Abstract
Lon protease, a member of the ATP-dependent protease family, regulates numerous cellular systems by degrading specific substrates. Here, we demonstrate that Lon is involved in the regulation of quorum-sensing (QS) signaling systems in Pseudomonas aeruginosa, an opportunistic human pathogen. The organism has two acyl-homoserine lactone (HSL)-mediated QS systems, LasR/LasI and RhlR/RhlI. Many reports have demonstrated that these two systems are regulated and interconnected by global regulators. We found that lon-disrupted cells overproduce pyocyanin, the biosynthesis of which depends on the RhlR/RhlI system, and show increased levels of a transcriptional regulator, RhlR. The QS systems are organized hierarchically: the RhlR/RhlI system is subordinate to LasR/LasI. To elucidate the mechanism by which Lon negatively regulates RhlR/RhlI, we examined the effect of lon disruption on the LasR/LasI system. We found that Lon represses the expression of LasR/LasI by degrading LasI, an HSL synthase, leading to negative regulation of the RhlR/RhlI system. RhlR/RhlI was also shown to be regulated by Lon independently of LasR/LasI via regulation of RhlI, an HSL synthase. In view of these findings, it is suggested that Lon protease is a powerful negative regulator of both HSL-mediated QS systems in P. aeruginosa.
Proteolysis in bacteria is important for maintaining cellular homeostasis. Most intracellular proteolysis is initiated by ATP-dependent proteases, including Lon, FtsH, ClpAP, ClpXP, and HslVU, which have been identified in Escherichia coli (8, 11, 18). These proteases harbor a domain conferring ATPase activity, characteristic of the ATPase family associated with diverse cellular activities (AAA+) (16). They are involved in protein quality control by degrading misfolded and denatured proteins. Among them, Lon and ClpXP are responsible for 70 to 80% of the energy-dependent degradation of proteins in vivo (22, 27). Furthermore, they perform important regulatory functions in bacterial cells by controlling the availability of critical regulatory proteins.
Lon, first identified in E. coli, has been shown to degrade specific regulatory proteins involved in a variety of biological processes: SulA, which regulates cell division (29); RcsA, a transcriptional activator for capsule synthesis (41); λ N, which antagonizes the termination of early transcription of λ DNA (26); and CcdA, the antitoxin of the ccd postsegregational killing system carried by the F plasmid (42). Lon has been shown to regulate virulence factors in some pathogenic bacteria. For instance, it specifically degrades HilC and HilD, which are transcriptional regulators for the expression of Salmonella pathogenicity island 1 in Salmonella enterica serovar Typhimurium (38). It recognizes YmoA, which regulates the Yop regulon in Yersinia pestis (17). Furthermore, it is involved in degrading HrpR, which regulates the expression of the Hrp system in Pseudomonas syringae (4). Proteolysis of these regulators by Lon is important for pathogenesis. In fact, we have previously demonstrated that it is essential for systemic infection with serovar Typhimurium in mice (39).
Pseudomonas aeruginosa is an opportunistic human pathogen that tends to infect individuals with cystic fibrosis or immunocompromised patients, such as those suffering burns or undergoing cytotoxic chemotherapy (24). A variety of virulence factors have been reported in P. aeruginosa, e.g., proteases, including elastase, alkaline protease, LasA protease, protease IV, and membrane protease (25). They interfere with the host immune response by degrading target cell components. In addition, extracellular virulence factors, such as pyocyanin, exotoxin, hemolycin, and rhamnolipids, are involved in the expression of P. aeruginosa virulence (5). In most cases, synthesis of these factors is controlled by a quorum-sensing (QS) system.
A QS system is a regulatory mechanism that allows bacteria to monitor their population size by responding to the extracellular concentration of a signal molecule. P. aeruginosa has two QS systems mediated by acyl-homoserine lactone (HSL) as the signal molecule. They are encoded by the lasR-lasI and rhlR-rhlI gene pairs. lasI and rhlI encode HSL synthases (LasI and RhlI), which are responsible for the synthesis of 3-oxo-C12-HSL and C4-HSL, respectively. lasR and rhlR encode the transcriptional activators (LasR and RhlR) that respond to their cognate signal molecules and activate transcription of lasI and rhlI, respectively. The two systems function sequentially: the RhlR/RhlI system is subordinate to the LasR/LasI system, because rhlI and rhlR are among the genes activated by LasR and LasI. The QS systems control over 200 genes, including those for the pathogenesis of P. aeruginosa infections (36, 44). The P. aeruginosa QS system is connected in complicated ways with other cellular regulatory networks. For instance, it is regulated by functions such as Vfr (1), GacA (32), RpoS (46), and RpoN (40).
In this study, we provide the first evidence that the Lon protease of P. aeruginosa is involved in negative regulation of the LasR/LasI and RhlR/RhlI QS systems. We identified a lon homologue in P. aeruginosa PAO1 and constructed a lon insertion mutant and characterized it. By these means, we have found that lon disruption results in LasR/LasI-dependent activation of the RhlR/RhlI system. We demonstrate that Lon regulates the expression of LasR/LasI by posttranslational control of LasI. It also regulates the RhlR level independently of LasR/LasI. We also suggest that Lon is involved in the regulation of RhlR through modulation of RhlI.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1 and Table 2, respectively. Bacteria were routinely grown in L broth (1% Bacto tryptone [Difco]-0.5% Bacto yeast extract [Difco]-0.5% sodium chloride, pH 7.4) and L agar at 37°C. When necessary, the medium was supplemented with gentamicin (25 μg ml−1), carbenicillin (100 μg ml−1), or ampicillin (25 μg ml−1).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant propertiesa | Reference or source |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Prototroph | 15 |
| CS9008 | Δlon::Gm in PAO1 | This study |
| CS9011 | ΔrhlR in PAO1 | This study |
| CS9013 | ΔlasR in PAO1 | This study |
| CS9027 | Δlon::Gm in CS9013 | This study |
| CS9028 | Δlon::Gm in CS9011 | This study |
| CS9038 | ΔlasI in PAO1 | This study |
| CS9044 | ΔrhlI in PAO1 | This study |
| CS9051 | ΔrhlI in CS9013 | This study |
| CS9053 | CS9008 harboring pTKY805 | This study |
| CS9062 | Δlon::Gm in CS9051 | This study |
| E. coli | ||
| CS5485 | DH5αZ1 harboring pTKY715 | This study |
| CS5487 | DH5αZ1 harboring pTKY714 | This study |
| DH5αZ1 | DH5α lacIq | Our collection |
| SM10(λpir) | thri thr leu tonA lacY supE recA::RP4-2Tc::MuKm λpir | Our collection |
| C. violaceum | ||
| CV026 | Mini-Tn5 mutant of ATCC 31532 | 23 |
Gm, gentamicin resistance.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant propertiesa | Reference or source |
|---|---|---|
| pTKY613 | pUC18 with 1,659-bp fragment containing a part of lon | This study |
| pTKY614 | pTKY613 carrying Δlon::Gm | This study |
| pTKY713 | pEX18 with HindIII-EcoRI fragment containing disrupted lon gene | This study |
| pTKY714 | pUHE212-1 with 788-bp rhlR fragment | This study |
| pTKY715 | pUHE212-1 with 719-bp rhlI fragment | This study |
| pTKY762 | pTKY714 carrying 574-bp rhlR NruI-NruI region-disrupted gene | This study |
| pTKY763 | pEX18 with EcoRI-HindIII fragment containing disrupted rhlR gene | This study |
| pTKY764 | pHSG399 with 1,936-bp fragment containing lasR gene | This study |
| pTKY765 | pTKY764 carrying 1,528-bp lasR PstI-PstI region-disrupted gene | This study |
| pTKY766 | pEX18 with EcoRI-HindIII fragment containing disrupted lasR gene | This study |
| pTKY788 | pHSG399 with 1,080-bp fragment containing lasI gene | This study |
| pTKY789 | pTKY788 carrying 942-bp lasI BssHII-BssHII region-disrupted gene | This study |
| pTKY790 | pEX18 with BamHI-HindIII fragment containing disrupted lasI gene | This study |
| pTKY791 | pTKY715 carrying 572-bp rhlI PvuII-NruI region-disrupted gene | This study |
| pTKY792 | pEX18 with BamHI-HindIII fragment containing disrupted rhlI gene | This study |
| pTKY805 | pME6032 with MluI-EcoRI fragment carrying the lon region including its own promoter | |
| pEX18 | Gene replacement vector with MCS from pUC18 | 14 |
| pHSG399 | Cloning vector | Our collection |
| pME6032 | Cloning vector | 30 |
| pMS255 | Gm cassette | 2 |
| pUC18 | Cloning vector | Our collection |
| pUHE212-1 | N-terminal His tag vector | 10 |
Gm, gentamicin resistance.
Construction of plasmids.
Plasmids pTKY714 and pTKY715, encoding N-terminally His-tagged rhlR and N-terminally His-tagged rhlI, respectively, were constructed by amplifying a BglII-HindIII fragment carrying rhlR with the primers His-rhlRF (5′-GGGGAAGCTTAAAGGAGGATGGAC-3′) and His-rhlRR (5′-GGCTTAGATCATGAGGATG-3′) and a BamHI-HindIII fragment carrying rhlI with the primers His-rhlIF (5′-GACGGATCCATGATCAATGC-3′) and His-rhlIR (5′-TCGGAAGCTTCGGCCAAATCC-3′) by PCR, after which the fragment was cloned into pUHE212-1.
Plasmid pTKY805 for complementation analysis of lon was constructed by cloning in pME6032 a 2,676-bp PCR fragment, bp −196 to +2480 relative to the lon translational start site.
Construction of Δlon::Gm, ΔrhlR, Δlon::Gm ΔrhlR, ΔlasR, Δlon::Gm ΔlasR, ΔlasI, ΔrhlI, ΔlasI ΔrhlI, and Δlon::Gm ΔlasI ΔrhlI mutants.
To construct the ΔrhlR mutant CS9011, the plasmid pTKY714 was cleaved at the two NruI sites to eliminate the 214-bp central fragment of rhlR. The resultant plasmid, pTKY762, was cleaved at the EcoRI and HindIII sites in the vector, and the rhlR-disrupted fragment was ligated to the vector pEX18, a transferable suicide vector (14), yielding pTKY763. This plasmid was then used in a marker exchange experiment in P. aeruginosa PAO1.
The ΔlasR mutant CS9013 was constructed as follows. We generated the plasmid pTKY764 by cloning in pHSG399 a 1,936-bp PCR fragment flanking bp −622 to +1215 relative to the lasR translational start site. This plasmid was cleaved at two PstI sites to eliminate the 408-bp central fragment of lasR, generating pTKY765. The lasR-disrupted fragment in this plasmid was ligated to the vector pEX18. The resultant plasmid, pTKY766, was used in a marker replacement experiment in strain PAO1.
The ΔlasI mutant CS9038 was constructed as follows. The plasmid pTKY788 was generated by cloning in pHSG399 a 1,080-bp PCR fragment, bp −99 to +981 relative to the lasI translational start site. This plasmid was cleaved at two BssHII sites to eliminate the 138-bp central fragment of lasI, yielding the plasmid pTKY789. The lasI-disrupted fragment in this plasmid was ligated to the vector pEX18. The resultant plasmid, pTKY790, was used in a marker exchange experiment in strain PAO1.
The ΔrhlI mutant CS9044 was constructed as follows. The plasmid pTKY715 was cleaved at PvuII and NruI sites to eliminate the 147-bp central fragment of rhlI, yielding the plasmid pTKY791. The rhlI-disrupted fragment in this plasmid was ligated to the vector pEX18. The resultant plasmid, pTKY792, was used in a gene replacement experiment in strain PAO1.
To construct the ΔrhlI ΔlasR double mutant CS9051, the plasmid pTKY792 was mobilized into the strain CS9013 (ΔlasR). The transconjugants were selected by resistance to carbenicillin based on a single-crossover event. A double-crossover event in the ΔlasR mutant was then assessed by its resistance to sucrose and sensitivity to carbenicillin.
To construct the Δlon::Gm (CS9008), Δlon::Gm ΔlasR (CS9027), Δlon::Gm ΔrhlR (CS9028), and Δlon::Gm ΔlasR ΔrhlI (CS9062) mutants, the DNA fragment between nucleotides 814 and 2484 in the lon coding region was initially amplified from the chromosome of strain PAO1 by PCR. The 1,659-bp fragment generated was cleaved with HindIII at the 5′ end and EcoRI at the 3′ end and then cloned into the vector pUC18. The resultant plasmid, pTKY613, was cleaved at the two HincII sites to eliminate the 370-bp central fragment of lon and then ligated to the fragment encoding the gentamicin resistance gene generated from SmaI-digested pMS255. The Δlon::Gm fragment obtained by digestion of the resultant plasmid, pTKY614, with HindIII and EcoRI was ligated to the vector pEX18. The resultant plasmid, pTKY713, was used to replace Δlon::Gm in strains PAO1, CS9011, CS9013, and CS9051.
Generation of the anti-RhlR, anti-RhlI and anti-LasI antisera.
To purify the N-terminally His-tagged RhlR and His-tagged RhlI, which had formed inclusion bodies, 3-liter cultures of the E. coli DH5αZ1 derivative harboring pTKY714 or pTKY715 were incubated at 37°C until the cell density reached an optical density at 600 nm (OD600) of 0.8. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM for 3 h, and the cells were collected by centrifugation. After one freeze-thaw cycle, the wet cell pastes were resuspended in lysis buffer (100 mM NaH2PO4, 10 mM Tris base, 8 M urea, pH 8.0) and sonicated. After centrifugation, the pellets were washed twice with 1% Triton X-100 and once with 0.1% sodium dodecyl sulfate (SDS). After further centrifugation, the precipitates were suspended in SDS sample buffer, following by SDS-polyacrylamide gel electrophoresis (PAGE). The eluates from the gel corresponding to each protein band were used to immunize a rabbit. To generate anti-LasI antiserum, the purified LasIΔG protein (12) (provided by M. Churchill) was used to immunize a rabbit. The resultant antisera were verified by reaction with the purified antigen proteins.
Comparative analysis of C4-HSL.
The detection of C4-HSL was based on the method of Shaw et al. (37). Bacterial cells were grown in L broth to stationary phase at 37°C and removed by centrifugation. Samples (80 ml) of the supernatants were filtered through a Millex-GV filter (Millipore) and extracted twice with 50 ml of ethyl acetate. HSL developed on the TLC plate was detected using an indicator bacterium, Chromobacterium violaceum mutant CV026, by the method of Latifi et al. (23).
SDS-PAGE and immunoblotting.
Gel electrophoresis was carried out according to the method of Laemmli (21) using 15% SDS-polyacrylamide gels. The separated proteins were transferred onto Immun-BlotPVDF membranes (Bio-Rad) and then incubated with anti-E.coli Lon serum (1:12,500), anti-Pseudomonas LasI and anti-RhlR sera (1:25,000), or anti-Pseudomonas RhlR serum (1:25,000), followed by alkaline phosphatase-conjugated anti-rabbit immunoglobulin G. The enzymatic reactions were performed in the presence of 0.3 mg ml−1 lysozyme nitroblue tetrazolium (Wako) and 0.15 mg ml−1 5-bromo-4-chloro-3-indolylphosphate (Sigma).
Pulse-labeling and coimmunoprecipitation.
Bacterial cells of strains PAO1 and CS9008 were grown in L broth at 37°C until the cell density reached an OD600 of 0.5. The cells were collected by centrifugation and were resuspended in M9 medium supplemented with 0.25% glucose, 40 μg ml−1 (each) of 18 amino acids (methionine and cysteine were excluded), and 2 μg ml−1 thiamine. They were then incubated at 37°C for 30 min, labeled with 3.7 Mbq ml−1 [35S]Met and [35S]Cys (Protein Labeling Mix; >37 Tbq mmol−1; GE Health Science) for 1 min, and chased with 200 μg ml−1 unlabeled methionine and cysteine. Aliquots of the cells were taken at appropriate intervals and mixed with trichloroacetic acid (final concentration, 5%), chilled on ice for 15 min, and centrifuged at 16,000 × g for 2 min. The pellets were washed with acetone and resuspended in 100 μl of SDS buffer (10% SDS, 1 mM EDTA, 50 mM Tris-HCl, pH 8.0). One milliliter of RIPA buffer (1% Triton X-100, 1% Na-deoxycholate, 0.1% SDS, 0.15 M NaCl, 50 mM Tris-HCl, pH 8.0, 0.1 mM EDTA) was added to each resuspended pellet, and the mixture was centrifuged. The supernatants were used for immunoprecipitation with anti-LasI and anti-RhlR sera by incubation overnight on ice. To collect the immunocomplexes, protein A-Sepharose beads (GE Health Science) were added, and the mixtures were incubated for 1 h at 4°C. After centrifugation, the pellets were washed twice with RIPA buffer and once with 10 mM Tris-HCl, pH 8.0, and finally dissolved in 40 μl of SDS sample buffer (21). A portion of each sample was analyzed by 15% SDS-PAGE. The radioactivity incorporated into LasI was visualized using Molecular Imager FX (Bio-Rad).
RNA extraction and quantitative, real-time reverse transcription (RT)-PCR.
Bacterial cells were grown in L broth at 37°C to mid-exponential phase (OD600, 1.0 to 1.4) and for 24 h. A 0.4-ml aliquot of these cultures was added to 0.8 ml of RNAprotect bacterial reagent (Qiagen). Total RNA was isolated by using an RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Residual DNA was removed from the RNase-Free DNase Set (Qiagen).
Differential expression of genes was examined by quantitative, real-time RT-PCR using a QuantiTect Probe RT-PCR kit (Qiagen) according to the manufacturer's instructions with TaqMan probes and primer pairs designed with the Primer Express software package (ABI). The following probe and primer sequences were used: for lasI, sense primer, 5′-GCCCCTACATGCTGAAGAACA-3′, antisense primer, 5′-CGAGCAAGGCGCTTCCT-3′, and probe 5′-(6-carboxyfluorescein [FAM])CTTCCCGGAGCTTCTGCACGGC(6-carboxytetramethylrhodamine [TAMRA])-3′; for rhlI, sense primer, 5′-GCAGCTGGCGATGAAGATATTC-3′, antisense primer, 5′-CGAACGAAATAGCGCTCCAT-3′, and probe, 5′-(FAM)AGCCTGCAATGCGCCTGGTACCT(TAMRA)-3′; for rhlR, sense primer, 5′-AACGCGAGATCCTGCAATG-3′, antisense primer, 5′-GCGCGTCGAACTTCTTCTG-3′, and probe 5′-(FAM)TGAGCATCTCCGAGAGCACGGT(TAMRA)-3′; and for rplU, sense primer, 5′-TCACCGAAGGCGAATTCCT-3′, antisense primer, 5′-TTCACGTCTTCGCCATTGG-3′, and probe, 5′-(VIC)ATTTCGACCGCGTCCTGCTGGTT(TAMRA)-3′. To check for residual contaminating genomic DNA, control reactions without the reverse transcriptase mixture were analyzed in the same way. Prior to comparative analysis, the relative efficiency of each probe and primer pair was tested and compared with that of the probe and primer pair for ribosomal rplU (referred to as the normalizer gene) to ensure that the threshold cycle (CT) data analysis approach could be employed. The absolute value of the slope of the log input amount versus ΔCT was less than 0.1 for all comparisons, which allowed us to use the ΔΔCT calculation to determine the relative levels of gene expression in all experimental cultures compared to the levels in controls. All reactions were performed in duplicate with an Mx3000P QPCR system (Stratagene), and the experiments were replicated at least three times.
RESULTS
Construction of a lon-disrupted mutant of P. aeruginosa PAO1.
DNA sequence analysis of the lon region of P. aeruginosa strain PAO1 (GenBank accession number NC_002516) showed that a lon homologue encodes a protein with 69.6% identity to the Lon protein of E. coli. The open reading frame for Lon is 2,397 bp long and encodes a peptide of 798 amino acid residues with a predicted molecular mass of 88.6 kDa, which is larger than E. coli Lon (784 amino acids; 87.4 kDa). The E. coli Lon subunit has been shown to comprise three functional domains (35): the N-terminal domain (N; also referred to as LAN), which interacts selectively with target proteins (16); the central ATPase (the AAA+ module, or A domain); and the C-terminal proteolytic (P) domain. The deduced amino acid sequence of P. aeruginosa Lon shows potential consensus regions in the A and P domains.
A genetically defined lon mutant was constructed by inserting a gentamicin resistance gene into lon. The disruption in the resultant mutant strain, CS9008, was confirmed by immunoblotting; an antiserum specific for the E. coli Lon protein failed to react with a cell lysate from the mutant strain, whereas a band corresponding to approximately 89 kDa was detected in a cell lysate from the wild-type strain (data not shown). Disruption of the lon gene did not affect the increase in OD used to monitor bacterial growth (data not shown). However, the numbers of viable cells were always lower for the lon-disrupted mutant than for the wild-type cells. The lon-disrupted mutant exhibited a long-filament phenotype. Because filamentous cells have larger volumes than normal cells, the lon-disrupted cells gave ODs similar to those of the wild-type cells despite their lower viability.
During further phenotypic characterization of the lon-disrupted mutant, we found that the mutant overproduced a blue-green phenazine pigment, pyocyanin. At 24 h of incubation, the level of pyocyanin in the lon-disrupted cells was 4.4-fold higher than in the wild-type cells (data not shown). Pyocyanin is a synthesized by-product of the phzA1-phzG1 operon, expression of which is under the control of the RhlR/RhlI circuit of the QS system. Many genes are regulated by the RhlR/RhlI system. One of them, rpoS, was also more highly expressed in the lon-disrupted cells than in the wild-type cells (data not shown). These findings suggest that Lon protease may be involved in the expression of the RhlR/RhlI circuit.
Effect of lon disruption on the RhlR/RhlI system.
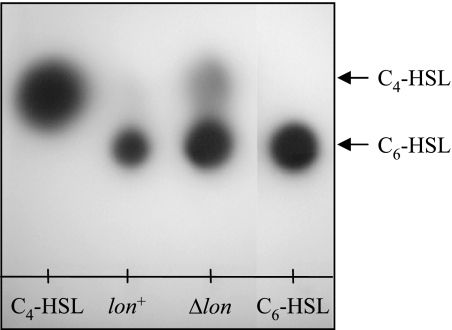
To determine whether Lon protease is involved in regulating the RhlR/RhlI system, we initially compared C4-HSL production directed by RhlI in the wild-type and lon-disrupted mutant strains. The lactones were extracted with ethyl acetate and detected by a bioassay that depended on the induction of violacein in the C. violaceum mutant CV026 (47). Besides C4-HSL, P. aeruginosa produces C6-HSL, which is also synthesized by RhlI and is involved in the induction of violacein in CV026 (47). These molecules were separated by C18 reversed-phase thin-layer chromatography. Whereas, only a trace of C4-HSL was detected in the wild-type strain, the amount of C4-HSL was dramatically increased in the lon-disrupted mutant (Fig. 1); also, more C6-HSL was detected.
FIG. 1.
Effect of lon disruption on C4-HSL production by P. aeruginosa. The extracts from the supernatants of cultures of bacterial strains PAO1 (lon+) and CS9008 (Δlon) were applied to C18 reversed-phase thin-layer plates and then developed with methanol/water (60:40 [vol/vol]). The spots were visualized with the C. violaceum reporter strain. C4-HSL (0.125 μmol) and C6-HSL (0.0125 μmol) were applied as HSL standards.
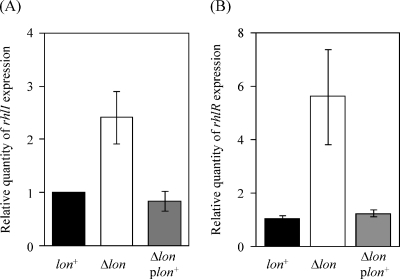
To determine the levels of RhlI in both the wild-type and lon-disrupted mutant cells, we initially constructed a plasmid encoding His-tagged RhlI, isolated the His-tagged protein, and used it to immunize rabbits as described in Materials and Methods. The anti-RhlI serum was verified by reaction with the purified RhlI proteins, but it could not detect the cellular level of RhlI in either the wild-type or the lon-disrupted cells, suggesting that the protein is short-lived and/or present in small amounts. Therefore, we decided to test the effect of the lon disruption on the level of the rhlI transcript by quantitative RT-PCR (Fig. 2A). The level of rhlI mRNA was approximately 2.5-fold higher in the lon-disrupted cells than in the lon+ cells. The increase in expression of the rhlI gene by lon disruption was fully suppressed by providing a functional copy of lon in trans, suggesting that Lon is involved in the expression of rhlI. The effect of lon disruption on the expression of rhlR was also examined by measuring the rhlR transcript level (Fig. 2B). The lon disruption increased rhlR transcription fourfold, and this increase was fully suppressed by lon. These results suggest that Lon is involved in the expression of both rhlI and rhlR.
FIG. 2.
Expression of rhlI (A) and rhlR (B) in wild-type and lon-disrupted cells. Total RNAs were prepared from strains PAO1 (lon+), CS9008 (Δlon), and CS9053 (Δlon plon+) grown in L broth for 24 h 37°C. The levels of rhlI and rhlR transcripts were measured by quantitative, real-time RT-PCR and then normalized to rplU expression. The values represent the means and standard deviations of changes in comparison with the transcription level in PAO1.
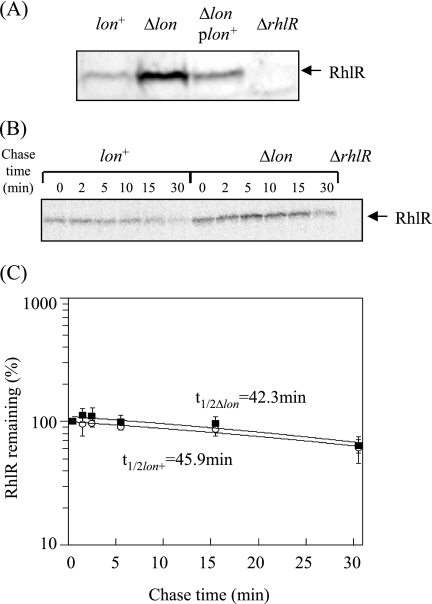
Using the anti-RhlR antiserum established in the present study, the levels of RhlR were determined in both wild-type and lon-disrupted cells (Fig. 3 A). The apparent levels of RhlR are consistent with the transcriptional-analysis results. That is, there was more RhlR in lon-disrupted cells than that in lon+ cells, and the accumulation of RhlR in the lon-disrupted cells was suppressed by transcomplementation with functional lon. A previous report demonstrated that E. coli Clp and Lon proteases are responsible for degrading apo-TraR, a transcriptional activator of the QS system in Agrobacterium tumefaciens (48). RhlR and TraR belong to the LuxR-like protein family. Therefore, we examined whether Lon protease degrades RhlR by determining the in vivo half-life of RhlR protein by pulse-labeling and chasing, followed by immunoprecipitation using the anti-RhlR antiserum. As shown in Fig. 3B and C, there was no significant difference between the half-lives of RhlR proteins in wild-type and lon-disrupted cells. This excluded the possibility that Lon may degrade the RhlR protein.
FIG. 3.
Cellular levels and in vivo stabilities of RhlR. (A) Whole-cell extracts were prepared from strains PAO1 (lon+ rhlR+), CS9008 (Δlon rhlR+), CS9011 (lon+ ΔrhlR), and CS9053 (Δlon plon+) grown in L broth for 24 h at 37°C and then separated on 15% SDS-polyacrylamide gels. The separated proteins were transferred to polyvinylidene difluoride membranes and then immunostained with anti-RhlR serum. (B) Cells of strains PAO1 (lon+) and CS9008 (Δlon) were grown to exponential phase. They were pulse-labeled with [35S]methionine and [35S]cysteine for 1 min at 37°C and chased with unlabeled methionine and cysteine. Samples were taken at the times indicated, followed by immunoprecipitation of RhlR. (C) Quantification of the precipitated RhlR protein relative to the value at 1 min. Mean values and standard deviations of at least three independent experiments are given.
Lon regulates the RhlR/RhlI system by a LasR/LasI-dependent pathway.
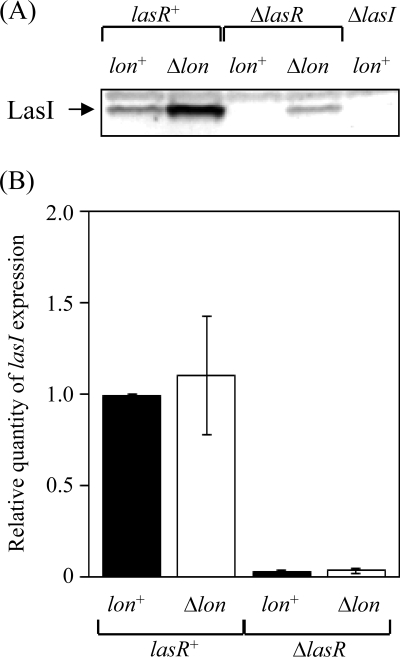
Regulation of the RhlR/RhlI system is intimately and hierarchically connected to the LasR/LasI system. The RhlR/RhlI system is subordinate to the LasR/LasI system, since expression of rhlR depends on the latter. Two key regulator proteins are involved in activating the LasR/LasI system: the transcriptional activator LasR and the autoinducer synthesis protein LasI. Expression of the hemolysin gene is known to be under the control of the LasR/LasI system. Since hemolytic activity was greater in lon-disrupted than in wild-type cells (data not shown), it was suggested that Lon might be involved in modulating the LasR/LasI system, decreasing the cellular level of RhlR. To examine this possibility, we determined the level of LasI protein in bacterial cells. To generate the anti-LasI antiserum, the purified LasIΔG protein provided by M. Churchill (12) was injected into a rabbit. The immunoblotting result is shown in Fig. 4A. Whereas a little LasI was present in the wild-type cells (lon+ lasR+), a large amount was detected in the lon-disrupted cells (Δlon lasR+), suggesting that Lon modulates the cellular level of LasI. The accumulation of LasI in the lon-disrupted cells was completely suppressed by providing a functional lon gene in trans (data not shown). Therefore, it is possible that Lon modulates the LasR/LasI system.
FIG. 4.
Cellular levels of LasI (A) and relative levels of lasI expression (B). (A) Whole-cell extracts were prepared from strains PAO1 (lon+ lasR+), CS9008 (Δlon lasR+), CS9013 (lon+ ΔlasR), CS9027 (Δlon ΔlasR), and CS9038 (lon+ ΔlasI) grown in L broth for 24 h at 37°C and then separated on 15% SDS-polyacrylamide gels. The separated proteins were transferred to polyvinylidene difluoride membranes and then immunostained with anti-LasI serum. (B) Total RNAs were prepared from strains PAO1 (lon+ lasR+), CS9008 (Δlon lasR+), CS9013 (lon+ ΔlasR), and CS9027 (Δlon ΔlasR) grown in L broth to exponential phase at 37°C. The levels of lasI transcript were measured by quantitative, real-time RT-PCR and then normalized to rplU expression. The values represent the means and standard deviations of changes in comparison with the transcription level in PAO1.
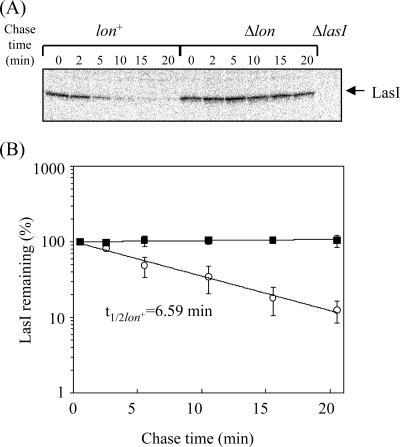
To explain the involvement of Lon in regulating the LasR/LasI system, we examined the cellular level of LasI in a lasR-disrupted background (Fig. 4A) because the expression of lasI is positively regulated by the LasR/3-oxo-C12-HSL complex. The increase in the cellular level of LasI effected by lon disruption persisted when lasR was additionally disrupted, suggesting that Lon controls the cellular level of LasI protein by a mechanism independent of LasR. The effect of lon disruption on lasI expression was examined in cells with the mutation background used for immunoblotting analysis (Fig. 4A) by quantitative RT-PCR. The results shown in Fig. 4B demonstrate that lon disruption did not affect lasI transcription in either lasR+ or ΔlasR cells. These results suggest that Lon may directly control the level of LasI by degrading it. To test whether Lon is involved in the turnover of LasI, we determined the in vivo half-life of this protein by pulse-labeling and chasing, followed by immunoprecipitation of LasI (Fig. 5A and B). Whereas LasI decayed with a half-life of 6.6 min in the wild-type cells, it did not seem to decay in the lon-disrupted cells even after a 20-min chase. The result indicates that Lon is involved in the turnover of LasI protein.
FIG. 5.
In vivo stabilities of LasI. (A) Cells of strains PAO1 (lon+) and CS9008 (Δlon) were grown to exponential phase and pulse-labeled with [35S]methionine and [35S]cysteine for 1 min at 37°C and then chased with unlabeled methionine and cysteine. Samples were taken at the times indicated, followed by immunoprecipitation of LasI. (B) Quantification of the precipitated LasI protein relative to the value at 1 min. Mean values and standard deviations of at least three independent experiments are given.
It is well known that rhlR expression depends mainly on the LasR/3-oxo-C12-HSL complex during growth in LB broth (31). To elucidate the role of the LasR/LasI system in the regulation of RhlR/RhlI by Lon, we measured the levels of RhlR in lon-disrupted cells after introducing a lasR disruption mutation (Fig. 6 A). The results showed that the lasR mutation decreased the cellular level of RhlR even in the lon-disrupted background. Quantitative RT-PCR analysis also demonstrated that the lasR-disrupted mutation decreased the level of rhlR transcripts (Fig. 6B), suggesting that the LasR/LasI system is involved in regulation of the RhlR/RhlI system by Lon. Taking these findings together, it is suggested that the RhlR/RhlI system is modulated by Lon protease through the LasR/LasI system, where the turnover of LasI is under the control of the Lon protease.
FIG. 6.
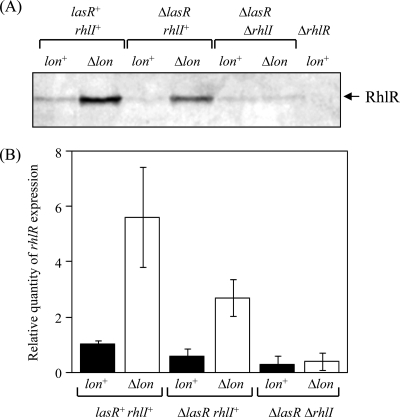
Cellular levels of RhlR (A) and relative levels of rhlR expression (B) in the absence of a lasR and/or rhlI gene in cells. (A) Whole-cell extracts were prepared from strains PAO1 (lon+ lasR+ rhlI+), CS9008 (Δlon lasR+ rhlI+), CS9013 (lon+ ΔlasR rhlI+), CS9027 (Δlon ΔlasR rhlI+), CS9051 (lon+ ΔlasR ΔrhlI), and CS9062 (Δlon ΔlasR ΔrhlI) grown in L broth for 24 h at 37°C and then separated on 15% SDS-polyacrylamide gel. The separated proteins were transferred to polyvinylidene difluoride membranes and then immunostained with anti-RhlR serum. (B) Total RNAs were prepared from strains PAO1 (lon+ lasR+ rhlI+), CS9008 (Δlon lasR+ rhlI+), CS9013 (lon+ ΔlasR rhlI+), CS9027 (Δlon ΔlasR rhlI+), CS9051 (lon+ ΔlasR ΔrhlI), and CS9062 (Δlon ΔlasR ΔrhlI) grown in L broth to exponential phase at 37°C. The levels of lasI transcript were measured by quantitative real-time RT-PCR and then normalized to rplU expression. The values represent the means and standard deviations of changes in comparison with the transcription level in PAO1.
LasR/LasI-independent regulation of the RhlR/RhlI system by Lon.
If the RhlR/RhlI system is down-regulated by Lon only through modulation of the LasR/LasI system, the increase in the cellular levels of RhlR due to stabilization of LasI in the lon-deficient mutant cells (Fig. 5) would be lost if lasR were disrupted. As shown by immunoblotting (Fig. 6A), there was a significant increase in the cellular level of RhlR in the lon-disrupted cells (Δlon ΔlasR), even in the absence of lasR, compared to lon+ cells (lon+ ΔlasR). Quantitative RT-PCR analysis of rhlR transcripts showed that the enhancement of rhlR transcription by lon disruption was not abolished by introducing the lasR-disrupted mutation (Fig. 6B). These results suggest that Lon may regulate expression of the RhlR/RhlI system via a LasR/LasI-independent pathway, in addition to the LasR/LasI-dependent pathway.
We then looked for the LasR/LasI-independent pathway for controlling rhlR expression. Since the expression of rhlI is known to be autoregulated by the RhlR/RhlI system (13), it is possible that the accumulation of RhlR protein in lon-disrupted cells may depend on the RhlI protein. To evaluate this suggestion, we analyzed the effect of lon disruption on the cellular level of RhlR in both lasR- and rhlI-disrupted backgrounds. The result in Fig. 6A shows that the increase in the RhlR level caused by lon disruption was suppressed by additional disruption of rhlI. A double mutation in rhlI and lasR simultaneously abolished the increase of rhlR transcription by lon disruption (Fig. 6B). These results suggest that Lon also regulates the expression of rhlR by modulating RhlI, independently of the LasR/LasI system.
DISCUSSION
In the present study, we have demonstrated that Lon protease in P. aeruginosa is involved in the regulation of two HSL-mediated QS systems, LasR/LasI and RhlR/RhlI. First, we showed that LasI accumulated dramatically in lon-disrupted cells independently of LasR, which is involved in the autoregulation of lasI expression (Fig. 4). Then, we found that LasI was significantly stabilized in lon-disrupted cells by determining its half-life in vivo (Fig. 5). We therefore proposed that Lon negatively controls the LasR/LasI system through degradation of LasI, which directs the synthesis of 3-oxo-C12-HSL. The LasR/3-oxo-C12-HSL complex positively regulates lasI expression, creating a positive regulatory feedback loop (43). It was therefore assumed that an elevated level of LasI could lead to increased lasI transcription. However, this was not actually the case, as indicated in Fig. 4B. It is also known that the complex activates the expression of rsaL (between lasR and lasI), the product of which negatively regulates lasI expression (34). Therefore, it is assumed that the accumulation of LasI caused by lon disruption could further activate the positive regulator, LasR, and increase the negative regulator, RsaL. This inverse regulation of lasI expression by LasR and RsaL could explain why lon disruption did not affect lasI transcription (Fig. 4) in spite of the accumulation of LasI. QS systems are organized hierarchically: RhlR/RhlI is subordinate to LasR/LasI (9). The present results suggest that Lon is involved in negative regulation of RhlR/RhlI expression through the degradation of LasI. On the other hand, the enhancement of the cellular level of RhlR by lon disruption was not completely abolished by the additional disruption of lasR. Interestingly, it was abolished by additional rhlI disruption (Fig. 6). These results suggest that an additional pathway, independent of the LasR/LasI system but dependent on RhlI, is involved in the regulation of rhlR expression by Lon. Taking these findings together, it is suggested that degradation of at least two positive regulators of QS by Lon protease leads to tight regulation of the expression of the two QS systems in P. aeruginosa.
A previous report demonstrated that the expression of rhlR is mainly induced by the LasR/3-oxo-C12-HSL complex (31). However, the results in Fig. 6 show that the rhlR transcript and RhlR accumulate sufficiently even in lasR-disrupted cells after lon disruption. rhlR contains at least four transcription start sites, P1 to P4 (28). Expression from P1 and P4 is dependent on the LasR/3-oxo-C12-HSL complex, and that from P2 and P3 depends on other factors (28). It has been demonstrated that expression of rhlR starts from P2 in stationary-phase P. aeruginosa PAO1 in LB medium (28). For this reason, sufficient rhlR transcripts are produced to lead to the accumulation of RhlR protein in the lasR-disrupted background. The result in Fig. 6 also shows that the enhancement of rhlR expression by lon disruption is abolished by introducing an rhlI mutation, suggesting that RhlI possibly activates rhlR expression. The pathway through which RhlI is involved in the activation of rhlR transcription has not been elucidated. The previous transcriptome analysis demonstrated that the level of rhlR transcription activated by both C4-HSL and 3-oxo-C12-HSL was higher than that activated by 3-oxo-C12-HSL alone (36). Therefore, it can be assumed that the excess levels of C4-HSL synthesized by RhlI due to lon disruption could activate the transcription of rhlR.
HSL levels are crucial for controlling QS systems and are therefore subject to regulation by a number of additional mechanisms. For example, the expression of HSL synthase genes is affected by various regulatory factors. The transcription of lasI is activated by the LasR/3-oxo-C12-HSL complex, VqsR (20), and PprB (7) and is repressed by RsaL (6). The LasR/3-oxo-C12-HSL complex and PprB (7) regulate rhlI transcription positively, but RpoS (46), RpoN (40), and Dks (19) regulate it negatively. RsmA could be involved in the stability of lasI mRNA (33). In addition to transcriptional control, our present study indicates that posttranslational control of LasI is important in regulating the HSL level, leading to negative control of QS system expression in P. aeruginosa.
Very recently, Bertani et al. reported that P. putida Lon protease is involved in regulating the HSL-mediated QS system PpuR/PpuI (3). They assumed that Lon was involved in regulating ppuI expression via the degradation of a LuxR-type protein, PpuR, on the basis of a previous report in which Lon and another ATP-dependent protease, ClpXP, were shown to be involved in degrading TraR, a LuxR-type protein of A. tumefaciens, in the absence of HSL (48). However, Lon could not degrade TraR in the presence of HSL. In our results, lon disruption did not affect the stability of the LuxR-type protein RhlR in the presence (Fig. 3) or absence (data not shown) of HSL. Furthermore, another LuxR-type protein from Erwinia carotovora was shown to be more sensitive to trypsin in vitro in the absence of HSL (45), suggesting that this protein might be nonspecifically degraded by proteases. Since ppuI expression is involved in the positive-induction loop of the PpuR/PpuI system, Lon in P. putida might possibly regulate ppuI expression by degrading PpuI. All HSL-mediated QS systems in many gram-negative bacteria are constructed by LuxR/I-type proteins. Since Lon protease is conserved, it might be generally involved in regulating all HSL-mediated QS systems.
Acknowledgments
We thank T. Tomoyasu for helpful discussions. We also thank T. Nemoto for technical assistance. We are grateful to M. E. A. Churchill for supplying the LasIΔG protein, to N. Gotoh for supplying the plasmid pME6032, to Y. Tsuda for supplying plasmids pEX18 and pMS255, and to T. Ikeda for supplying the C. violaceum strain CV026 and C4-HSL.
This research was supported in part by grants-in-aid for scientific research 16790252 and 19790313 to A.T. from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese government.
Footnotes
Published ahead of print on 11 April 2008.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1791113-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, A., M. Schmidt, W. Jager, and A. Puhler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 16237-39. [DOI] [PubMed] [Google Scholar]
- 3.Bertani, I., G. Rampioni, L. Leoni, and V. Venturi. 2007. The Pseudomonas putida Lon protease is involved in N-acyl homoserine lactone quorum sensing regulation. BMC Microbiol. 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretz, J., L. Losada, K. Lisboa, and S. Hutcheson. 2002. Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae. Mol. Microbiol. 45397-409. [DOI] [PubMed] [Google Scholar]
- 5.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 684839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Kievit, T. R., P. C. Seed, J. Nezezon, L. Passador, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 1812175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong, Y. H., X. F. Zhang, H. M. Soo, E. P. Greenberg, and L. H. Zhang. 2005. The two-component response regulator PprB modulates quorum-sensing signal production and global gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 561287-1301. [DOI] [PubMed] [Google Scholar]
- 8.Dougan, D., A. Mogk, K. Zeth, K. Turgay, and B. Bukau. 2002. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 5296-10. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35439-468. [DOI] [PubMed] [Google Scholar]
- 10.Gamer, J., H. Bujard, and B. Bukau. 1992. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor sigma 32. Cell 69833-842. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman, S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19565-587. [DOI] [PubMed] [Google Scholar]
- 12.Gould, T. A., H. P. Schweizer, and M. A. Churchill. 2004. Structure of the Pseudomonas aeruginosa acyl-homoserine synthase LasI. Mol. Microbiol. 531135-1146. [DOI] [PubMed] [Google Scholar]
- 13.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 341082-1093. [DOI] [PubMed] [Google Scholar]
- 14.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 21277-86. [DOI] [PubMed] [Google Scholar]
- 15.Holloway, B. W. 1969. Genetics of Pseudomonas. Bacteriol. Rev. 33419-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer, L. M., D. D. Leipe, E. V. Koonin, and L. Aravind. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 14611-31. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, M., E. Silva-Herzog, and G. Plano. 2005. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 541364-1378. [DOI] [PubMed] [Google Scholar]
- 18.Jenal, U., and R. Hengge-Aronis. 2003. Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 6163-172. [DOI] [PubMed] [Google Scholar]
- 19.Jude, F., T. Kohler, P. Branny, K. Perron, M. P. Mayer, R. Comte, and C. van Delden. 2003. Posttranscriptional control of quorum sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J. Bacteriol. 1853558-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juhas, M., L. Wiehlmann, B. Huber, D. Jordan, J. Lauber, P. Salunkhe, A. S. Limpert, F. von Gotz, I. Steinmetz, L. Eberl, and B. Tummler. 2004. Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa. Microbiology 150831-841. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 22.Laskowska, E., D. Kuczynska-Wisnik, J. Skorko-Glonek, and A. Taylor. 1996. Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol. Microbiol. 22555-571. [DOI] [PubMed] [Google Scholar]
- 23.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17333-343. [DOI] [PubMed] [Google Scholar]
- 24.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 21051-1060. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto, K. 2004. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol. Chem. 3851007-1016. [DOI] [PubMed] [Google Scholar]
- 26.Maurizi, M. R. 1987. Degradation in vitro of bacteriophage lambda N protein by Lon protease from Escherichia coli. J. Biol. Chem. 2622696-2703. [PubMed] [Google Scholar]
- 27.Maurizi, M. R. 1992. Proteases and protein degradation in Escherichia coli. Experientia 48178-201. [DOI] [PubMed] [Google Scholar]
- 28.Medina, G., K. Juarez, R. Diaz, and G. Soberon-Chavez. 2003. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology 1493073-3081. [DOI] [PubMed] [Google Scholar]
- 29.Mizusawa, S., and S. Gottesman. 1983. Protein degradation in Escherichia coli the lon gene controls the stability of the SulA protein. Proc. Natl. Acad. Sci. USA 80358-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murata, T., N. Gotoh, and T. Nishino. 2002. Characterization of outer membrane efflux proteins OpmE, OpmD, and OpmB of Pseudomonas aeruginosa: molecular cloning and dvelopment of specific antisera. FEMS Microbiol. Lett. 21757-63. [DOI] [PubMed] [Google Scholar]
- 31.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1793127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pessi, G., and D. Haas. 2001. Dual control of hydrogen cyanide biosynthesis by the global activator GacA in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 20073-78. [DOI] [PubMed] [Google Scholar]
- 33.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. Holden, M. Camara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 1836676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rampioni, G., M. Schuster, E. P. Greenberg, I. Bertani, M. Grasso, V. Venturi, E. Zennaro, and L. Leoni. 2007. RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 661557-1565. [DOI] [PubMed] [Google Scholar]
- 35.Rotanova, T. V., E. E. Melnikov, A. G. Khalatova, O. V. Makhovskaya, I. Botos, A. Wlodawer, and A. Gustchina. 2004. Classification of ATP-dependent proteases Lon and comparison of the active sites of their proteolytic domains. Eur. J. Biochem. 2714865-4871. [DOI] [PubMed] [Google Scholar]
- 36.Schuster, M., P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 1852066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. J. Cronan, K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 946036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaya, A., Y. Kubota, E. Isogai, and T. Yamamoto. 2005. Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to down-regulation of Salmonella pathogenicity island 1 gene expression. Mol. Microbiol. 55839-852. [DOI] [PubMed] [Google Scholar]
- 39.Takaya, A., M. Suzuki, H. Matsui, T. Tomoyasu, H. Sashinami, A. Nakane, and T. Yamamoto. 2003. Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar Typhimurium infection of mice. Infect. Immun. 71690-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, L. S., J. S. Webb, S. A. Rice, and S. Kjelleberg. 2003. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 220187-195. [DOI] [PubMed] [Google Scholar]
- 41.Torres-Cabassa, A., and S. Gottesman. 1987. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J. Bacteriol. 169981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Melderen, L., M. H. Thi, P. Lecchi, S. Gottesman, M. Couturier, and M. R. Maurizi. 1996. ATP-dependent degradation of CcdA by Lon protease. Effects of secondary structure and heterologous subunit interactions. J. Biol. Chem. 27127730-27738. [DOI] [PubMed] [Google Scholar]
- 43.Venturi, V. 2006. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30274-291. [DOI] [PubMed] [Google Scholar]
- 44.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 1852080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welch, M., D. E. Todd, N. A. Whitehead, S. J. McGowan, B. W. Bycroft, and G. P. C. Salmond. 2000. N-Acyl homoserine lactone binding to the CarA receptor determines quorum-sensing specificity in Erwinia. EMBO J. 19631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 1824356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winson, M. K., M. Camara, A. Latifi, M. Foglino, S. R. Chhabra, M. Daykin, M. Bally, V. Chapon, G. P. Salmond, B. W. Bycroft, A. Lazdunski, S. S. A. B. Gordon, and P. Williams. 1995. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 929427-9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, J., and S. C. Winans. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA 981507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]