Abstract
The DevR-DevS two-component system of Mycobacterium tuberculosis mediates bacterial adaptation to hypoxia, a condition believed to be associated with the initiation and maintenance of dormant bacilli during latent tuberculosis. The activity of the Rv3134c-devRS operon was studied in M. tuberculosis using several transcriptional fusions comprised of promoter regions and the gfp reporter gene under inducing and aerobic conditions. Aerobic transcription was DevR independent, while hypoxic induction was completely DevR dependent. The hypoxia transcriptional start point, TH, was mapped at −40 bp upstream of Rv3134c. In contrast, the divergently transcribed Rv3135 gene was not induced under hypoxic conditions. DNase I footprinting and mutational analyses demonstrated that induction required the interaction of DevR∼P with binding sites centered at bp −42.5 and −63.5 relative to TH. Binding to the distal site (D) was necessary to recruit another molecule of DevR∼P to the proximal site (P), and interaction with both sequences was essential for promoter activation. These sites did not bind to either unphosphorylated or phosphorylation-defective DevR protein, which was consistent with an essential role for DevR∼P in activation. Phosphorylated DevR also bound to three copies of the motif at the hspX promoter. The Rv3134c and hspX promoters have a similar architecture, wherein the proximal DevR∼P binding site overlaps with the promoter −35 element. A model for the likely mode of action of DevR at these promoters is discussed.
Mycobacterium tuberculosis is a remarkable human pathogen whose success is attributed in no small measure to its ability to establish an asymptomatic latent infection. Latently infected individuals comprise approximately one-third of the world's population and serve as a vast reservoir of bacilli for future reactivation of disease. Existing therapies are not very effective against metabolically sluggish dormant bacilli, and prolonged treatment is required to virtually eradicate them. One of the important aspects of understanding latent tuberculosis is to learn about the molecular basis of adaptation of tubercle bacilli to a dormant state in response to environmental stresses. In mice, latent tuberculosis is believed to result from bacterial adaptation to a dormant state in response to hypoxia and nitric oxide (18, 35). Both of these stimuli induce the expression of M. tuberculosis devR (sometimes called dosR) and its target genes referred to as the dormancy regulon (21, 23, 27, 32). Induction of some or all of these genes also occurs in an in vivo dormancy model and in gamma-interferon (IFN-γ)-activated mouse macrophages (11, 26). A recent report demonstrated the generation of a robust IFN-γ response in latently infected individuals to dormancy regulon antigens, suggesting that these antigens may contribute to the control of latent infection (15).
The most well-characterized in vitro model of latency and persistence was pioneered by Wayne et al., who used hypoxic culture conditions to transform growing bacteria into nonreplicating persistent organisms (33, 34, 35). This model and its variations have contributed substantially to our understanding of the molecular mechanisms underlying the dormancy response. One variation that is frequently used is the standing of aerobic cultures, which creates a local hypoxic environment in settled bacteria and strongly induces devR and devR regulon genes (8, 12, 23, 27). DevR-DevS (Rv3133c-Rv3132c) is a two-component system of M. tuberculosis. The devRS genes are cotranscribed along with Rv3134c, a gene which codes for an alanine-valine-rich protein of unknown function that belongs to the universal stress protein family of proteins found in many bacteria, including M. tuberculosis (6). DevR activation occurs through phosphorylation by histidine kinases, DevS and Rv2027c/DosT (22, 23, 24), and induces the expression of several genes in M. tuberculosis cultures exposed to hypoxia (21). Most of the induced genes of the devR regulon contain one or more copies of a 20-bp palindromic sequence in their upstream regions (21). It was reported earlier that (i) unphosphorylated DevR interacted with devR, Rv3134c, and hspX promoters (2, 21); (ii) phosphorylation-defective DevR D54E protein bound nearly as well as phosphorylated DevR to a 20-bp double-stranded oligonucleotide bearing the consensus binding motif (22); and (iii) M. bovis BCG cultures expressing DevR D54E protein failed to support hspX induction during hypoxia (21). These results were puzzling since it was difficult to reconcile the near equivalent binding of phosphorylated wild-type and mutant DevR proteins to a consensus binding sequence in vitro and the requirement for phosphorylated protein to activate hspX expression. One possibility was that there may be some difference(s) in regulation between BCG and M. tuberculosis. A second possibility was that the in vitro binding that was noted to occur in reactions containing an excess of protein (2, 22) or DNA (22) may not mirror the scenario in axenically cultured bacteria. Therefore, it was imperative to resolve these inconsistencies in order to clearly understand the mechanism of DevR-mediated gene activation in M. tuberculosis.
The Rv3134c-devRS operon is transcribed in aerobic cultures of M. tuberculosis from two regions upstream of Rv3134c and devR, respectively (2). We performed a detailed analysis of the transcriptional regulation of this operon under aerobic and hypoxic conditions in M. tuberculosis. The aerobic promoters were DevR-independent, while positive autoregulation was noted under hypoxic conditions from a newly defined promoter located upstream of Rv3134c. Phosphorylated DevR bound to two sequences in the Rv3134c promoter. Binding to the distal site (D) preceded binding to the proximal site (P) that overlapped with the −35 promoter element and protein binding to both the motifs was necessary and sufficient for induction of the Rv3134c-devRS operon. DevR∼P bound cooperatively to three sites in the promoter of hspX (Rv2031c/acr), one of the best-studied members of the DevR regulon. We show that positive autoregulation is mediated by the cooperative binding of phosphorylated DevR at the hypoxia-inducible Rv3134c promoter, answer an important question that DevR∼P (and not unphosphorylated protein) is the binding and activating species, and provide an explanation for the inability of unphosphorylated DevR to support DevR regulon induction in aerobic cultures despite being present at measurable levels. Both the Rv3134c and hspX promoters have a similar architecture comprised of multiple DevR binding sites and an overlap of the proximal binding site with the −35 promoter element. A model for DevR-mediated activation at these promoters is discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
M. tuberculosis H37Rv and ΔdevR (complete deletion) bacteria were cultured in Dubos medium containing 0.05% Tween 80 plus 0.5% albumin, 0.75% dextrose, and 0.085% NaCl (AD complex). M. tuberculosis H37Rv and ΔdevR (20) strains were a generous gift from N. G. Stoker. For reporter assays, −80°C frozen stock cultures of M. tuberculosis were aerobically subcultured twice to mid-logarithmic phase (A595 ∼ 0.3) and then diluted to an A595 of 0.05. Standing cultures were established by dispensing 200-μl culture aliquots in triplicate into 96-well black, clear-bottom microtiter plates (Becton Dickinson, United Kingdom) and incubating the plates at 37°C. The aerobic promoter activity was measured in cultures that were simultaneously grown in 50-ml tubes (10 ml of culture) placed in racks in a shaker incubator at 220 rpm. A shaking speed of 220 rpm was necessary to prevent promoter induction in aerobic cultures in order to distinguish between aerobic and hypoxia-inducible transcription. Under these conditions the A595 of cultures typically increased to ∼0.15 in the wells and to ∼0.25 under shaking conditions over a 48-h period. Culture aliquots of 200 μl were sampled at 48 h, and the green fluorescent protein (GFP) fluorescence was measured as described previously (2). Escherichia coli strains and culture conditions were as described earlier (2). When needed, antibiotics were used at the following concentrations: ampicillin at 100 μg/ml; kanamycin at 50 μg/ml in E. coli and at 25 μg/ml in M. tuberculosis; and hygromycin at 200 μg/ml in E. coli and at 50 μg/ml in M. tuberculosis. All of the plasmids used in the present study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant featurea | Source or reference |
|---|---|---|
| pSC-DevR | pGEX4T1 overexpressing DevR with GST N-terminal tag | 2 |
| pDevR D54V | pSC-DevR containing aspartate to valine mutation at amino acid residue 54 of DevR | K. Kaur and J. S. Tyagi, unpublished data |
| pFPV27 | E. coli-mycobacterial shuttle plasmid with promoterless gfp, Kmr | 31 |
| pOperon | pFPV27 containing operon promoter (−608 to +998), Hygr | This study |
| p3134c-1 | pFPV27 containing Rv3134c promoter (−608 to +90), Hygr | This study |
| p3134c-2 | pFPV27 containing Rv3134c promoter (−608 to −89), Hygr | This study |
| p3134c-3 | pFPV27 containing Rv3134c promoter (−348 to +90), Hygr | This study |
| p3134c-4 | pFPV27 containing Rv3134c promoter (−348 to −33), Hygr | This study |
| p3134c-5 | pFPV27 containing Rv3134c promoter (−348 to −89), Hygr | This study |
| p3134c-6 | pFPV27 containing Rv3134c promoter (−348 to −118), Hygr | This study |
| p3134c-7 | pFPV27 containing Rv3134c promoter (−173 to +90), Hygr | This study |
| pdevR-1 | pFPV27 containing devR promoter (−532 to +164), Hygr | This study |
| pdevR-2 | pFPV27 containing devR promoter (−390 to +164), Hygr | This study |
| pdevR-3 | pFPV27 containing devR promoter (−237 to +164), Hygr | This study |
| p3135-1 | pFPV27 containing Rv3135 promoter (−674 to +24), Kmr | This study |
| p3135-2 | pFPV27 containing Rv3135 promoter (−466 to +24), Kmr | This study |
| p3135-3 | pFPV27 containing Rv3135 promoter (−257 to +24), Kmr | This study |
| pmut-P | p3134c-6 containing mutated proximal hypoxia motif, Hygr | This study |
| pmut-D | p3134c-6 containing mutated distal hypoxia motif, Hygr | This study |
| pmut-PD | p3134c-6 containing mutated proximal and distal hypoxia motif, Hygr | This study |
| psigA | pFPV27 containing sigA promoter (−238 to +80), Hygr | N. K.Taneja and J. S. Tyagi, unpublished data |
| prrn | pFPV27 containing rrn promoter (−321 to +48), Hygr | N. K. Taneja and J. S. Tyagi, unpublished data |
| phspX | pFPV27 containing hspX promoter (−132 to +48), Hygr | D. K. Saini and J. S. Tyagi, unpublished data |
The coordinates are with reference to the translational start site of the respective M. tuberculosis gene, except for hspX, where the coordinates are with reference to its TSP as defined earlier (21), and for rrn, where coordinates are with reference to the +1 nucleotide of mature 16S rRNA. Hygr, hygromycin resistance; Kmr, kanamycin resistance.
RNA isolation and primer extension.
RNA was isolated from M. tuberculosis H37Rv cultures grown in Dubos medium (see above) under aerobic (shaking) and hypoxic (48-h standing) conditions using TRI reagent (Molecular Research Center) according to the manufacturer's protocol. Briefly, a 40-ml culture of M. tuberculosis was grown in a 250-ml flask in a shaker incubator at 220 rpm. At an A595 of ∼0.2, a 20-ml aliquot was centrifuged immediately at 5,000 rpm for 10 min at 4°C. The remaining culture aliquot was kept standing (10 ml in 50-ml tubes) for 48 h at 37°C, the harvested cell pellets were each resuspended in 1 ml of TRI reagent, and the cells were lysed in a mini bead beater using 0.1-mm zirconium/silica beads (Biospec). After phase separation by bromochloropropane, the RNA in the aqueous phase was precipitated with isopropanol, treated with RNase-free DNase I (Promega), and purified using RNeasy mini column (Qiagen, Germany). The Rv3134c and Rv3135 transcription start points (TSPs) were mapped by primer extension as described previously (25), using the 32P-labeled primers R9c and 3135r (see Table S1 in the supplemental material), respectively, and 10 or 20 μg of RNA (two or three separate lots). The primers were annealed at 60 and 55°C for 45 min, and the cDNA was synthesized at 42°C for 60 min using StrataScript reverse transcriptase (Stratagene). The sequencing ladders were generated using M. tuberculosis DNA and the same primers that were used in the primer extension analysis.
Reporter plasmids construction.
GFP reporter plasmids were constructed as described previously (2). In brief, M. tuberculosis H37Rv DNA was amplified with Pfu DNA polymerase (Stratagene) using specific primers to generate various promoter fragments (Fig. 1 and see Tables S1 and S2 in the supplemental material). The fragments were first cloned in pGEM-T Easy (Promega) and then in the reporter plasmid pFPV27 (31) at the EcoRI site. The identity of the cloned fragments was verified by DNA sequencing. The constructs were modified by the introduction of a hygromycin resistance cassette into each plasmid (except Rv3135 constructs) and electroporated in wild-type H37Rv and ΔdevR strains of M. tuberculosis.
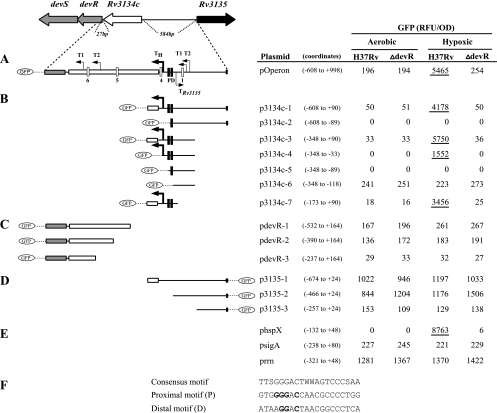
FIG. 1.
Promoter activity under aerobic and hypoxic conditions. The genomic organization of the Rv3134c-devRS operon and the Rv3135 gene in M. tuberculosis is shown at the top. TSPs are indicated by bent arrows. Boxes P and D refer to proximal and distal binding sites, respectively; white boxes indicate low-scoring putative binding sequences (2, 21). The regions contained within individual GFP transcriptional fusion constructs are indicated alongside each plasmid. (A) Operon promoters; (B) Rv3134c upstream promoters; (C) devR upstream promoters; (D) Rv3135 upstream promoter; (E) control promoters. (F) Sequences of the proximal (P) and distal (D) motifs, along with the proposed consensus sequence (21); W, A/T. Bases in the P and D motifs that were mutated (G→C, etc.) for interaction experiments with DevR are indicated in boldface. For each construct the solid line and the boxes depict the intergenic region and coding sequences contained therein, respectively. The coordinates are with reference to the translational start site of the respective M. tuberculosis gene. The GFP fluorescence (RFU/OD) produced within M. tuberculosis strains (H37Rv and ΔdevR mutant) grown under aerobic and hypoxic conditions is shown. GFP fluorescence is shown from one representative experiment out of three independent experiments. Fluorescence values from promoters that were induced under hypoxic conditions are underlined. The promoter coordinates are indicated in parentheses alongside each plasmid.
Mutated versions of the proximal (P) and distal (D) binding sites (the −348 to +90 positions in p3134c-3) were generated by using the mega primer-based method (25). Briefly, the first PCR was carried out with an internal mutagenic primer and a vector-specific primer, Fpv27a. The resulting product was purified and used as a mega primer with Gfpseq, a vector-specific primer from the opposite end, for the second PCR. For the construction of pmutP/pmutD plasmids the template used in the first PCR was p3134c-3, and for constructing pmutPD the plasmid pmutD was used as the template. The final products were purified, digested with EcoRI, and cloned in pFPV27 at the EcoRI site. The hygromycin resistance cassette was inserted as described above. The mutations were confirmed by DNA sequencing, and these plasmids were electroporated in M. tuberculosis H37Rv.
Gel shift assay and DNase I footprinting.
M. tuberculosis DevR with an N-terminal glutathione S-transferase (GST) tag (DevR-GST; henceforth referred to as DevR) was overexpressed in pSCDevR-containing cultures of E. coli and purified as described previously (2). Plasmid pSCDevR was used as a PCR template to construct pDevR D54V by site-directed mutagenesis (Table 1). The DevR D54V-GST mutant protein (henceforth referred to as DevR D54V) was overproduced from pDevR D54V in E. coli. The mutant protein is defective in phosphorylation as described earlier (21). Gel shift and DNase I footprinting assays were performed with purified DevR (2) and DevR D54V mutant proteins. Electrophoretic mobility shift assays (EMSAs) were carried out as described previously (2). Many response regulators can be phosphorylated, albeit less efficiently, by low-molecular-weight phosphor donors such as acetyl phosphate (29). Since phosphorylated DevR rapidly lost its phosphosignal in the presence of DevS and Rv2027c kinases (23, 24), DevR was phosphorylated by incubating it with 50 mM acetyl phosphate for 30 min at 30°C in 40 mM Tris-Cl (pH 8.0) and 5 mM MgCl2. DevR was incubated without acetyl phosphate (unphosphorylated protein), and DevR D54V was incubated with acetyl phosphate in parallel for use in gel shift assays and DNase footprinting. The phosphorylation defect in DevR D54V was confirmed by failure to be phosphorylated with acetyl phosphate (not shown). Radiolabeled DNA fragments were generated by PCR using appropriate primers (see Table S1 in the supplemental material), one of which was end labeled with [γ-32P]ATP (∼3,000 Ci/mmol; BRIT, Hyderabad, India). After phosphorylation, binding to DevR was performed in a 20-μl reaction with 1 to 2 ng of 32P-labeled DNA (10,000 cpm) for 20 min at 30°C in binding buffer [25 mM Tris-HCl (pH 8.0), 0.5 mM EDTA, 20 mM KCl, 6 mM MgCl2, 5% glycerol, and 1 μg of poly(dI-dC)]. The reaction was electrophoresed on a 5% nondenaturing gel at 120 V (constant) in 0.5× Tris-borate-EDTA buffer at 4°C after the gel was prerun for 30 min under similar conditions. The gel was dried and analyzed by using a phosphorimager with QuantityOne software (Bio-Rad).
For DNase footprinting assays, DevR phosphorylation and DNA-protein interaction were performed as described above except that 10 to 15 ng (75,000 to 100,000 cpm) of labeled DNA was used in a reaction volume of 50 μl. After DNase I treatment (0.2 U; Promega) for 4 min at 22°C in the presence of 2.5 MgCl2 and 5 mM CaCl2, the reaction was stopped by the addition of 90 μl of 2× stop solution (200 mM NaCl, 30 mM EDTA, 1% sodium dodecyl sulfate, and 66 μg of yeast tRNA/ml). The reaction products were extracted with phenol-chloroform, precipitated with 3 volumes of ethanol at −80°C for 1 h, washed with 70% ethanol, and air dried. DNA was dissolved in formamide-urea loading dye, loaded onto 6% denaturing polyacrylamide gel alongside a DNA ladder (generated by using the fmol DNA cycle sequencing system [Promega]), and run at 70 W. The gel was dried and visualized by using a phosphorimager.
RESULTS
We showed earlier that the Rv3134c-devRS operon is transcribed from promoters located upstream of Rv3134c and devR in aerobic cultures of M. tuberculosis (2). Here we carried out a detailed analysis of the induction mechanism under hypoxia in order to begin to understand the critical regulatory features of DevR and DevR regulon expression.
Transcription and autoregulation of DevR.
We constructed 11 gfp transcriptional fusion reporter plasmids containing various lengths of devR and Rv3134c upstream sequences and measured their GFP fluorescence in wild-type and ΔdevR M. tuberculosis cultures under shaking and standing conditions (Fig. 1). It was previously shown that transcription of devR was upregulated rapidly on standing (12). The induction was probably due to the development of hypoxia in the vicinity of cells settled at the bottom of the wells (8), and the term “hypoxic” is used here to refer to these cultures. Since GFP requires oxygen for maturation to the fluorescent form, the 96-well plate format reporter assay was first standardized under various concentrations of oxygen (0.2, 2, 6, and 10% and atmospheric). After 48 h of standing, the GFP fluorescence derived from the hypoxia-inducible hspX promoter rose proportionately with an increase in oxygen concentration and was maximal at an ambient atmosphere (data not shown). GFP fluorescence was not detected in the presence of 0.2% oxygen (data not shown). Therefore, an ambient atmosphere was used to incubate the 96-well plates. After 48 h, the cultures were sufficiently hypoxic to induce the Rv3134c-devRS operon, and an adequate level of oxygen was present for GFP maturation. The reporter activity measurements were rapid, convenient, and reproducible under these conditions.
The pOperon reporter construct (positions −608 to +998, with reference to the translational start site of Rv3134c) contains both devR and Rv3134c upstream promoters and provided us with an overview of operon transcription (Fig. 1A). The promoter activity was moderate under aerobic conditions, equivalent to that of sigA promoter (psigA, Fig. 1E), and independent of DevR. Promoter activity was strongly induced in standing cultures (∼28-fold) in a DevR-dependent manner (Fig. 1A), which demonstrated that M. tuberculosis DevR activates its own expression under hypoxic but not in aerobic conditions. Next, constructs containing various deletions of the Rv3134c upstream region were assessed for GFP activity (Fig. 1B. The complete upstream region in p3134c-1 (positions −608 to +90) supported modest aerobic activity and was highly induced under hypoxia (∼83-fold) in a DevR-dependent manner. This fragment contains the aerobic TSPs, T1Rv3134c and T2Rv3134c, and four putative DevR-binding sites that include the proximal (P) and distal (D) motifs plus two other consensus sequences (sequences 1 and 4, Fig. 1) described previously (2). Deletion of the P motif and downstream sequences abolished both aerobic activity and hypoxic induction (p3134c-2), which indicated the importance of the sequence. To assess the usage of aerobic TSPs and the influence of the putative DevR-binding sites on inducible transcription, constructs deleted of 260 bp from the far-upstream region of Rv3134c were analyzed (constructs p3134c-3 to p3134c-6). The region from positions −348 to +90 (in p3134c-3) conferred aerobic activity and DevR-dependent hypoxic inducibility (174-fold) to the same extent as observed with p3134c-1 and defined the minimal region required for basal and inducible operon activity. Since plasmid p3134c-4 (positions −348 to −33) displayed no aerobic activity but was inducible, albeit not to the same extent as p3134c-3, we concluded that the region from positions −33 to +90 was necessary for both aerobic expression and full induction. Further sequence deletion (p3134c-5), which removed the P sequence also, eliminated aerobic activity and inducibility completely (as in p3134c-2), indicating the importance of the deleted region (between positions −32 and −88) for aerobic transcription and hypoxic induction. Surprisingly, when the deletion extended to both putative binding sequences P and D (p3134c-6, positions −348 to −118), moderate promoter activity was detected (equivalent to that of the psigA promoter) under both aerobic and hypoxic growth conditions, and in both wild-type H37Rv and the devR mutant, and in each of these conditions, the expression of GFP was essentially the same. Interestingly, a 263-bp region between positions −173 and +90 (in plasmid p3134c-7) deleted of the aerobic TSPs, supported rather low aerobic expression but a high DevR-dependent hypoxic induction (∼192-fold), allowing us to conclude that the aerobic TSP was not used in hypoxia and that additional TSP was active under inducing conditions.
We had previously shown that aerobic transcription also initiated upstream of devR within Rv3134c coding sequences (2). To further analyze this activity, deletion constructs pdevR-1 to pdevR-3 were studied under aerobic and hypoxic conditions (Fig. 1C). The region from position −532 upstream of devR to position +164 inside the devR coding region (pdevR-1) contains the previously reported aerobic TSPs (T1devR and T2devR) and also two putative binding sites (sites 5 and 6). It displayed moderate aerobic transcriptional activity (quite comparable to psigA) that was not induced on standing. A further deletion of 142-bp from the 5′ end supported a similar level of transcription (pdevR-2), while further deletion (region having T2devR) resulted in a significant reduction in activity in both of the strains (pdevR-3). This shows that T2devR might be major aerobic TSP in this region. Collectively, the data indicate that the devR upstream region plays no role in hypoxic induction and that putative binding sites 5 and 6 do not interact with DevR. Considering the GFP activities of pOperon, p3134c-1, and pdevR-1 together, we conclude that the low-strength promoters located upstream of devR and Rv3134c are involved in DevR-independent aerobic basal transcription, whereas the TH promoter upstream of Rv3134c contributes to DevR-dependent hypoxic inducibility of the operon. sigA and rrn promoter GFP fusions served as independent controls since they are not known to be induced during hypoxia; their activities remained unchanged in the presence or absence of DevR (Fig. 1E).
Transcription of divergent Rv3135 (orfX) gene is not regulated by DevR.
Rv3135 and Rv3134c genes are divergently transcribed from an intergenic region of 584 bp. Rv3135 (orfX [5]) codes for PPE50 protein and belongs to the PPE gene family which, along with the PE gene family, was proposed to contribute to antigenic diversity in M. tuberculosis (4). We examined the influence of DevR on Rv3135 promoter activity. A few fragments upstream of Rv3135 carrying progressive 5′ deletions were cloned in pFPV27 reporter plasmid, and their promoter activity was assessed in M. tuberculosis and ΔdevR strains (Fig. 1D). The complete intergenic region present in p3135-1 (from positions −674 to +24, with reference to translational start site of Rv3135) exhibited very high aerobic activity that was not altered under standing conditions in both the strains, indicating that Rv3135 was not regulated by DevR. Deletion of far upstream sequences from −674 to −466 (p3135-2) did not affect promoter activity, but deletion of the sequence from positions −466 to −257 (p3135-3) greatly reduced activity establishing that the major promoter was present in this region. The TSP was mapped at position −428 with reference to the Rv3135 translational start site and was preceded by strong matches with the SigA −10 and −35 promoter elements (Fig. 2B and C).
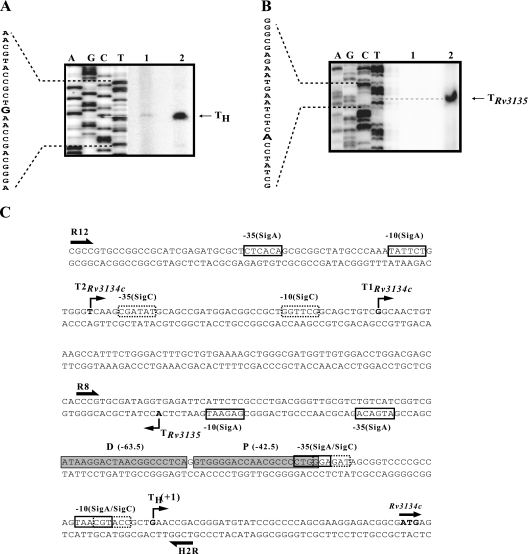
FIG. 2.
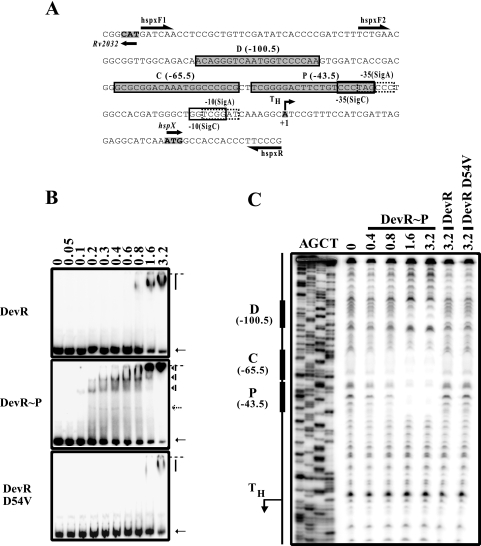
Primer extension analysis. (A and B) Primer extension with M. tuberculosis H37Rv RNA from aerobic (lane 1) and 48-h standing cultures (lane 2) using primer R9c (A) and no RNA control (lane 1) and aerobic culture (lane 2) using primer 3135r (B). The sequence of the noncoding strand was generated by using the respective primers. G (in boldface), whose complementary C sequence comigrates with the extended RNA, is designated TH for Rv3134c. Likewise, A is the Rv3135 TSP. (C) Salient features of the Rv3134c-Rv3135 promoter. The start codon of Rv3134c is shaded. TH, hypoxia Rv3134c TSP determined in Fig. 2A. Putative −10 and −35 elements of SigC and SigA are indicated by dashed and solid boxes, respectively; for P and D, shaded boxes indicate proximal and distal binding sites, respectively. Rv3134c aerobic TSPs are described elsewhere (2).
Rv3134c TSP mapping.
A distinct hypoxia-specific TSP, TH (Fig. 2A), was mapped at the G nucleotide located at −40 bp with reference to the Rv3134c translational start site by primer extension using RNA extracted from hypoxic cultures of M. tuberculosis (+1, Fig. 2C). Its location was consistent with the inducible GFP expression observed with p3134c-4 and p3134c-7 constructs that carried minimal upstream sequences. Sequences having partial matches with −10 and −35 elements of SigA and SigC sigma factors were noted (1, 30). The −35 sequences partially overlapped with the P sequence centered at −42.5 bp (Fig. 2A). From TSP mapping of the divergently placed genes it is evident that aerobic transcripts from Rv3134c and Rv3135 overlapped by 84 bp (Fig. 2C).
Binding of DevR in gel shift assays.
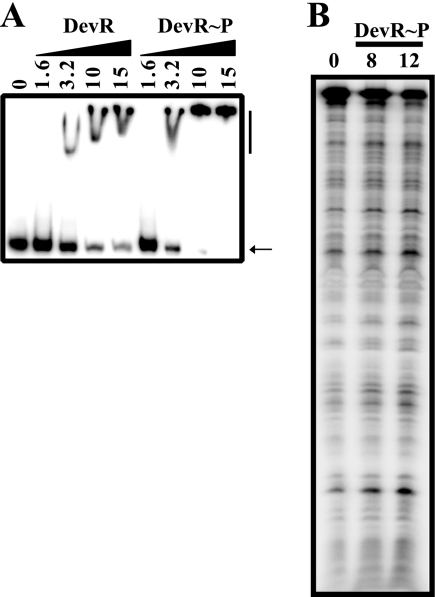
We showed previously that devR upstream sequences interacted weakly with DevR (at high concentration [2]), which could not be rationalized in terms of its constitutive promoter activity and suggested that the interaction may be nonspecific. Therefore, we first assessed the interaction of DevR with the sigA and rrn control promoters. Let us consider the case of sigA. Three features of the sigA promoter established that its activity was not DevR dependent; first, the promoter was equally active in H37Rv and ΔdevR strains; second, it was not induced during hypoxia; and third, it lacked a potential DevR binding site. However, at ≥3.2 μM, both unphosphorylated and phosphorylated DevR interacted with the sigA promoter (Fig. 3A). On DNase footprinting with the sigA promoter fragment, no protection was observed with DevR∼P (Fig. 3B). On the basis of these observations, we conclude that, at high concentrations, unphosphorylated DevR interacts with DNA in a sequence-independent manner. Nonspecific binding of unphosphorylated protein to DNA has also been reported with ArcA at high protein concentrations by the gel shift assay (10).
FIG. 3.
The sigA promoter interacts nonspecifically with DevR. (A) EMSA was performed with 32P-labeled sigA promoter fragment (positions −238 to +80). Lane 1, no protein; lanes 2 to 9, DevR (lanes 2 to 5) or DevR∼P (lanes 6 to 9) at various concentrations (μM). The arrow indicates free DNA and the vertical line, bound complex. (B) DNase footprinting was performed with the same fragment.
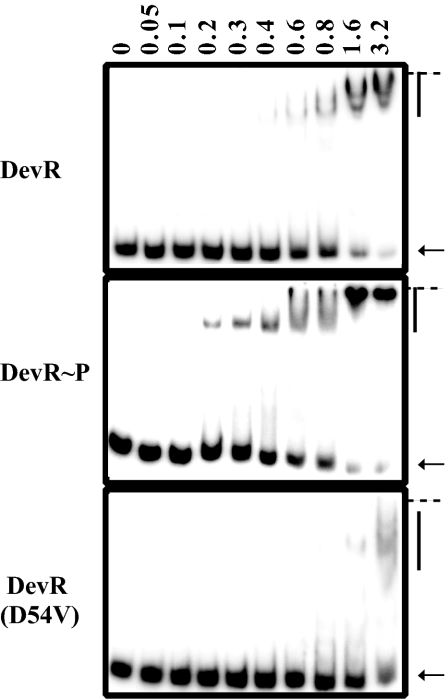
We examined DNA-protein interactions between the Rv3134c promoter and unphosphorylated DevR, DevR∼P, or phosphorylation-defective DevR D54V mutant protein at various protein concentrations (Fig. 4). DevR D54V protein bound poorly and that, too, was only at 3.2 μM. Unphosphorylated DevR appeared to bind slightly better than mutant DevR protein, perhaps because it was partially phosphorylated in E. coli (by acetyl phosphate or cross talk) from which it was purified, as reported for other response regulator proteins (7, 14). Phosphorylation increased DevR binding to the promoter region compared to only a minimal amount of binding observed with unphosphorylated DevR and none at all with DevR D54V protein. At higher concentrations, the faster-moving complex appeared to decrease, and a broad protein-DNA complex of lower mobility was seen (denoted by arrowheads, Fig. 4). At 1.6 μM protein concentration, most of the DNA was bound.
FIG. 4.
DevR∼P binds with enhanced affinity to the Rv3134c promoter. 32P-labeled Rv3134c promoter DNA (positions −173 to +90) was incubated in the absence (lane 1) or presence (lanes 2 to 10) of increasing concentrations of DevR protein (μM). The arrow indicates the position of free DNA, and the vertical line indicates liganded DNA. The well position is marked by a horizontal line.
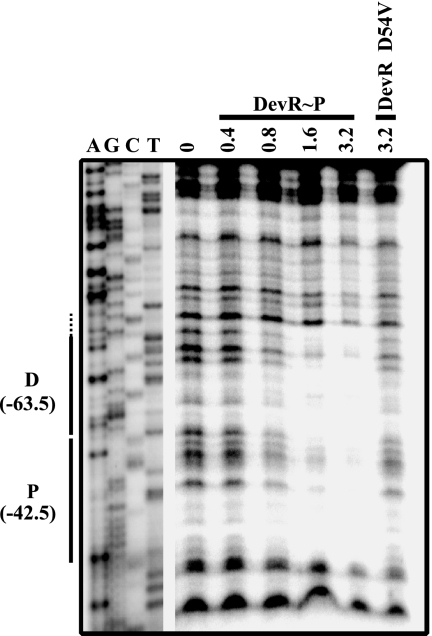
Phosphorylated DevR binds to P and D binding sites in the Rv3134c promoter.
In silico analysis predicted the presence of four putative binding sites including the P and D motifs in the Rv3134c promoter region (Fig. 1A) (2, 21). To determine which of them interact with DevR, DNase footprinting was first performed with the complete Rv3134c promoter fragment (positions −348 to +90). Phosphorylated DevR protected the two high-scoring tandemly placed P and D motifs centered at base pair positions −42.5 and −63.5 relative to the hypoxia tsp, TH (data not shown), while protection was not observed with unphosphorylated DevR even at a 3.2 μM protein concentration (data not shown). The putative sites 1 and 4 (Fig. 1A) did not bind to DevR∼P (data not shown). Using a shorter DNA fragment (bp −173 to −33), a progressive increment in the specific protection of D and P sites was noted with an increasing concentration of DevR∼P but not with 3.2 μM DevR D54V protein (Fig. 5). Based on the results of EMSA and DNase footprinting, we conclude that DNase footprinting is a reliable measure of sequence-specific interaction even at high DevR∼P concentrations and that the binding observed in EMSAs with unphosphorylated or mutant DevR proteins was nonspecific.
FIG. 5.
DNase I protection of the Rv3134c promoter. DNase footprinting was performed with a 140-bp (positions −173 to −33) labeled antisense strand of Rv3134c promoter region (amplified with R8 and H2R primers, see Table S1 in the supplemental material) and either DevR∼P, DevR, or DevR D54V protein (μM). D and P represent distal and proximal binding sites, respectively, with reference to hypoxic TSP, TH. Protection beyond the binding site is depicted by a dotted line. Dideoxy sequencing reactions using the same primers are also shown.
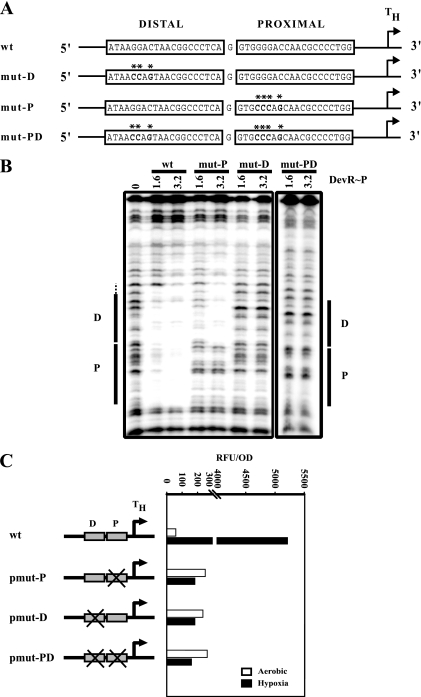
Effect of binding site mutations on in vitro binding and in vivo promoter activity.
The crystal structure of the C-terminal domain of DevR revealed that a tetramer (a dimer of dimers) bound to two 20-bp palindromic consensus sequences to form a complex (36). Within the 20-bp motif, the four bases, G4G5G6 and C8, per half-palindrome were highly conserved in putative DevR-binding sites, and it was proposed that these conserved nucleotides may provide contact points for DevR (36). In the present study, we demonstrated that DevR∼P interacts with the tandem P and D sites located upstream of Rv3134c. So, next, in order to understand the specific roles of DevR interaction with these sites in transcription activation, the P and D sequences were mutated individually and in combination. Four bases in the P site (G4G5G6 and C8) and three bases in the D site (G5G6 and C8 since G4 is not conserved in this motif) were mutated to their complementary bases in the first half of the palindrome (Fig. 6A) and assessed for their effect on DevR-DNA interaction and on promoter activity. DNase footprinting with the mut-P fragment (having a P-site mutation) showed protection of the D motif only, indicating that the mutated residues were required for DevR∼P binding to the P site. Surprisingly, and interestingly, the mut-D fragment (having the mutated D site) was not protected at either the distal or the proximal binding site (Fig. 6B). To confirm the results, four residues (C4C5C6 and G8) in the second-half of the palindrome were mutated in both motifs, and similar footprinting results were obtained as with the previous mutations (data not shown). The doubly mutated mut-PD fragment showed no protection at either of the altered motifs. These results established the distal D site to be the primary binding site and binding to the proximal P site to be dependent on protein occupancy at the D site. These results suggest that (i) since sequence-specific interaction of DevR∼P was abolished upon mutating either half of the palindromic motif, both halves of the motif were important for binding, (ii) mutation in any one-half of the palindromic sequence abrogated protein binding to the motif, which suggested DevR∼P dimer (not a monomer) to be the interacting species, and (iii) the ability of the protein to bind independently to the D site (when the P site was mutated) further suggests that DevR∼P may interact with the motif as a dimer.
FIG. 6.
In vitro and in vivo mutational analysis of the P and D binding sites at the Rv3134c promoter. (A) Sequence of wild-type (wt) and mutated binding sites (mut-P, mut-D, and mut-PD). Mutations are indicated by asterisks. (B) DNase footprinting with DevR∼P (μM) and 32P-labeled Rv3134c promoter (positions −173 to −33) containing wild-type, mut-P, mut-D, and mut-PD sequences (C) GFP fluorescence of wild-type and mutated site-containing plasmids. “X” indicates the mutated box. Representative data from one of two experiments, each of which was performed in triplicate, are shown.
The mutated fragments were cloned in gfp reporter plasmids (pmut-P, pmut-D, and pmut-PD) and introduced in M. tuberculosis H37Rv. In 48-h standing cultures, hypoxic induction of GFP fluorescence was completely abrogated in all of the mutants but, interestingly, aerobic activity increased by ∼5-fold compared to the wild-type promoter (Fig. 6C). The lack of hypoxic induction in pmut-D and pmut-PD strains can be explained by the inability to bind DevR at the P and D sites (Fig. 6B). The P-site mutant fragment bound DevR∼P at the distal D site but failed to support hypoxic induction in the pmut-P strain. These studies established that DevR∼P binding to both of these sites is necessary and sufficient for hypoxic induction in vivo.
DevR∼P interacts with three binding sites in the hspX promoter.
We assessed the interaction of DevR with the hspX promoter that is very highly induced and is a marker for the M. tuberculosis dormancy response (8, 12, 21, 27). The hspX promoter was inactive in aerobic conditions and very highly induced on standing in a DevR-dependent manner (Fig. 1E). The hspX promoter contains three putative binding sites, namely, proximal (P), central (C), and distal (D) sites (Fig. 7A). DevR∼P bound more strongly to the hspX promoter compared to either unphosphorylated DevR or DevR D54V protein in EMSA. Three bound species with retarded mobility were observed and, with increasing protein concentration, a greater proportion of DNA was bound in the two larger complexes. At 1.6 μM, almost all of the DNA was liganded to protein (Fig. 7B).
FIG. 7.
DevR∼P binds cooperatively to the hspX promoter. (A) DNA sequence of the hspX promoter. The start codons of hspX and the divergent gene Rv2032 are shaded and in boldface. TH, hypoxia-specific TSP. Proximal (P), central (C), and distal (D) binding sites are as described previously (21); putative −10 and −35 promoter elements are boxed; arrows indicate the primers used in EMSA and footprinting assays. (B) 32P-labeled hspX promoter DNA fragment (bp −132 to +48) was incubated in the absence (lane 1) or presence (lanes 2 to 10) of increasing concentrations of DevR (μM) as indicated. The arrow indicates the position of free DNA, and the vertical arrowheads indicate DNA-protein complexes. A faint band migrating just behind the free DNA is a minor contaminant in the probe. Another faint band in the gel shift assay with DevR∼P was not observed consistently (dotted arrow). The well position is marked by a horizontal line. (C) DNase protection with a 215-bp hspX promoter fragment (bp −167 to +48) and DevR protein as indicated.
By DNase footprinting DevR binding was mapped to the P (−43.5 bp), C (−65.5 bp), and D (−100.5 bp) sites relative to the hypoxia TSP, TH. Binding to the P and C sites was first noted at 0.8 μM, and all of the three sites were occupied at 1.6 μM protein concentration (Fig. 7C). The binding pattern suggests that DevR∼P binds more strongly to the C and P sequences and less well to sequence D. No binding was noted with either unphosphorylated DevR or DevR D54V proteins. The arrangement and distances of the C and P sites from the hspX hypoxic TSP are much more similar to the arrangement and distances of the D and P sites relative to the Rv3134c hypoxic TSP than the hspX D and P sites are. It was shown earlier in M. bovis BCG that when either the P or the D motif was mutated, hspX promoter induction was reduced but not abrogated (21). Mutation of the P site affected hspX induction more severely compared than mutation of the D site (21) and is consistent with the overlap of the P site with the −35 promoter element. On the basis of the data presented here, it appears that the C and P sites may be the primary binding sites for DevR∼P at the hspX promoter and that the D site may play a compensatory role or be needed for optimal gene induction. Rv2032 (acg) is divergently transcribed and coinduced with hspX during hypoxia (8). It will be useful to analyze the mechanism of DevR activation of these and other such divergently organized and coactivated genes.
DISCUSSION
We analyzed the transcriptional activity and the mechanism of activation of the Rv3134c-devRS operon, which encodes the DevR-DevS two-component system of M. tuberculosis. devR and Rv3134c upstream sequences direct aerobic transcription in a DevR-independent manner, while specific sequences located upstream of Rv3134c control hypoxic transcription in a strictly DevR-dependent fashion. These combined functions are reflected in the activity of the complete operon promoter under both conditions. Positive autoregulation is mediated by a direct interaction between phosphorylated DevR and tandemly placed D and P binding sites located upstream of TSP, TH. The reporter constructs that contained this region (pOperon and p3134c-1, -3, -4, and -7) were highly induced on standing in a DevR-dependent manner. The Rv3134c upstream region contains both aerobic and hypoxia-responsive promoters, and various deletion constructs had various expression of GFP. The deletion of downstream (p3134c-4) or upstream (p3134c-7) sequences retained the hypoxia motifs but prevented full activation. Further experiments are required to understand their role in complete activation. Aerobic Rv3134c promoter activity was abolished completely when P and Tsp TH sequences were deleted (p3134c-2 and p3134c-5) and restored at an ∼5-fold-higher level when the binding sites were either singly or doubly mutated (pmut constructs) or deleted (p3134c-6). One possible explanation is that this region intrinsically folded into an inhibitory structure that was disrupted by sequence mutation or deletion. Second, interaction with another regulatory factor may be abolished, or transcript stabilization may be affected. Recently, it was noted that aerobic transcription of Rv3134c was elevated in an mprA mutant. Since MprA-binding to this region was not observed, it was suggested that the MprA-mediated inhibitory effect on promoter activity was indirect (19). A third possibility was that the divergent Rv3135 and Rv3134c aerobic promoters overlap, and sequence disruption or deletion leads to an increase in Rv3134c promoter activity. Although it is evident from TSP mapping that these promoters do overlap, the Rv3135 TSP was located within p3134c-6 and not within the inhibitory sequence. It was also noted that fairly good GFP expression (equivalent to sigA promoter activity) occurred under hypoxic conditions from p3134c-6 or pmut plasmids. Since TH is not utilized in these constructs, it is likely that the expression is derived from the upstream promoter(s) (T1 or T2), which is DevR independent. Taken together, these results suggest that the operon is under complex regulation, and further studies are necessary in order to understand the regulatory role of other factors and interacting sequences in transcribing this operon.
Weak interaction was noted between devR promoter sequences and unphosphorylated DevR at a high concentration (12.5 μM [2]) by EMSA. Based on our present findings, that interaction does not appear to be sequence specific. Similarly, the rather weak interaction between the consensus sequence and a high concentration of unphosphorylated and/or phosphorylation defective DevR (25 μM [22]) appears to be uncharacteristic of interaction between a transcriptional regulator and its consensus element. Therefore, these results indicate that DNase footprinting, and not EMSA, is a reliable measure of sequence-specific binding of DevR. The G4G5G6 nucleotide sequence in the consensus motif was proposed as a basis for recognition, and the G4 nucleotide appears to be essential for DevR-DNA interaction (36). The D site at the Rv3134c promoter (primary binding site) contains an A nucleotide at position 4 in place of the G nucleotide. Likewise, the C site in the hspX promoter contains a C nucleotide at position 4. Since both of these sequences bound to DevR∼P, we conclude that the G4 nucleotide was not essential for specific DevR∼P DNA interaction at these promoters; however, our results confirm the importance of G5, G6, and C8, as proposed elsewhere (36), since mutation in these sequences abolished protein binding.
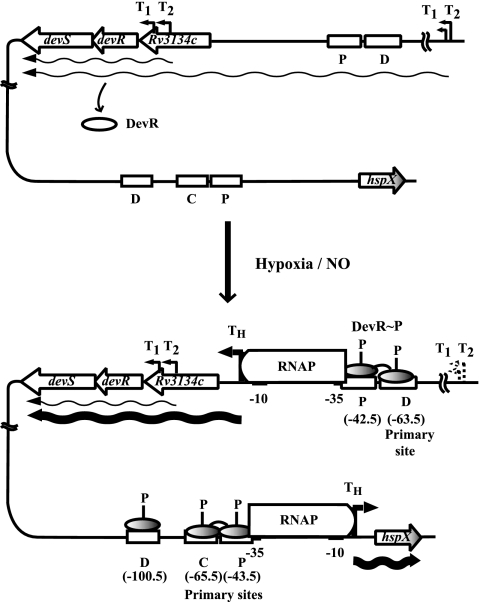
DevR belongs to the NarL/UhpA family of response regulators (6). Like DevR, phosphorylated but not unphosphorylated NarL interacted specifically with its cognate DNA motif (17). Phosphorylation of NarL releases the DNA binding determinants and may also facilitate its oligomerization (16). With respect to DevR, we know now that phosphorylated protein binds first to the D site (−63.5 bp) and cooperatively recruits another molecule of DevR∼P to the secondary binding P site (−42.5 bp) that overlaps the promoter −35 element of the hypoxia-inducible Rv3134c promoter, TH (Fig. 8). Putative promoter −10 and −35 elements with 3/6 and 4/6 matches for SigA sigma factor (1) and 5/6 and 4/6 matches for SigC (30) were detected upstream of TH (Fig. 2). Binding of DevR∼P to the sites may be facilitated through DNA bending by the occurrence of DNA kinking induced at each half-site bound to DevR (36). The hspX upstream sequences have a similar layout of DevR binding sites and overlap with the promoter −35 element (Fig. 7 and 8). Our results demonstrate that DevR∼P binding to both of the sites in the Rv3134c promoter is necessary for gene activation under hypoxia. Similarly, DevR∼P binding to at least two sites is necessary for optimal induction of hspX. The architecture of both the hypoxia-inducible promoters resembles CRP-activated class III promoters (3) where one CRP-binding site overlaps the −35 promoter element (as in class II promoters) and a second binding site is located further upstream (as in class I promoters). In the DevR regulon, other genes, namely, Rv2626c and ctpF, have potential binding sites in a arrangement similar to that in Rv3134c (P and D sites) and hspX (P and C sites). Rv2626c codes for a conserved hypothetical protein, and ctpF codes for a cation transporter (21). Further experimental analysis is necessary in order to understand the mechanism of phosphorylation-mediated DevR activation; determine the nature of interactions, if any, between DevR and RNA polymerase; and understand their role in the activation of these promoters.
FIG. 8.
Transcriptional regulation of Rv3134c and hspX promoters. (Top panel) Under normoxia, devRS operon is transcribed from aerobic promoters (thin wavy arrows) to provide basal levels of DevR and DevS. Unphosphorylated DevR does not bind or induce the operon or hspX gene. (Bottom panel) Upon induction, DevR∼P species is generated. It binds first to the D site and cooperatively recruits another DevR∼P molecule to the P site to induce the operon (thick wavy line). At the hspX promoter, DevR∼P binds first with the P and C sites and then with the D site to induce its transcription. At both promoters, DevR∼P binds to the P site overlapping the −35 promoter element and also to one or two additional upstream sites. The oligomeric status (monomer or dimer) of free and DNA-bound DevR is not known and, for simplicity, DevR is represented as an oval (white for unphosphorylated and shaded for phosphorylated).
We had previously reported that the M. tuberculosis Rv3134c and devR promoters were modulated by DevR in aerobic cultures of M. smegmatis and E. coli (2). Those observations are not consistent with the present findings in M. tuberculosis, wherein the promoters are expressed in a DevR-independent manner under aerobic conditions. There could be several reasons for the observed differences. First, in our previous study, GFP fluorescence was measured in tube cultures of M. smegmatis shaken at 190 rpm. At this shaking speed, DevR was observed to be upregulated even under aerobic conditions in M. tuberculosis (possibly due to the partial settling of cells) but not at a shaking speed was 220 rpm. Indeed, the reporter activity in aerobic cultures of M. smegmatis/pSC1 was much lower at a shaking speed of 220 rpm versus a shaking speed of 190 rpm (data not shown). Since, in M. smegmatis/pSC2, M. tuberculosis DevR was produced from its constitutive promoter and endogenous DevR was produced from its native promoter (2), it is possible that the DevR orthologues could compete and result in a marginal repression. Second, the detection of five TSPs in the Rv3134c and devR promoter regions and an additional TSP for Rv3135 points to a complex regulation in this region and, furthermore, the regulatory mechanisms may not be completely conserved between the species. For example, three different promoters, namely, the σ70/SigA-like aerobic promoter (T2Rv3134c [2]), the SigC-like aerobic promoter (T1Rv3134c [2]), and the hypoxia-inducible TH promoter (the present study), are located upstream of Rv3134c, suggesting the possibility of σ70/SigA-like promoter being recognized in M. smegmatis and E. coli but not the SigC-like promoter since the SigC orthologue is absent in these bacteria. Third, DevR motifs are located downstream of the σ70-like aerobic promoter, T2Rv3134c (Fig. 1), and binding to these sites may lower promoter activity in E. coli and M. smegmatis. These observations suggest that experiments of this nature are best performed in a homologous system.
The transcriptional activity of the Rv3134c-devRS operon in aerobic and induced cultures is consistent with a model wherein phosphorylated DevR (not unphosphorylated protein) is the transcriptional activator of the operon and of DevR target genes (Fig. 8). In aerobic cultures, DevR is expressed at basal levels from DevR-independent promoters located upstream of Rv3134c and devR. Upon stimulation by, say, hypoxia, nitric oxide, and/or redox changes (9, 13, 28), activated DevR∼P is generated from preexisting DevR. A positive feedback loop stimulates TH-directed transcription and leads to increased levels of DevR and DevR regulon genes products. Generally, the presence of cooperativity associated with multiple binding sites lowers the protein concentration at which an effect is seen. The cooperativity in DevR interaction at both the Rv3134c and the hspX promoters is explained by the spacing of motifs, which allows two or more DevR dimers to bind on the same face of the DNA helix. Both the motifs, P and D, located at the −42.5 and −63.5 nucleotide positions, respectively, are crucial for in vivo activation of Rv3134c. At present, we do not know the mechanism by which the DevR regulon reverts to a basal level of expression. One possibility is that DevR dephosphorylation activity associated with DevS (23) might be regulated by disappearance of the stimulatory signal. Alternatively, phosphorylated DevR maybe intrinsically unstable, and its continued synthesis may be required to mediate its activating functions.
Supplementary Material
Acknowledgments
This study was financially supported by grant to J.S.T. from the Department of Biotechnology, Government of India. S.C. is grateful to the CSIR for a Senior Research Fellowship. We also acknowledge the facilities of the Biotechnology Information Systems, Department of Biotechnology, and the Government of India.
We are grateful to present and past members of the laboratory for lively and stimulating discussions. We acknowledge A. Baisantry and D. Bakshi for preliminary EMSA analysis of DevR interaction with the sigA and rrn promoters. The technical assistance of Sanjay Kumar in routine laboratory work is also acknowledged.
Footnotes
Published ahead of print on 21 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agarwal, N., and A. K. Tyagi. 2006. Mycobacterial transcriptional signals: requirements for recognition by RNA polymerase and optimal transcriptional activity. Nucleic Acids Res. 344245-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagchi, G., S. Chauhan, D. Sharma, and J. S. Tyagi. 2005. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology 1514045-4053. [DOI] [PubMed] [Google Scholar]
- 3.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293199-213. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 5.Dasgupta, N., and J. S. Tyagi. 1998. Identification of a RFLP associated with a deletion in M. tuberculosis Erdman that maps in a transcriptionally active open reading frame, orfX, in Mycobacterium tuberculosis Erdman. Tuberc. Lung Dis. 7975-81. [DOI] [PubMed] [Google Scholar]
- 6.Dasgupta, N., V. Kapur, K. K. Singh, T. K. Das, S. Sachdeva, K. Jyothisri, and J. S. Tyagi. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuberc. Lung Dis. 80141-159. [DOI] [PubMed] [Google Scholar]
- 7.Emmerich, R., K. Panglungtshang, P. Strehler, H. Hennecke, and H. M. Fischer. 1999. Phosphorylation, dephosphorylation, and DNA-binding of the Bradyrhizobium japonicum RegSR two-component regulatory proteins. Eur. J. Biochem. 263455-463. [DOI] [PubMed] [Google Scholar]
- 8.Florczyk, M. A., L. A. McCue, A. Purkayastha, E. Currenti, M. J. Wolin, and K. A. McDonough. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 715332-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ioanoviciu, A., E. T. Yukl, P. Moenne-Loccoz, and P. R. de Montellano. 2007. DevS, a heme-containing two-component oxygen sensor of Mycobacterium tuberculosis. Biochemistry 464250-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon, Y., Y. S. Lee, J. S. Han, J. B. Kim, and D. S. Hwang. 2001. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J. Biol. Chem. 27640873-40879. [DOI] [PubMed] [Google Scholar]
- 11.Karakousis, P. C., T. Yoshimatsu, G. Lamichhane, S. C. Woolwine, E. L. Nuermberger, J. Grosset, and W. R. Bishai. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J. Exp. Med. 200647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kendall, S. L., F. Movahedzadeh, S. C. Rison, L. Wernisch, T. Parish, K. Duncan, J. C. Betts, and N. G. Stoker. 2004. The Mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis 84247-255. [DOI] [PubMed] [Google Scholar]
- 13.Kumar, A., J. C. Toledo, R. P. Patel, J. R. Lancaster, Jr., and A. J. Steyn. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. USA 10411568-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ladds, J. C., K. Muchova, D. Blaskovic, R. J. Lewis, J. A. Brannigan, A. J. Wilkinson, and I. Barak. 2003. The response regulator Spo0A from Bacillus subtilis is efficiently phosphorylated in Escherichia coli. FEMS Microbiol. Lett. 223153-157. [DOI] [PubMed] [Google Scholar]
- 15.Leyten, E. M., M. Y. Lin, K. L. Franken, A. H. Friggen, C. Prins, K. E. van Meijgaarden, M. I. Voskuil, K. Weldingh, P. Andersen, G. K. Schoolnik, S. M. Arend, T. H. Ottenhoff, and M. R. Klein. 2006. Human T-cell responses to 25 novel antigens encoded by genes of the dormancy regulon of Mycobacterium tuberculosis. Microbes Infect. 82052-2060. [DOI] [PubMed] [Google Scholar]
- 16.Maris, A. E., M. Kaczor-Grzeskowiak, Z. Ma, M. L. Kopka, R. P. Gunsalus, and R. E. Dickerson. 2005. Primary and secondary modes of DNA recognition by the NarL two-component response regulator. Biochemistry 4414538-14552. [DOI] [PubMed] [Google Scholar]
- 17.Maris, A. E., M. R. Sawaya, M. Kaczor-Grzeskowiak, M. R. Jarvis, S. M. Bearson, M. L. Kopka, I. Schroder, R. P. Gunsalus, and R. E. Dickerson. 2002. Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 9771-778. [DOI] [PubMed] [Google Scholar]
- 18.Nathan, C., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 978841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pang, X., P. Vu, T. F. Byrd, S. Ghanny, P. Soteropoulos, G. V. Mukamolova, S. Wu, B. Samten, and S. T. Howard. 2007. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 1531229-1242. [DOI] [PubMed] [Google Scholar]
- 20.Parish, T., D. A. Smith, S. Kendall, N. Casali, G. J. Bancroft, and N. G. Stoker. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 711134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts, D. M., R. P. Liao, G. Wisedchaisri, W. G. Hol, and D. R. Sherman. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 27923082-23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saini, D. K., V. Malhotra, D. Dey, N. Pant, T. K. Das, and J. S. Tyagi. 2004. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150865-875. [DOI] [PubMed] [Google Scholar]
- 24.Saini, D. K., V. Malhotra, and J. S. Tyagi. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 56575-80. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 26.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 987534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sousa, E. H., J. R. Tuckerman, G. Gonzalez, and M. A. Gilles-Gonzalez. 2007. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 161708-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction system: structure-function relationships and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction systems: structure-function relationships and mechanism of catalysis. American Society for Microbiology, Washington, DC.
- 30.Sun, R., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 5225-38. [DOI] [PubMed] [Google Scholar]
- 31.Valdivia, R. H., A. E. Hromockyj, D. Monack, L. Ramakrishnan, and S. Falkow. 1996. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene 17347-52. [DOI] [PubMed] [Google Scholar]
- 32.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wayne, L. G., and G. A. Diaz. 1967. Autolysis and secondary growth of Mycobacterium tuberculosis in submerged culture. J. Bacteriol. 931374-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 642062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 55139-163. [DOI] [PubMed] [Google Scholar]
- 36.Wisedchaisri, G., M. Wu, A. E. Rice, D. M. Roberts, D. R. Sherman, and W. G. Hol. 2005. Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency. J. Mol. Biol. 354630-641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.