsin, X. Veaute, L. Ielpi, A. Labigne, and J. P. Radicella. 2005. Suppression of homologous and homeologous recombination by the bacterial MutS2 protein. Mol. Cell 17113-120. [DOI] [PubMed] [Google Scholar]
- 39.Rocha, E. P., E. Cornet, and B. Michel. 2005. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 1e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt, W., S. Odenbreit, D. Heuermann, and R. Haas. 1995. Cloning of the Helicobacter pylori recA gene and functional characterization of its product. Mol. Gen. Genet. 248563-572. [DOI] [PubMed] [Google Scholar]
- 41.Sharples, G. J., S. M. Ingleston, and R. G. Lloyd. 1999. Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA. J. Bacteriol. 1815543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shea, M. E., and H. Hiasa. 2003. The RuvAB branch migration complex can displace topoisomerase IV·quinolone·DNA ternary complexes. J. Biol. Chem. 27848485-48490. [DOI] [PubMed] [Google Scholar]
- 43.Shinagawa, H., K. Makino, M. Amemura, S. Kimura, H. Iwasaki, and A. Nakata. 1988. Structure and regulation of the Escherichia coli ruv operon involved in DNA repair and recombination. J. Bacteriol. 1704322-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shurvinton, C. E., and R. G. Lloyd. 1982. Damage to DNA induces expression of the ruv gene of Escherichia coli. Mol. Gen. Genet. 185352-355. [DOI] [PubMed] [Google Scholar]
- 45.Singleton, M. R., S. Scaife, and D. B. Wigley. 2001. Structural analysis of DNA replication fork reversal by RecG. Cell 10779-89. [DOI] [PubMed] [Google Scholar]
- 46.Suerbaum, S., and C. Josenhans. 2007. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat. Rev. Microbiol. 5441-452. [DOI] [PubMed] [Google Scholar]
- 47.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 9512619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388539-547. [DOI] [PubMed] [Google Scholar]
- 49.Wang, G., M. Z. Humayun, and D. E. Taylor. 1999. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol. 7488-493. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Y., K. P. Roos, and D. E. Taylor. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 1392485-2493. [DOI] [PubMed] [Google Scholar]
- 51.West, S. C. 1997. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31213-244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.