Abstract
The ability of Escherichia coli to colonize both intestinal and extraintestinal sites is driven by the presence of specific virulence factors, among which are the autotransporter (AT) proteins. Members of the trimeric AT adhesin family are important virulence factors for several gram-negative pathogens and mediate adherence to eukaryotic cells and extracellular matrix (ECM) proteins. In this study, we characterized a new trimeric AT adhesin (UpaG) from uropathogenic E. coli (UPEC). Molecular analysis of UpaG revealed that it is translocated to the cell surface and adopts a multimeric conformation. We demonstrated that UpaG is able to promote cell aggregation and biofilm formation on abiotic surfaces in CFT073 and various UPEC strains. In addition, UpaG expression resulted in the adhesion of CFT073 to human bladder epithelial cells, with specific affinity to fibronectin and laminin. Prevalence analysis revealed that upaG is strongly associated with E. coli strains from the B2 and D phylogenetic groups, while deletion of upaG had no significant effect on the ability of CFT073 to colonize the mouse urinary tract. Thus, UpaG is a novel trimeric AT adhesin from E. coli that mediates aggregation, biofilm formation, and adhesion to various ECM proteins.
Pathogenic bacteria often interact with their host through surface proteins referred to as adhesins. Two major classes of adhesins have been described, and both utilize vastly different mechanisms for their assembly at the cell surface. Fimbrial adhesins such as the prototypical type 1 fimbriae of Escherichia coli are composed primarily of a major repeating subunit protein and often contain minor-subunit proteins (including the adhesin) at the tip of the organelle. The biogenesis of these fimbriae is dependent on a highly conserved chaperone-usher system (20, 29). Nonfimbrial adhesins are a second class of adherence factors and encompass a diverse range of adhesins that includes the autotransporter (AT) family of proteins. AT proteins are particularly unique in that the information required for receptor recognition and routing and anchorage to the outer membrane is provided by the protein itself (25). Recent studies have identified a new group of nonfimbrial adhesins that have the capacity to form stable trimeric structures on the bacterial cell surface. These adhesins are characterized by a membrane-anchored C-terminal domain that forms a trimeric β-barrel pore and facilitates the translocation of a passenger domain (consisting of an extended stalk and an N-terminal head) to the bacterial cell surface via the type V secretion pathway (54, 67).
Trimeric AT proteins have been identified from a range of different gram-negative bacterial pathogens. Where characterized, the function of trimeric AT proteins is universally associated with bacterial adherence, and thus they constitute an important group of virulence factors (14). The YadA adhesin from Yersinia enterocolitica represents the best-characterized trimeric AT protein. YadA mediates adherence to host epithelial cells and extracellular matrix (ECM) proteins, is crucial for the colonization of the intestinal mucosa by Yersinia enterocolitica, and contributes to the serum resistance of Y. enterocolitica by inhibiting complement activation (3, 24, 68-70). Two trimeric AT adhesins have been described for Neisseria meningitidis: NadA, which mediates the binding to and invasion of epithelial cells, and NhhA, which mediates binding to human epithelial cells and ECM components, such as laminin and heparan sulfate (12, 59). The BadA protein from Bartonella henselae is a giant trimeric AT protein of more than 3,000 amino acids that plays a role in binding to ECM proteins and in cell host infection (52, 53). Hia and Hsf are trimeric AT proteins from Haemophilus influenzae that have been characterized and that mediate high-affinity adherence to respiratory epithelial cells (33). Trimeric AT proteins from Moraxella catarrhalis (UspA1 and UspA2) (27, 34) and plant pathogens such as Xanthomonas campestris (XadA) (50) have also been identified.
Escherichia coli is a highly diverse bacterial species that includes both harmless gut commensal strains and virulent intestinal and extraintestinal pathogens. In contrast to intestinal pathogens, extraintestinal pathogenic E. coli (ExPEC) can inhabit the intestinal tract as part of the normal flora and infect extraintestinal sites such as the urinary tract, the bloodstream, and the central nervous system (30). The range of infections caused by E. coli can be attributed to the extensive variation in DNA content (up to 1 Mb) that exists between different strain types. These differences reflect variation in gene content and are typically associated with pathogenicity islands. For example, a genome comparison analysis of the uropathogenic E. coli (UPEC) strain CFT073 with the E. coli K-12 strain MG1655 showed that they share only 41.7% of their encoded proteins; 14.3% of the proteins encoded by the E. coli K-12 MG1655 genome are absent from E. coli CFT073, while 23.8% of the proteins encoded by the E. coli CFT073 genome are absent from E. coli K-12 MG1655 (76). It is therefore reasonable to assume that part of the CFT073 genome encodes currently uncharacterized proteins that play a role in virulence.
Previous in silico analysis of the E. coli CFT073 genome sequence identified the presence of 10 putative proteins potentially secreted via the AT pathway (46). Two of the predicted proteins are highly homologous to E. coli K-12 antigen 43 (Ag43) protein, Ag43a (c3655), and Ag43b (c1273). We recently demonstrated that Ag43a and Ag43b possess functional differences; Ag43a promotes strong biofilm growth, and its expression is associated with long-term persistence in the urinary bladder (71). The protein encoded by one of the remaining putative AT-encoding genes from CFT073 (c4424/upaG) is highly homologous to members of the surface-exposed trimeric AT adhesins. UpaG was recently identified through a reverse vaccinology approach as a potentially protective antigen against ExPEC (17), suggesting the need for further analysis of its structural and functional properties. In this study, we performed an in-depth molecular analysis of UpaG. We have demonstrated that UpaG is exported to the cell surface by virtue of a C-terminal β domain and that it mediates the aggregation of E. coli as well as its adhesion to abiotic surfaces, T24 bladder epithelial cells, and ECM proteins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Cells were routinely grown at 37°C in M63B1-0.2% glucose minimal medium (M63B1glu) unless otherwise specified and supplemented with the appropriate antibiotic: kanamycin (Km; 50 mg ml−1), chloramphenicol (Cm; 25 mg ml−1), ampicillin (Amp; 100 mg ml−1), apramycin (30 mg ml−1), or zeocin (50 mg ml−1). HeLa epithelial cells (ATCC number CCL-2), T24 cells (Marinpharm), T24-red epithelial cells (Marinpharm), and BSC1 cells (ATCC number CCL-26) were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco).
TABLE 1.
Bacterial strains and plasmids used in this study
| E. coli strain(s) or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| OS56 | MG1655 flu Gfp+ Ampr | 62 |
| OS56(pBAD/Myc-HisA-Kan) | OS56 with plasmid pBAD/Myc-HisA-Kan, Kmr | 71 |
| OS56(pupaG) | OS56 with plasmid pupaG, Kmr | This study |
| CFT073 | Wild-type UPEC isolate | 42 |
| CFT073 ΔupaG | CFT073 ΔupaG::Cm; upaG mutant, Cmr | This study |
| CFT073 PcL upaG | CFT073 Km PcL upaG, constitutively expressed upaG, Kmr | This study |
| CFT073 RExBAD upaG | CFT073 Cm RExBAD upaG, arabinose-inducible upaG, Cmr | This study |
| UT5600 | ΔompT proC leu-6 trpE38 entA | 21 |
| UT5600(pUpaGL2-L1-β) | UT5600 with plasmid pUpaGL2-L1-β, Cmr | This study |
| UT5600(pUpaGL1-β) | UT5600 with plasmid pUpaGL1-β, Cmr | This study |
| UT5600(pUpaGβ) | UT5600 with plasmid pUpaGβ, Cmr | This study |
| U6 | UPEC | 73 |
| U15 | UPEC | 73 |
| E. coli collection | 96 commensal E. coli isolates constituted from several animal populations | Reference 64 and Table S2 in the supplemental material |
| ECOR collection | E. coli isolates of different origins | 45 |
| UPEC collection | UPEC clinical isolates | 73 |
| 1094 | Commensal E. coli isolate from a healthy patient | C. Le Bouguenec |
| 1102 | Commensal E. coli isolate from a healthy patient | C. Le Bouguenec |
| 1103 | Commensal E. coli isolate from a healthy patient | C. Le Bouguenec |
| 1110 | Commensal E. coli isolate from a healthy patient | C. Le Bouguenec |
| 1125 | Commensal E. coli isolate from a healthy patient | C. Le Bouguenec |
| 1127 | Commensal E. coli isolate from a healthy patient | C. Le Bouguenec |
| 536 | UPEC isolate (O6:K15:H31) | 8 |
| J96 | UPEC isolate (O4:K6) | 22 |
| IHE3034 | E. coli isolate (O18:K1:H7/9) | 32 |
| 55989 | Enteroaggregative E. coli | C. Le Bouguenec |
| O42 | Enteroaggregative E. coli | 75 |
| Plasmids | ||
| pJunβ | Plasmid with a hybrid outer membrane protein containing the β-AT domain of the IgA protease of N. gonorrhoeae and the leucine zipper of c-Jun, Cmr | 74 |
| pUpaGL2-L1-β | Plasmid with a hybrid protein containing the leucine zipper of c-Jun and the L2 and L1 linker and β domains of UpaG of E. coli CFT073, Cmr | This study |
| pUpaGL1-β | Plasmid with a hybrid protein containing the leucine zipper of c-Jun and the L1 linker and β domains of UpaG of E. coli CFT073, Cmr | This study |
| pUpaGβ | Plasmid with a hybrid protein containing the leucine zipper of c-Jun and the β domain of UpaG of E. coli CFT073, Cmr | This study |
| pupaG | upaG gene from CFT073 in pBAD/Myc-HisA-Kan, Kmr | This study |
| pET22b-F2 | pET22b with 281 amino acids of the N terminus of UpaG (amino acids 943 to 1224), Ampr | This study |
DNA manipulations and genetic techniques.
DNA techniques were performed as described by Sambrook et al. (58). Isolation of plasmid DNA was carried out using the QIAprep Spin Miniprep kit (Qiagen). Restriction endonucleases were used according to the manufacturer's specifications (New England Biolabs). Oligonucleotides were purchased from Sigma (France or Australia) and are listed in Table S1 in the supplemental material. DNA sequencing was performed by MWG Services. The construction of mutants and upaG-controlled-expression strains (PcL and RExBAD) (15, 57) was performed using a three-step PCR procedure described previously (36; http://www.pasteur.fr/recherche/unites/Ggb/matmet.html).
Construction of plasmids used in this study.
The upaG gene was amplified from CFT073 by PCR using primers 110 and 139 and ligated into the XhoI site of pBAD/Myc-HisA to generate plasmid pupaG. Plasmids pUpaGL2-L1-β, pUpaGL1-β, and pUpaGβ were constructed as follows. Plasmid pJunβ was digested by BamHI to remove the region encoding the β-AT domain of the immunoglobulin A (IgA) protease from Neisseria gonorrhoeae. The region encoding the translocator unit of UpaG (including the L2, L1, and β domains) was amplified using primer BamH-4424-3 in combination with primer BamHTag2.β-5, BamHTag2.L1-5, or BamHTag2.L2-5, respectively. These primers included a sequence encoding the DPLEPI E-tag epitope. Amplified fragments were cloned into pJun to generate the plasmids pUpaGL2-L1-β, pUpaGL1-β, and pUpaGβ. All plasmid constructs were verified by sequence analysis and were used to transform E. coli UT5600 for functional expression analysis.
Structural analysis and 3D molecular modeling.
The SignalP 3.0 program was used to predict the signal peptide of UpaG (7), and the SMART program was used to detect protein domains in UpaG (61). A three-dimensional (3D) model of UpaG was constructed using the Protein Data Bank files for one outer membrane translocator domain of the Haemophilus influenzae Hia trimeric AT protein (Hia shares 20% identity and 30% similarity with UpaG; these percentages increased to 37% identity and 55% similarity when the last 107 amino acids were compared): 2GR7.pdb (Hia amino acids 992 to 1098) and 2GR8.pdb (Hia amino acids 1022 to 1098). The structural alignment of the two polypeptide chains was obtained using the combinatorial extension algorithm (63), which determines an optimal alignment of multiple protein structures based on a Monte Carlo optimization method. Multiple-sequence alignment of these proteins with UpaG was performed with Clustal W. The homology model of UpaG was then generated with the program MODELLER (40). Finally, the SCWRL 3.0 program (11) and energy minimizations were used for protein side chain conformational predictions. The model generated was subjected to a series of tests to assess its internal consistency and reliability. Evaluations were performed using energy criteria and root mean square deviation, Ramachandran plot (inspection of ψ/φ angles), and Procheck programs. The Ramachandran plot statistics were as follows: for residues in most-favored regions [A,B,L], 94%; for residues in additional allowed regions [a,b,l,p], 6%; for residues in generously allowed regions [∼a,∼b,∼l,∼p], 0%; and for residues in disallowed regions, 0%. The root mean square deviation calculated using backbone atoms (N, Ca, C′, O) for the Gln9-Gln105 region was 0.83 Å.
Cell aggregation assay.
Liquid cultures of E. coli CFT073, CFT073 ΔupaG, CFT073 PcL upaG, CFT073 RExBAD upaG (supplemented with 0.2% arabinose), and CFT073 RExBAD upaG (supplemented with 0.2% glucose) were grown separately overnight in M63B1. Cultures were adjusted to an optical density at 600 nm (OD600) of 2.5 (in 3.0 ml) by dilution with M63B1 medium and left to stand at room temperature. At regular time intervals, the OD600 of the upper part of the culture was measured. Cultures of E. coli strains bearing plasmids pUpaGL2-L1-β, pUpaGL1-β, and pUpaGβ were grown separately in lysogeny broth (LB) to an OD600 of 0.5 and then induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C under vigorous shaking conditions. Following this, 100-ml samples were withdrawn at 20-min intervals from the upper part of the culture and the OD600 was measured. All assays were performed in triplicate.
Biofilm assays.
Biofilm formation on polystyrene surfaces was measured using 96-well microtiter plates (Iwaki) as previously described (57). Briefly, cells were grown for 24 h in M63B1glu (containing 0.2% arabinose for induction of upaG gene expression) at 37°C, washed to remove unbound cells, and stained with 0.1% crystal violet. Quantification of bound cells was performed by the addition of acetone-ethanol (20:80 [vol/vol]) and measurement of the dissolved crystal violet at an absorbance of 570 nm. All experiments were performed in triplicate.
Generation of anti-UpaG antibodies.
A fragment of the upaG gene (encoding 281 amino acids of the passenger domain of UpaG) was amplified using the primers c4424.943-5 and c4424.1224-3 and cloned into plasmid pET22b to generate pET22b-F2. The resulting construction was verified by sequencing and introduced by electroporation into E. coli BL21(DE3). A total of 500 ml of LB was inoculated with 2 ml of an overnight culture of BL21(pET22b-F2). The cells were grown to an OD600 value of 0.4 and induced with 1 mM IPTG for 4 h. The His-tagged UpaG943-1224 protein (a region of UpaG comprising amino acids 943 to 1224 of the N-terminal passenger domain) was purified by affinity chromatography on dry-silica-based resin precharged with Ni2+ ions (Protino Ni-TED resin; Macherey-Nagel). The purity was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Rabbit polyclonal antibodies were conventionally prepared using purified His-tagged UpaG943-1224. Prior to immunodetection, the serum was absorbed against a crude protein extract of CFT073 ΔupaG.
Protein localization and Western blot analysis.
Outer membrane extractions were performed as previously described (16). Briefly, CFT073, CFT073 ΔupaG, CFT073 PcL upaG, and CFT073 RExBAD upaG (with or without arabinose) cells were harvested by centrifugation, washed in phosphate-buffered saline (PBS), and resuspended to an OD600 of 20. The cell suspension was centrifuged, and the pellet was snap-frozen on dry ice. Cells were then resuspended in sonication buffer (10% sucrose, 50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 5 mM dithiothreitol, 1 mg ml−1 lysozyme) and incubated at 37°C for 10 min and then on ice for 20 min. Complete cell lysis was achieved by sonication, and cellular debris were removed by centrifugation (10,000 rpm for 5 min). The supernatant was then incubated with 0.5% Sarkosyl for 30 min at room temperature on a rotating platform. The insoluble outer membrane fraction was harvested by centrifugation at 18,000 rpm for 45 min at 4°C, resuspended in 100 ml of 1× SDS final sample buffer, and boiled for 3 min prior to gel electrophoresis. A volume of cells corresponding to an OD600 value of 2.0 was examined by electrophoresis on an 8% SDS-polyacrylamide gel containing 8 M urea. Immunodetection was performed following transfer onto nitrocellulose membranes (Protran; Schleicher & Schuell), using a 1:1,000 dilution of polyclonal rabbit antiserum raised against the UpaG protein.
To monitor the presence of hybrid proteins, a volume of UT5600 cells (containing plasmid pUpaGL2-L1-β, pUpaGL1-β, or pUpaGβ) corresponding to an OD600 value of 2.0 was harvested by centrifugation following induction with 1 mM IPTG. Pellets were resuspended and boiled for 10 min prior to gel electrophoresis, and in each case, an amount equivalent to an OD600 value of 2.0 was loaded onto the gel. Proteins were transferred onto nitrocellulose membranes (Protran; Schleicher & Schuell), and immunodetection was performed using a 1:10,000 dilution of anti-E-tag monoclonal antibody conjugated with peroxidase (Amersham Pharmacia).
To determine the levels of fibronectin in T24 and HeLa cells, the cells were suspended in 250 μl of the lysis buffer (20 mM Tris-HCl, 1% [vol/vol] NP-40, 137 mM NaCl, 5 mM Na3VO4, 30 mM NaF, protease inhibitor cocktail). Cells were mixed for 15 min at 4°C and pelleted by centrifuged for 15 min. Proteins contained in the supernatants were examined by SDS-PAGE. Proteins were transferred onto nitrocellulose membranes (Protran; Schleicher & Schuell), and immunodetection was performed using a 1:2,000 dilution of antifibronectin antiserum conjugated with peroxidase (Sigma).
Immunofluorescence microscopy.
Immunofluorescence microscopy analysis was performed as follows. Overnight cultures of the different strains were grown at 37°C in M63B1glu (in the presence of 0.2% arabinose, where indicated below). Cells were loaded onto 0.1%-poly-l-lysine-treated immunofluorescence microscope slides. Slides were washed three times with PBS. Cells were fixed with 3% paraformaldehyde for 10 min before being quenched with 50 mM NH4Cl. Slides were saturated for 15 min with 0.5% bovine serum albumin (BSA) before incubation with a 1:1,000 dilution of the primary polyclonal rabbit antiserum raised against UpaG and next with a 1:300 dilution of the secondary polyclonal goat anti-rabbit serum coupled to Alexa 488 (Molecular Probes-Invitrogen) along with 10 mg ml−1 of 4′,6-diamidino-2-phenylindole (DAPI). Finally, the slides were mounted in Mowiol 4088 (Calbiochem) and observed by epifluorescence microscopy with green fluorescent protein and DAPI filters.
Immunogold electron microscopy.
Cells for immunoelectron microscopy were prepared from overnight cultures of wild-type CFT073, CFT073 ΔupaG, and CFT073 PcL upaG strains grown at 37°C in M63B1glu. Cells were fixed with freshly prepared 8% paraformaldehyde at 4°C, embedded in 10% gelatin, and cryoprotected in 15% polyvinyl pyrrolidone-1.7% sucrose. Blocks were mounted on aluminum pins and frozen in liquid nitrogen before being cryosectioned with a cryoultramicrotome (model UC6 FCS; Leica). Thin cryosections (70 nm) were placed on glow-discharged carbon-coated copper grids, washed twice with PBS, and quenched with 50 mM NH4Cl before being blocked with 0.5% BSA for 15 min. Samples were reacted with the primary polyclonal rabbit antiserum raised against UpaG (1:100 dilution in PBS containing 0.5% BSA) for 30 min and washed four times in PBS (5 min). Samples were then incubated with anti-rabbit IgG gold conjugate (10-nm diameter, diluted 1:50 in PBS containing 0.5% BSA) for 30 min and subsequently washed four times (5 min) in PBS followed by four washes (2 min each) in sterile ultrapure water. Cryosections were embedded in 2% methyl cellulose and stained with 4% uranyl acetate before being examined under a Jeol JEM1010 transmission electron microscope operated at 80 kV. Images were captured using an analySIS Megaview III digital camera.
Epithelial cell adhesion assay.
The interaction of CFT073, CFT073 ΔupaG, and CFT073 PcL upaG with cultured T24, HeLa, and BSC1 epithelial cells was studied essentially as previously described (39). Briefly, wells were seeded with 1.5 ×105 cells in six-well tissue culture plates. Once cells were confluent (1.2 ×106 cells per well), the culture medium was removed and cells were washed once with DMEM plus 10% fetal bovine serum. The use of confluent cells resulted in a reduction in the amount of plastic surface available for bacterial binding. Overnight bacterial cultures were mixed vigorously by vortexing them in order to disperse any bacterial cell clumps. Bacteria were added to the monolayers at a multiplicity of infection of 10 in DMEM. Incubation was carried out for 1 h at 37°C in 5% CO2 (longer incubation times resulted in killing of the eukaryotic cells by CFT073). The inoculating dose of bacteria was confirmed by serial dilution and plating. The absence of bacterial clumps attached to the eukaryotic cells was verified with phase-contrast microscopy. Monolayers were washed three times with DMEM plus 10% fetal bovine serum to remove nonadherent bacteria. The remaining bacteria were released by eukaryotic cell lysis with 0.1% Triton X-100. The number of adherent bacteria was determined by serial dilution and plating. All experiments were performed in triplicate.
Mixed-monolayer cell adhesion assay.
T24-red cells expressing the red fluorescent protein and HeLa cells were mixed in different proportions and seeded on six-well tissue culture plates to obtain a mixed cell culture monolayer. Once cells were confluent, the proportion of HeLa and T24-red cells was checked by fluorescence microscopy. Wells with approximately the same proportion of HeLa and T24-red cells were infected with CFT073 ΔupaG or CFT073 PcL upaG at a multiplicity of infection of 50. Bacterial cultures were dispersed by vigorous vortexing prior to infection. After 1 h of incubation at 37°C in 5% CO2, the mixed cell monolayers were washed with PBS in order to eliminate nonadherent bacteria. Cells were fixed with a paraformaldehyde solution (3.5% in PBS) for 15 to 30 min and then permeabilized with 0.1% Triton X-100 for 5 min in PBS. Actin was labeled by phalloidin Alexa 647 incubated for 30 min at room temperature in blocking solution, and this permitted the identification of both types of eukaryotic cells. Images were acquired on an inverted fluorescence microscope (Axiovert 135; Carl Zeiss MicroImaging, Inc.) equipped with a cooled, charge-coupled-device camera (MicroMax, 5 MHz; Princeton Instruments) driven by Metamorph Imaging System software (Universal Imaging Corp.). Bacteria were observed by phase-contrast microscopy and were colored green using the Adobe Photoshop CS program.
Binding assays (enzyme-linked immunosorbent assays [ELISA]).
Microtiter plates (Maxisorb; Nunc) were coated overnight at 4°C with 10 mg of the following ECM proteins ml−1: heparan (from bovine kidney; Sigma), fibronectin (from human plasma; Sigma), laminin (from human placenta; Sigma), and human collagen type I, type III, and type IV (Sigma). BSA (Sigma) was used as a negative control. Wells were washed twice with TBS (150 mM NaCl, 20 mM Tris, pH 7.5) and then blocked with TBS-2% milk for 1 h. After being washed with TBS, 200 μl of CFT073, CFT073 ΔupaG, CFT073 PcL upaG, CFT073 RExBAD upaG (with arabinose), or CFT073 RExBAD upaG (without arabinose) (OD600 = 0.1) was added, and the mixtures were incubated for 2 h. After being washed to remove nonadherent bacteria, adherent cells were fixed with 4% paraformaldehyde, washed a further three times, and incubated for 1 h with anti-E. coli serum (diluted 1:500 in TBS-Tween 0.5%) and then for 1 h with a secondary anti-rabbit horseradish peroxidase antibody (diluted 1:1,000). Finally, cells were washed and adherent bacteria were detected by adding 150 μl of 1-Strep ABTS [2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid diammonium salt); Pierce] and measurement of absorbance at 405 nm.
Mouse model of UTI.
The mouse model of urinary tract infections (UTI) described previously was used for this study (56). Female C57BL/6 and C3H/HeJ mice (8 to 10 weeks old) were purchased from the Animal Resources Center, Western Australia, and housed in sterile cages with ad libitum access to sterile water. Urine was collected from each mouse 24 h prior to challenge and examined using a microscope with a hemocytometer and by culture. Mice with a combination of >5 × 102 CFU of bacteria per ml of urine and >2 × 105 white blood cells in urine were defined as having a preexisting condition and were excluded from the study. Mice were anesthetized by brief inhalation exposure to isoflurane, and the periurethral area was sterilized by swabbing it with 10% povidone-iodine solution, which was removed with sterile PBS. Mice were catheterized using a sterile Teflon catheter (0.28-mm internal diameter, 0.61-mm outer diameter, and 25-mm length; Terumo) by inserting the device directly into the bladder through the urethra. An inoculum of 25 μl, containing 5 × 108 CFU of bacteria in PBS containing 0.1% India ink was instilled directly into the bladder using a 1-ml tuberculin syringe attached to the catheter. The catheter was removed immediately after the challenge, and mice were returned to their cages. Urine was collected from each mouse at 18 h after inoculation for quantitative colony counts. Groups of mice were euthanized at 18 h and 5 days after challenge by cervical dislocation; bladders were then excised aseptically, weighed, and homogenized in PBS. Bladder homogenates were serially diluted in PBS and plated onto LB agar for colony counts. Data are expressed as the mean total numbers of CFU per 0.1 g of bladder tissue ± standard errors of the means. All animal experiments were repeated at least twice with a minimum group size of eight, and the data shown are the composite of all independent experiments.
RESULTS
Structural analysis of the putative trimeric AT UpaG.
The 5,337-bp upaG gene from E. coli CFT073 encodes a protein that shares many structural features with the trimeric YadA and NhhA AT proteins. UpaG possesses a characteristically long putative signal sequence with a predicted cleavage site after amino acid 53 (Fig. 1A), as well as a large N-terminal passenger domain (amino acids 54 to 1685) with 14 repeats of both Hep-Hag and Him motifs often found in bacterial invasins and hemagglutinins (Fig. 1A). Furthermore, the 89-C-terminal-amino-acid sequence of UpaG contains a putative translocator domain homologous to a C-terminal region of YadA and other trimeric AT adhesins (Fig. 1B).
FIG. 1.
In silico analysis of the UpaG protein. (A) Schematic illustration of the domain organization of UpaG from E. coli CFT073, YadA from Yersinia enterocolitica, and NhhA from Neisseria meningitidis. Indicated are the signal peptide (S.P.) and the localizations of the Hep-Hag and Him domains (invasin and hemagglutinin domains). Alignments were generated using Clustal W. (B) Multiple-sequence alignment of the translocation units from UpaG, YadA, and NhhA. Identical residues are indicated by shaded boxes, whereas conservative substitutions are indicated by unshaded boxes. The L2, L1, and β subdomains of UpaG are aligned with the corresponding regions from YadA and NhhA. (C) Computer model of the putative 3D structure of UpaG. (a) Monomeric structure; (b) trimeric structure; (c) upper view. The different subregions are in green (β), red (L1), and yellow (L2).
The amino acid sequence of the putative translocator unit of UpaG contains three subdomains; L2 (amino acids 1690 to 1707), L1 (amino acids 1708 to 1724), and β (amino acids 1725 to 1778), all of which are highly conserved in members of the trimeric AT adhesin family (37). Computer modeling of the putative 3D structure of the last 107 amino acids of UpaG generated and refined by energy minimization predicts an N-terminal α-helix of 37 amino acids (9-45) followed by four antiparallel β-sheets of 8, 9, 9, and 7 amino acids connected by short turns (Fig. 1C). By analogy with other members of the trimeric AT family, the trimerization of this region may produce a 12-strand β-barrel that may form in the outer membrane and facilitate the exposure of the N-terminal part of the protein at the cell surface (Fig. 1C). The strong amino acid sequence similarity of UpaG to other trimeric AT proteins, in combination with these structural predictions, is compatible with a model predicting a trimeric organization of the protein.
UpaG is located at the cell surface.
Our structural predictions of the C-terminal part of UpaG suggested that UpaG might localize to the outer membrane. To test this tenet, we raised a polyclonal rabbit antiserum against a region of UpaG comprising amino acids 943 to 1224 of the N-terminal passenger domain. Although we were able to detect very weak transcription of upaG in CFT073 by quantitative reverse transcriptase real-time PCR (see Fig. S1 in the supplemental material), immunodetection employing anti-UpaG antibodies failed to detect the UpaG protein in CFT073 outer membrane preparations (Fig. 2A). To circumvent this expression problem, we constructed a unique set of CFT073 UpaG overexpression strains by inserting a chromosomally located, arabinose-inducible (pBAD) or constitutive (PcL) promoter upstream of upaG to generate the strain CFT073 RExBAD upaG or CFT073 PcL upaG, respectively (15, 57). A band of the expected apparent molecular mass (178 kDa) that reacted specifically with antibodies directed against the N-terminal region of UpaG could be detected in outer membrane preparations of the CFT073 RExBAD upaG cells in the presence of arabinose and from CFT073 PcL upaG cells. In addition, bands with a higher molecular mass that may represent UpaG multimers (including trimers) were also observed (Fig. 2A). To demonstrate the surface localization of UpaG, we performed immunofluorescence microscopy (Fig. 2B). UpaG antiserum readily reacted with intact cells expressing upaG, confirming that the N-terminal region of UpaG was effectively translocated to the cell surface. No reaction was seen with CFT073 and CFT073 ΔupaG cells. The surface location of UpaG was confirmed using immunogold labeling and electron microscopy employing cell sections prepared from CFT073 PcL upaG (which constitutively produces UpaG). In contrast, no evidence of UpaG production was observed on the surfaces of CFT073 (Fig. 2C) or CFT073 ΔupaG (data not shown) cells. Examination of the immunogold micrographs indicated that UpaG could form structures extending approximately 100 nm from the outer membrane (Fig. 2C); this is in agreement with our estimates of the size of the exposed domain.
FIG. 2.
Surface localization of the UpaG passenger domain. (A) Western blot analysis of UpaG performed using outer membrane fractions from CFT073, CFT073 ΔupaG, CFT073 PcL upaG, and CFT073 RExBAD upaG in the presence (+) or absence (−) of 0.2% arabinose. Outer membrane fractions were resolved by SDS-PAGE in the presence of 8 M urea. The monomeric form of UpaG is indicated (m), as are possible dimers (d) and trimers (t). (B) Immunofluorescence assays of UpaG from CFT073, CFT073 ΔupaG, CFT073 PcL upaG, and CFT073 RExBAD upaG in the presence (+) or absence (−) of 0.2% arabinose. Overnight cultures were fixed and incubated with anti-UpaG serum, followed by incubation with a secondary polyclonal goat anti-rabbit serum coupled to Alexa 488 and DAPI. (C) Immunogold electron microscopy of cell sections of CFT073 and CFT073 PcL upaG probed with anti-UpaG and then with gold-labeled anti-rabbit IgG. Anti-UpaG-labeled gold particles were observed on the surface of CFT073 PcL upaG (but not CFT073) up to a distance of approximately 100 nm (arrows). One CFT073 cell and two individual CFT073 PcL upaG cells are shown as representatives of the average labeling observed for cells in several fields of view.
The C-terminal L1-β region of UpaG is a functional transporter domain.
Trimeric AT proteins are characterized by their short C-terminal translocator domain. To verify that the C-terminal region of UpaG possessed translocation activity, we assessed whether it could export a heterologous passenger domain to the cell surface. We constructed a hybrid protein combining the L2, L1, and β domains of the C-terminal region of UpaG and, in place of the UpaG passenger domain, the leucine zipper domain of the c-Jun protein (Fig. 3A). c-Jun is a eukaryotic transcriptional factor that has the ability to dimerize through strong interactions imparted by its leucine zipper fold (1, 2). It has been shown that the export of this c-Jun leucine zipper domain to bacterial cell surfaces can promote, by virtue of its dimerization properties, cell-cell interactions and thus bacterial aggregation (74). Therefore, we hypothesized that the production and export of the heterologous passenger domain of the hybrid protein c-Jun-UpaGL2-L1-β on the surfaces of E. coli cells should confer novel autoaggregation traits via c-Jun leucine zipper dimerization. The hybrid protein was cloned into the pJunβ plasmid, in which the c-Jun leucine zipper is fused to the PelB signal sequence and placed under the control of the lac promoter (74) to generate the plasmid pUpaGL2-L1-β. The production of this hybrid protein was demonstrated using an antibody directed against an E-tag epitope located between the c-Jun leucine zipper and the UpaG C-terminal potential translocator unit (Fig. 3B). To test whether the translocator unit of UpaG could present the leucine zipper sequence of c-Jun to the surfaces of upaG-negative E. coli UT5600 cells, we monitored cell-to-cell adhesion in an aggregation assay. In contrast to noninduced cells which remained in suspension, E. coli cells induced for the expression of UpaGL2-L1-β aggregated strongly and sedimented rapidly to the bottom of the tube (Fig. 3C and D). These results suggest that the c-Jun leucine zipper domain was, when fused to the UpaGL2-L1-β domain, effectively transported to the bacterial cell surface and could facilitate cell-to-cell aggregation (74).
FIG. 3.
Surface localization of the c-Jun leucine zipper in a c-Jun-UpaG chimeric protein. (A) Schematic illustration of recombinant c-Jun-UpaG proteins constructed in this study. The position of the PelB signal sequence (SS), the E-tag, the leucine zipper of c-Jun (noted as Jun), and the L2, L1, and β subdomains are indicated, as is the lac promoter (plac). (B) Immunoblots of whole-cell protein extracts from induced cultures expressing the different chimeric constructions. The blot was probed with anti-E-tag MAb. The UpaGL2-L1-β protein migrated as a polypeptide of 23 kDa despite its calculated mass of 19.8 kDa. The molecular masses of UpaGβ (14.6 kDa) and UpaGL1-β (16.3 kDa) were as calculated. (C) Cell-cell aggregation driven by the interaction of the surface-exposed leucine zipper dimerization domain in the different constructions in the presence (black) or absence (gray) of the inducer IPTG. E. coli UT5600 was used as the control. IPTG-induced and uninduced cultures were left to stand without being shaken. Samples of 100 μl were taken from the top of the cultures (1 cm from the surface) at regular time intervals, and the OD600 was measured. The degree of aggregation is inversely proportional to the turbidity. The OD600 just after the induction was taken as 100%. Data represent the means from three independent experiments. Standard deviations are indicated by error bars. (D) Representative photographs of induced (+) and uninduced (−) E. coli cultures taken after 120 min without shaking.
In order to identify the minimal C-terminal part of UpaG able to efficiently export the heterologous passenger leucine zipper of c-Jun, we also constructed hybrid proteins containing the β domain of UpaG lacking the L2 subdomain or lacking both the L2 and L1 subdomains (Fig. 1A). The hybrid proteins were cloned into the pJunβ plasmid to generate the plasmids pUpaGL1-β and pUpaGβ, respectively (Fig. 3A). Immunodetection using anti E-tag antibodies performed on whole extracts of IPTG-induced E. coli cells transformed with the plasmids pUpaGL2-L1-β, pUpaGL1-β, and pUpaGβ allowed the detection of the chimeric E-tag/c-Jun/UpaG proteins. All of the hybrid proteins were produced in equivalent amounts and migrated as monomeric polypeptides under denaturing conditions (Fig. 3B). To compare the functional activities of these hybrid proteins, we examined their ability to mediate bacterial cell aggregation. Cells expressing pUpaGL2-L1-β aggregated more strongly and sedimented more rapidly to the bottom of the tube than cells expressing UpaGL1-β. Whereas the L2 domain was not fully required for transport of the passenger domain, its presence appeared necessary for optimal transport, indicating that it may contribute to the correct folding and activity of the C-terminal transporter domain of UpaG. No significant aggregation was observed from cells expressing UpaGβ, suggesting that the L1 subdomain was absolutely required for efficient export of the N-terminal passenger domain. Taken together, these results suggest that the UpaG C-terminal region is a functional translocator domain and that the UpaGL1-β domain is necessary and sufficient to direct the translocation of the hybrid passenger domain. These results, combined with our immunoelectron microscopy experiments, confirm that UpaG is located in the outer membrane.
Expression of UpaG leads to increased aggregation and biofilm formation.
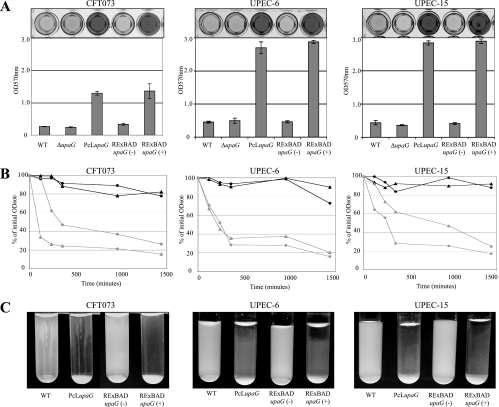
To investigate the functional role of UpaG at the bacterial cell surface, we tested whether the expression of upaG could enhance biofilm formation by CFT073. As shown in Fig. 4A, the expression of upaG in CFT073 RExBAD upaG (following arabinose induction) and CFT073 PcL upaG led to a significantly increased capacity to form a biofilm. In contrast, the CFT073 ΔupaG mutant strain did not display any difference in biofilm growth from wild-type CFT073. To eliminate the possibility that this observation was limited to CFT073, we expressed upaG in two other UPEC strains (U6 and U15) from our laboratory collection using the same constitutive and inducible promoter system. As observed for the CFT073 RExBAD upaG and CFT073 PcL upaG strains, the expression of upaG dramatically increased biofilm formation by both strains (Fig. 4A). In addition, the upaG gene was PCR amplified from CFT073 and cloned as a transcriptional fusion behind the tightly regulated araBAD promoter in the pBAD/Myc-HisA expression vector. The resulting plasmid (pupaG) was used to transform the upaG-negative E. coli K-12 strain OS56 (62). Under inducing conditions, OS56(pupaG) produced a strong biofilm (data not shown).
FIG. 4.
Biofilm formation and aggregation phenotypes. (A) Static biofilm formation in polystyrene microtiter plates by CFT073, UPEC-6, and UPEC-15 (and their upaG derivatives) in the presence (+) or absence (−) of 0.2% arabinose, where indicated. Biofilm growth was quantified by the solubilization of crystal violet-stained cells with ethanol-acetone and the determination of the absorbance at 570 nm. WT, wild type. (B) Autoaggregation assay demonstrating the settling profiles from liquid suspensions of CFT073/UPEC-6/UPEC-15 (black circles), CFT073/UPEC-6/UPEC-15 PcL upaG derivatives (gray circles), and CFT073/UPEC-6/UPEC-15 RExBAD upaG derivatives in the presence (gray triangles) or absence (black triangles) of 0.2% arabinose. Cells were diluted to an OD600 of 2.5 in a 3.0-ml volume. One-hundred-microliter samples were taken from the top of the cultures (1 cm from the surface) at regular time intervals, and the OD600 was measured. (C) Photographs of E. coli cultures taken after 24 h without shaking.
The enhanced biofilm growth mediated by UpaG either could be caused by direct adhesion or may occur as a result of intercellular reactions that promote cell aggregation. To explore these possibilities, we examined the primary adherence capacity of cells overexpressing UpaG to a glass surface within a short (2-h) time span. The expression of upaG did not result in an increase in the number of isolated adherent bacteria, suggesting that UpaG production does not promote initial cell-to-surface adhesion (data not shown). However, examination of the glass surface by phase-contrast microscopy revealed the presence of large cell aggregates indicative of bacterial clumping (data not shown). To further demonstrate the role of UpaG in promoting cell-cell aggregation, we performed an assay to examine the settling kinetics of CFT073, U6, and U15 in association with their respective derivative RExBAD upaG and PcL upaG strains. As shown in Fig. 4B and C, while neither CFT073, U6, nor U15 formed aggregates, the expression of UpaG in all of these E. coli backgrounds resulted in strong cell aggregation. Taken together, these results demonstrate that the production of UpaG induces biofilm formation primarily through the promotion of cell-cell aggregation.
UpaG also plays a role in bacterial adherence to epithelial cells.
The strong similarity between UpaG, NhhA, and YadA (Fig. 1) suggested that UpaG might also play a role in adhesion to epithelial cells. To test this, T24 bladder epithelial cell monolayers were infected with CFT073, CFT073 PcL upaG, or CFT073 ΔupaG. The enumeration of cell-adherent bacteria revealed that the CFT073 PcL upaG strain adhered to T24 cells 1.5-fold more efficiently than CFT073 or CFT073 ΔupaG (P = 0.005) (Fig. 5A). In contrast, CFT073, CFT073 PcL upaG, and CFT073 ΔupaG did not display significant differences in adherence to HeLa cervical epithelial cells or BSC1 kidney epithelial cells (P was 0.265 or 0.0796, respectively) (Fig. 5A). This suggested that UpaG may exhibit a specific affinity for T24 cells. Consistent with this hypothesis, adherence assays employing a coculture of mixed T24 cells expressing the red fluorescence protein and HeLa cells demonstrated that the majority of CFT073 PcL upaG cells bound to T24 cells (red cells) (Fig. 5B). This specificity for T24 cells was not observed for CFT073 ΔupaG. Taken together, these results indicate that the production of UpaG by CFT073 specifically enhances its capacity to adhere to T24 bladder epithelial cells.
FIG. 5.
Adhesive properties of UpaG. (A) Adherence of CFT073, CFT073 ΔupaG, and CFT073 PcL upaG strains to T24, HeLa, and BSC1 epithelial cells. Values correspond to the means ± standard deviations from three independent experiments (*, denotes a P of <0.01). WT, wild type. (B) Adherence of CFT073 ΔupaG and CFT073 PcL upaG cells to a mixed monolayer of T24 cells expressing the red fluorescence protein (red cells) and HeLa cells. Actin was labeled by phalloidin Alexa 647 (in blue), revealing all eukaryotic cells. Bacteria were observed using phase-contrast microscopy and colored green using Adobe Illustrator CS software. CFT073 PcL upaG adhered in significantly greater numbers to T24 bladder epithelial cells than to HeLa epithelial cells (left panel). In contrast, CFT073 ΔupaG showed no difference in adherence to T24 bladder epithelial cells and HeLa epithelial cells (right panel).
UpaG binds to the ECM proteins laminin and fibronectin.
A property of many trimeric AT proteins is their ability to mediate adherence to a variety of substrates, including ECM components. The ability of UpaG to mediate binding to selected human ECM components was examined by in vitro binding assays. CFT073, CFT073 ΔupaG, CFT073 PcL upaG, and CFT073 RExBAD upaG cells were added to wells of 96-well plates coated with purified plasma laminin, heparan, fibronectin, and collagens I, III, and IV. BSA was used as negative control in the assay. Cell adherence to ECM proteins was detected with a polyclonal serum raised against E. coli and quantified by ELISA. As shown in Fig. 6, CFT073 PcL upaG and arabinose-induced CFT073 RExBAD upaG cells adhered strongly to fibronectin and laminin. This binding was not type 1 fimbria dependent, as the same adhesion profile was observed in the presence of 0.1 M α-d-mannose (data not shown). No adherence to these ECM proteins was observed for CFT073 or CFT073 ΔupaG. These results support our previous findings indicating the cell surface location of UpaG and demonstrate that UpaG can promote significant adherence of E. coli to fibronectin and laminin (P < 0.001). Interestingly, immunodetection experiments using antifibronectin antibodies showed that T24 cells contained at least a 10-fold-higher quantity of fibronectin than HeLa cells (Fig. S2). The capacity of UpaG to bind to fibronectin could therefore explain the preferential adhesion of CFT073 PcL upaG to T24 bladder epithelial cells.
FIG. 6.
Adherence to ECM proteins. Adherence of CFT073, CFT073 ΔupaG, CFT073 PcL upaG, and CFT073 RExBAD upaG in the presence of 0.2% of arabinose (+) or glucose (−) to heparan, laminin, fibronectin, and collagens I, II, and IV. Cells that adhered to ECM proteins were detected with a polyclonal serum raised against E. coli and quantified by ELISA. Data are means (± standard deviations) from two independent experiments. WT, wild type.
Prevalence of upaG in UPEC.
The widespread occurrence of trimeric AT adhesins in gram-negative pathogens prompted us to investigate the prevalence of upaG in a well-characterized collection of 191 UPEC and commensal E. coli isolates of human and animal origins (45). For this purpose, we employed primers designed to amplify the region encompassing nucleotides 943 to 1224 of upaG. We found that a base pair product of the correct size was amplified from 40 of the 191 strains tested (21%). Further analysis revealed that 93% (37 of these 40 upaG-positive strains) of the PCR-positive strains belonged to the E. coli B2 and D phylogenetic groups, both of which are experimentally and epidemiologically associated with extraintestinal infections (9, 10, 48), and that 7% (3 of the 40 upaG-positive strains) belonged to the A or B1 phylogenetic group. In total, the percentage of B2 and D strains positive for upaG was 35%, compared to less than 4% for the remaining phylogenetic groups. These results are in good agreement with a recent report that examined the prevalence of upaG in a small selection of ExPEC strains (17) and suggest that upaG is frequently associated with ExPEC.
Evaluation of the role of UpaG in the colonization of the mouse urinary tract.
To study the role of UpaG in virulence, we examined the ability of CFT073, CFT073 ΔupaG, and CFT073 PcL upaG to survive in the mouse urinary tract. There was no significant difference in the abilities of the three strains to colonize the C57BL/6 mouse bladder on day 1 (short term) or day 5 (longer term) following infection (Fig. 7A). No colonization of the kidneys was observed for any of the strains; this is consistent with previous data from our laboratory using C57BL/6 mice (71). To address the effect of upaG deletion or constitutive expression on kidney colonization, we repeated the same experiment using C3H/HeJ mice. C3H/HeJ mice are defective in the Toll-like receptor 4 (TLR-4) protein, do not respond to lipopolysaccharide, and are highly susceptible to E. coli-induced pyelonephritis (44). At day 1 and day 5 following the infection of C3H/HeJ mice, there was no significant difference in the abilities of the three strains to colonize the bladder or kidneys (Fig. 7B).
FIG. 7.
Role of UpaG in virulence. (A) Persistence of CFT073, CFT073 ΔupaG, and CFT073 PcL upaG in the bladders of C57BL/6 mice following intraurethral challenge. At days 1 and 5 following infection, all three strains were recovered in equivalent numbers. (B) Persistence of CFT073, CFT073 ΔupaG, and CFT073 PcL upaG in the bladders and kidneys of C3H/HeJ mice following intraurethral challenge. At day 1 and day 5 following infection, there was no significant difference in the abilities of the three strains to colonize the bladder or kidneys.
DISCUSSION
AT proteins represent one of the major families of secreted proteins in gram-negative bacteria. Recently, a specialized class of AT adhesins that adopt a trimeric protein conformation and remain anchored to the cell surface were described (12, 43, 47, 77). In this study, we characterized UpaG, a new functional member of the trimeric AT proteins. UpaG is the first trimeric AT protein to be characterized in detail from E. coli, and its prevalence is strongly associated with E. coli strains from the B2 or D clonal group.
One of the most conserved features of the trimeric AT family is the translocator unit. Studies of the YadA and Hia adhesins have revealed that the 70 and 76 C-terminal residues of these proteins, respectively, are sufficient for the translocation of a heterologous passenger domain to the bacterial surface (54). In the case of the NhhA trimeric AT protein from N. meningitidis, the 72 C-terminal residues are sufficient for trimerization and localization of the N-terminal protein domain to the bacterial surface (59). Here, we examined the region of UpaG that could act as a functional translocation domain by constructing a series of fusions to the c-Jun leucine zipper hybrid domain. The 89 C-terminal amino acids of UpaG are predicted to contain four β-strands and a linker region with two α-helices (L1) and (L2); our data suggest that the last 71 amino acids of UpaG, corresponding to the L1-β subdomain, represent the translocator unit of UpaG. The reduced level of aggregation observed in E. coli cells expressing a truncated translocator unit in pUpaGL1-β compared to that of a full-length translocator unit in pUpaGL2-L1-β could be due to a slightly reduced efficiency of translocation. Alternatively, we cannot exclude the possibility that the reduced size of the linker between the β domain and the passenger domain might result in the decreased stability of the protein and therefore the reduced interaction between the leucine zipper domains of c-Jun.
We recently characterized the Ag43a and Ag43b AT adhesins from UPEC CFT073 and demonstrated that the Ag43a variant contributes to long-term persistence in the urinary bladder (71). This study identified the existence of pathogenicity-adapted variants of Ag43 with distinct virulence-related functions. Ag43-mediated aggregation and biofilm formation were not observed in CFT073 when the Ag43a and Ag43b proteins were constitutively produced, suggesting that additional structures at the cell surface may mask their function. Unlike with these observations, the expression of UpaG in CFT073 produced a strong aggregation phenotype and promoted significant biofilm growth. We attribute these different phenotypes to the size difference between Ag43 and UpaG. Whereas the passenger domain of Ag43 comprises 498 amino acids and is predicted to extend approximately 10 nm from the bacterial outer membrane (31), 3D modeling suggested that the passenger domain of UpaG that is exposed to the surface comprises around 1,700 amino acids, and we estimate from immunogold EM images that it is approximately 100 nm in size. This is consistent with size predictions for the passenger domain of other members of the trimeric AT family. For example, the size of the exposed domain of YadA, a 455-amino-acid protein, was measured at about 23 nm (27), whereas the exposed domain of BadA, a giant 3,082-amino-acid protein, was estimated to be about 300 nm (52).
While conventional AT proteins have diverse effector functions, including cytotoxicity, lipase or protease activity, and adhesin properties, it appears that all trimeric AT proteins identified so far display an adhesive activity mediating bacterial interactions with either host cells or ECM proteins (4, 12, 13, 55, 59, 66). Consistent with these data, the results reported here demonstrate that UpaG mediates the adherence of E. coli to T24 bladder epithelial cells. In addition, we also observed that the expression of UpaG mediated cell-to-cell aggregation and biofilm formation by E. coli on abiotic surfaces. While we have no information with regard to the structural domains of UpaG responsible for these different functions, binding to eukaryotic cells/ECM proteins might be due to Him and Hep-Hag motifs found in the passenger domain of the protein. These domains are known to contribute to the binding activity of other proteins from the invasin/hemagglutinin family. Crystal structure determinations from other trimeric AT proteins have demonstrated that in each case the passenger domain has threefold symmetry and three identical faces. This structure provides the potential for three binding pockets to adhere with strong affinity to receptor targets (14). Moreover, many Hia-like and YadA-like AT proteins possess predicted coiled-coiled motifs, raising the possibility of multimerization of the passenger domain. This may provide one explanation for the ability of UpaG to promote bacterial cell aggregation (14).
Adherence to human ECM proteins is an important aspect of host colonization for many bacterial pathogens, including N. meningitidis, H. influenzae, Helicobacter pylori, Streptococcus pyogenes, and Staphylococcus aureus (18, 19, 38, 49). The trimeric YadA, UspA2, and NhhA AT proteins are all associated with high-affinity bacterial binding to ECM proteins (41, 59, 69). Our data revealed that UpaG mediates the binding of E. coli to laminin and fibronectin and that this binding is independent of type 1 fimbriae. The UpaG-specific adhesion to T24 bladder cells could be explained by the differential expression of eukaryotic cell surface ligands. Indeed, we observed that T24 bladder cells possess a higher level of fibronectin than HeLa cells. Since ECM proteins become exposed following epithelial cell damage, adhesins such as UpaG that mediate binding to fibronectin and laminin may contribute to colonization by promoting adherence at such sites. However, we note that this does not exclude the possibility of the existence of a specific host receptor for UpaG. Interestingly, the YadA protein has been shown to mediate tissue-specific colonization by Yersinia pseudotuberculosis (28), implying a possible differential affinity of this protein for different cellular types, as was observed for UpaG.
Based on the E. coli genome sequences available within the NCBI database, the chromosomal location of upaG appears to be conserved; upaG is located upstream of lldP, a gene encoding an l-lactate permease. Interestingly, the presence of upaG orthologs seems to be restricted to some closely related enterobacteriaceae, including E. coli, Shigella species, and Salmonella species (5, 35).
Orthologs of upaG are absent from genera such as Yersinia and other proteobacteria from the gamma subdivision. This suggests either that upaG acquisition occurred after the separation of E. coli and related enterobacteria or that there is a correlation between UpaG and the environment encountered by enterobacteria, such as in the animal gut. However, the existence of paralog proteins containing typical Hep-Hag/Him and YadA-like motifs in various bacterial species suggests that this type of adhesin has been acquired independently to fulfill similar functions. E. coli strains fall into four main phylogenetic groups (A, B1, B2, and D) (26). Strains associated with invasive extraintestinal clinical syndromes, such as bacteremia, pyelonephritis, prostatitis, and meningitis, belong mainly to group B2 and to a lesser extent to group D. In contrast, most commensal strains and other strains associated with less invasive syndromes, such as cystitis, generally belong to group A (48). Our epidemiological data provide molecular evidence to suggest that upaG is significantly more frequently associated with E. coli strains from phylogenetic groups B2 and D. These data are consistent with a previous report that demonstrated an association between upaG and ExPEC strains that cause bacteremia as well as the absence of upaG in certain E. coli strains, such as members of the K-12 lineage (17).
The apparent lack of production of UpaG in E. coli CFT073 in vitro correlates with a previous report that examined UpaG production in the neonatal meningitis E. coli strain S26 (17). Those authors demonstrated that immunization of mice with UpaG provided significant protective immunity against a lethal intraperitoneal challenge with E. coli S26. Although UpaG expression could not be detected during the laboratory growth of E. coli S26, pooled human sera collected from patients suffering from bacteremia due to ExPEC infection reacted positively with UpaG in immunoblot experiments, suggesting that UpaG is produced during bacteremic infection and can induce a host antibody response. We detected weak transcription of upaG by quantitative reverse transcription-PCR following the in vitro growth of CFT073. However, we were unable to detect the expression of the corresponding protein by Western blotting using UpaG-specific antiserum on outer membrane preparations of CFT073 grown under the same conditions. Furthermore, the upaG mutant of CFT073 showed no statistically significant difference from the parent strain in its ability to colonize the mouse urinary tract in both C57BL/6 and C3H/HeJ mouse infection models. The lack of expression of UpaG by CFT073 appears to be consistent with other studies, since UpaG was not detected among outer membrane proteins prepared from CFT073 following colonization of the mouse bladder (65) or after CFT073 infection of the mouse peritoneal cavity (51). However, the fact that passive immunization of mice with UpaG could protect them from a lethal systemic challenge, combined with the observation that an antibody response to UpaG can be detected in patients with bacteremia, suggests that the expression of upaG may be enhanced during infection.
The CFT073 PcL upaG strain was constructed to examine the effect of UpaG constitutive expression in a defined UPEC background. CFT073 PcL upaG aggregated strongly, formed an excellent biofilm and adhered to laminin, fibronectin, and T24 bladder epithelial cells. The direct association between UpaG expression and these phenotypes was confirmed by the construction and examination of two additional UpaG-producing strains (U6 and U15). When CFT073 PcL upaG was examined in the C57BL/6 mouse UTI model, no significant difference was observed in the levels of colonization of the bladder. The same result was obtained using the C3H/HeJ mouse infection model, although there was a trend in the data to suggest that CFT073 PcL upaG colonized the bladder and kidney less efficiently that the parent CFT073 strain. We speculate that this may be due to the forced expression of UpaG, which may interfere with the functions of other surface adhesins, as has been described previously (6, 23, 60, 62, 71, 72).
While UpaG has recently been found to be a promising ExPEC vaccine candidate (17), an understanding of the functions associated with UpaG expression as well as an investigation of its regulation under in vitro and in vivo conditions is required to gain a better understanding of the arsenal of adhesins used by E. coli to colonize the urinary tract and other human sites.
Supplementary Material
Acknowledgments
We thank D. Skurnik and E. Denamur for providing commensal E. coli strains, L. A. Fernandez for providing plasmid pJunβ, E. Veiga for helpful discussions, and Rick Webb for help with electron microscopy. We are grateful to N. Wolff for helping with UpaG 3D modeling, for helpful discussions, and for critical reading of the manuscript. We thank P. Cossart and M. Delepierre for kindly providing their laboratory facilities for some of the analyses presented herein.
J.V. is a Marie Curie Fellow. A.T.-A. is an EMBO Fellow. This work was supported by grants from the Institut Pasteur, CNRS URA 2172, the Network of Excellence EuroPathoGenomics, the European community (LSHB-CT-2005-512061), the Fondation BNP PARIBAS, the Australian National Health and Medical Research Council (455914), the Australian Research Council (DP0557615), and the University of Queensland.
Footnotes
Published ahead of print on 18 April 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abate, C., D. Luk, E. Gagne, R. G. Roeder, and T. Curran. 1990. Fos and Jun cooperate in transcriptional regulation via heterologous activation domains. Mol. Cell. Biol. 105532-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abate, C., D. Luk, R. Gentz, F. J. Rauscher III, and T. Curran. 1990. Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc. Natl. Acad. Sci. USA 871032-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balligand, G., Y. Laroche, and G. Cornelis. 1985. Genetic analysis of virulence plasmid from a serogroup 9 Yersinia enterocolitica strain: role of outer membrane protein P1 in resistance to human serum and autoagglutination. Infect. Immun. 48782-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barenkamp, S. J. 1996. Immunization with high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae modifies experimental otitis media in chinchillas. Infect. Immun. 641246-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belda, E., A. Moya, and F. J. Silva. 2005. Genome rearrangement distances and gene order phylogeny in gamma-Proteobacteria. Mol. Biol. Evol. 221456-1467. [DOI] [PubMed] [Google Scholar]
- 6.Beloin, C., K. Michaelis, K. Lindner, P. Landini, J. Hacker, J. M. Ghigo, and U. Dobrindt. 2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J. Bacteriol. 1881316-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 8.Berger, H., J. Hacker, A. Juarez, C. Hughes, and W. Goebel. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J. Bacteriol. 1521241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bingen, E., B. Picard, N. Brahimi, S. Mathy, P. Desjardins, J. Elion, and E. Denamur. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177642-650. [DOI] [PubMed] [Google Scholar]
- 10.Bonacorsi, S., O. Clermont, V. Houdouin, C. Cordevant, N. Brahimi, A. Marecat, C. Tinsley, X. Nassif, M. Lange, and E. Bingen. 2003. Molecular analysis and experimental virulence of French and North American Escherichia coli neonatal meningitis isolates: identification of a new virulent clone. J. Infect. Dis. 1871895-1906. [DOI] [PubMed] [Google Scholar]
- 11.Canutescu, A. A., A. A. Shelenkov, and R. L. Dunbrack, Jr. 2003. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 122001-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Arico, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 1951445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, G. H. McCracken, Jr., and E. J. Hansen. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 1814026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13199-205. [DOI] [PubMed] [Google Scholar]
- 15.Da Re, S., B. Le Quere, J. M. Ghigo, and C. Beloin. 2007. Tight modulation of Escherichia coli bacterial biofilm formation through controlled expression of adhesion factors. Appl. Environ. Microbiol. 733391-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deighan, P., A. Free, and C. J. Dorman. 2000. A role for the Escherichia coli H-NS-like protein StpA in OmpF porin expression through modulation of micF RNA stability. Mol. Microbiol. 38126-139. [DOI] [PubMed] [Google Scholar]
- 17.Durant, L., A. Metais, C. Soulama-Mouze, J. M. Genevard, X. Nassif, and S. Escaich. 2007. Identification of candidates for a subunit vaccine against extraintestinal pathogenic Escherichia coli. Infect. Immun. 751916-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberhard, T., R. Virkola, T. Korhonen, G. Kronvall, and M. Ullberg. 1998. Binding to human extracellular matrix by Neisseria meningitidis. Infect. Immun. 661791-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finlay, B. B., and M. Caparon. 2006. Bacterial adherence to cell surface and extracellular matrix, p. 105-120. In P. Cossart, P. Bouquet, S. Normark, and R. Rappuoli (ed.), Cellular microbiology, 2nd ed. ASM Press, Washington, DC.
- 20.Gerlach, R. G., and M. Hensel. 2007. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297401-415. [DOI] [PubMed] [Google Scholar]
- 21.Grodberg, J., and J. J. Dunn. 1988. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol. 1701245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hacker, J., L. Bender, M. Ott, J. Wingender, B. Lund, R. Marre, and W. Goebel. 1990. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb. Pathog. 8213-225. [DOI] [PubMed] [Google Scholar]
- 23.Hasman, H., T. Chakraborty, and P. Klemm. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 1814834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heesemann, J., C. Eggers, and J. Schroder. 1987. Serological diagnosis of yersiniosis by immunoblot technique using virulence-associated antigen of enteropathogenic Yersiniae. Contrib. Microbiol. Immunol. 9285-289. [PubMed] [Google Scholar]
- 25.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 1726175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 195989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudson, K. J., and A. H. Bouton. 2006. Yersinia pseudotuberculosis adhesins regulate tissue-specific colonization and immune cell localization in a mouse model of systemic infection. Infect. Immun. 746487-6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung, D. L., and S. J. Hultgren. 1998. Pilus biogenesis via the chaperone/usher pathway: an integration of structure and function. J. Struct. Biol. 124201-220. [DOI] [PubMed] [Google Scholar]
- 30.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 31.Klemm, P., L. Hjerrild, M. Gjermansen, and M. A. Schembri. 2004. Structure-function analysis of the self-recognizing antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 51283-296. [DOI] [PubMed] [Google Scholar]
- 32.Korhonen, T. K., M. V. Valtonen, J. Parkkinen, V. Vaisanen-Rhen, J. Finne, F. Orskov, I. Orskov, S. B. Svenson, and P. H. Makela. 1985. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 48486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laarmann, S., D. Cutter, T. Juehne, S. J. Barenkamp, and J. W. St. Geme. 2002. The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol. Microbiol. 46731-743. [DOI] [PubMed] [Google Scholar]
- 34.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 1821364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lerat, E., V. Daubin, and N. A. Moran. 2003. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-Proteobacteria. PLoS Biol. 1E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lesic, B., S. Bach, J. M. Ghigo, U. Dobrindt, J. Hacker, and E. Carniel. 2004. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol. Microbiol. 521337-1348. [DOI] [PubMed] [Google Scholar]
- 37.Linke, D., T. Riess, I. B. Autenrieth, A. Lupas, and V. A. Kempf. 2006. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 14264-270. [DOI] [PubMed] [Google Scholar]
- 38.Ljungh, A., A. P. Moran, and T. Wadstrom. 1996. Interactions of bacterial adhesins with extracellular matrix and plasma proteins: pathogenic implications and therapeutic possibilities. FEMS Immunol. Med. Microbiol. 16117-126. [DOI] [PubMed] [Google Scholar]
- 39.Martinez, J. J., M. A. Mulvey, J. D. Schilling, J. S. Pinkner, and S. J. Hultgren. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 192803-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marti-Renom, M. A., A. C. Stuart, A. Fiser, R. Sanchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29291-325. [DOI] [PubMed] [Google Scholar]
- 41.McMichael, J. C., M. J. Fiske, R. A. Fredenburg, D. N. Chakravarti, K. R. VanDerMeid, V. Barniak, J. Caplan, E. Bortell, S. Baker, R. Arumugham, and D. Chen. 1998. Isolation and characterization of two proteins from Moraxella catarrhalis that bear a common epitope. Infect. Immun. 664374-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 581281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen, B. B., J. S. Kastrup, H. Rasmussen, T. L. Holtet, J. H. Graversen, M. Etzerodt, H. C. Thogersen, and I. K. Larsen. 1997. Crystal structure of tetranectin, a trimeric plasminogen-binding protein with an alpha-helical coiled coil. FEBS Lett. 412388-396. [DOI] [PubMed] [Google Scholar]
- 44.Nowicki, B., J. Singhal, L. Fang, S. Nowicki, and C. Yallampalli. 1999. Inverse relationship between severity of experimental pyelonephritis and nitric oxide production in C3H/HeJ mice. Infect. Immun. 672421-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parham, N. J., U. Srinivasan, M. Desvaux, B. Foxman, C. F. Marrs, and I. R. Henderson. 2004. PicU, a second serine protease autotransporter of uropathogenic Escherichia coli. FEMS Microbiol. Lett. 23073-83. [DOI] [PubMed] [Google Scholar]
- 47.Pepe, J. C., M. R. Wachtel, E. Wagar, and V. L. Miller. 1995. Pathogenesis of defined invasion mutants of Yersinia enterocolitica in a BALB/c mouse model of infection. Infect. Immun. 634837-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Preissner, K. T., and G. S. Chhatwal. 2004. Extracellular matrix and host cell surfaces: potential sites of pathogen interaction, p. 87-104. In P. Cossart, P. Bouquet, S. Normark, and R. Rappuoli (ed.), Cellular microbiology, 2nd ed. ASM Press, Washington, DC.
- 50.Ray, S. K., R. Rajeshwari, Y. Sharma, and R. V. Sonti. 2002. A high-molecular-weight outer membrane protein of Xanthomonas oryzae pv. oryzae exhibits similarity to nonfimbrial adhesins of animal pathogenic bacteria and is required for optimum virulence. Mol. Microbiol. 46637-647. [DOI] [PubMed] [Google Scholar]
- 51.Redford, P., P. L. Roesch, and R. A. Welch. 2003. DegS is necessary for virulence and is among extraintestinal Escherichia coli genes induced in murine peritonitis. Infect. Immun. 713088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riess, T., S. G. Andersson, A. Lupas, M. Schaller, A. Schafer, P. Kyme, J. Martin, J. H. Walzlein, U. Ehehalt, H. Lindroos, M. Schirle, A. Nordheim, I. B. Autenrieth, and V. A. Kempf. 2004. Bartonella adhesin a mediates a proangiogenic host cell response. J. Exp. Med. 2001267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riess, T., G. Raddatz, D. Linke, A. Schafer, and V. A. Kempf. 2007. Analysis of Bartonella adhesin A expression reveals differences between various B. henselae strains. Infect. Immun. 7535-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 1853735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roggenkamp, A., H. R. Neuberger, A. Flugel, T. Schmoll, and J. Heesemann. 1995. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol. Microbiol. 161207-1219. [DOI] [PubMed] [Google Scholar]
- 56.Roos, V., G. C. Ulett, M. A. Schembri, and P. Klemm. 2006. The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect. Immun. 74615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roux, A., C. Beloin, and J. M. Ghigo. 2005. Combined inactivation and expression strategy to study gene function under physiological conditions: application to identification of new Escherichia coli adhesins. J. Bacteriol. 1871001-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Scarselli, M., D. Serruto, P. Montanari, B. Capecchi, J. Adu-Bobie, D. Veggi, R. Rappuoli, M. Pizza, and B. Arico. 2006. Neisseria meningitidis NhhA is a multifunctional trimeric autotransporter adhesin. Mol. Microbiol. 61631-644. [DOI] [PubMed] [Google Scholar]
- 60.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 1861249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 1868058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shindyalov, I. N., and P. E. Bourne. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11739-747. [DOI] [PubMed] [Google Scholar]
- 64.Skurnik, M., and P. Toivanen. 1992. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Bacteriol. 1742047-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 726373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.St. Geme, J. W., III, and D. Cutter. 1995. Evidence that surface fibrils expressed by Haemophilus influenzae type b promote attachment to human epithelial cells. Mol. Microbiol. 1577-85. [DOI] [PubMed] [Google Scholar]
- 67.Surana, N. K., D. Cutter, S. J. Barenkamp, and J. W. St. Geme III. 2004. The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. J. Biol. Chem. 27914679-14685. [DOI] [PubMed] [Google Scholar]
- 68.Tahir, Y. E., P. Kuusela, and M. Skurnik. 2000. Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification of eight NSVAIG-S motifs in the amino-terminal half of the protein involved in collagen binding. Mol. Microbiol. 37192-206. [DOI] [PubMed] [Google Scholar]
- 69.Tamm, A., A. M. Tarkkanen, T. K. Korhonen, P. Kuusela, P. Toivanen, and M. Skurnik. 1993. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol. 10995-1011. [DOI] [PubMed] [Google Scholar]
- 70.Tertti, R., M. Skurnik, T. Vartio, and P. Kuusela. 1992. Adhesion protein YadA of Yersinia species mediates binding of bacteria to fibronectin. Infect. Immun. 603021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ulett, G. C., J. Valle, C. Beloin, O. Sherlock, J. M. Ghigo, and M. A. Schembri. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 753233-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ulett, G. C., R. I. Webb, and M. A. Schembri. 2006. Antigen-43-mediated autoaggregation impairs motility in Escherichia coli. Microbiology 1522101-2110. [DOI] [PubMed] [Google Scholar]
- 73.Valle, J., S. Da Re, N. Henry, T. Fontaine, D. Balestrino, P. Latour-Lambert, and J. M. Ghigo. 2006. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. USA 10312558-12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Veiga, E., V. de Lorenzo, and L. A. Fernandez. 2003. Autotransporters as scaffolds for novel bacterial adhesins: surface properties of Escherichia coli cells displaying Jun/Fos dimerization domains. J. Bacteriol. 1855585-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vial, P. A., R. Robins-Browne, H. Lior, V. Prado, J. B. Kaper, J. P. Nataro, D. Maneval, A. Elsayed, and M. M. Levine. 1988. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J. Infect. Dis. 15870-79. [DOI] [PubMed] [Google Scholar]
- 76.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9917020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, P., B. B. Chomel, M. K. Schau, J. S. Goo, S. Droz, K. L. Kelminson, S. S. George, N. W. Lerche, and J. E. Koehler. 2004. A family of variably expressed outer-membrane proteins (Vomp) mediates adhesion and autoaggregation in Bartonella quintana. Proc. Natl. Acad. Sci. USA 10113630-13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.