Abstract
Enzymatic colicins such as colicin E9 (ColE9) bind to BtuB on the cell surface of Escherichia coli and rapidly recruit a second coreceptor, either OmpF or OmpC, through which the N-terminal natively disordered region (NDR) of their translocation domain gains entry into the cell periplasm and interacts with TolB. Previously, we constructed an inactive disulfide-locked mutant ColE9 (ColE9s-s) that binds to BtuB and can be reduced with dithiothreitol (DTT) to synchronize cell killing. By introducing unique enterokinase (EK) cleavage sites in ColE9s-s, we showed that the first 61 residues of the NDR were inaccessible to cleavage when bound to BtuB, whereas an EK cleavage site inserted at residue 82 of the NDR remained accessible. This suggests that most of the NDR is occluded by OmpF shortly after binding to BtuB, whereas the extreme distal region of the NDR is surface exposed before unfolding of the receptor-binding domain occurs. EK cleavage of unique cleavage sites located in the ordered region of the translocation domain or in the distal region of the receptor-binding domain confirmed that these regions of ColE9 remained accessible at the E. coli cell surface. Lack of EK cleavage of the DNase domain of the cell-bound, oxidized ColE9/Im9 complex, and the rapid detection of Alexa Fluor 594-labeled Im9 (Im9AF) in the cell supernatant following treatment of cells with DTT, suggested that immunity release occurred immediately after unfolding of the colicin and was not driven by binding to BtuB.
During times of nutrient stress, many bacteria synthesize and release a range of bacteriocins that target and kill related organisms. Presumably, this gives them greater access to the limited food supply and improves their chances of survival over “unarmed” neighbors occupying the same ecological niche (36). Escherichia coli is no exception and produces a range of toxic bacteriocins, called colicins, that target other enterobacteriaceae (9). Colicins are bactericidal antibiotics that (i) bind to one or more β-barrel-shaped outer membrane receptors of target cells (that are principally involved with the uptake of small metabolite growth factors), (ii) translocate across the cell envelope via protein-protein interactions with host periplasmic proteins, and (iii) kill the host bacterium by either depolarizing the cytoplasmic membrane, inhibiting murein biosynthesis or degrading the host's nucleic acids.
A combination of deletion mapping and structural studies (12, 21, 31, 32, 39, 42) have shown that each step in the colicin killing process is accomplished by one of three separate domains of the colicin that tend to be separated by a short stretch of residues that act as a linker. Receptor binding occurs via a centrally located receptor-binding domain (i.e., the R domain). Translocation is via a N-terminal natively disordered region (NDR) joined to an ordered region making up the translocation domain (i.e., the T domain), while cell killing is achieved by a C-terminally located cytotoxic domain. Enzymatic colicins differ from pore-forming colicins because they are coexpressed with a neutralizing immunity protein that binds to and inactivates the cytotoxic nuclease domain on synthesis.
Structures of the cocrystal complexes of colicins E3, E2, B, and Ia with their outer membrane receptor have shown that it is the tips of the receptor-binding domain that make contact with the β-barrel with a relatively small buried surface area that partially overlaps with the binding site of the receptors with their physiologically cognate small metabolite growth factor (7, 26, 29, 38). Once bound to an outer membrane receptor, each colicin crosses the bacterial cell envelope through the assistance of one of two different groups of periplasmic proteins. Group A colicins co-opt the tol-pal translocation system, which consists of five proteins, TolA, TolB, TolR, TolQ, and Pal (27), whereas the group B colicins, which include the pore-forming colicins B, D, Ia, 5, and 10, utilize the ton translocation system, which consists of the TonB, ExbB, and ExbD proteins (6). The translocation domains of most group A colicins, which include the enzymatic colicins E2 to E9, colicin A, and colicin E1, recruit a second coreceptor shown to be an outer membrane porin, principally OmpF or TolC, but potentially OmpC and PhoE (28). Group B colicins do not recruit a coreceptor at the cell surface (9).
Site-directed mutagenesis and biophysical analyses have demonstrated the importance of a small peptide sequence, called the TolB box, located at the N terminus of the E group colicins E2 to E9 and colicin A that interacts with the β-propeller domain of TolB (8, 17) and thus displaces the TolB-Pal interactions of the untreated cells (3). Analysis of this N-terminal region of colicin E9 by nuclear magnetic resonance spectroscopy showed that the region is natively disordered and highly flexible due to a high occupancy of glycine residues (10). It has been shown that some flexibility in the arms of the coiled-coil R domain of ColE9 are necessary for killing activity (34), which presumably facilitates the entry of the NDR and the cytotoxic domain into the periplasm. However, the final cellular location of any region of a colicin molecule, with the exception of the cytotoxic domain and the TolB box region of the T domain are not known. Recent data showed that the R domain of ColE2 is still in contact with its receptor after cell killing (14). Since the ordered region of the T domain would have to be unfolded to pass through the OmpF lumen (46), this perhaps suggests that regions of both the T and the R domains may remain at the cell surface.
Interaction of the T domain with the translocation proteins has been shown to be necessary for immunity release into the extracellular milieu (15), but the precise sequence and timing of events with respect to receptor binding and interaction with Tol proteins has not been determined. In the present study we introduced unique enterokinase (EK) cleavage sites at a number of locations within a disulfide-locked colicin E9 (ColE9s-s) (34) to study the surface accessibility of regions of the colicin molecule shortly after receptor binding. We show that the first 61 residues of the NDR of the T domain are not surface exposed immediately after binding of the R domain to BtuB and demonstrate that the extreme distal region of the NDR of the T domain is surface exposed and not occluded by OmpF, which has implications for the cellular localization of the ordered T and R domains. We use a sensitive fluorescence assay to demonstrate for the first time that immunity release from enzymatic colicins occurs immediately after reduction of the disulfide bond of ColE9s-s leading to unfolding of the helical hairpin.
MATERIALS AND METHODS
Plasmids, bacterial strains, and media.
E. coli DH5α was used as the host strain for cloning and mutagenesis. E. coli BL21(DE3) (Novagen) was used as the host strain for the expression vector pET21a (Novagen), which has a strong IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible T7 polymerase promoter and a C-terminal hexahistidine tag (His tag) to facilitate the purification of overexpressed proteins. E. coli DPD1718 contains a fusion of the E. coli recA promoter region to the Photorhabdus luminescens luxCDABE reporter integrated into the lacZ locus of E. coli DPD1692 (11). All cultures were routinely grown in Luria-Bertani (LB) broth or on plates of LB agar, supplemented where required with 100 μg of ampicillin/ml or 30 μg of chloramphenicol/ml. For the synchronization and protection experiments, we used a disulfide-locked colicin E9 construct, BH29, encoded by plasmid pBH29. This protein contains two cysteine mutations in the ColE9 receptor-binding domain that, in the reduced protein, do not affect its colicin activity. The formation of a disulfide bond under oxidizing conditions leads to a loss of biological activity, as previously described (34). Plasmids pYZ23, pYZ25, pYZ39, pYZ49, and pYZ52 were derived from pBH29 and engineered a unique, EK recognition site (DDDK) at different locations in the BH29 protein (Table 1).
TABLE 1.
Colicin activities of various ColE9 mutants containing unique EK sites
| Colicin | EK sitea | % lux inductionb |
|---|---|---|
| YZ23/Im9 | 61 | 53.5 |
| YZ49/Im9 | 82 | 53.2 |
| YZ52/Im9 | 287 | 75.5 |
| YZ39/Im9 | 441 | 6.1 |
| YZ25/Im9 | 478 | 41.3 |
| Free YZ25 | 478 | 41.3 |
The position of the engineered EK site (i.e., the residue number) is indicated.
lux induction is a measure of DNA damage and hence colicin activity and is expressed for each colicin construct relative to the ColE9 BH29/Im9, which is regarded as having 100% activity.
Protein expression and purification.
Recombinant proteins were expressed from recombinant pET21a clones with 1 mM IPTG and purified from BL21(DE3) cells by using metal chelate chromatography as described previously (33, 34).
Free colicin E9 protein purification.
ColE9/Im9 or ColE9/Im9 mutant complexes were dialyzed overnight against denaturing buffer (6 M guanidine [Gn]-HCl, 20 mM Tris-HCl [pH 8.0], 50 mM NaCl). Samples were applied to a 5-ml HiTrap chelating HP column charged with 25 ml of charge buffer (50 mM NiSO4) and equilibrated with 25 ml of denaturant buffer (6 M Gn-HCl, 20 mM Tris-HCl [pH 8.0], 50 mM NaCl) at 5 ml/min. The His-tagged immunity proteins are retained on the column, and the flowthrough, which contains free colicin E9 or free ColE9 mutant protein, was collected. The column was washed with 25 ml of denaturant buffer (6 M Gn-HCl, 20 mM Tris-HCl [pH 8.0], 50 mM NaCl) at 2 ml/min and then washed with 25 ml of 20 mM Tris-HCl (pH 8.0)-50 mM NaCl. The immunity protein was eluted from the column with elute buffer (1 M imidazole, 50 mM NaCl, 20 mM Tris-HCl [pH 7.5]) by using a 0 to 1 M imidazole gradient at 5 ml/min. The elution was monitored at an optical density of 280 nm (OD280) and by subjecting samples of the fractions to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein-containing fractions were pooled and then dialyzed extensively in 5 l potassium phosphate buffer (50 mM K2HPO4, 50 mM KH2PO4 [pH 7.4]) at 4°C.
Pentapeptide scanning mutagenesis.
Random pentapeptide insertions into the DNase domain of ColE9 were engineered as described previously (19), using the plasmid pCS4, which encodes both the ceaI and ceiI genes (18). Transposon insertions into ceaI were identified as inactive mutants in a stab test assay of the colicin activity of several hundred transconjugant colonies. The locations of the insertions were determined by restriction enzyme analysis and DNA sequencing. Plasmids encoding ColE9 harboring pentapeptide insertions in the DNase domain were transformed into E. coli BL21(DE3) and tested for colicin activity.
In vitro EK cleavage assay.
LB medium was pretreated with 2 ng of EK/ml overnight at 37°C to remove any EK cleavage sites in the yeast extract present in the growth medium, and then the medium was autoclaved to inactivate the EK. Incubation of the colicins with EK was performed in EK-pretreated LB at a substrate/enzyme ratio of 500:1 (4 μM to 8 nM) at 37°C in 20 mM Tris-Cl-20 mM NaCl-2 mM CaCl2 (pH 8.0). Samples (10 μl) were obtained at the desired time points and subjected to SDS-PAGE on 10 or 16% gels.
Diamide oxidation and DTT reduction.
For diamide oxidation, the colicin E9 protein samples were dialyzed overnight against phosphate-buffered saline (pH 7.4; PBS) to remove the dithiothreitol (DTT). Protein samples were then incubated with 1 mM diamide (N,N,N′,N′-tetramethylazodicarbamide) for 30 min in the dark at room temperature before extensive dialysis against PBS. For reduction, the protein samples were incubated with 10 mM DTT for 1 h at room temperature, followed by extensive dialysis against degassed PBS. To avoid the possibility of spontaneous oxidation during certain assays, reduced protein was alkylated by using iodoacetamide (50 mM final concentration), followed by extensive dialysis.
In vivo EK cleavage assay.
An overnight culture of E. coli DPD1718 cells was diluted 1:50 with EK-pretreated LB broth and grown for 3 h at 37°C in the presence of 30 μg of chloramphenicol/ml, until reaching an OD600 of 0.3 to 0.4. The cells were then diluted twofold with LB broth to 100 μl in black 96-well optical bottom microtiter plates (Nunc), and 4 nM oxidized colicin, followed by 0.4 nM EK, was added to the wells. The microtiter plates were incubated at 37°C for 30 and 60 min before the addition of 1 mM DTT. The induction of luminescence was monitored over a period of 200 min, with readings taken every 300 s.
In vivo capture of cleaved protein.
E. coli DPD1718 cells were grown in 5 ml as described above and incubated with either 4 nM free YZ23 or YZ49, followed immediately with or without 0.5 nM EK for 1 h. The cells were boiled to deactivate the EK, sonicated, and mixed with 8 nM Im9 for 30 min to bind to any free colicin. The sample was concentrated to 1 ml and incubated with 200 μl of MagneHis Ni-Particles (Promega) for 2 h at room temperature with shaking to capture any Im9 complexed proteins. The beads were centrifuged and washed five times in PBS, and the proteins were eluted by using 20 μl of PBS containing 1 M imidazole.
Luminescence reporter assay.
This assay makes use of an SOS-inducible chromosomal lux operon to detect DNA damage induced by colicin E9 in E. coli reporter cells and was carried out as previously described (40). All assays were performed in a microtiter plate luminometer (Lucy 1; Anthos Labtech, Salzburg, Austria) at 37°C. The luminometer, plates, and media were prewarmed to 37°C to prevent induction of a stress response due to cooling.
Data analysis.
Luminescence values are mostly presented as relative luminescence units (RLU). The gamma value is defined as the luminescence induced for any given sample concentration minus the luminescence of the control cells at the same time point divided by the luminescence of the control cells at that time point: (Lsample − Lcontrol)/Lcontrol (11). The activity of mutant colicin proteins was calculated by dividing the gamma value of the mutant by the gamma value of BH29 and expressed as a percentage. EK cleavage was assessed by dividing the gamma value of the EK-treated sample with that of the untreated control. All assays were performed at least twice, with three to six replicates for each condition.
Immunity protein labeling.
Im9 (C23S/S6C) was dialyzed against PBS and unfolded in 3 M Gn-HCl in the same buffer. It was then incubated with an ∼25-fold molar excess of Alexa Fluor 594-labeled C5-maleimide and incubated for 2 h at room temperature, followed by extensive dialysis against PBS. The extent of labeling was then checked via comparison to the unlabeled protein at OD587 (ɛAlexa Fluor 594 = 97,700 cm−1 M−1) and OD280 (ɛIm9(C23S/S6C) = 9,970 cm−1 M−1, prelabeling).
Fluorescence assay for immunity release.
E. coli DH5α cells from overnight cultures were grown to mid-log phase (OD600 of ≥0.4) in LB medium. Alexa Fluor 594-labeled BH29/Im9 was formed by preincubation of oxidized free BH29 with Alexa Fluor 594-labeled Im9 (C23S/S6C) at a molar ratio of 1:1.5 and was subsequently added to 1 ml of cells to a final concentration of 10 nM. Receptor binding was allowed for 5 min at 37°C (it was usually complete within 2 min), after which cells were spun for 2 min at 6,000 rpm and resuspended in 100 μl of prewarmed LB medium to remove unbound colicin. At this stage the cell-bound relative fluorescence units (RFU) were measured, and then DTT to a final concentration of 2 mM or nonreducing buffer was added to the cells; the cells were then incubated for an additional 30 min at 37°C. The cells were then spun for 1 min at 10,000 rpm, and the RFU of the cell supernatant and cell pellet (resuspended in prewarmed LB medium) were measured in 96-well plates (optical bottom) in triplicate by using a Victor2 1420 multilabel plate reader (Wallac) controlled by the Wallac 1420 software.
The values of DTT-induced release were plotted after background release in the absence of DTT had been subtracted.
RESULTS
Early events in translocation: differences in EK cleavage of the NDR of ColE9.
ColE9s-s, containing a disulfide bond in the distal arms of the coiled-coil R domain, was used to study some of the early events in colicin killing (Fig. 1). It has been reported that the T domain of the locked colicin makes contact with OmpF prior to any movement of the alacoil of the R domain (23), but the extent of the interaction of the TolB extensor region (37) with OmpF at this stage has not been determined. We engineered an EK cleavage site (DDDK) into the NDR of the T domain between residues 61 and 62 of the disulfide-locked ColE9, and we named it YZ23. The lux reporter assay (40) showed that reduced YZ23 was still active and retained 53.5% of the activity of BH29 (Table 1). Cleavage of free YZ23 and YZ23/Im9 complex with EK in vitro showed that the protein was fully cleaved after approximately 30 min, and biologically inactive (Fig. 2A and B) demonstrating (i) that the EK cleavage site of this mutant colicin was fully accessible in solution and (ii) that the presence of the immunity protein did not affect the efficiency of the cleavage.
FIG. 1.
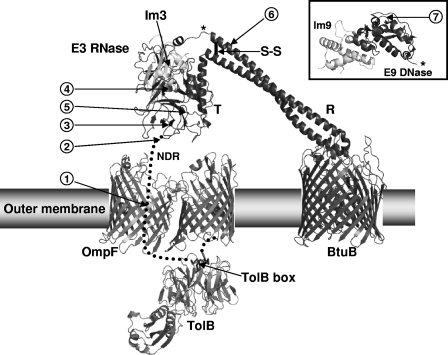
Structural model of the assembly of the ColE9S-S translocon shortly after binding to the BtuB receptor. The model is based on the structure of ColE3/Im3 modeled on that of the BtuB-ColE3 R domain complex (26, 39). The figure highlights the position of the disulfide bond (S-S) within the R domain (R), the NDR, and the TolB box relative to the ordered T domain (T), and the E3 RNase bound to Im3 in relation to the BtuB, OmpF outer membrane receptors, and periplasmic TolB. The locations of the EK cleavage sites are shown by numbered arrows 1 to 7, where arrow 1 indicates an insertion of DDDK after residue 61; arrow 2 indicates an insertion of DDDK after residue 82; arrow 3 indicates an insertion of DDDK after G101; arrow 4 indicates a mutation of P161-ADDI-T166 to P161-DDDK-T166; arrow 5 indicates a mutation of S287-VSDV-L292 to S287-DDDK-L292; arrow 6 indicates a mutation of K441-DAKDK-L447 to K441-ADDDK-L447; and arrow 7 indicates an insertion of DDDK after S478. The asterisk shows the position of attachment of the ColE9 DNase/Im9 domains (inset) on the full-length colicin. For clarity, OmpF is shown as a dimer rather than the physiologically relevant trimeric arrangement. (Adapted from reference 3 with permission of the publisher.)
FIG. 2.
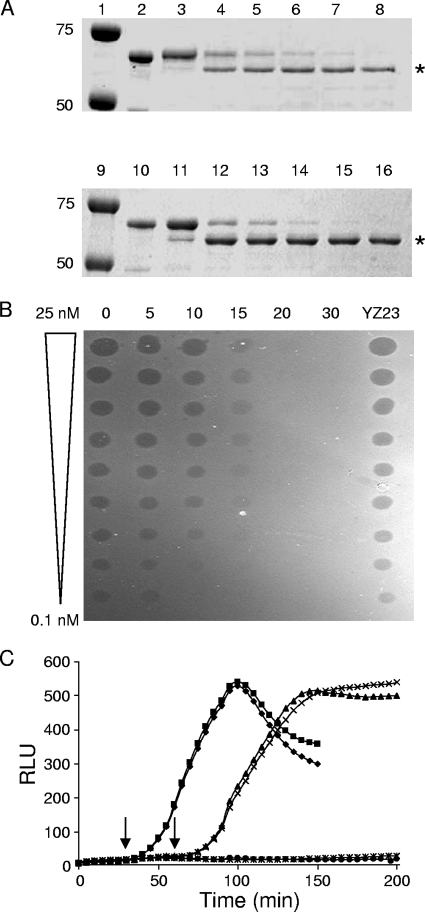
Proteolytic cleavage of an EK cleavage site at residue 61 of ColE9 (YZ23). (A) SDS-PAGE of free YZ23 (lanes 2 to 8) and YZ23/Im9 (lanes 10 to 16) containing an engineered EK cleavage site at residue 61, incubated with EK in vitro for 0 min (lanes 3 and 11), 5 min (lanes 4 and 12), 10 min (lanes 5 and 13), 15 min (lanes 6 and 14), 20 min (lanes 7 and 15), and 30 min (lanes 8 and 16). Untreated controls are shown in lanes 2 and 10, and protein molecular mass markers of 50 and 75 kDa in lanes 1 and 9 are indicated. The asterisk marks the cleavage products. (B) Spot test activity assay of the EK cleaved YZ23. Twofold doubling dilutions of 25 nM YZ23 cleaved with EK for 0, 5, 10, 15, 20, or 30 min were spotted as 2-μl aliquots onto a freshly grown lawn of E. coli DH5α cells. Untreated YZ23 were spotted as a control. (C) Luminescence (RLU) generated by YZ23 in the lux reporter assay showing protection of YZ23 from EK cleavage. E. coli DPD1718 cells were incubated with 4 nM oxidized YZ23, followed by incubation with EK for 30 min (⧫) or 60 min (▴) before being reduced with 1 mM DTT. E. coli DPD1718 cells that were not treated with EK but reduced with 1 mM DTT after 30 min (▪) or 60 min (×) were used as positive controls for DNA damage, while E. coli DPD1718 cells that were treated with trypsin for 30 min prior to 1 mM DTT treatment (✳) or untreated DPD1718 (•) were grown as negative controls of DNA damage. Arrows indicate the addition of DTT at 30 and 60 min, respectively.
To determine the accessibility of the first 61 residues of the extensor TolB box region after receptor binding, E. coli DPD1718 cells were challenged with oxidized free YZ23 or YZ23/Im9 complex and then treated with EK for up to 60 min to facilitate the cleavage of any surface-accessible colicin. The disulfide-locked colicin was then reduced with DTT, and the residual activity of the EK-treated colicin was then determined by using the sensitive lux reporter assay for DNA damage (40). Incubation with EK had very little effect on the activity of both the free YZ23 and the YZ23/Im9 complex after incubation for up to 60 min before DTT reduction (Fig. 2C). The increase in luminescence in this in vivo EK cleavage experiment indicates substantial DNA damage and shows that YZ23 was not significantly cleaved by EK while it was bound to the BtuB outer membrane receptor. The difference in the in vitro and in vivo cleavage experiments with YZ23 indicate that the EK recognition site in YZ23 was not accessible to EK after binding to the BtuB receptor. When considered along with previous studies (23), these results strongly suggest that the first 61 residues of the T domain containing the TolB box of enzymatic colicins rapidly interacts with their coreceptor, OmpF, and that the initial interaction with OmpF occurs before any movement of the alacoil of the R domain has occurred.
The region of the T domain that interacts with OmpF has been shown to include residues 5 to 9 of both colicins E3 and E9 (23, 45) and, in addition, residues between 60 and 80 of ColE9 (23). The TolB box of ColE9 maps between residues 32 and 46 (20), which suggests that the NDR interacts with OmpF at two positions either side of the TolB box, looping into the periplasm to interact with TolB according to the model proposed by Bonsor et al. (3) and shown in Fig. 1. This suggests that the entire NDR region of the T domain of ColE9 encompassing residues 1 to 83 might be involved in the early interactions with OmpF, but whether this occurs before any unfolding of the R domain is currently unknown. We tested this by introducing an EK site (DDDK) between residues 82 and 83 of BH29 to form YZ49. lux reporter (Table 1) and activity spot assays (data not shown) showed that YZ49 was active against sensitive E. coli cells. Incubation with EK in vitro demonstrated that cleavage of YZ49 occurred after 5 min and continued accumulatively over the next 30 min but, unlike the cleavage of YZ23, cleavage of YZ49 at the EK site was slower and did not proceed to completion (Fig. 3A). This demonstrates that the EK cleavage site engineered into BH29 between residues 82 and 83 is accessible to EK, but that cleavage in vitro is not as efficient at this site as it is at an EK site introduced between residues 61 to 62. In the in vivo EK digestion experiment, luminescence induced by EK-treated YZ49 was reduced compared to that of untreated YZ49, demonstrating that some protection had resulted after cleavage of the colicin at the EK site introduced between residues 82 and 83 (Fig. 3B). Calculation of the gamma values demonstrated that up to 50% protection from lux induction was provided by EK digestion of cell-bound YZ49 after 30 or 60 min (Table 2).
FIG. 3.
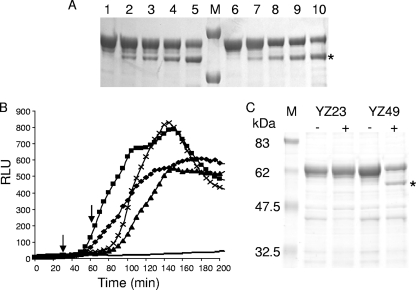
Proteolytic cleavage of an EK cleavage site at residue 82 of ColE9 (YZ49). (A) Im9-free YZ49 (lanes 1 to 5) and YZ49/Im9 (lanes 6 to 10) were incubated with EK in vitro for 10 min (lanes 2 and 7), 15 min (lanes 3 and 8), 20 min (lanes 4 and 9), or 30 min (lanes 5 and 10). Untreated controls are shown in lanes 1 and 6. The asterisk marks the cleavage products, and protein molecular mass markers of 50 and 75 kDa are shown in lane M. (B) Luminescence (RLU) generated by YZ49 after incubation with EK in the lux reporter assay. E. coli DPD1718 cells were treated with 4 nM oxidized YZ49, followed by incubation with EK for 30 min (⧫) or 60 min (▴) before being reduced with 1 mM DTT. E. coli DPD1718 cells incubated with YZ49 that were not treated with EK but reduced with 1 mM DTT after 30 (▪) or 60 min (×) were used as positive controls for DNA damage, while untreated DPD1718 (−) was grown as negative controls for DNA damage. The arrows indicate the addition of DTT at 30 or 60 min, respectively. (C) Capture of colicin proteins in vivo showing specific cleavage of YZ49 at the EK cleavage site. E. coli DPD1718 cells were treated with oxidized free YZ49 or YZ23 and then incubated with (+) or without (−) EK for 60 min. Bound colicin was rescued from heat-inactivated cells after binding to exogenously added Im9 using nickel-treated magnetic beads, concentrated, and subjected to SDS-12% PAGE. Molecular mass markers (M) are indicated on the left in kilodaltons. The asterisk marks the specific cleavage product of 54.2 kDa.
TABLE 2.
Percent lux induction of DPD1718 cells after incubation of YZ23/Im9, YZ49/Im9, YZ52/Im9, YZ39/Im9, YZ25/Im9, and free YZ25 with EK for 30 or 60 min
| Colicin | EK sitea | % lux induction after EK treatmentb
|
|
|---|---|---|---|
| 30 min | 60 min | ||
| YZ23/Im9 | 61 | 100* | 100* |
| YZ49/Im9 | 82 | 49.4 | 52.9 |
| YZ52/Im9 | 287 | 73.5 | 61.9 |
| YZ39/Im9 | 441 | 90.0 | 78.7 |
| YZ25/Im9 | 478 | 95.4 | 100 |
| Free YZ25 | 478 | 94.6 | 76.4 |
The position of the engineered EK site (i.e., the residue number) is indicated.
The percent lux induction after EK treatment was expressed as a percentage of the lux induction of the untreated control and calculated by dividing the gamma value (see Materials and Methods) of a selected time point by that of the EK-untreated control at the same time point. Incubations of all colicins in Table 2 with trypsin for 30 min resulted in no lux induction. *, Values extrapolated from the gamma values calculated at one time point only, in this case 60 min after the addition of DTT. The data presented are representative of data generated from similar experiments repeated on at least three occasions.
To demonstrate that the observed inhibition of lux induction occurred due to specific cleavage at the engineered EK cleavage site rather than by nonspecific in vivo degradation, we sought to identify the expected specific cleavage product of 54.2 kDa. E. coli DPD1718 cells were incubated with free YZ49 and either treated with EK or not. Proteins were extracted from the cells, and full-size YZ49 or the 54.2-kDa cleaved product of YZ49 were fished out with nickel beads after binding to exogenously added Im9, concentrated, and subjected to SDS-PAGE. Figure 3C shows the appearance of the 54.2-kDa cleaved product in the EK-treated sample and the absence of any cleaved product in the sample without EK treatment. The experiment was conducted with free YZ23 in which the EK cleavage site is rapidly occluded by OmpF (Fig. 2C) to demonstrate that no cleavage of YZ23 occurred in either EK-treated or untreated samples (Fig. 3C).
Early events in translocation: proteolytic cleavage of the ordered T domain of ColE9.
The role of the ordered T domain from residues 84 to 315 of enzymatic E colicins remains undetermined. Even though the distal end of the disordered region of the T domain of colicin E9 was accessible to EK cleavage while bound to BtuB, we sought to determine the accessibility of the ordered T domain. From an analysis of the crystal structure of ColE3 T84-315, we engineered an EK site between S287 and L292 that changed the sequence of residues from S287-VSDV-L292 to S287-DDDK-L292, and we named the mutant colicin YZ52. Biological spot test activity and lux reporter assay (Table 1) showed that YZ52 was biologically active. Exposure to EK for 60 min in vitro showed that YZ52 was cleaved (data not shown) but less efficiently than either YZ23 or YZ49, a finding indicative of the structural differences between the NDR and the ordered T domain (Fig. 1). lux induction from cells incubated with EK- treated YZ52 was reduced compared to the untreated control, showing that the cleavage site remained accessible to the action of the protease (Table 2). Two additional mutations were made in external loops of the ordered T domain. One was an insertion of the EK site (DDDK) between residues G101 and G102, and the other was a conservative mutation changing the residues P161-ADDI-T166 to P161-DDDK-T166. Both mutants were cleaved with EK in vitro but were biologically inactive and thus were not tested in vivo.
The distal arm of the R domain is accessible to cleavage by EK.
The crystal structure of a 135-residue receptor-binding domain of ColE3 in complex with BtuB has shown that 27 residues at the apex of the coiled-coil of the colicin are responsible for this interaction (26). Since the T+R domains of ColE3 and ColE9 are 96% identical, it is reasonable to assume that the interaction between the ColE9 R domain and BtuB will be identical. We generated an EK site at the top of the distal arm of the R domain of BH29 by changing the residues K441-DAKDK-L447 to K441-ADDDK-L447 (Fig. 1), and we named the mutant YZ39. The spot test assay showed that YZ39 retained activity against sensitive E. coli DH5α cells (data not shown), but the lux reporter assay indicated that the activity was only 6.1% of that of reduced BH29 (Table 1). Free and Im9-complexed YZ39 was incubated with EK in vitro for 30 min, and the efficiency of cleavage was assessed by using SDS-PAGE, the spot test activity assay, and the lux reporter assay. The activity of free and Im9-complexed YZ39 was reduced compared to untreated YZ39 (data not shown), and the presence of EK cleavage products of 47 and 16 kDa with the concomitant decrease in concentration of YZ39 confirmed that specific cleavage had occurred at the EK site (data not shown). EK inhibited lux induction in vivo by YZ39/Im9 after incubations of 30 and 60 min (Table 2). Similar data were found for free YZ39 (data not shown).
Receptor binding does not trigger DNase entry or loss of immunity protein.
For full activity, the DNase domain of colicin E9 must traverse the cell envelope of the bacterial cell and enter the cytoplasm. We used the EK cleavage assay to determine the status of the DNase domain immediately after receptor binding. Pentapeptide scanning mutagenesis (19) was used to determine regions of the DNase that would tolerate the insertion of an EK site without affecting the function of the DNase (data not shown). Two potential permissive insertion sites were determined at Ser478 and Ser528. We choose to insert the EK cleavage site at Ser478 of the DNase domain of ColE9 forming the mutant strain, YZ25, because this region represents an exposed loop on the opposite side of the DNase domain to the Im9 binding site. EK cleavage in vitro showed that the cleavage site is accessible, and the presence of the immunity protein did not significantly alter the cleavage reaction (data not shown). E. coli DPD1718 cells were treated with oxidized free YZ25 or YZ25/Im9 complex and then incubated with EK for 30 and 60 min, followed immediately by reduction of the disulfide bond with DTT. Figure 4 shows that incubation with EK for 30 or 60 min resulted in a reduction in lux induction by free YZ25, but not by the YZ25/Im9 complex. This suggests that the DNase domain of free YZ25 was accessible to EK cleavage and thus had not begun the translocation process through the outer membrane. Analysis of the gamma values 60 min after DTT reduction showed that lux induction was less in the YZ25-treated cells compared to the YZ25/Im9-treated cells (Table 2). The only difference between these two experiments is the presence of the immunity protein. Since there is no difference in the dynamics of killing between free and complexed ColE9 (40), this implies that the observed protection against cleavage in vivo directly resulted from the presence of Im9 bound to YZ25, perhaps as a result of steric hindrance to the DNase domain created by the immunity protein and the rigidity enforced on YZ25/Im9 through its complex with BtuB.
FIG. 4.
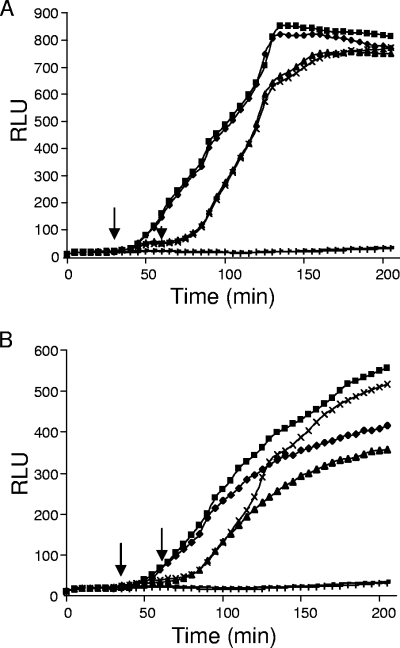
Protection from proteolytic cleavage of the DNase by Im9. The lux induction assay was used to show that Im9-complexed YZ25 was not cleaved (A), whereas Im9-free YZ25 was cleaved by EK after binding to BtuB (B). The luminescence of cultures of E. coli DPD1718 treated with oxidized YZ25/Im9 (A) or free YZ25 (B), followed by EK treatment for 30 min (⧫) or 60 min (▴), and then immediately reduced with 1 mM DTT was compared to that of E. coli DPD1718 treated with oxidized YZ25/Im9 or free YZ25 without EK treatment but reduced with 1 mM DTT after 30 min (▪) or 60 min (×). The luminescence of E. coli DPD1718 cells treated with oxidized YZ25/Im9 (A) or YZ25 (B) and then trypsin treatment for 30 min, followed by reduction with 1 mM DTT (+), and cells with no addition (−) is shown. Arrows indicate the addition of DTT at 30 and 60 min, respectively.
Immunity protein is released on reduction of the disulfide bond.
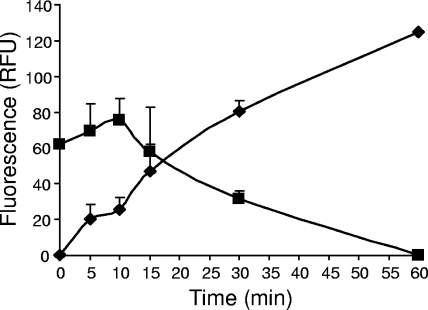
In order to confirm that Im9 is still bound to colicin E9 after binding of disulfide-locked BH29 to E. coli DPD1718 cells, we introduced C23S and S6C mutations to create a unique cysteine residue in helix one of Im9 that did not affect the activity of the mutant immunity protein. The Im9 C23S/S6C mutant protein was fluorescently labeled with Alexa Fluor 594 (Im9AF) and then incubated with free BH29 to reconstitute the ColE9/Im9AF complex. Figure 5 shows an increase in fluorescence in the culture supernatant, and a concomitant reduction of cell-bound fluorescence, after the addition of DTT to BH29/Im9AF-treated E. coli DH5α cells. This finding is in agreement with data presented by Duche et al. (15), who suggested that the trigger for immunity release of enzymatic colicins is an interaction of colicin with the Tol proteins.
FIG. 5.
Immunity protein is released from the colicin after reduction of the disulfide bond. DH5α(pET21a) cells were preincubated with 10 nM BH29-Im9AF. After the removal of unbound BH29-Im9AF, activity was initiated by the addition of 2 mM DTT. The release of fluorescently labeled Im9 in the cell supernatant (RFU) is shown at set time intervals over a 60-min period as values plotted after subtraction of background release from samples that were not reduced with DTT (⧫). The coinciding reduction in cell-bound fluorescence (RFU) over the same period is also shown (▪). The means ± the standard errors of the mean are shown for two independent experiments.
Because of the sensitive nature of our experimental setup, we were able to observe the dynamics of immunity release quantitatively. It has been shown previously that significant immunity release can be detected 20 min after the addition of ColE2/Im2 to sensitive cells by using Western blot analysis (15). By monitoring the increase in Im9AF-generated fluorescence in the cell supernatant of cell-bound oxidized BH29 after reduction by DTT, we showed, using this “synchronized” system, that immunity release begins immediately upon reduction and continues to rise over the 60-min period (Fig. 5). The slight lag in the loss of fluorescence from cell-bound BH29/Im9AF over the first 10 min is due to the relatively high fluorescent signals produced at these time points in combination with the fluorescence-quenching effects of the cells. At this stage of the experiment, the increase of fluorescence in the supernatant is much more sensitive, does not involve any quenching effects, and clearly shows that immunity is lost from the ColE9/Im9 complex immediately after DTT is added to the cells.
DISCUSSION
It is generally accepted that after binding to the BtuB outer membrane receptor the initial stages of translocation involve the establishment of a colicin translocon (45) in which an outer membrane protein (OmpF) coreceptor is rapidly recruited, leading to the interaction of the N-terminal T domain of the colicin with the Tol proteins, principally TolB. A number of models have been devised to explain colicin translocation and the subsequent loss of the immunity protein, leading to entry of the cytotoxic nuclease domain (3, 37, 38, 46).
Using a disulfide-locked colicin to synchronize cell killing through DTT-induced reduction of the disulfide bond, we determined the accessibility of unique, EK sites in colicin E9 to EK cleavage in order to extend our knowledge of some of the early spatial and temporal events that occur immediately after binding of the colicin to BtuB. Although enzymatic colicins require some flexibility and movement of the R-domain helical hairpin to allow translocation of the T domain, recruitment of OmpF occurs before any unfolding of the colicin. It has been proposed that OmpF recruitment by enzymatic E colicins requires residues 1 to 7 (37, 45) and 60 to 80 (23) of the unstructured TolB extensor region. Our data show that the first 61 residues of the T domain are not accessible to EK cleavage once bound to a bacterial cell and are most probably trapped within OmpF or located within the periplasm. Housden et al. (23) predicted that the binding site extends to at least residue 64 but may not include residue 75. Our data provide experimental evidence that the extreme distal region of the NDR around serine 82 of the TolB extensor region is not occluded by OmpF prior to R domain unfolding.
The role of the ordered T domain from residue 84 onward of enzymatic colicins is currently unknown. It is predominantly β-sheet in structure, with a number of adjoining external loops and two alpha helices at the extreme distal end immediately before the R domain (39), and would need an energy-dependent unfolding for it to enter the OmpF lumen (46). It is known that ColA is still in contact with its receptor when pore formation occurs (2), and it has recently been shown for colicin E2 that both the receptor binding and the translocation domains remain attached to BtuB and the Tol proteins, respectively, after the colicin has initiated cell killing (14). However, the cellular localization of the ordered T domain is not known. We engineered EK cleavage sites into the ordered T domain at three different locations between G101 and L292 and showed in vitro that all three regions were accessible to EK cleavage. Two of the insertions, located in different externally located loops mapping on opposite sides of the domain, rendered the colicin inactive, and therefore cleavage could not be tested in vivo. The third mutation of S287-VSDV-L292 to S287-DDDK-L292 was biologically active and was cleaved in vivo by EK, showing that the ordered T domain is not involved in the initial insertion of the T domain into the coreceptor.
It is of interest that two of the mutants generated by the incorporation of EK cleavage sites in the ordered T domain were inactive. The first of these, a DDDK insertion between residues G101 and G102, placed the EK cleavage site within an external loop that we deemed ideally suited to any potential cleavage by EK. However, it became apparent through in vitro biophysical analysis of the mutated colicin that the insertion of the EK cleavage site caused premature breakdown of the polypeptide and destabilized the interaction of ColE9 with TolB (Y. Zhang and C. Penfold, unpublished data). The second inactive mutation involved the mutagenesis of an alanine to aspartic acid and an isoleucine-to-lysine substitution within the peptide sequence PADDIT. This peptide sequence is also in an exposed loop and is conserved among a variety of colicins. These two regions may be important, for example, to allow the T domain of colicins flexibility to cross the periplasmic space if, as has been suggested, ColA and the enzymatic colicins interact with TolR and the ton-dependent colicins interact with ExbB and ExbD near the inner membrane (5, 24). The ordered T domain of ColE9 has two highly conserved sequence motifs across colicin groups. Both motifs, named the T1 box (residues 202 to 214) and T2 box (residues 272 to 276), contain primarily hydrophobic residues and were suggested as being potentially important for interactions with host proteins during a common translocation pathway (13), although there is no direct experimental evidence to support this. We have used site-directed mutagenesis of several hydrophobic residues of both the T1 box and the T2 box extended to include Tyr285 of ColE9 to show the importance of three residues of the T2 box to the activity of ColE9 (22). One of these mutants, Y285A was completely inactive. Although the ColE9 Y285A mutant displayed full DNase activity and also interacted, in part, with TolB in an surface plasmon resonance experiment, receptor-binding activity was affected, and there was significant breakdown of the purified protein, suggesting that ColE9 Y282A is substantially less stable than ColE9 (22). Restoration of activity to the alanine mutant was only observed with hydrophobic substitutions, which when added to the fact that the T2 box is buried and not solvent exposed, suggests that this instability is most likely the result of a loss of hydrophobic packing around this residue induced by the creation of an internal cavity surrounding the smaller alanine residue. Similar destabilization has been reported in hemoglobin (44) and T4 lysozyme (16, 43) caused by the generation of an internal cavity when a much larger hydrophobic residue was replaced by a smaller nonhydrophobic residue. Xu et al. (43) investigated the effect of substituting a leucine residue for an alanine residue at a number of sites within the hydrophobic core of T4 lysozyme, determined the crystal structure of each mutant protein, and showed an approximate relationship between the increase in cavity size associated with the substitution and the reduction in protein stability.
We have made use of our disulfide-locked colicin E9/Im9 complex to monitor the dynamics of immunity protein release. Although an EK cleavage site was engineered into the DNase domain away from the immunity binding site, the presence of the immunity protein prevented EK-induced cleavage of the ColE9/Im9 complex in vivo. This was in contrast to the results with free colicin, without the immunity protein, which was cleaved by EK after binding to BtuB. Since Im9 did not prevent cleavage of the DNase domain by EK in vitro, and loss of lux induction in vivo is due to specific cleavage of the EK cleavage sites, we speculate that the presence of the immunity protein in combination with the rigidity enforced on YZ25/Im9 by its interaction with the outer membrane affected the access of EK to the cleavage site in the DNase domain. Loss of immunity was then confirmed after reduction of the disulfide bond with DTT using loss of cell-bound Im9AF fluorescence and an increase of fluorescence in the cell supernatant showing that immunity protein is definitely released before the DNase domain of enzymatic colicins enter the cell.
By using a more sensitive assay, we showed that immunity release is not driven by binding to BtuB but occurs immediately after DTT-induced restoration of flexibility to the R domain and interaction of the T domain with the Tol proteins. This is in contrast to previous studies that suggest that immunity release from enzymatic bacteriocins occurs at least 20 min after the colicin/immunity complex binds to the cell surface receptor (15, 25), which may be due to differences in the detection limits of each experimental system, and reflects the value of our cell synchronization system and the sensitivity of fluorescence detection of released Im9AF.
Even though cells are synchronized for killing, not all cells are in the same growth phase, nor are all BtuB receptor proteins in the same functional state (1). Therefore, immunity release occurs over a longer time frame than perhaps anticipated with this system. Because we are able to synchronize the start of cell killing and use a very sensitive fluorescent detection method, we can detect immunity loss much earlier than previously detectable (15, 25).
These data add to the growing evidence that the time required for DNase colicins to kill sensitive cells is similar to the rapid killing seen with pore-forming colicins (4) and is much faster than previously predicted (35, 40). Notably, the receptor binding and translocation of the T domain of ColE2 occurs in seconds (14), while the interaction of the DNase domains of ColE9 and colicins E2 and E8 with a lipid membrane has also been shown to occur in seconds (30). The recent use of trypsin protection against killing by the DNase colicins E2, E7, E8, and E9 has shown that trypsin cannot prevent DNA damage caused by ColE7 even if added immediately after ColE7 (41).
The fluorescence assay for immunity release will provide a solid platform for investigating the role of individual Tol proteins and of specific colicin mutations in the release of Im9 and entry of the DNase domain into the cell envelope.
Acknowledgments
We are extremely grateful to Nick Housden (University of York, York, United Kingdom) for the construction of Fig. 1. Figure 1 was adapted from Fig. 8 of Bonsor et al. (3), and we gratefully acknowledge Nick Housden and Colin Kleanthous (University of York, York, United Kingdom) and the American Chemical Society for their permission to adapt this figure.
We are grateful to the BBSRC, United Kingdom (studentships to L.E.H. and D.C.W.), and the University of Nottingham, Nottinghamshire, United Kingdom (studentship to Y.Z.), for financial support.
Footnotes
Published ahead of print on 11 April 2008.
REFERENCES
- 1.Bassford, P. J., R. J. Kadner, and C. A. Schnaitman. 1977. Biosynthesis of the outer membrane receptor for vitamin B12, E colicins, and bacteriophage BF23 by Escherichia coli: kinetics of phenotypic expression after the introduction of bfe+ and bfe alleles. J. Bacteriol. 129265-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bénédetti, H., R. Lloubès, C. Lazdunski, and L. Letellier. 1992. Colicin A unfolds during its translocation in Escherichia coli cells and spans the whole-cell envelope when its pore has formed. EMBO J. 11441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonsor, D. A., I. Grishkovskaya, E. J. Dodson, and C. Kleanthous. 2007. Molecular mimicry enables competitive recruitment by a natively disordered protein. J. Am. Chem. Soc. 1294800-4807. [DOI] [PubMed] [Google Scholar]
- 4.Bourdineaud, J. P., P. Boulanger, C. Lazdunski, and L. Letellier. 1990. In vivo properties of colicin A: channel activity is voltage dependent but translocation may be voltage independent. Proc. Natl. Acad. Sci. USA 871037-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouveret, E., L. Journet, A. Walburger, E. Cascales, H. Bénédetti, and R. Lloubès. 2002. Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 84413-421. [DOI] [PubMed] [Google Scholar]
- 6.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84365-380. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan, S. K., P. Lukacik, S. Grizot, R. Ghirlando, M. Ali, T. J. Barnard, K. S. Jakes, P. K. Kienker, and L. Esser. 2007. Structure of colicin I receptor bound to the R-domain of colicin Ia: implications for protein import. EMBO J. 262594-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr, S., C. N. Penfold, V. Bamford, R. James, and A. M. Hemmings. 2000. The structure of TolB, an essential component of the tol-dependent translocation system, and its protein-protein interaction with the translocation domain of colicin E9. Struct. Fold. Des. 857-66. [DOI] [PubMed] [Google Scholar]
- 9.Cascales, E., S. K. Buchanan, D. Duche, C. Kleanthous, R. Lloubes, K. Postle, M. Riley, S. Slatin, and D. Cavard. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71158-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, E. S., S. B. Whittaker, K. Tozawa, C. MacDonald, R. Boetzel, C. N. Penfold, A. Reilly, N. J. Clayden, M. J. Osborne, A. M. Hemmings, C. Kleanthous, R. James, and G. R. Moore. 2002. Structural dynamics of the membrane translocation domain of colicin E9 and its interaction with TolB. J. Mol. Biol. 318787-804. [DOI] [PubMed] [Google Scholar]
- 11.Davidov, Y., R. Rozen, D. R. Smulski, T. K. Van Dyk, A. C. Vollmer, D. A. Elsemore, R. A. LaRossa, and S. Belkin. 2000. Improved bacterial SOS promoter::lux fusions for genotoxicity detection. Mutat. Res. 46697-107. [DOI] [PubMed] [Google Scholar]
- 12.de Graaf, F. K., M. J. Stukart, F. C. Boogerd, and K. Metselaar. 1978. Limited proteolysis of cloacin DF13 and characterization of the cleavage products. Biochemistry 171137-1142. [DOI] [PubMed] [Google Scholar]
- 13.de Zamaroczy, M., and R. H. Buckingham. 2002. Importation of nuclease colicins into Escherichia coli cells: endoproteolytic cleavage and its prevention by the immunity protein. Biochimie 84423-432. [DOI] [PubMed] [Google Scholar]
- 14.Duche, D. 2007. Colicin E2 is still in contact with its receptor and import machinery when its nuclease domain enters the cytoplasm. J. Bacteriol. 1894217-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duche, D., A. Frenkian, V. Prima, and R. Lloubes. 2006. Release of immunity protein requires functional endonuclease colicin import machinery. J. Bacteriol. 1888593-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eriksson, A., W. Baase, X. Zhang, D. Heinz, M. Blaber, E. Baldwin, and B. Matthews. 1992. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science 255178-183. [DOI] [PubMed] [Google Scholar]
- 17.Garinot-Schneider, C., C. N. Penfold, G. R. Moore, C. Kleanthous, and R. James. 1997. Identification of residues in the putative TolA box which are essential for the toxicity of the endonuclease toxin colicin E9. Microbiology 1432931-2938. [DOI] [PubMed] [Google Scholar]
- 18.Garinot-Schneider, C., A. J. Pommer, G. R. Moore, C. Kleanthous, and R. James. 1996. Identification of putative active-site residues in the DNase domain of colicin E9 by random mutagenesis. J. Mol. Biol. 260731-742. [DOI] [PubMed] [Google Scholar]
- 19.Hallet, B., D. J. Sherratt, and F. Hayes. 1997. Pentapeptide scanning mutagenesis: random insertion of a variable five amino acid cassette in a target protein. Nucleic Acids Res. 251866-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hands, S. L., L. E. Holland, M. Vankemmelbeke, L. Fraser, C. J. Macdonald, G. R. Moore, R. James, and C. N. Penfold. 2005. Interactions of TolB with the translocation domain of colicin E9 require an extended TolB box. J. Bacteriol. 1876733-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilsenbeck, J. L., H. Park, G. Chen, B. Youn, K. Postle, and C. Kang. 2004. Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 Å resolution. Mol. Microbiol. 51711-720. [DOI] [PubMed] [Google Scholar]
- 22.Holland, L. E. 2003. Investigating the translocation mechanism of colicin E9, and identification of USP as an HNH endonuclease. Ph.D. thesis, University of Nottingham, Nottinghamshire, United Kingdom.
- 23.Housden, N. G., S. R. Loftus, G. R. Moore, R. James, and C. Kleanthous. 2005. Cell entry mechanism of enzymatic bacterial colicins: porin recruitment and the thermodynamics of receptor binding. Proc. Natl. Acad. Sci. USA 10213849-13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Journet, L., E. Bouveret, A. Rigal, R. Lloubès, C. Lazdunski, and H. Bénédetti. 2001. Import of colicins across the outer membrane of Escherichia coli involves multiple protein interactions in the periplasm. Mol. Microbiol. 42331-344. [DOI] [PubMed] [Google Scholar]
- 25.Krone, W. J., P. de Vries, G. Koningstein, A. J. de Jonge, F. K. de Graaf, and B. Oudega. 1986. Uptake of cloacin DF13 by susceptible cells: removal of immunity protein and fragmentation of cloacin molecules. J. Bacteriol. 166260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurisu, G., S. D. Zakharov, M. V. Zhalnina, S. Bano, V. Y. Eroukova, T. I. Rokitskaya, Y. N. Antonenko, M. C. Wiener, and W. A. Cramer. 2003. The structure of BtuB with bound colicin E3 R-domain implies a translocon. Nat. Struct. Biol. 10948-954. [DOI] [PubMed] [Google Scholar]
- 27.Lazzaroni, J. C., J. F. Dubuisson, and A. Vianney. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 84391-397. [DOI] [PubMed] [Google Scholar]
- 28.Mock, M., and A. P. Pugsley. 1982. The BtuB group col plasmids and homology between the colicins they encode. J. Bacteriol. 1501069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohanty, A. K., C. M. Bishop, T. C. Bishop, W. C. Wimley, and M. C. Wiener. 2003. Enzymatic E-colicins bind to their target receptor BtuB by presentation of a small binding epitope on a coiled-coil scaffold. J. Biol. Chem. 27840953-40958. [DOI] [PubMed] [Google Scholar]
- 30.Mosbahi, K., C. Lemaitre, A. H. Keeble, H. Mobasheria, B. Morel, R. James, G. R. Moore, E. J. A. Lea, and C. Kleanthous. 2002. The cytotoxic domain of colicin E9 is a channel-forming endonuclease. Nat. Struct. Biol. 9476-484. [DOI] [PubMed] [Google Scholar]
- 31.Ohno-Iwashita, Y., and K. Imahori. 1980. Assignment of the functional loci in colicin E2 and E3 molecules by the characterization of their proteolytic fragments. Biochemistry 19652-659. [DOI] [PubMed] [Google Scholar]
- 32.Ohno-Iwashita, Y., and K. Imahori. 1982. Assignment of the functional loci in the colicin E1 molecule by characterization of its proteolytic fragments. J. Biol. Chem. 2576446-6451. [PubMed] [Google Scholar]
- 33.Penfold, C. N., C. Garinot-Schneider, A. M. Hemmings, G. R. Moore, C. Kleanthous, and R. James. 2000. A 76-residue polypeptide of colicin E9 confers receptor specificity and inhibits the growth of vitamin B12-dependent Escherichia coli 113/3 cells. Mol. Microbiol. 38639-649. [DOI] [PubMed] [Google Scholar]
- 34.Penfold, C. N., B. Healy, N. G. Housden, R. Boetzel, M. Vankemmelbeke, G. R. Moore, C. Kleanthous, and R. James. 2004. Flexibility in the receptor-binding domain of the enzymatic colicin E9 is required for toxicity against Escherichia coli cells. J. Bacteriol. 1864520-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds, B. L., and P. R. Reeves. 1969. Kinetics of adsorption of colicin CA42-E2 and reversal of its bactericidal activity. J. Bacteriol. 100301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riley, M. A., and D. M. Gordon. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7129-133. [DOI] [PubMed] [Google Scholar]
- 37.Sharma, O., and W. A. Cramer. 2007. Minimum length requirement of the flexible N-terminal translocation subdomain of colicin E3. J. Bacteriol. 189363-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma, O., E. Yamashita, M. V. Zhalnina, S. D. Zakharov, K. A. Datsenko, B. L. Wanner, and W. A. Cramer. 2007. Structure of the complex of the colicin E2 R-domain and its BtuB receptor: the outer membrane colicin translocon. J. Biol. Chem. 28223163-23170. [DOI] [PubMed] [Google Scholar]
- 39.Soelaiman, S., K. Jakes, N. Wu, C. Li, and M. Shoham. 2001. Crystal structure of colicin E3: implications for cell entry and ribosome inactivation. Mol. Cell 81053-1062. [DOI] [PubMed] [Google Scholar]
- 40.Vankemmelbeke, M., B. Healy, G. R. Moore, C. Kleanthous, C. N. Penfold, and R. James. 2005. Rapid detection of colicin E9 induced DNA damage using Escherichia coli cells carrying SOS promoter::lux fusions. J. Bacteriol. 1874900-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker, D., K. Mosbahi, M. Vankemmelbeke, R. James, and C. Kleanthous. 2007. The role of electrostatics in colicin nuclease domain translocation into bacterial cells. J. Biol. Chem. 28231389-31397. [DOI] [PubMed] [Google Scholar]
- 42.Wiener, M., D. Freymann, P. Ghosh, and R. M. Stroud. 1997. Crystal structure of colicin Ia. Nature 385461-464. [DOI] [PubMed] [Google Scholar]
- 43.Xu, J., W. A. Baase, E. Baldwin, and B. W. Matthews. 1998. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 7158-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yutani, K., K. Ogasahara, and Y. Sugino. 1985. Effect of amino acid substitutions on conformational stability of a protein. Adv. Biophys. 2013-29. [DOI] [PubMed] [Google Scholar]
- 45.Zakharov, S. D., V. Y. Eroukova, T. I. Rokitskaya, M. V. Zhalnina, O. Sharma, P. J. Loll, H. I. Zgurskaya, Y. N. Antonenko, and W. A. Cramer. 2004. Colicin occlusion of OmpF and TolC channels: outer membrane translocons for colicin import. Biophys. J. 873901-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zakharov, S. D., M. V. Zhalnina, O. Sharma, and W. A. Cramer. 2006. The colicin E3 outer membrane translocon: immunity protein release allows interaction of the cytotoxic domain with OmpF porin. Biochemistry 4510199-10207. [DOI] [PubMed] [Google Scholar]