Abstract
Bacteria of the order Rhizobiales are able to establish nitrogen-fixing symbioses with legumes. Commonly, genes for symbiosis are harbored on large symbiotic plasmids. Although the transfer of symbiotic plasmids is commonly detected in nature, there are few experimentally characterized examples. In Rhizobium etli, the product of rctA inhibits the conjugation of the symbiotic plasmid by reducing the transcription of the virB operon. rctA is transcribed divergently from this operon, and its product is predicted to have a DNA binding domain. In the present study, using DNase I footprinting and binding assays, we demonstrated the specific binding of RctA to the virB operon promoter. A 9-bp motif in the spacer region of this promoter (the rctA binding motif box) and the presence of a functional −10 region were critical elements for RctA binding. Transcriptional fusion analyses revealed that the elimination of either element provoked a relief of RctA-mediated repression. These data support a model in which RctA inhibits the access of the RNA polymerase to the virB promoter. Interestingly, rctA expression levels were modulated by transcriptional interference from transcripts emanating from the virB promoter. This phenomenon adds another level of regulation for this system, thus revealing a novel mechanism of plasmid transfer regulation in the Rhizobiales.
The ability to establish nitrogen-fixing symbioses is prevalent in bacteria of the order Rhizobiales. Commonly, most of the genes needed to establish symbiosis are either harbored on the so-called symbiotic plasmids (pSyms) or restricted to symbiosis islands (SI) located on the bacterial chromosome. As befits a trait that confers niche extension, there is evidence for the mobility of these genomic compartments. Indeed, sequence analyses of pSyms, including pRetCFN42d of Rhizobium etli (17), pNGR234a of Rhizobium sp. strain NGR234 (13), and pSymA of Sinorhizobium meliloti (1, 15), as well as of the SI of Bradyrhizobium japonicum (23, 18) and Mesorhizobium loti (22, 43), have lead to the identification of conjugation-related genes, mainly the virB1-to-virB11 and traA-traCDG systems, carried by these elements. Moreover, a common feature of these genetic compartments is that the GC contents of these elements differ significantly from those of the rest of the genomes. These data suggest that these gene clusters originated out of, and were transmitted to, other genetic systems. It is likely that these compartments may still be prone to lateral transfer.
Evidence for the movement of pSyms among naturally occurring rhizobial populations has been inferred through phylogenetic and/or population genetics analyses of a variety of systems (45, 38). The transfer of SI, initially detected in field experiments investigating the SI of M. loti (41), was recently demonstrated for the SI of B. japonicum (16). Direct experimental evidence for lateral transfers has also been obtained, albeit such transfers have been found to occur at various rates (ranging from 10−3 to 10−9 transconjugants per receptor cell) for the SI of M. loti (42), the pSym pNGR234a of Rhizobium sp. NGR234 (20), and pRL1JI, the pSym of Rhizobium leguminosarum bv. viciae (10). Conjugational transfer in these three systems is regulated in part by quorum sensing (10, 20, 31), a common strategy used by other rhizobial nonsymbiotic plasmids, such as pTi of Agrobacterium tumefaciens (3) and pRetCFN42a of R. etli CFN42 (44). Thus, although pSym and SI transfer is widely detectable in nature, there are few examples in which mobilization and its regulation have been experimentally characterized.
R. etli CFN42 is a gram-negative bacterium capable of establishing a nitrogen-fixing symbiosis with the common bean (Phaseolus vulgaris). It contains six plasmids, with sizes ranging from 184 to 642 kb. One of them, pRetCFN42d (371 kb), is the pSym. A sequence analysis revealed that this plasmid also possesses a full set of genes involved in conjugation, comprising genes for a mating pair formation (Mpf) type IV secretion system (2) and a DNA transfer and replication system (Dtr) (24). The genes for the Mpf system are arranged as a virB1-to-virB11 operon with the peculiarity of possessing an additional gene, yhd0053, prior to virB1. The genes for the Dtr system include a traA gene, a functional relaxase gene (29), and a traCDG operon featuring genes for two accessory proteins (traC and traD) and a conjugative coupling protein (traG); interestingly, quorum sensing-related genes are absent from this plasmid.
Even though the automobilization of a pSym under laboratory conditions has never been detected, pSym transfer through cointegration with pRetCFN42a, a different automobilizable plasmid regulated by quorum sensing, was observed previously (44). The natural cointegration of these two plasmids occurs at a relatively high frequency and is mediated by both site-specific and homologous recombination (6).
Our previous work suggested that the pSym has an intrinsic ability for conjugal transfer, independent of pRetCFN42a, although this ability is tightly repressed (27). By various genetic strategies, two genes that participate in the regulation of the pRetCFN42d conjugational transfer were identified previously (28). The first one, named rctA (for regulation of conjugal transfer), is transcribed divergently from the virB operon, and it was determined previously by transcriptional fusion analyses to be a repressor of the virB genes. Consistent with the possible role of rctA, an in silico analysis of the predicted sequence of the corresponding protein revealed the presence of a winged-helix DNA binding domain. The second gene found, rctB, is located downstream of traA, and it appears to act as an inhibitor of the repressor activity of rctA. Functional homologues of all these genes also exist on plasmids pAtC58 of A. tumefaciens (46) and pSme1021a of S. meliloti (15), indicating that this model also applies to these organisms (28). Interestingly, this system represents a different alternative for the regulation of conjugal transfer in the Rhizobiales in which tight control is achieved by two novel regulator proteins in a quorum sensing-independent manner.
In the present study, using electrophoretic mobility shift assays (EMSA), DNase I footprinting, and transcriptional fusions, we characterized the mechanism by which rctA represses virB operon transcription. Our data demonstrate the specific binding of RctA to DNA and identify the specific sequence to which RctA binds in order to exert its repressor activity. Moreover, our work reveals the occurrence of transcriptional interference between the rctA and virB transcriptional units, a mechanism that conceivably allows the fine-tuning of conjugational activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Rhizobium strains were grown at 30°C in PY rich medium (26) or in Y minimal medium containing 10 mM succinate and 10 mM ammonium chloride (5). Escherichia coli strains were grown at 37°C in Luria-Bertani medium. Antibiotics were added, when required, at the following concentrations (in micrograms per milliliter): carbenicillin, 100 (E. coli); chloramphenicol, 15 (E. coli); gentamicin, 15 (R. etli) or 30 (E. coli); kanamycin, 15 (R. etli) or 30 (E. coli); nalidixic acid, 20 (R. etli); spectinomycin, 100 (E. coli); and tetracycline, 5 (R. etli) or 10 (E. coli). For the detection of β-galactosidase activity on agar plates, 30 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) ml−1 was used. For fusion analyses, cells were grown until mid-exponential phase in minimal medium. β-Glucuronidase activities in 1-ml culture samples were measured with p-nitrophenyl glucuronide as the substrate (8) and normalized according to the cell protein concentration.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant feature(s) | Source or reference(s) |
|---|---|---|
| Strains | ||
| S. meliloti | ||
| 1021 | Wild-type strain; Smr | 15 |
| A. tumefaciens | ||
| C58 | Wild-type nopaline-resistant strain | 46 |
| R. etli | ||
| CFN2001 | CFN42 derivative lacking p42a and p42d | 6 |
| CFN2001 Tn5.C | CFN2001 derivative with pRetCFN42d harboring a Tn5 insertion in an unmapped location | 28 |
| CFN2001 Tn5.2 | CFN2001 derivative with pRetCFN42d; rctA::Tn5 | 28 |
| CFN2001 Tn5.6 | CFN2001 derivative with pRetCFN42d; rctB::Tn5 | 28 |
| E. coli | ||
| S17.1 | thi pro recA hsdR hsdM RP4-2-Tc::Mu-Km::Tn7 | 39 |
| DH5α | supE44 ΔlacU169 φ80dlacZΔM15 hsdR171 recA1 endA1 gyrA96 thi-1 relA1 | 19 |
| BL21(DE3)/pLysS | F−ompT hsdSB(rB− mB−) gal dcm (DE3) pLysS (CamR) | Novagen |
| Plasmids | ||
| p53Gus | pBBR1MCS5 derivative with a uidA gene of pWM5 (pBBR1MCS5::uidA) | L. Girard |
| p53virB::Gus | Transcriptional fusion of virB promoter in p53Gus using fragment pVT | 28 |
| p53rctA::Gus | Transcriptional fusion of rctA promoter in p53Gus using fragment pVT | 28 |
| p53virB-10m::Gus | Transcriptional fusion of virB promoter in p53Gus using fragment pVT-10m | This work |
| p53rctA-10m::Gus | Transcriptional fusion of rctA promoter in p53Gus using fragment pVT-10m | This work |
| p53virB-rbm::Gus | Transcriptional fusion of virB promoter in p53Gus using fragment pVT-RBM | This work |
| p53rctA-rbm::Gus | Transcriptional fusion of rctA promoter in p53Gus using fragment pVT-RBM | This work |
| pTE::rctB | pTE3 with rctB cloned in front of the trp promoter | 27 |
| pTE::rctA | pTE3 with rctA cloned in front of the trp promoter | 28 |
| pCR2.1-TOPO | PCR direct-cloning vector | Invitrogen |
| pET-16B | Protein His tag fusion and expression vector | Novagen |
| pRK404 | Broad-host-range vector; Tcr | 11, 36 |
| pSSH01 | pCR2.1-TOPO with fragment pVT cloned | This work |
| pSSH02 | pCR2.1-TOPO with fragment pVT-RBM cloned | This work |
| pSSH03 | pCR2.1-TOPO with fragment pVT-10m cloned | This work |
| pSSH04 | pET-16B with rctA cloned in NdeI/BamHI region | This work |
| pSSH05 | pSSH04 fused to pRK404 | This work |
Microbiological and DNA manipulations.
Plasmids were isolated with the AquaPlasmid kit (MultiTarget Pharmaceuticals, Salt Lake City, UT). Plasmid transfer from E. coli to Rhizobium was done by biparental mating by using E. coli S17.1 with the appropriate plasmid as a donor. Rhizobium plasmids were visualized by the Eckhardt procedure (12). Plasmid transformation of E. coli was done using CaCl2-competent cells (33).
Recombinant-DNA techniques were carried out using standard procedures (33). The primers used for PCR amplification are shown in Table 2. PCR amplifications were carried out with Pfu DNA polymerase (Altaenzymes, Alberta, Canada) in a TC-312 thermocycler (Techgene, Burlington, NJ). The DNA amplification regime consisted of 30 cycles comprising 94°C for 1 min, 1 min at variable temperatures, and 72°C for 1 min. For all PCR products cloned with the TOPO TA cloning kit (Invitrogen, Carlsbad, CA), 3′ A overhangs were added. For ligations, T4 polynucleotide ligase (Amersham Biosciences, Piscataway, NJ) was used.
TABLE 2.
Oligonucleotides used in this work
| Name | Sequencea | Locationb |
|---|---|---|
| 38Tl | 5′ TCCCGCCACAGCTTC 3′ | 161542 RetlpSym |
| 38Tu | 5′ TGCCGATCTGCTTCAGC 3′ | 161142 RetlpSym |
| Pv35u | 5′ TTAATCGCGCTTGTCATGTCATTTA 3′ | 161256 RetlpSym |
| Pv10u | 5′ CATTTAACTGTGTTATATACGGCAGTA 3′ | 161265 RetlpSym |
| Pv1u | 5′ TATACGGCAGTATGAATGCGGGCAG 3′ | 161280 RetlpSym |
| rbml | 5′ CCGTATATAACCACTGGCCCTGACATGACAAGC 3′ | 161286 RetlpSym |
| rbmu | 5′ GCTTGTCATGTCAGGGCCAGTGGTTATATACGG 3′ | 161254 RetlpSym |
| 10Ml | 5′ TTCATACTGCCGAGACATACACAGTTAAAT 3′ | 161295 RetlpSym |
| 10Mu | 5′ ATTTAACTGTGTATGTCTCGGCAGTATGAA 3′ | 161266 RetlpSym |
| 38SmTu | 5′ CACGCGCCAGAGCTTCTC 3′ | 721157 SmelpSymA |
| 38SmTl | 5′ TCCCGAGCTGCTTCAGCC 3′ | 721556 SmelpSymA |
| 38AtTl | 5′ ACCACCTCCAAAGCTTCTC 3′ | 160059 AtumpTA |
| 38AtTu | 5′ ATGCCGATCTGCTTGATGC 3′ | 159661 AtumpTA |
| RctAl | 5′ AGGAATACATATGACAAGCGCGATTAAAACGC 3′ | 161262 RetlpSym |
| RctAu | 5′ AAAGGATCCCACTAAAGGCCGAAAAATCAGTC 3′ | 160872 RetlpSym |
| 38Race | 5′ CAACGGATGGTCGAGGATCTC 3′ | 161182 RetlpSym |
| 37Race | 5′ CGGGATTGGAAAGGCATAGGA 3′ | 161487 RetlpSym |
Restriction sites are underlined; noncomplementary bases are in italics; noncomplementary bases used for mutation are in bold italics.
The location is indicated by the first 5′ nucleotide and the replicon where the sequence is located. RetlpSym, pSym of R. etli; SmelpSym, pSymA of S. meliloti; AtumpTA, plasmid A of A. tumefaciens.
Promoter mapping.
To map the transcriptional start sites of rctA and virB, 5-ml cultures of the appropriate strains expressing either rctA (CFN 2001 Tn5.C and CFN 2001 Tn5.C/p53rctA::Gus) or virB (strains CFN 2001 Tn5.2 and CFN 2001 Tn5.C/p53virB::Gus) were grown in PY medium and RNA was isolated using the High Pure RNA isolation kit (Roche, Nutley, NJ). Transcription initiation sites were mapped with a kit for the rapid amplification of cDNA 5′ ends (version 2.0; Invitrogen, Carlsbad, CA) using oligonucleotides 38Race and 37Race (Table 2) for rctA and virB, respectively. The products were sequenced to identify the transcription start sites. Promoter regions were predicted based on the R. etli promoter consensus (30).
Plasmid construction.
Fragment pVT, encompassing the whole regulatory region comprising the promoters of both rctA and the virB operon (see Fig. 1), was amplified using primers 38Tu and 38Tl and cloned into pCR2.1-TOPO, yielding plasmid pSSH01. The mutant virB promoters were constructed with overlapping mutagenic oligonucleotides (34) by using primer pairs 38Tu/rbml and 38Tl/rbmu for the pVT-derivative fragment pVT-RBM and 38Tu/10Ml and 38Tl/10Mu (Table 2) for the pVT-derivative fragment pVT-10m. Both fragments were cloned into pCR2.1-TOPO, generating plasmids pSSH02 and pSSH03, respectively.
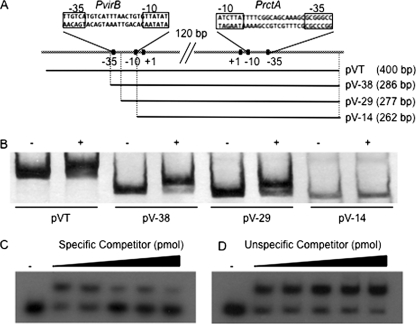
FIG. 1.
Specific binding of RctA to the virB promoter region. (A) Scheme of the mapped promoters of rctA and the virB operon and the regions used in EMSA and transcriptional fusions. Fragment pVT encompasses the whole regulatory region comprising the promoters of both rctA and the virB operon, while fragments pV-38, pV-29, and pV-14 are shortened derivatives of pVT (the final number in each fragment designation indicates the terminal nucleotide, with respect to the transcriptional start site of virB). (B) Results of EMSA using fragments depicted in panel A. The DNA concentration was adjusted for homogeneity with the concentration (measured as counts per minute) of the probe. −, RctA not added; +, RctA added. The DNA/RctA molar ratio was always 1/1. (C and D) Results of competitive EMSA using fragment pV-38. The probed DNA concentration in each lane was 1.5 pmol; the RctA concentration was 1.5 pmol; specific (C) and nonspecific (D) unlabeled probes were added in increasing concentrations (0, 0.5, 1, 1.5, and 3 pmol). −, RctA not added.
To construct the β-glucuronidase transcriptional fusions with the mutant virB promoters, plasmids pSSH02 and pSSH03 were cut with XbaI and KpnI. The resulting fragments were cloned separately into p53Gus restricted with XbaI-KpnI, generating plasmids p53virB-rbm::Gus and p53virB-10m::Gus. To construct transcriptional fusions of these fragments with the rctA promoter, we repeated the same procedure but using SpeI and XhoI, yielding plasmids p53rctA-rbm::Gus and p53rctA-10m::Gus.
To generate an amino-terminally His-tagged RctA derivative, the rctA coding sequence was amplified using primers RctAl and RctAu (Table 2), which contain custom-made NdeI and BamHI sites, respectively. After digestion with the appropriate enzymes, the PCR product was ligated into pET16b (40), which was cut similarly, giving rise to plasmid pSSH04. For introduction into R. etli, pSSH04 was digested with BamHI and ligated with BamHI-restricted pRK404 (11, 36) to yield pSSH05. All constructs were verified by DNA sequencing.
Overproduction and purification of RctA in E. coli.
For the overproduction of RctA, cells of E. coli BL21(DE3)/pLysS/pSSH04 were grown in 100 ml of Luria-Bertani medium at 30°C to an A620 of 0.4. At this point, 100 μM IPTG (isopropyl-β-D-thiogalactopyranoside) was added; cells were harvested 2 h later, and the cell pellet was resuspended in 5 ml of ice-cold extraction buffer (20 mM sodium phosphate, 0.5 M NaCl, pH 7.4). Cells were broken by three cycles of thawing and freezing, followed by three passages through a French press (Thermo Spectronic Instruments, Rochester, NY). The extract was centrifuged at 10°C for 10 min at 7,800 × g to obtain the cell-free fraction. To purify His-tagged RctA, a 1-ml Ni2+ affinity column (Pharmacia Biotech, Uppsala, Sweden) was equilibrated with extraction buffer containing 100 mM imidazole. Five milliliters of cell extract containing the His-tagged RctA was added to the column, the column was washed with the same buffer, and His-tagged RctA was batch eluted with extraction buffer containing 200 mM imidazole. Proteins were analyzed by sodium dodecyl sulfate-16.5% polyacrylamide gel electrophoresis as described previously (25, 35).
EMSA analyses.
DNA regions were amplified by PCR using the following oligonucleotide pairs: for R. etli CFN42 genomic DNA, 38Tu/38Tl (fragment pVT), Pv35u/38Tl (fragment pV-38), Pv10u/38Tl (fragment pV-29), and Pv1u/38Tl (fragment pV-14); for S. meliloti 1021 genomic DNA, 38SmTu/38SmTl (fragment pVT-Sm); for A. tumefaciens C58 genomic DNA, 38AtTu/38AtTl (fragment pVT-At); and for purified pSSH02 and pSSH03, 38Tu/38Tl (fragments pVT-RBM and pVT-10m, respectively). Products were electrophoresed on a 1.5% agarose gel and purified by band slicing (4). Fragments were 5′ end labeled with [γ-33P]ATP by using T4 polynucleotide kinase (USB Corporation, Cleveland, OH). Unincorporated ATP was removed by gel filtration using Centri-Sep spin columns (Applied Biosystems, Foster City, CA). Labeling efficiency was measured by liquid scintillation analysis using an LS6500 counter (Beckman Coulter, Fullerton, CA).
His-tagged RctA was incubated with the desired fragments for 30 min at room temperature in binding buffer (20 mM Tris-HCl [pH 8.5], 10% glycerol, 50 mM KCl, 3 mM MgCl2, 0.5 mg of bovine serum albumin/ml). For competition assays, the unlabeled fragment was added to the binding reaction mixture and the mixture was incubated for 10 min prior to the addition of the labeled fragment. Binding reaction mixtures were electrophoresed on a 6% TB-EDTA (Tris base, 40 mM; boric acid, 40 mM; EDTA, 1 mM)-polyacrylamide gel at 60 V for 1.5 h. The gel was dried on top of a Whatman filter paper and autoradiographed.
DNase I protection assay.
Fragment pVT was 32P labeled at the 5′ end of the bottom strand. A probe concentration equivalent to about 100,000 cpm was preincubated at room temperature with increasing concentrations of His-tagged RctA in the same binding buffer used for EMSA analyses. After 20 min, 0.003 U of DNase I (Roche, Nutley, NJ) in dilution buffer (8 mM Tris-HCl [pH 7.9], 40 mM MgSO4, 4 mM CaCl2, 40 mM KCl, 2 mM EDTA [pH 8.0], 24% glycerol) was added to the mixture and the mixture was incubated at room temperature for 2 min. The reaction was stopped by adding 300 μl of stop solution (570 mM ammonium acetate, 80% ethanol, 50 μg of carrier tRNA ml−1). The DNA was precipitated, dried, and dissolved in 8 μl of loading buffer (45 mM Tris-borate [pH 8.0], 1 mM EDTA, 80% formamide). Samples were denatured at 85°C for 5 min and resolved by electrophoresis through an 8% polyacrylamide sequencing gel. Gels were vacuum dried and visualized with a PhosphorImager (Molecular Dynamics). Sequencing reactions were included for size markers.
RESULTS
rctA and virB are transcribed from convergent promoters.
Given the close proximity of rctA and the virB operon, a prerequisite to understanding their relationship was to map their promoters. To do so, we identified the start sites for each transcriptional unit by nucleotide sequencing of the products obtained in assays for the rapid amplification of cDNA 5′ ends (see Materials and Methods). To enhance the sensitivity of these assays, we employed mRNA for which the corresponding gene was transcribed either in cis (from the promoter in the pSym) or in trans (from the promoter cloned in the plasmid p53Gus). The methods produced identical results (data not shown) and allowed us to determine the transcription initiation sites and to predict the location of the promoter for each gene. As shown in Fig. 1A, the genes are transcribed from promoters facing each other (i.e., convergent promoters): while the rctA promoter is located within the coding sequence of yhd0053 (between nucleotides 55 and 84 of the predicted coding sequence), the virB operon promoter lies in the intergenic region between rctA and yhd0053 (the first gene of the virB operon). The location of the virB promoter was verified by mutagenesis (see below), while the position of the rctA promoter was further confirmed by deletion analysis (see Fig. S1 in the supplemental material).
RctA binds specifically to the virB operon promoter.
As reported previously (28), RctA is predicted to have a winged-helix DNA binding domain (14, 32). This prediction suggests that the repressor activity of RctA may be due to direct binding to a regulatory region involved in the transcription of the virB operon. To test this hypothesis, we generated a His-tagged RctA derivative for use in EMSA (see Materials and Methods). To verify that this His-tagged derivative was functional in vivo, plasmid pSSH05 was introduced by conjugation into an R. etli rctA mutant derivative (see Materials and Methods) and the expression of both rctA and the virB operon was analyzed using the appropriate β-glucuronidase transcriptional fusions. As shown in Table 3, the His-tagged RctA derivative retained its biological activity, being able to complement an rctA mutant strain, as evidenced by the shutting off of the expression of the virB operon and the simultaneous activation of the transcription of rctA.
TABLE 3.
Activities of transcriptional fusions of rctA and virB promoters in different genetic backgrounds
| Strain (relevant genotype)a | Sp act (nmol min−1 mg of protein−1)b from the indicated promoter in β-glucuronidase fusion with:
|
|||||
|---|---|---|---|---|---|---|
| WT virB
|
virB- RBM
|
virB-10m
|
||||
| PvirB | PrctA | PvirB | PrctA | PvirB | PrctA | |
| CFN2001 Tn5.C (rctA+ rctB+) | 59 ± 9 | 240 ± 18 | 220 ± 72 | 39 ± 13 | ND | 215 ± 9 |
| CFN2001 Tn5.2 (rctA rctB+) | 257 ± 18 | 48 ± 15 | 237 ± 109 | 32 ± 22 | ND | 221 ± 16 |
| CFN2001 Tn5.6 (rctA+ rctB++) | 283 ± 58 | 67 ± 24 | 188 ± 56 | 29 ± 11 | ND | 229 ± 16 |
| CFN2001 Tn5.2/pSSH05 [rctA+(k) rctB+] | 87 ± 14 | 323 ± 55 | NA | NA | NA | NA |
| CFN2001 (rctA rctB) | 225 ± 57 | 33 ± 7 | 213 ± 68 | 55 ± 30 | ND | 220 ± 7 |
| CFN2001/pTErctA [rctA+(tr) rctB] | 2 ± 2 | 405 ± 100 | 247 ± 104 | 55 ± 30 | ND | 208 ± 24 |
| CFN2001/pTErctB [rctA rctB+(tr)] | 471 ± 51 | 4 ± 3 | 232 ± 77 | 24 ± 7 | ND | 230 ± 20 |
+, wild-type expression; ++, unregulated expression; (tr), expression from the trp promoter; (k) expression from the pET16B promoter.
Specific β-glucuronidase activities are expressed as means ± standard deviations of results from at least three independent experiments. WT virB, wild-type virB (on fragment pVT); virB-RBM, virB with a mutated RBM box (on fragment pVT-RBM); virB-10m, virB with a mutated −10 box (on fragment pVT-10m); ND, not determined; NA, not applicable.
The His-tagged RctA derivative (molecular mass, 15.34 kDa) was purified to homogeneity by Ni affinity chromatography and then used to set up EMSA with different radiolabeled fragments encompassing the putative regulatory regions of rctA and the virB operon (Fig. 1A). For these experiments, fragment pVT encompassed the whole regulatory region comprising both the rctA and virB promoters, while fragments pV-38, pV-29, and pV-14 were shortened derivatives of pVT (the final number in each fragment designation indicates the terminal nucleotide, with respect to the transcriptional start site of virB). As shown in Fig. 1B, fragments pVT, pV-38, and pV-29 clearly formed RctA-dependent, retarded complexes, while the migration of fragment pV-14 was unaffected upon RctA addition. The main difference between the smallest retarded fragment (pV-29) and pV-14 was the absence in the latter of 15 bp, spanning part of the spacer region between the −10 and −35 boxes of the virB operon promoter (Fig. 1A). Thus, this region of the virB operon promoter is critical for the binding of RctA to DNA.
To verify if the binding of RctA to fragment pV-38 was specific, competitive EMSA (see Materials and Methods) were set up. In these assays, when the binding of RctA to pV-38 was challenged by the prior addition of increasing amounts of unlabeled fragment pV-38 as a specific competitor, the amount of the retarded complex was reduced (Fig. 1C). In contrast, when an unlabeled competitor fragment from S. meliloti (a PCR product from nucleotides 1453158 to 1453424 of S. meliloti pSymB) was used, no decrease in the amount of retarded DNA complexes was seen (Fig. 1D).
These results clearly show that (i) RctA is able to bind specifically to DNA and (ii) a region located between the −10 and −35 regions of the virB promoter is needed for specific binding.
The binding of RctA depends on a conserved nucleotide sequence.
As reported previously, rctA homologues negatively control the conjugative transfer of plasmids pAtC58 of A. tumefaciens and pSymA of S. meliloti (28). These rctA homologues can functionally substitute for rctA from R. etli. Therefore, it was reasonable to expect that RctA from R. etli should recognize similar sequences in R. etli, A. tumefaciens, and S. meliloti. Aiming to identify the nucleotides recognized by RctA, we made an alignment of the putative promoters of the virB operons from these three species. This alignment revealed the presence of nine nearly invariable nucleotides between the −10 and the −35 boxes of the virB promoter (Fig. 2A).
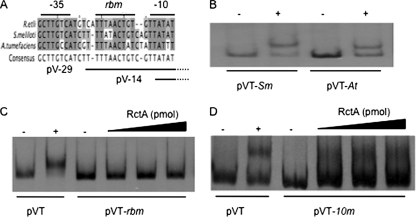
FIG. 2.
The binding of RctA requires an RBM box and an active −10 region. (A) Multiple-sequence alignment of the virB promoters of three rhizobiales showing the nucleotide conservation that delineates the RBM box. Solid horizontal lines below the alignment mark the limits of regions pV-29 and pV-14 with reference to the virB promoter. (B) Results of EMSA using fragments pVT-Sm and pVT-At; the DNA concentration was 1 pmol. −, RctA not added; +, 2 pmol of RctA added. (C and D) Results of EMSA using fragments pVT-RBM and pVT-10m, respectively. The DNA concentration in each lane was 1 pmol; RctA was added in increasing concentrations (0, 2, 4, and 8 pmol). −, RctA not added; +, 2 pmol of RctA added.
To verify that RctA of R. etli is able to bind to the virB promoter regions of S. meliloti and A. tumefaciens, we used specific oligonucleotides to amplify the equivalents of fragment pVT from each species (pVT-Sm and pVT-At, respectively) to use them in EMSA. As shown in Fig. 2B, retarded complexes with both fragments were found upon the addition of similar proportions of RctA from R. etli.
To demonstrate the role of the conserved nine base pairs in the binding of RctA to DNA, we constructed a mutant version of fragment pVT in which these nucleotides were changed from TTT AAC TGT to GGG CCA GTG, generating fragment pVT-RBM. When EMSA was performed with this fragment, RctA from R. etli was unable to bind, even upon the addition of an eightfold molar excess of RctA versus pVT-RBM (Fig. 2C). These results demonstrate that nucleotides within a conserved 9-bp sequence, termed the rctA binding motif (RBM) box, are required for the binding of RctA to DNA.
The binding of RctA protects a zone encompassing the RBM box and the −10 region.
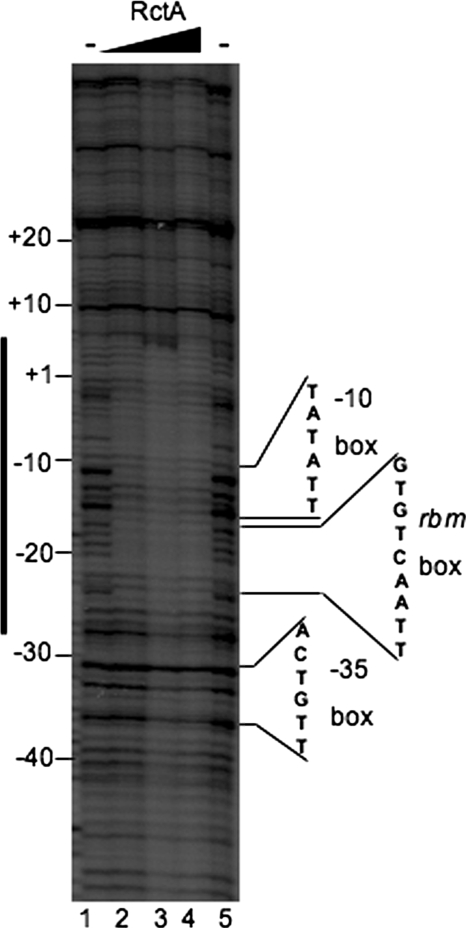
The identification of a motif in the spacer region of the virB promoter needed for the binding of RctA is fully consistent with the proposed role of this protein as a transcriptional repressor. To ascertain if the binding of RctA obliterates the access to other transcriptional elements, we performed a DNase I protection assay of the pVT fragment in the presence of increasing amounts of RctA (Fig. 3). Our results show a well-delineated protection zone in the virB promoter upon the addition of RctA, even at the lowest protein concentration tested. This region encompassed nucleotides −26 to +5 of the virB promoter region relative to the transcription start site and, as expected, included the RBM box (nucleotides −17 to −25). Interestingly, although the −35 box (nucleotides −32 to −37) was not within the protected region, the −10 box (nucleotides −10 to −15) of the promoter was significantly protected. This result further supports the idea that the interaction of RctA with the virB promoter may impair the transcription of the virB operon.
FIG. 3.
DNase I protection of the virB operon regulatory region by RctA. Increasing amounts of His-tagged RctA were mixed with a 32P-end-labeled DNA fragment corresponding to fragment pVT and treated with DNase I. Samples were subjected to electrophoresis on an 8% polyacrylamide sequencing gel. The −10 and −35 promoter sequences and the RBM box are indicated on the right; they were determined by running sequencing reactions with the same fragments in parallel (data not shown). The protected regions are indicated by a vertical black bar. The black triangle above the autoradiogram represents increasing amounts of RctA. Lanes: 1 and 5, DNA alone; 2 to 4, 2, 4, and 8 pmol of RctA, respectively. −, RctA not added.
Moreover, the finding that the −10 region was also protected in the presence of RctA opens up the possibility that this sector is also needed for the binding of RctA. To test this possibility, we constructed a pVT mutant fragment in which the −10 region was changed from TTA TAT to AGA CAT. This mutant fragment (pVT-10m) was then used for EMSA in the presence of various amounts of RctA. As shown in Fig. 2D, only a scarce amount of retarded complexes with the pVT-10m fragment was seen, even at an eightfold molar excess of RctA versus DNA. Interestingly, the amount of these retarded complexes was not increased when larger amounts of RctA were added, suggesting that these complexes were unstable in vitro. Thus, these results indicate that the binding of RctA to DNA requires elements located both in the RBM box and in the −10 region.
The binding of RctA to the virB promoter represses virB operon transcription.
Since fragment pVT harbors the promoters for both the rctA gene and the virB operon, the introduction of this fragment into a promoterless uidA reporter plasmid allows an evaluation of the expression of both promoters, depending on the orientation of the insert. To explore the functional consequences of the mutation in the RBM box for the expression of virB and rctA, we constructed two transcriptional fusions with the fragment pVT-RBM, one in the direction of the rctA promoter (p53rctA-rbm::Gus) and the other in direction of the virB operon promoter (p53virB-rbm::Gus).
The introduction of fusions with wild-type promoters into an otherwise wild-type background confirmed, as previously reported (28), low-level expression from the virB promoter but high-level expression from the rctA promoter (Table 3). In contrast, when RBM mutant fusions were introduced into a wild-type background, we found that the level of expression from the virB promoter was high but that expression from the rctA promoter was diminished (Table 3). The expression patterns obtained with these mutant fusions closely matched the one found with a wild-type-promoter fusion in an rctA mutant background (Table 3). In fact, the expression pattern seen for the RBM mutant fusions (a high expression level for the virB promoter and a low expression level for the rctA promoter) was maintained in backgrounds lacking rctA or overexpressing rctB (Table 3). Given the location of the RBM sequence and the inability of RBM mutant constructs to bind RctA, these results are fully consistent with the interpretation that the binding of RctA to the virB promoter represses the transcription of this operon.
The transcription of the virB operon interferes with rctA expression.
It has been reported previously (28) that mutations in rctA have the interesting effect of provoking a reduction of rctA expression (Table 3). This effect was also seen under conditions that conceivably interfered with RctA function, such as the overexpression of RctB (Table 3). These observations were explained by invoking the hypothesis of positive autoregulation for this gene (28). However, the convergent organization of the virB and rctA promoters, coupled with the presence of a single RctA binding site far from the rctA promoter, raises the alternative possibility that transcription from the virB promoter interferes with rctA expression. In this view, the loss of the repressor (as in an rctA mutant) or the blocking of its activity (as in a strain overexpressing rctB) should allow transcription from the virB promoter, which may structurally interfere with expression from the rctA promoter.
These two hypotheses (positive autoregulation and transcriptional interference) can be distinguished by studying the expression patterns of both rctA and virB genes in a mutant affected in the −10 box of the virB promoter. According to the positive-autoregulation hypothesis, the loss of virB expression should have no effect on rctA expression, which would remain high in a wild-type background or low in either an rctA mutant strain or a strain overexpressing rctB. In contrast, according to the transcriptional-interference hypothesis, the loss of transcription from the virB promoter would provoke a high level of constitutive transcription from the rctA promoter. To discern between these alternatives, we constructed two transcriptional fusions with the fragment pVT-10m, one in the direction of the rctA promoter (p53rctA-10m::Gus) and the other in the direction of the virB operon promoter (p53virB-10m::Gus). As shown in Table 3, virB expression was completely abolished when this mutant fragment was used. Notably, high-level constitutive expression of rctA from this mutant fusion was observed even in an rctA mutant and in a strain overexpressing rctB (Table 3). Interestingly, high-level constitutive expression of rctA was observed despite the fact that in the virB −10 box mutant fragment (pVT-10m), RctA binding was severely reduced (Fig. 2D). These observations are fully consistent with the expectations of the transcriptional-interference model.
To further substantiate this point, all the transcriptional fusions were introduced into genetic backgrounds lacking the pSym and, hence, both rctA and rctB (Table 3). In one of these strains, rctA was supplied on a separate plasmid under the control of the strong tryptophan promoter, while in another rctB was overexpressed (Table 3). Interestingly, the high-level constitutive expression of rctA from the virB −10 box mutant gene-rctA::Gus fusion was maintained even under circumstances in which rctA expression should have increased or decreased (Table 3) according to the positive-autoregulation hypothesis. Thus, these results clearly reveal that the expression of rctA is modulated by transcriptional interference emanating from the virB promoter.
DISCUSSION
In this work, we have provided direct evidence for the role of RctA as a transcriptional repressor of conjugational transfer genes in Rhizobium, based on structural and functional data. Using site-specific mutagenesis and EMSA, we have identified a 9-nucleotide motif in the spacer sequence of the virB operon promoter that is required for specific binding, which we have named the RBM box. As shown by DNase I footprinting assays, the binding of RctA protects a region that encompasses not only the RBM box, but also the −10 region of the virB promoter. This protection pattern fully explains the fivefold reduction in virB expression previously observed, since RctA binding should hinder the access of the RNA polymerase to the virB promoter. The binding of RctA to the virB promoter regions of both A. tumefaciens pAtC58 and S. meliloti pSymA, as demonstrated here, indicates that the regulatory characteristics described here should extend to these homologous systems (28).
Interestingly, although the RBM box is the main determinant for RctA binding, it is not the sole factor. As shown in Fig. 2D, the elimination of the promoter −10 box near the RBM box significantly reduced the binding of RctA. In this sense, the −10 box played an important role, albeit an ancillary one, in the binding of RctA. The requirement for a functional −10 box adjacent to the RBM box has important consequences for recognition. This requirement ensures that the binding of RctA should be targeted to active promoters. In this regard, it is germane to mention that two potential RBM boxes have been located, by sequence analysis, near the traCDG region. Only one of these boxes, the one that has a recognizable −10 box nearby, is bound by RctA (unpublished data).
A second important regulatory aspect that emerges from our data is the presence of transcriptional interference. As mentioned before, the convergent organization of the virB and rctA promoters generates the possibility of interference between them. Transcriptional interference has been defined as the suppressive influence of one transcriptional process, directly and in cis, on a second transcriptional process (37) and has been observed previously for artificially convergent promoters (21, 9) and bacteriophage promoters (7). As revealed by data from the transcriptional fusions, the rctA-virB region in R. etli shows all the hallmarks of transcriptional interference. When either RctA or the RBM box was absent, the transcription of the virB operon was activated, simultaneously reducing the transcription of the rctA promoter. Our data show that the reduction in rctA transcription was due most likely to transcriptional interference and not to autoregulation by RctA, as previously thought. Support for this conclusion comes from the fivefold increase in rctA transcriptional activity upon the elimination of transcription from the virB promoter. This effect was observed even in the absence of the whole pSym, thus ruling out any potential influence in trans as an explanation for this phenomenon.
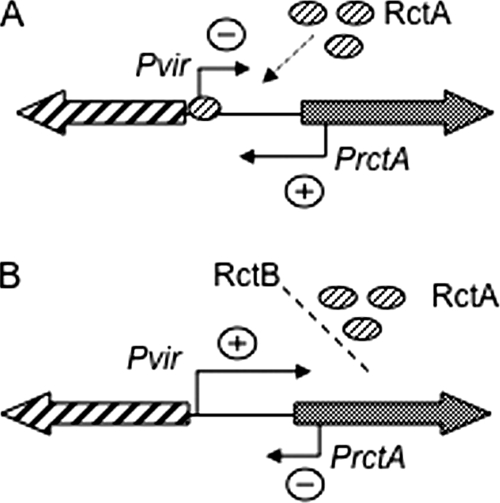
The finding of transcriptional interference adds another level for the regulation of this system. Under conditions that limit conjugative transfer, the expression of the virB operon is repressed by the binding of RctA to the virB promoter; this binding provokes high-level expression of rctA due to the lack of transcriptional interference, thus ensuring tight repression of the system (Fig. 4A). Whenever RctA binding is diminished, virB expression is activated, thus ensuring the establishment of transcriptional interference with rctA. This last effect warrants the full expression of the conjugative system (Fig. 4B).
FIG. 4.
Role of transcriptional interference in the regulation of the virB operon. (A) The virB operon promoter is blocked by the binding of RctA to the RBM box, repressing the expression of the promoter and allowing rctA transcription without interference. (B) RctB blocks RctA access to the RBM box, allowing virB operon transcription, which in turn interferes with rctA expression. +, activation; −, repression or inhibition.
A critical aspect of this system relates to the conditions that allow the elimination of repression by RctA. Thus far, the activation of conjugation has been seen upon the inactivation of rctA or the overexpression of rctB. There are two common mechanisms that regulate the conjugational transfer of automobilizable plasmids, quorum sensing (3, 6) and peptide signaling (3), but neither of them seems to participate in pSym transfer in the absence of pRet42a (unpublished results). Future work will be devoted to identifying environmental signals that preclude RctA functioning and the detailed role of rctB in this process.
Supplementary Material
Acknowledgments
We are indebted to José Luis Puente for critical and constructive discussions and Miguel Ángel Cevallos for useful scientific advice. We are grateful to Laura Cervantes and Javier Rivera for skillful technical assistance, to José Espíritu for computer support, to Verónica Martínez for help with the DNase I footprinting experiments, to Ana Laura Ramos for providing the nonspecific competitor PCR product, to Patricia Bustos, Rosa Isela Santamaría, and Jorge Yáñez for DNA sequencing, and to Paul Gaytán and Eugenio López for oligonucleotide synthesis.
Partial financial support was provided by grant IN226802 (Dirección General de Asuntos del Personal Académico, UNAM). E.S. was supported during the Ph.D. program (Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México) by scholarships from Consejo Nacional de Ciencia y Tecnología (México) and Dirección General de Estudios de Posgrado (UNAM).
Footnotes
Published ahead of print on 18 April 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K. C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 989883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 431359-1365. [DOI] [PubMed] [Google Scholar]
- 3.Beck von Bodman, S., G. T. Hayman, and S. K. Farrand. 1992. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc. Natl. Acad. Sci. USA 89643-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle, J. S., and A. M. Lew. 1995. An inexpensive alternative to glassmilk for DNA purification. Trends Genet. 118. [DOI] [PubMed] [Google Scholar]
- 5.Bravo, A., and J. Mora. 1988. Ammonium assimilation in Rhizobium phaseoli by the glutamine synthetase-glutamate synthase pathway. J. Bacteriol. 170980-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brom, S., L. Girard, C. Tun-Garrido, A. García-de los Santos, P. Bustos, V. González, and D. Romero. 2004. Transfer of the symbiotic plasmid of Rhizobium etli CFN42 requires cointegration with p42a, which may be mediated by site-specific recombination. J. Bacteriol. 1867538-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callen, B. P., K. E. Shearwin, and J. B. Egan. 2004. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol. Cell 14647-656. [DOI] [PubMed] [Google Scholar]
- 8.Corvera, A., D. Promé, J. C. Promé, E. Martínez-Romero, and D. Romero. 1999. The nolL gene from Rhizobium etli determines nodulation efficiency by mediating the acetylation of the fucosyl residue in the nodulation factor. Mol. Plant-Microbe Interact. 12236-246. [DOI] [PubMed] [Google Scholar]
- 9.Crampton, N., W. A. Bonass, J. Kirkham, C. Rivetti, and N. H. Thomson. 2006. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 345416-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danino, V. E., A. Wilkinson, A. Edwards, and J. A. Downie. 2003. Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol. Microbiol. 50511-525. [DOI] [PubMed] [Google Scholar]
- 11.Ditta, G., T. Schmidhauser, E. Yakobson, P. Lu, X. W. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13149-153. [DOI] [PubMed] [Google Scholar]
- 12.Eckhardt, T. 1978. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid 1584-588. [DOI] [PubMed] [Google Scholar]
- 13.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387394-401. [DOI] [PubMed] [Google Scholar]
- 14.Gajiwala, K. S., and S. K. Burley. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10110-116. [DOI] [PubMed] [Google Scholar]
- 15.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernández-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293668-672. [DOI] [PubMed] [Google Scholar]
- 16.Gomes-Barcellos, F., P. Menna, J. S. da Silva Batista, and M. Hungría. 2007. Evidence of horizontal transfer of symbiotic genes from a Bradyrhizobium japonicum inoculant strain to indigenous diazotrophs Sinorhizobium (Ensifer) fredii and Bradyrhizobium elkanii in a Brazilian Savannah soil. Appl. Environ. Microbiol. 732635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González, V., P. Bustos, M. A. Ramírez-Romero, A. Medrano-Soto, H. Salgado, I. Hernández-González, J. C. Hernández-Celis, V. Quintero, G. Moreno-Hagelsieb, L. Girard, O. Rodríguez, M. Flores, M. A. Cevallos, J. Collado-Vides, D. Romero, and G. Dávila. 2003. The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 4R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Göttfert, M., S. Rothlisberger, C. Kundig, C. Beck, R. Marty, and H. Hennecke. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 1831405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 20.He, X., W. Chang, D. L. Pierce, L. O. Seib, J. Wagner, and C. Fuqua. 2003. Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 185809-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horowitz, H., and T. Platt. 1982. Regulation of transcription from tandem and convergent promoters. Nucleic Acids Res. 105447-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7331-338. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9189-197. [DOI] [PubMed] [Google Scholar]
- 24.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 451-8. [DOI] [PubMed] [Google Scholar]
- 25.Nesterenko, M. V., M. Tilley, and S. J. Upton. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 28239-242. [DOI] [PubMed] [Google Scholar]
- 26.Noel, K. D., A. Sanchez, L. Fernández, J. Leemans, and M. A. Cevallos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez-Mendoza, D., A. Domínguez-Ferreras, S. Muñoz, M. J. Soto, J. Olivares, S. Brom, L. Girard, J. A. Herrera-Cervera, and J. Sanjuan. 2004. Identification of functional mob regions in Rhizobium etli: evidence for self-transmissibility of the symbiotic plasmid pRetCFN42d. J. Bacteriol. 1865753-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Mendoza, D., E. Sepúlveda, V. Pando, S. Muñoz, J. Nogales, J. Olivares, M. J. Soto, J. A. Herrera-Cervera, D. Romero, S. Brom, and J. Sanjuan. 2005. Identification of the rctA gene, which is required for repression of conjugative transfer of rhizobial symbiotic megaplasmids. J. Bacteriol. 1877341-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez-Mendoza, D., M. Lucas, S. Muñoz, J. A. Herrera-Cervera, J. Olivares, F. de la Cruz, and J. Sanjuan. 2006. The relaxase of the Rhizobium etli symbiotic plasmid shows nic site cis-acting preference. J. Bacteriol. 1887488-7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramírez-Romero, M. A., I. Masulis, M. A. Cevallos, V. González, and G. Dávila. 2006. The Rhizobium etli sigma70 (SigA) factor recognizes a lax consensus promoter. Nucleic Acids Res. 341470-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsay, J. P., J. T. Sullivan, G. S. Stuart, I. L. Lamont, and C. W. Ronson. 2006. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol. Microbiol. 62723-734. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, V. A., D. A. Case, and V. Tsui. 2004. Predicting interactions of winged-helix transcription factors with DNA. Proteins 57172-187. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Santoyo, G., J. M. Martínez-Salazar, C. Rodríguez, and D. Romero. 2005. Gene conversion tracts associated with crossovers in Rhizobium etli. J. Bacteriol. 1874116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 36.Scott, H. N., P. D. Laible, and D. K. Hanson. 2003. Sequences of versatile broad-host-range vectors of the RK2 family. Plasmid 5074-79. [DOI] [PubMed] [Google Scholar]
- 37.Shearwin, K. E., B. P. Callen, and J. B. Egan. 2005. Transcriptional interference—a crash course. Trends Genet. 21339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva, C., P. Vinuesa, L. E. Eguiarte, E. Martinez-Romero, and V. Souza. 2003. Rhizobium etli and Rhizobium gallicum nodulate common bean (Phaseolus vulgaris) in a traditionally managed milpa plot in Mexico: population genetics and biogeographic implications. Appl. Environ. Microbiol. 69884-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol. Gen. Genet. 196413-420. [DOI] [PubMed] [Google Scholar]
- 40.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 18560-89. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan, J. T., H. N. Patrick, W. L. Lowther, D. B. Scott, and C. W. Ronson. 1995. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. USA 928985-8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 955145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan, J. T., J. R. Trzebiatowski, R. W. Cruickshank, J. Gouzy, S. D. Brown, R. M. Elliot, D. J. Fleetwood, N. G. McCallum, U. Rossbach, G. S. Stuart, J. E. Weaver, R. J. Webby, F. J. de Bruijn, and C. W. Ronson. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 1843086-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tun-Garrido, C., P. Bustos, V. González, and S. Brom. 2003. Conjugative transfer of p42a from Rhizobium etli CFN42, which is required for mobilization of the symbiotic plasmid, is regulated by quorum sensing. J. Bacteriol. 1851681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wernegreen, J. J., and M. A. Riley. 1999. Comparison of the evolutionary dynamics of symbiotic and housekeeping loci: a case for the genetic coherence of rhizobial lineages. Mol. Biol. Evol. 1698-113. [DOI] [PubMed] [Google Scholar]
- 46.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 2942317-2323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.