Abstract
Bacteria growing in biofilms are more resistant to antibiotics than their planktonic counterparts. How this transition occurs is unclear, but it is likely there are multiple mechanisms of resistance that act together in order to provide an increased overall level of resistance to the biofilm. We have identified a novel efflux pump in Pseudomonas aeruginosa that is important for biofilm-specific resistance to a subset of antibiotics. Complete deletion of the genes encoding this pump, PA1874 to PA1877 (PA1874-1877) genes, in an P. aeruginosa PA14 background results in an increase in sensitivity to tobramycin, gentamicin, and ciprofloxacin, specifically when this mutant strain is growing in a biofilm. This efflux pump is more highly expressed in biofilm cells than in planktonic cells, providing an explanation for why these genes are important for biofilm but not planktonic resistance to antibiotics. Furthermore, expression of these genes in planktonic cells increases their resistance to antibiotics. We have previously shown that ndvB is important for biofilm-specific resistance (T. F. Mah, B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole, Nature 426:306-310, 2003). Our discovery that combining the ndvB mutation with the PA1874-1877 gene deletion results in a mutant strain that is more sensitive to antibiotics than either single mutant strain suggests that ndvB and PA1874-1877 contribute to two different mechanisms of biofilm-specific resistance to antibiotics.
Bacteria growing in biofilms are more resistant to antimicrobial agents than their planktonic counterparts are (16). Various hypotheses have been put forward to explain biofilm resistance, but to date, there are no data that entirely explain this phenomenon (24). For instance, it has been suggested that the exopolysaccharide matrix that surrounds the cells in the biofilm prevents diffusion of the antimicrobial agents through the biofilm, thus preventing access of the antimicrobial agent to the cells. While this may be the case for some antimicrobial agents, for others it has been shown that they can penetrate the matrix but still cannot kill the cells in the biofilm (2, 39). It has also been suggested that cells within the biofilm grow slowly in response to oxygen, nutrient deprivation, or environmental stress. While a number of studies support the idea that a lower growth rate can explain some level of biofilm-specific resistance, other studies have suggested that the full extent of resistance cannot be accounted for by this mechanism (7, 12, 39). Furthermore, it has been suggested that high cell density signaling or quorum sensing plays a role in resistance to antimicrobial agents, but again, their exact role is not clear (6).
Conversely, resistance mechanisms utilized by planktonic cells have been well studied. These mechanisms, such as expression of antibiotic-degrading enzymes, decrease in antibiotic influx or increase in antibiotic efflux, are often encoded by transmissible elements and can spread rapidly through different species of bacteria (20). In particular, ATP-binding cassette (ABC) transporters and resistance-nodulation-division (RND) efflux pumps contribute to planktonic resistance to a variety of antibiotics and biocides (21, 35). While no ABC transporters have been identified in Pseudomonas aeruginosa that specifically contribute to drug resistance (34), nine RND efflux pumps have been identified in P. aeruginosa that account for this organism's high intrinsic resistance to antibiotics (22, 28, 35). Like ABC transporters, RND transporters in P. aeruginosa also have a wide range of substrates (10). For instance, MexAB-OprM, MexCD-OprJ, and MexXY extrude quinolones, macrolides, tetracyclines, β-lactams, and more, while aminoglycosides are more specific to MexXY-OprM (26).
Since RND multidrug efflux pumps contribute to antibiotic resistance in planktonic cells, it has been suggested that expression of efflux pumps in biofilms might lead to antibiotic resistance in these surface-attached communities (11). It was found that four of the best-studied RND efflux pumps in Pseudomonas aeruginosa, MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY had no impact on biofilm-specific resistance when mature biofilms were tested (11). However, a recent study suggested that MexAB-OprM and MexCD-OprJ are involved in biofilm resistance to the macrolide azithromycin (14). Furthermore, a putative multidrug efflux pump, YhcQ, has been implicated in Escherichia coli biofilm antibiotic resistance (23).
In order to identify antibiotic resistance mechanisms utilized by biofilm bacteria, we developed a high-throughput system to search for Tn5 insertion mutants of P. aeruginosa that do not develop the characteristic increase in resistance to antimicrobial agents when grown in a biofilm (25). This method allowed us to previously identify ndvB as a gene important for biofilm-specific resistance and describe a novel mechanism of resistance whereby ndvB-derived glucans interact with the antibiotic tobramycin and prevent it from accessing its cellular target (25). Here we present the characterization of another mutant from this screen, which has allowed us to identify a new, putative efflux pump with a role in biofilm antibiotic resistance.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and chemicals.
P. aeruginosa PAO1 (17) and PA14 (36) and E. coli DH5α (3) were grown in rich medium (Luria-Bertani [LB]) or minimal medium at 37°C. The minimal medium was minimal M63 salts (33) supplemented with arginine (0.4%) and MgSO4 (1 mM). The P. aeruginosa PAO1 transposon insertion mutants were obtained from the University of Washington Genome Center (17). The P. aeruginosa PA14 deletion strains ΔPA1874, ΔPA1875, ΔPA1876, ΔPA1877, and ΔPA1874-1877 were constructed as reported previously (25) using the following primers: P504 (5′-GGCGGGATCCGACCACGTCGTTGACCACTG-3′) and P455 (5′-GGCGGCGCATATGGATAGGGGTAACTTTCGCCTG-3′) for the 5′ PA1874 fragment, P456 (5′-GGCGGCGCATATGCGTTTCGGTTTCAGCGGAACC-3′) and P505 (5′-GGCGGAATTCGTAATGCCGTTGATCAGTTGC-3′) for the 3′ PA1874 fragment, P506 (5′-GGCGGGATCCGAACATCACCGACATCCTCAAC-3′) and P459 (5′-GGCGGCGCATATGCCGTCCCTTGCGCGCGTACTG-3′) for the 5′ PA1875 fragment, P460 (5′-GGCGGCGCATATGCGCGAGGCCAAGGCGTCGCTG-3′) and P507 (5′-GGCGGAATTCCGTCGTCGAGCAGCAGCAG-3′) for the 3′ PA1875 fragment, P462 (5′-GGCGGGATCCCATCCGCGAGATGACCCAGGC-3′) and P463 (5′-GGCGGCGCATATGGAGGATCACGACGTTATCCGG-3′) for the 5′ PA1876 fragment, P464 (5′-GGCGGCGCATATGCAGCGCGGTATCGGCTACCTG-3′) and P465 (5′-GGCGGAATTCGTTCGCCGACTCCTGGAAGTTG-3′) for the 3′ PA1876 fragment, P466 (5′-GGCGGGATCCCAACTGATGAGCGCCGAGCAG-3′) and P467 (5′-GGCGGCGCATATGCAGCAACGGGTCGGCCAGTTG-3′) for the 5′ PA1877 fragment, P468 (5′-GGCGGCGCATATGGAGGTATTGCCGATCATTCCG-3′) and P469 (5′-GGCGGAATTCCGTCGCCGCGGATATTTTATC-3′) for the 3′ PA1877 fragment, PA1874F2 (5′-GATCAAGCTTACATGGTGCGGCCGTTCG-3′) and PA1874R2 (5′-GGAATCTAGAGCCGTTGCTGGCATTGACCT-3′) for the 5′ PA1874 to PA1877 (PA1874-1877) fragment, and PA1877F2 (5′-GGAGTCTAGATCCAGCGGCAGATCCACCT-3′) and PA1877R2 (5′-CACAGAATTCGAAGCGATCCACGGCGACAT-3′) for the 3′ PA1874-1877 fragment. Tobramycin, gentamicin, ciprofloxacin, nalidixic acid, and rifampin were purchased from Sigma-Aldrich Co. (St. Louis, MO).
Biofilm formation assay.
Biofilm formation was measured by using the microtiter dish assay system described previously (31, 32). Biofilms were grown for 24 h before they were stained with 0.1% crystal violet.
MBC-P and MBC-B assays.
To determine the minimal bactericidal concentration for biofilms (MBC-B) (25), bacterial strains were inoculated into the wells of a 96-well microtiter dish and biofilms were allowed to form over 24 h. Serial dilutions of the antibiotic of choice were added into the wells, and the biofilms were exposed to the drug for 24 h. The medium containing the antibiotic was replaced with fresh medium so that after 24 additional hours, bacteria from the biofilm that survived antibiotic treatment would detach from the biofilm and repopulate the planktonic phase. These bacteria were transferred using a multipronged device (Dan-Kar Corp., Woburn, MA) onto an agar growth plate in order to determine the concentration of the drug that killed the biofilm cells. To determine the minimal bactericidal concentration for planktonic cells (MBC-P) (25), bacterial strains were inoculated into the wells of a 96-well microtiter dish at the same time as serial dilutions of the antibiotic of choice were added. The planktonic bacteria were exposed to drug for 24 h, and viability was assessed by transferring a small amount of the culture onto an agar growth plate using a multipronged device.
Tobramycin accumulation assay.
Biofilms were formed over 24 h and then exposed to tobramycin for 8 h. After exposure to tobramycin, the wells containing the biofilms were rinsed with phosphate-buffered saline to remove the external tobramycin and the medium was replaced with a glycine lysis buffer (0.1 M glycine [pH 3.0]). The biofilms were left overnight at 37°C to allow for complete lysis of the biofilm cells. After ∼16 h, the biofilm/glycine mixture was evaporated to dryness and resuspended in water. To measure the amount of tobramycin that had accumulated in the biofilms, the resuspended biofilm/glycine mix was spotted onto a lawn of sensitive E. coli. The presence of tobramycin was noted as a zone of clearing on the lawn of E. coli (25).
RT-PCR.
Total bacterial RNA was isolated from the planktonic or biofilm cells. Briefly, the planktonic cells were prepared from the log-phase cells subcultured at 37°C in LB broth (optical density at 600 nm of ca. 0.7), while the biofilm cells were prepared from colony biofilms grown on agar plates overnight at 37°C followed by another night at room temperature as described previously (8). The RNA samples were prepared by the Trizol method using Trizol reagent (Invitrogen). After treatment of the RNA samples with RNase-free DNase I (Sigma), DNase I was inactivated according to the manufacturer's instructions. The RNA samples were further purified by using the PureLink Micro-to-Midi total RNA purification system (Invitrogen) and then quantitated using the SmartSpec Plus spectrophotometer (Bio-Rad) with the confirmation on agarose gel. Equal amounts of DNase-treated RNA samples were used as templates for reverse transcription with the iScript cDNA synthesis kit (Bio-Rad), followed by PCR amplification of the cDNA samples with Taq DNA polymerase (Invitrogen). A pair of primers specific for and internal to the PA1874 gene (i.e., PA14_40260 in PA14) (5′-GCGTCGGCATCGATACCAAT-3′ [forward] and 5′-ACGATCACCGTCACCGTCTC-3′ [reverse]) was used to amplify mRNA as a measure of PA1874 expression. For a control, the mRNA of the constitutively expressed rpoD gene was amplified and quantitated by the two-step reverse transcription-PCR (RT-PCR) with the primers (5′-GATCCGGAACAGGTGGAAGAC-3′ [forward] and 5′-TCAGCAGTTCCACGGTACCC-3′ [reverse]). RT was performed in a volume of 50 μl with the following incubations: 5 min at 25°C, 40 min at 42°C, and 5 min at 85°C. PCR was subsequently carried out in a 50-μl reaction mixture containing the primers and the cDNA samples incubated for 1 min at 94°C, followed by 35 cycles (with 1 cycle consisting of 1 min at 94°C, 30 s at 56°C, and 20 s at 70°C) and a final step of 10 min at 70°C. The RT-PCR products were analyzed by agarose gel electrophoresis for the expected products (PA1874, 382 bp; rpoD, 226 bp). To control for DNA contamination of RNA samples, non-RT reactions (i.e., PCR) were carried out. In no instance was a product obtained in the absence of an RT reaction. We estimate the difference in expression between the planktonic and biofilm cells to be 10-fold on the basis of the fact that dilution of the reaction mixture by 1/10 reduced the planktonic PA1874 gene signal to zero yet reduced the biofilm PA1874 signal to a level equal to the undiluted planktonic PA1874 signal.
Cloning of PA1875-1877 genes.
A ca. 4.7-kb fragment containing the putative operon of the PA1875-1877 genes (beginning immediately after the ATG start codon of PA14_40250) was amplified from the genomic DNA of P. aeruginosa PA14 using Platinum Taq HF DNA polymerase (Invitrogen) with primers (forward primer, 5′-ACAAGAGCTCATGCGCGGGCGCAGGCAGTACGCG-3′ [the SacI site is underlined]; reverse primer, 5′-GACCGAATTCCTGTCCTGCACGCCTCAGTA-3′ [the EcoRI site is underlined]). PCR mixtures were subjected to 35 cycles of 1 min at 94°C, 1 min at 60°C, and 6 min at 70°C before finishing with a 10-min incubation at 72°C. Following digestion of the two purified PCR products with SacI and EcoRI enzymes, the fragment was cloned into the SacI/EcoRI-restricted pJB866 vector (5) to yield pLZ866A. Subsequently, the plasmid (pLZ866A) obtained in E. coli DH5α was used to transform E. coli S17-1, from which pLZ866A and pJB866 (empty vector) were mobilized into P. aeruginosa PA14 via conjugation (43) by selection of the transconjugants on LB agar containing tetracycline (100 μg/ml) and ciprofloxacin (0.1 μg/ml; for counterselection). The presence of the appropriate plasmids in either E. coli or P. aeruginosa was confirmed by plasmid isolation, PCR amplification, and/or restriction enzyme digestion. The role of the plasmid-borne PA14_40250/40240/40230 operon in antibiotic resistance was assessed in LB broth by MIC determination, in which the cells carrying pJB866 or pLZ866A were pregrown in the presence or absence of 2 mM m-toluic acid.
RESULTS
Identification of genes important for biofilm-specific resistance to antibiotics.
We carried out a genetic screen to identify mutants of P. aeruginosa that are more sensitive to the antibiotic tobramycin, specifically when this microbe is growing in a biofilm (25). One of the mutants from the screen had a Tn5 insertion in the first gene of a predicted four-gene operon PA14_40260/40230 locus in P. aeruginosa PA14, which corresponds to PA1874-1877 genes in P. aeruginosa PAO1 (Fig. 1). The probability that these genes are cotranscribed is high (between 0.94 and 0.99) (40, 42). On the basis of the published annotation, the PA1874 gene is predicted to encode a large outer membrane protein with sequence similarity to LapA of Pseudomonas putida and Bap of Staphylococcus aureus (42). LapA is important for biofilm formation in Pseudomonas fluorescens, and Bap was identified in a screen for mutants of S. aureus unable to make a biofilm (9, 15).
FIG. 1.
Genomic context of the PA1874-1877 operon. P. aeruginosa PAO1 gene designations are indicated in the figure. P. aeruginosa PA14 gene designations are as follows: PA1874 (1874) = PA14_40260, PA1875 = PA14_40250, PA1876 = PA14_40240, and PA1877 = PA14_40230. The predicted location or function of the protein product is listed below the gene designation (42).
The other genes in this operon are predicted to encode an outer membrane protein with homology to an RND efflux pump outer membrane protein (PA1875), an ABC transporter protein (PA1876), and a membrane fusion protein predicted to be part of an ABC transporter (PA1877) (Fig. 1) (42). Given the predicted functions of the gene products of the PA1874-1877 operon and their phenotype in the initial genetic screen, we initiated this study in order to determine whether these gene products are important for biofilm-specific efflux of antibiotics.
Confirmation of biofilm-specific resistance defect.
To confirm the defect in biofilm-specific resistance to tobramycin that we observed in the original transposon insertion mutant from our screen, we obtained individual transposon insertion mutants in PA1874, PA1875, PA1876, and PA1877 genes from the P. aeruginosa PAO1 transposon insertion library at the University of Washington Genome Center (17). Using these mutants, we performed biofilm assays and growth assays and determined the MBC-P for tobramycin for each strain and found that the mutant strains all behaved like the wild-type strain in these assays (Table 1). However, the MBC-B values for tobramycin for these mutant strains were lower (Table 1) than the MBC-B value for the wild-type strain, suggesting that these genes are important for biofilm-specific resistance to tobramycin and confirming our original observation with the original transposon insertion mutant described above.
TABLE 1.
Phenotypes of strain PAO1 and PA1874-1877 operon transposon insertion mutants of P. aeruginosa
| Straina | Tobramycin
|
Biofilm formation | |
|---|---|---|---|
| MBC-P (μg/ml) | MBC-B (μg/ml) | ||
| PAO1 | 16 | 200 | Yes |
| PA1874tn | 16 | 100 | Yes |
| PA1875tn | 16 | 100 | Yes |
| PA1876tn | 16 | 100 | Yes |
| PA1877tn | 16 | 100 | Yes |
Transposon insertion mutants are indicated by tn at the end of the strain designation.
Because we did not know whether the phenotypes of the transposon insertion mutants (Table 1) were due to the transposon insertion in the PA1874 gene or the polar effect of this transposon insertion on the downstream genes in the operon, we constructed in-frame deletion mutations in each individual gene in the operon in an P. aeruginosa PA14 background and tested each knockout mutant for its biofilm-specific sensitivity phenotype (Table 2). Interestingly, while knockouts of the three genes predicted to encode components of an efflux pump, PA1875, PA1876, and PA1877, were more sensitive to tobramycin in the MBC-B assay, the PA1874 mutation showed a sensitivity profile identical to that of the wild-type strain, suggesting that the large, outer membrane protein is not important for biofilm-specific resistance to tobramycin (Table 2).
TABLE 2.
Phenotypes of PA14 and PA1874-1877 knockout mutants of P. aeruginosa
| Strain | Tobramycin
|
Biofilm formation | |
|---|---|---|---|
| MBC-P (μg/ml) | MBC-B (μg/ml) | ||
| PA14 | 32 | 100 | Yes |
| ΔPA1874 mutant | 32 | 100 | Yes |
| ΔPA1875 mutant | 32 | 50 | Yes |
| ΔPA1876 mutant | 32 | 25 | Yes |
| ΔPA1877 mutant | 32 | 25 | Yes |
| ΔPA1874-1877 mutant | 32 | 25 | Yes |
To confirm that the knockout mutant phenotypes were specific to biofilm resistance, we tested the growth, biofilm formation (using a static 96-well microtiter dish assay), and planktonic sensitivity phenotypes for each mutant. For all three assays, there was no defect in any of the mutants compared to the wild type; the knockout mutants were able to grow as well as the wild type (data not shown) and form a biofilm and had a planktonic resistance level (MBC-P) equivalent to that of the wild-type strain (Table 2).
Deletion of the PA1874-1877 operon affects biofilm-specific resistance to several antibiotics.
Tobramycin, a key aminoglycoside antibiotic for therapy of P. aeruginosa infection, was used in the screen and the initial experiments. In order to assess the specificity of the biofilm-specific sensitivity phenotype, we determined the MBC-B values of the whole-operon deletion (PA1874-1877) for the following antibiotics: gentamicin (aminoglycoside), ciprofloxacin (fluoroquinolone) (Table 3), nalidixic acid (quinolone), and rifampin. None of the MBC-P values for the various antibiotics differed from that of the wild-type strain, but the MBC-B values for each individual PA1874-1877 deletion mutant were two- to threefold lower than the MBC-B values for the wild type for gentamicin and ciprofloxacin. In the MBC-B assay, there was no loss of viability observed for either the wild-type or mutant strains for the maximum deliverable concentration of nalixidic acid and rifampin (data not shown). Furthermore, using a Kadouri drip-fed biofilm reactor (27), we confirmed the tobramycin biofilm sensitivity phenotype of the PA1874-1877 deletion mutant (data not shown). These data suggest that PA1874-1877 is acting like a multidrug efflux pump rather than a specific transporter for one antibiotic or one class of antibiotics.
TABLE 3.
MBC-Ps and MBC-Bs of tobramycin, gentamicin, and ciprofloxacin for PA14 and mutant strains
| Strain | Tobramycin
|
Gentamicin
|
Ciprofloxacin
|
|||
|---|---|---|---|---|---|---|
| MBC-P (μg/ml) | MBC-B (μg/ml) | MBC-P (μg/ml) | MBC-B (μg/ml) | MBC-P (μg/ml) | MBC-B (μg/ml) | |
| PA14 | 32a | 100a | 64b | 400b | 4b | 40b |
| ΔPA1874-1877 mutant | 32a | 25a | 64 | 200 | 4 | 20 |
| ΔndvB mutant | 32b | 25b | 64b | 200b | 4b | 10b |
| ndvB transposon insertion ΔPA1874-1877 double mutant | 16 | 12.5 | 32 | 50 | 2 | 5 |
Loss of the PA1874-1877 operon results in the accumulation of tobramycin in the mutant biofilm.
Because the gene products encoded by the PA1874-1877 operon are predicted to function as a transporter, we wanted to determine whether the loss of the PA1874-1877 operon had an effect on the ability of the mutant to pump tobramycin out of the cells of the biofilm. Because the PA1874-1877 operon deletion showed no impairment in planktonic resistance, we devised a biofilm-specific accumulation bioassay that was adapted from other efflux/accumulation assays used for planktonic cells (4, 30). Biofilms of the wild type and PA1874-1877 knockout mutant were formed over 24 h and then exposed to tobramycin for 8 h. After exposure to tobramycin, the wells containing the biofilms were rinsed with phosphate-buffered saline to remove the external tobramycin, and the medium was replaced with a glycine lysis buffer. The biofilms were left overnight at 37°C to allow for complete lysis of the biofilm cells. After ∼16 h, the biofilm/glycine mixture was evaporated to dryness and resuspended in water.
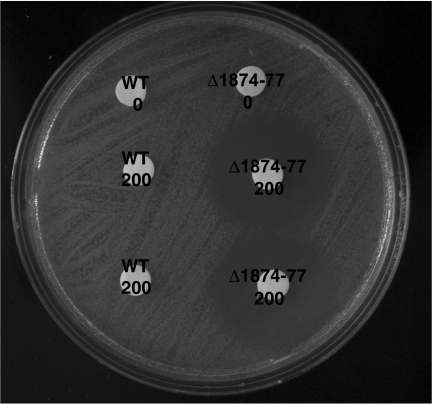
To measure the amount of tobramycin that had accumulated in the biofilms, the resuspended biofilm/glycine mix was spotted onto a lawn of sensitive E. coli (Fig. 2). The presence of tobramycin was noted as a zone of clearing on the lawn of E. coli (25). As predicted, the PA1874-1877 knockout mutant biofilm accumulated more tobramycin than the wild-type biofilm, implying that the loss of this predicted efflux system results in an increase in the accumulation of an antibiotic that is normally an efflux substrate.
FIG. 2.
Tobramycin accumulates in the PA1874-1877 deletion mutant biofilm but not the wild-type biofilm. Wild-type (WT) and mutant (Δ1874-1877) biofilms were exposed to 200 μg/ml tobramycin. The control biofilms were not exposed to tobramycin. The amount of tobramycin that had accumulated in the biofilms was observed as a zone of clearing on the lawn of E. coli, using the white disks as focal points.
The PA1874 gene is more highly expressed in biofilm bacteria.
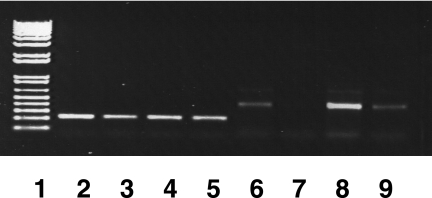
Deleting the entire PA1874-1877 operon or each individual gene in the operon had no effect on the planktonic resistance phenotypes of these mutants; in all cases except for the individual PA1874 knockout, the biofilm resistance of the mutant was reduced compared to the wild type (Table 2). In an effort to understand this biofilm-specific phenotype, we studied the expression profile of the PA1874 gene in planktonic and biofilm cells, by semiquantitative RT-PCR (Fig. 3). We observed that the PA1874 gene was ∼10-fold more highly expressed in biofilm cells than in planktonic cells (Fig. 3). The control for the experiment, rpoD, displayed equal expression levels in planktonic and biofilm cells, as expected. These data suggest that the PA1874-1877 mutants are specifically impaired in biofilm-specific resistance because the PA1874-1877 operon is preferentially expressed in the biofilm.
FIG. 3.
Analysis of PA1874 gene expression by RT-PCR. Expression of the PA1874 gene in planktonic (lanes 6 and 7) and biofilm (lanes 8 and 9) cells of P. aeruginosa PA14 measured by RT-PCR amplification of RNA with primers specific to PA1874 gene (lanes 6 to 9) and rpoD gene (control) (lanes 2 to 5). The RNA templates were isolated from planktonic and biofilm cells, and the different amounts of the templates were used (undiluted in lanes 2, 4, 6, and 8; 1:10 dilution in lanes 3, 5, 7, and 9; data with 1:100 dilution not shown). DNA size markers (0.1 to 12 kb) (1 kb plus DNA ladder; Invitrogen) are shown in lane 1.
Cloned PA1875-1877 genes increase the resistance of planktonic cells.
Intact PA1875-1877 genes from P. aeruginosa PA14 were cloned into a broad-host-range vector, pJB866, which carries the Pm promoter by induction of m-toluic acid (5), resulting in the creation of pLZ866A. Compared with planktonic PA14 cells carrying the vector only, the pLZ866A-containing cells (preinduced with m-toluic acid) showed decreased susceptibility to tobramycin (fourfold MIC change from 0.5 to 2 μg/ml), gentamicin (twofold MIC change from 4 to 8 μg/ml), and ciprofloxacin (twofold MIC change from 1 to 2 μg/ml). Consistent with these observations, when preinduced with m-toluic acid, E. coli DH5α with pLZ866A also showed decreased susceptibility to tobramycin and gentamicin (two- to fourfold MIC increase), but not to ciprofloxacin.
Multiple resistance mechanisms in biofilm-specific resistance.
Previously, we showed that ndvB-derived glucans are important for biofilm-specific resistance to tobramycin through a proposed mechanism involving sequestration of the tobramycin by the glucans (25). The glucans are produced in the cytoplasm of the cell yet function in the periplasm and possibly in the extracellular medium. Since we have not identified the glucan transporter and we have not identified the whole range of substrates of the PA1875-1877 transporter, it is formally possible that the products of the PA1874-1877 operon transport the ndvB-derived glucans, in addition to antibiotics, thus contributing to a documented mechanism of biofilm-specific resistance.
To test whether ndvB and PA1875-1877 function in the same pathway, we constructed a mutant combining the ndvB mutation and the PA1874-1877 deletion and tested the antibiotic resistance of this mutant strain in the 96-well microtiter dish assay (Table 3). The MBC-P values for tobramycin, gentamicin, and ciprofloxacin for this mutant were twofold lower than those for the wild-type strain, indicating a small role for these genes in planktonic resistance. However, the MBC-B of the ndvB and PA1875-1877 mutant to tobramycin, gentamicin, and ciprofloxacin were eightfold lower than the wild-type strain and twofold lower than each individual mutation. This cumulative effect of the combined ndvB and PA1875-1877 mutations over the phenotypes of the single mutants (Table 2) (25) implies that these two genes contribute to two different mechanisms of resistance in biofilms, supporting the hypothesis that there are multiple mechanisms of resistance in these surface-attached communities.
DISCUSSION
Biofilm cells are notably more resistant to antimicrobial agents than their planktonic counterparts. The protective mechanisms are not well understood. We present the identification of a novel biofilm-specific efflux pump in P. aeruginosa. Our conclusions are based on the gene annotations of PA1874-1877 provided by the Pseudomonas genome database (42), confirmation of the importance of these genes in biofilm-specific resistance to antibiotics (Tables 1 and 2), and demonstration of the accumulation of tobramycin in the PA1874-1877 operon knockout mutant biofilm (Fig. 2). Furthermore, MIC results from the planktonic cells carrying the cloned genes have suggested that the PA1874-1877 operon contributes resistance to multiple antibiotics.
The increased accumulation of tobramycin in the PA1874-1877 operon knockout mutant could be due to either an increase in permeability or a decrease in efflux. We favor the idea that the increase in accumulation is due to a decrease in efflux because deletion of the PA1874 gene has no effect on biofilm resistance to tobramycin, while deletion of the individual components of the predicted efflux pump, the PA1875, PA1876, and PA1877 genes, reduces biofilm resistance to two classes of antibiotics (Table 2). If the increase in accumulation of tobramycin in the mutant biofilm were due to altered permeability, we would expect that deletion of the PA1874 gene, encoding a predicted large outer membrane protein, would have the greatest effect on biofilm-specific resistance to tobramycin.
On the basis of the annotations compiled by the Pseudomonas aeruginosa Community Annotation project, it is likely that the PA1875-1877 operon encodes an ABC transporter. The PA1876 gene is predicted to encode an ATP-binding protein due to the presence of several conserved motifs, including Walker boxes A and B, suggesting that the pump should be classified as an ABC transporter (10, 42). Interestingly, there has not been an antibiotic-specific ABC transporter identified in P. aeruginosa. The PA1875 protein has homology to OprM, an outer membrane component of at least two different RND efflux systems in P. aeruginosa (1, 29). In fact, the PA1875 gene has been named opmL (42) for outer membrane protein M family, protein L (18). Additionally, the PA1875 gene is homologous to tolC, the E. coli oprM homolog. TolC is the outer membrane component for several RND and ABC transporters in E. coli (19). Finally, the protein product of the PA1877 gene is a predicted membrane fusion protein of the ABC transporter family that is homologous to HlyD, the membrane fusion protein of the E. coli α-hemolysin secretion system (13).
We have shown that the PA1874 gene is not required for biofilm-specific resistance to tobramycin, gentamicin, and ciprofloxacin (Tables 1 and 2). However, the protein product of PA1874 may be one of the substrates of this efflux system, as it is predicted to encode an outer membrane protein (42). Interestingly, PA1875-1877 genes show strong sequence similarity to the genes that encode the ABC transporter system for LapA, another large outer membrane protein with homology to PA1874. LapB is an ATPase with 58% identity to the PA1876 protein, LapC is a membrane fusion protein with 57% identity to the PA1877 protein, and LapE is an outer membrane protein with 42% similarity to the PA1875 protein (15).
The demonstration of the biofilm-specific expression of the PA1874-1877 operon (Fig. 3) provides an explanation for the observation that mutations of this operon affect biofilm resistance to antibiotics but not planktonic resistance. This type of biofilm-specific expression was also observed for ndvB, another gene shown to be important for biofilm-specific resistance to tobramycin (25). None of these genes have been identified as biofilm-specific genes in genomic and proteomic studies (37, 38, 41), likely due to differences between P. aeruginosa strains and experimental conditions used in these studies, but they may represent a class of genes that are regulated by a common regulatory system that is turned on at different stages in the biofilm life cycle.
Several factors, such as the extracellular polysaccharide matrix, high cell density, and slow growth, have been implicated in the increase in resistance that biofilm cells exhibit over their planktonic counterparts (24). Due to the success of a genetic screen that we carried out, some additional mechanisms are being proposed (25). Individually, these factors and mechanisms might add only a modest level of resistance, as evidenced by the two- or threefold reduction in resistance that we have observed in our studies (Table 2) (25). However, when added together, they contribute to the overall level of increased resistance that allows biofilms to survive in conditions where their planktonic counterparts do not.
Acknowledgments
This work was supported by a grant from the Canadian Cystic Fibrosis Foundation.
We thank Xian-Zhi Li, Peter Newell, and Sandeep Tamber for critical review of the manuscript.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 432624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderl, J. N., M. J. Franklin, and P. S. Stewart. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 441818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, NY.
- 4.Berlanga, M., N. Ruiz, J. Hernandez-Borrell, T. Montero, and M. Vinas. 2000. Role of the outer membrane in the accumulation of quinolones by Serratia marcescens. Can. J. Microbiol. 46716-722. [PubMed] [Google Scholar]
- 5.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, P. Karunakaran, and S. Valla. 1997. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 3835-53. [DOI] [PubMed] [Google Scholar]
- 6.Brooun, A., S. Liu, and K. Lewis. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M. R., D. G. Allison, and P. Gilbert. 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J. Antimicrob. Chemother. 22777-780. [DOI] [PubMed] [Google Scholar]
- 8.Caiazza, N. C., J. H. Merritt, K. M. Brothers, and G. A. O'Toole. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 1893603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 1832888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73241-268. [DOI] [PubMed] [Google Scholar]
- 11.De Kievit, T. R., M. D. Parkins, R. J. Gillis, R. Srikumar, H. Ceri, K. Poole, B. H. Iglewski, and D. G. Storey. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 451761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, D. J., D. G. Allison, M. R. W. Brown, and P. Gilbert. 1991. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. J. Antimicrob. Chemother. 27177-184. [DOI] [PubMed] [Google Scholar]
- 13.Gentschev, I., G. Dietrich, and W. Goebel. 2002. The E. coli α-hemolysin secretion system and its use in vaccine development. Trends Microbiol. 1039-45. [DOI] [PubMed] [Google Scholar]
- 14.Gillis, R. J., K. G. White, K.-H. Choi, V. E. Wagner, H. P. Schweizer, and B. H. Iglewski. 2005. Molecular basis of azithromycin-resistant Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 493858-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49905-918. [DOI] [PubMed] [Google Scholar]
- 16.Hoyle, B. D., and W. J. Costerton. 1991. Bacterial resistance to antibiotics: the role of biofilms. Prog. Drug Res. 3791-105. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 10014339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jo, J. T. H., F. S. L. Brinkman, and R. E. Hancock. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 471101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koronakis, V., J. Eswaran, and C. Hughes. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73467-489. [DOI] [PubMed] [Google Scholar]
- 20.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10S122-S129. [DOI] [PubMed] [Google Scholar]
- 21.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64159-204. [DOI] [PubMed] [Google Scholar]
- 22.Li, Y., T. Mima, Y. Komori, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. A new member of the tripartite multidrug efflux pumps, MexVW-OprM, in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 52572-575. [DOI] [PubMed] [Google Scholar]
- 23.Lynch, S. V., L. Dixon, M. R. Benoit, E. L. Brodie, M. Keyhan, P. Hu, D. F. Ackerley, G. L. Andersen, and A. Matin. 2007. Role of the rapA gene in controlling antibiotic resistance of Escherichia coli biofilms. Antimicrob. Agents Chemother. 513650-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 934-39. [DOI] [PubMed] [Google Scholar]
- 25.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426306-310. [DOI] [PubMed] [Google Scholar]
- 26.Masuda, N., E. Sakagowa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 443322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merritt, J. H., D. E. Kadouri, and G. A. O'Toole. 2005. Growing and analyzing static biofilms, p. 1B.1.1-1B.1.17. In Current protocols in microbiology, vol. 1. J. Wiley & Sons, Hoboken, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mima, T., H. Sekiya, T. Mizushima, T. Kuroda, and T. Tsuchiya. 2005. Gene cloning and properties of the RND-type multidrug efflux pumps MexPQ-OpmE and MexMN-OprM from Pseudomonas aeruginosa. Microbiol. Immunol. 49999-1002. [DOI] [PubMed] [Google Scholar]
- 29.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression of Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortimer, P. G. S., and L. J. V. Piddock. 1991. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 28639-653. [DOI] [PubMed] [Google Scholar]
- 31.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30295-304. [DOI] [PubMed] [Google Scholar]
- 32.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to the study of biofilms. Methods Enzymol. 31091-109. [DOI] [PubMed] [Google Scholar]
- 33.Pardee, A. B., F. Jacob, and J. Monod. 1959. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase in E. coli. J. Mol. Biol. 1165-178. [Google Scholar]
- 34.Poole, K. 2005. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 5620-51. [DOI] [PubMed] [Google Scholar]
- 35.Poole, K. 2004. Efflux-mediated multiresistance in gram-negative bacteria. Clin. Microbiol. Infect. 1012-26. [DOI] [PubMed] [Google Scholar]
- 36.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 2681899-1902. [DOI] [PubMed] [Google Scholar]
- 37.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 1841140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waite, R. D., A. Papakonstantinopoulou, E. Littler, and M. A. Curtis. 2005. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J. Bacteriol. 1876571-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walters, M. C., III, F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westover, B. P., J. D. Buhler, J. L. Sonnenburg, and J. I. Gordon. 2005. Operon prediction without a training set. Bioinformatics 21880-888. [DOI] [PubMed] [Google Scholar]
- 41.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413860-864. [DOI] [PubMed] [Google Scholar]
- 42.Winsor, G. L., R. Lo, S. J. Ho Sui, K. S. E. Ung, S. Huang, D. Cheng, W. H. Ching, R. E. W. Hancock, and F. S. L. Brinkman. 2005. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 33D338-D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, L., X.-Z. Li, and K. Poole. 2000. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob. Agents Chemother. 44287-293. [DOI] [PMC free article] [PubMed] [Google Scholar]