Abstract
Three active-site cysteine l,d-transpeptidases can individually anchor the Braun lipoprotein to the Escherichia coli peptidoglycan. We show here that two additional enzymes of the same family form peptide bonds between the third residues of peptidoglycan stems, generating meso-DAP3→meso-DAP3 unusual cross-links. This activity partially replaces the d,d-transpeptidase activity of penicillin-binding proteins.
The peptidoglycan of Escherichia coli results from the polymerization of a disaccharide-peptide subunit containing a linear stem pentapeptide (Fig. 1). The final polymerization steps involve the formation of the glycan strands by glycosyltransferases and cross-linking of peptide stems by transpeptidases. The latter reaction is mainly catalyzed by d,d-transpeptidases that form d-Ala4→meso-DAP3 cross-links (4-3 cross-links; Fig. 1A). The d,d-transpeptidases belong to the penicillin-binding protein (PBP) family and are the targets of β-lactams. A small proportion of the cross-links are unlikely to be generated by PBPs since they involve two meso-DAP residues (meso-DAP3→meso-DAP3 or 3-3 cross-links) (5). The nature of the corresponding transpeptidase and its relationship to PBPs has remained speculative for several decades (6). Recently, l,d-transpeptidases that catalyze formation of 3-3 peptidoglycan cross-links have been identified in gram-positive bacteria (7, 9). In Enterococcus faecium, the enzyme can bypass the PBPs, resulting in high-level resistance to β-lactam antibiotics (11). The catalytic domain of the l,d-transpeptidases displays a new fold (1, 2) and contains an active-site Cys residue (9, 10) instead of Ser in PBPs (4).
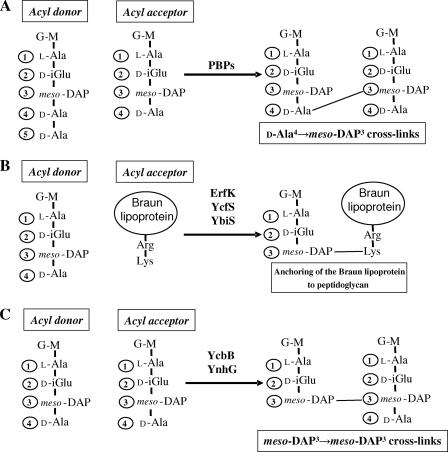
FIG. 1.
Reaction catalyzed by the d,d-transpeptidases (PBPs) and the l,d-transpeptidases. (A) The disaccharide-peptide subunit is composed of β1-4-linked N-acetylglucosamine (G) and N-acetylmuramic acid (M) and a linear stem pentapeptide linked to the d-lactoyl group of N-acetylmuramic acid by an amide bond (5). The third position of the pentapeptide stem is occupied by a meso-diaminopymelyl residue (meso-DAP) linked to the γ-carboxyl of d-glutamic acid (l-Ala1-d-iGlu2-meso-DAP3-d-Ala4-d-Ala5). PBPs cleave the d-Ala4-d-Ala5 peptide bond of a peptidoglycan donor stem and link the carbonyl of d-Ala4 to the side chain amine of meso-DAP3 in a peptidoglycan acceptor stem to form d-Ala4→meso-DAP3 cross-links. (B) The ErfK, YcfS, and YbiS l,d-transpeptidases anchor the Braun lipoprotein to peptidoglycan. These enzymes cleave the meso-DAP3-d-Ala4 peptide bond of a peptidoglycan donor stem and link the carbonyl of meso-DAP3 to the side chain amine of the C-terminal Lys residue of the Braun lipoprotein. (C) The YcbB and YnhG l,d-transpeptidases generate the meso-DAP3→meso-DAP3 peptidoglycan cross-links. These enzymes cleave the meso-DAP3-d-Ala4 peptide bond of a peptidoglycan donor stem and link the carbonyl of meso-DAP3 to the side chain amine of meso-DAP3 in a peptidoglycan acceptor stem.
In a recent attempt to identify the mode of synthesis of meso-DAP3→meso-DAP3 cross-links in E. coli, we deleted from the chromosome of strain BW25113 four genes encoding the proteins ErfK, YcfS, YbiS, and YnhG that display sequence similarity with the catalytic domain of l,d-transpeptidases from gram-positive bacteria (8). Unexpectedly, peptidoglycan analyses of the quadruple mutant, BW25113Δ4, and of its derivatives obtained by transcomplementation with each of the four genes have led to the identification of three l,d-transpeptidases that anchor the Braun lipoprotein to the peptidoglycan (ErfK, YcfS, and YbiS; Fig. 1B) (8). In contrast, the enzymes for synthesis of meso-DAP3→meso-DAP3 cross-links were not fully identified since overexpression of the fourth gene, ynhG, increased the abundance of 3-3 cross-links, whereas deletion of ynhG did not abolish their formation. In the present study, we identified a fifth l,d-transpeptidase, YcbB, and showed that this enzyme and YnhG are the only l,d-transpeptidases for synthesis of meso-DAP3→meso-DAP3 peptidoglycan cross-links in E. coli (Fig. 1C).
The ycbB gene was deleted from the chromosome of E. coli BW25113 using the procedure described by Datsenko and Wanner (3). Briefly, the linear PCR product used for gene replacement was obtained by amplification of the kanamycin resistance gene cassette of plasmid pKD4 with two primers (5′-TAAAATAACAGCCTGGCTATTCAGAGTATGATAAAAACAGGGGGCGTGTAGGCTGGAGCTGCTTC-3′ and 5′-AATCGCCCCCAATCATGCTAATTATTACGACAACTGATTTCCCCGCATATGAATATCCTCCTTAG-3′), which contained sequences flanking ycbB (underlined). Phage P1 was used to transduce the kanamycin cassette from the ycbB locus of E. coli BW25113 to that of the quadruple mutant E. coli BW25113Δ4 (8). The resulting mutant, BW25113Δ5, lacked ycbB in addition to erfK, ycfS, ybiS, and ynhG. Deletion of the five genes did not alter the growth characteristics in brain heart infusion broth (Difco). For transcomplementation analysis, the ycbB gene of E. coli BW25113 was amplified with the primers 5′-CGGAATTCTAAGGAGATACTA GATGTTGCTTAATATGATGTGTG-3′ and 5′-CCTCTAGATTACCTGATTAATTGTTCCGC-3, digested with EcoRI and XbaI (the sites are underlined), and cloned into the vector pTRC99a (Pharmacia) under the control of the plac promoter. The resulting plasmid, pTRC99aΩycbB, was introduced into BW25113Δ5 using ampicillin (100 μg/ml) for selection. Expression of ycbB was induced with IPTG (isopropyl-β-d-thiogalactopyranoside; 0.02 mM) at an optical density of 0.8 at 600 nm in cultures performed in brain heart infusion broth at 37°C. The peptidoglycan was extracted at the stationary phase and digested with muramidases (8). The resulting muropeptides were separated by reversed-phase high-performance liquid chromatography (rp-HPLC) (Fig. 2A) and identified by mass spectrometry (Fig. 2B), as previously described (8).
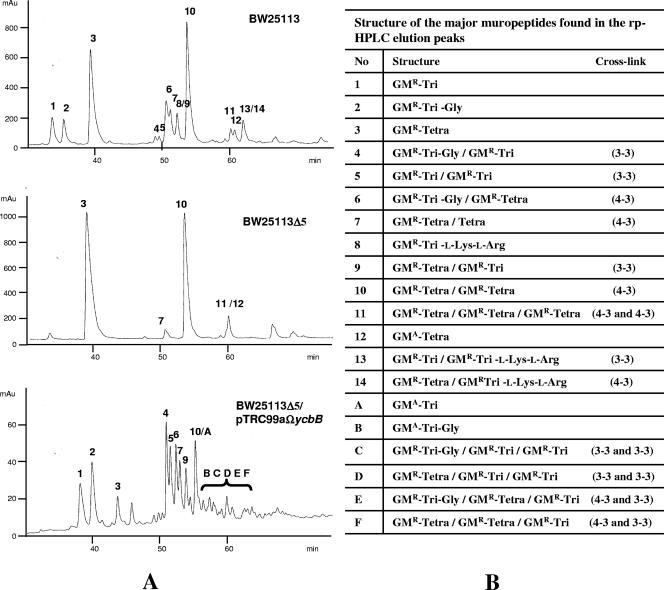
FIG. 2.
Structure of the peptidoglycan of E. coli BW25113, BW25113Δ5, and BW25113Δ5/pTRC99aΩycbB. (A) rp-HPLC profiles of muropeptides obtained by digestion of the peptidoglycan by muramidases. Muropeptides 1 to 14 and A have been assigned to specific peaks detected by the absorbance at 210 nm. Muropeptides B to F could not be assigned to any specific absorbance peaks due to their low abundance; mAU, absorbance units (103). (B) Identification of muropeptides in the main peaks by mass spectrometry. GMR, N-acetyl-glucosamine linked to reduced N-acetyl-muramic acid; GMA, N-acetyl-glucosamine linked to anhydro-N-acetyl-muramic acid; Tri, tripeptide l-Ala-d-iGlu-meso-DAP; Tri-Gly, tetrapeptide l-Ala-d-iGlu-meso-DAP-Gly; Tetra, tetrapeptide l-Ala-d-iGlu-meso-DAP-d-Ala. The type of cross-links (3-3 or 4-3) is indicated in parentheses for dimers and trimers.
Deletion of the five genes encoding l,d-transpeptidases in E. coli BW25113Δ5 resulted in the disappearance of 9 out of the 14 muropeptides identified by rp-HPLC and mass spectrometry in the parental strain E. coli BW25113 (Fig. 2). The nine missing muropeptides included all of the muropeptides (8, 13, and 14) that contained a tripeptide stem substituted by a fragment of the Braun lipoprotein. This result was expected since BW25113Δ5 did not produce the l,d-transpeptidases previously shown to anchor the Braun lipoprotein to peptidoglycan (ErfK, YcfS, and YbiS; Fig. 1B) (8). The nine missing muropeptides also comprised all peptidoglycan dimers containing meso-DAP3→meso-DAP3 cross-links (muropeptides 4, 5, 9, and 13). This observation shows that E. coli does not produce any l,d-transpeptidase for formation of 3-3 cross-links in addition to YcbB (the present study) and YnhG (8). The quintuple deletion also led to the disappearance of muropeptides containing a free tripeptide stem (muropeptides 1, 5, and 9), indicating that the l,d-transpeptidases cleaved the meso-DAP3-d-Ala4 peptide bonds (l,d-carboxypeptidase activity). Finally, muropeptides containing a modified tetrapeptide stem ending in Gly instead of d-Ala4 (muropeptides 2, 4, and 6) were absent, showing that Gly was used as an acyl acceptor resulting in the exchange of d-Ala4 by Gly.
Expression of ycbB in BW25113Δ5 restored production of all missing muropeptides except those resulting from the anchoring of the Braun lipoprotein, revealing that the YcbB l,d-transpeptidase is sufficient for the formation of 3-3 cross-links, hydrolysis of d-Ala4, and exchange of d-Ala4 by Gly. In comparison to the parental strain, the abundance of these muropeptides was increased due to high-level expression of ycbB cloned into the expression vector pTRC99a. For example, the relative abundance of cross-links generated by l,d-transpeptidation was estimated to increase from 5 to 50% based on integration of the rp-HPLC peak areas (Fig. 2). Overexpression of ycbB also led to the formation of six additional muropeptides (A to F) due to an increase in the l,d-transpeptidase, l,d-carboxypeptidase, and exchange activities. For example, the additional muropeptides included a trimer containing two 3-3 cross-links (muropeptide C). This trimer was not detectable in the parental strain in which the substantial majority of the cross-links were generated by PBPs (4-3 cross-links).
In conclusion, we have shown that E. coli produces five l,d-transpeptidases with two distinct functions (Fig. 1). ErfK, YcfS, and YbiS anchor the Braun lipoprotein to the peptidoglycan, whereas YcbB and YnhG form the meso-DAP3→meso-DAP3 peptidoglycan cross-links. In addition, all five l,d-transpeptidases appear to hydrolyze d-Ala4 and to exchange this residue with Gly since overexpression of the five l,d-transpeptidase genes individually led to the accumulation of tripeptide stems and of tetrapeptide stems ending in Gly.
Acknowledgments
This study was supported by the European Community (COBRA, contract LSHM-CT-2003-503335, 6th PCRD), the National Institute of Allergy and Infectious Diseases (grant R01 AI45626), and the Fondation pour la Recherche Médicale (Equipe FRM 2006, DEQ200661107918).
Footnotes
Published ahead of print on 2 May 2008.
REFERENCES
- 1.Biarrotte-Sorin, S., J. E. Hugonnet, V. Delfosse, J. L. Mainardi, L. Gutmann, M. Arthur, and C. Mayer. 2006. Crystal structure of a novel beta-lactam-insensitive peptidoglycan transpeptidase. J. Mol. Biol. 359533-538. [DOI] [PubMed] [Google Scholar]
- 2.Bielnicki, J., Y. Devedjiev, U. Derewenda, Z. Dauter, A. Joachimiak, and Z. S. Derewenda. 2006. B. subtilis YkuD protein at 2.0 Å resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes. Proteins 62144-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frere, J. M., and B. Joris. 1985. Penicillin-sensitive enzymes in peptidoglycan biosynthesis. Crit. Rev. Microbiol. 11299-396. [DOI] [PubMed] [Google Scholar]
- 5.Glauner, B., J. V. Holtje, and U. Schwarz. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 26310088-10095. [PubMed] [Google Scholar]
- 6.Goffin, C., and J. M. Ghuysen. 2002. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 66702-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magnet, S., A. Arbeloa, J. L. Mainardi, J. E. Hugonnet, M. Fourgeaud, L. Dubost, A. Marie, V. Delfosse, C. Mayer, L. B. Rice, and M. Arthur. 2007. Specificity of l, d-transpeptidases from gram-positive bacteria producing different peptidoglycan chemotypes. J. Biol. Chem. 28213151-13159. [DOI] [PubMed] [Google Scholar]
- 8.Magnet, S., S. Bellais, L. Dubost, M. Fourgeaud, J. L. Mainardi, S. Petit-Frere, A. Marie, D. Mengin-Lecreulx, M. Arthur, and L. Gutmann. 2007. Identification of the l, d-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J. Bacteriol. 1893927-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mainardi, J. L., M. Fourgeaud, J. E. Hugonnet, L. Dubost, J. P. Brouard, J. Ouazzani, L. B. Rice, L. Gutmann, and M. Arthur. 2005. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J. Biol. Chem. 28038146-38152. [DOI] [PubMed] [Google Scholar]
- 10.Mainardi, J. L., J.-E. Hugonnet, F. Rusconi, M. Fourgeaud, L. Dubost, A. Nguekam Moumi, V. Delfosse, C. Mayer, L. Gutmann, L. Rice, and M. Arthur. 2007. Unexpected inhibition of peptidoglycan l, d-transpeptidase from Enterococcus faecium by the β-lactam imipenem. J. Biol. Chem. 28230414-30422. [DOI] [PubMed] [Google Scholar]
- 11.Mainardi, J. L., R. Legrand, M. Arthur, B. Schoot, J. van Heijenoort, and L. Gutmann. 2000. Novel mechanism of beta-lactam resistance due to bypass of dd-transpeptidation in Enterococcus faecium. J. Biol. Chem. 27516490-16496. [DOI] [PubMed] [Google Scholar]