Abstract
Biofilms are polymicrobial, with diverse bacterial species competing for limited space and nutrients. Under healthy conditions, the different species in biofilms maintain an ecological balance. This balance can be disturbed by environmental factors and interspecies interactions. These perturbations can enable dominant growth of certain species, leading to disease. To model clinically relevant interspecies antagonism, we studied three well-characterized and closely related oral species, Streptococcus gordonii, Streptococcus sanguinis, and cariogenic Streptococcus mutans. S. sanguinis and S. gordonii used oxygen availability and the differential production of hydrogen peroxide (H2O2) to compete effectively against S. mutans. Interspecies antagonism was influenced by glucose with reduced production of H2O2. Furthermore, aerobic conditions stimulated the competence system and the expression of the bacteriocin mutacin IV of S. mutans, as well as the H2O2-dependent release of heterologous DNA from mixed cultures of S. sanguinis and S. gordonii. These data provide new insights into ecological factors that determine the outcome of competition between pioneer colonizing oral streptococci and the survival mechanisms of S. mutans in the oral biofilm.
Bacterial biofilms are complex microbial communities found on virtually all mucosal surfaces in humans and animals (8). Bacterial communities indigenous to humans consist of dozens to hundreds of different species or phylotypes, as has been shown in gut, vaginal, and oral biofilms (11, 12, 46). Changes in biofilm ecology can lead to diseases such as inflammatory bowel disease (28), vaginitis (12), and caries (46). Interactions between the bacterial species that reside in the biofilm influence the composition of the communities (21, 45), a process termed bacterial interference (16). As a consequence, changes in the populating species influence whether the biofilm favors either a benign or otherwise beneficial homeostatic state or the development of disease. In the beneficial state, the bacterial community restricts potentially harmful bacteria from colonizing, protecting the host from disease. However, some disease-causing microorganisms are present under healthy conditions, and shifts in the population may affect the virulence of a species; e.g., virulence genes that require diffusible intraspecies signal molecules for expression might only be transcribed when a certain bacterial density is archived. For example, caries-causing Streptococcus mutans can become a predominant member of the community under appropriate conditions, leading to dental caries formation (24). How changes in the species composition of dental biofilms occur is not well understood.
The polymicrobial community dental plaque is one of the best-studied biofilms (for a recent review, see reference 22). In the oral biofilm, more than 500 species or phylotypes have been identified (20, 36). The cell density can reach 1011 CFU ml−1 (11). The biofilm is challenged by frequent changes in environmental conditions, e.g., food intake, temperature, pH change, and salivary flow. Perhaps as a response to environmental challenges, the oral biofilm community evolved with individual members assuming specialized functions, e.g., primary and secondary colonizers (reviewed in reference 15), including members that can metabolize excreted products (such as lactic acid) produced by other species (35).
Approximately 20% of the oral bacteria are streptococci (27, 47). The oral streptococci pioneer early dental plaque formation and have a specific temporal and spatial distribution that is crucial for the development of oral biofilms (44). Reflecting their genetic similarity, the oral streptococci have similar metabolic activities, which include the ability to metabolize a wide variety of carbohydrates (50). The streptococci compete for adhesion-binding sites on the saliva-coated tooth surface (31) and are able to produce antimicrobial compounds, such as bacteriocins and hydrogen peroxide (H2O2) (18). When present in the oral biofilm in high numbers, the pioneer colonizers Streptococcus gordonii and Streptococcus sanguinis can antagonize S. mutans, as suggested by clinical studies (5) and sequential inoculation experiments in germfree rats (5, 29).
S. mutans can, however, become dominant in oral biofilms, leading to dental caries development. S. mutans dominance depends on competition with S. sanguinis and is influenced by the production of antimicrobial compounds (18). For example, S. mutans shows cell density-dependent expression of bacteriocins mutacin I and mutacin IV when inoculated before S. sanguinis in an in vitro plate assay. Conversely, S. sanguinis produces H2O2 when inoculated before S. mutans (19). These compounds are able to inhibit the competing species to keep their numbers low.
In the present study, we tested the hypothesis that S. sanguinis and S. gordonii grown under aerobic conditions are better able to inhibit S. mutans than when grown under anaerobic conditions. Our results show that the interspecies interactions were influenced by the availability of oxygen and the differential production of H2O2 by S. sanguinis and S. gordonii. Furthermore, in the presence of oxygen, S. sanguinis and S. gordonii released more DNA into the growth medium. Stimulation of the competence system of S. mutans leads to the production of higher levels of antistreptococcal bacteriocins. Hence, oxygen availability appears to be a crucial factor in the interspecies competition between S. mutans and S. sanguinis and S. gordonii in the oral biofilm.
MATERIALS AND METHODS
Bacterial strains and media.
S. mutans UA140 (40), S. sanguinis SK36 (54), and S. gordonii DL1 (34) and their derivatives were routinely grown at 37°C anaerobically (90% N2, 5% CO2, 5% H2) or aerobically (5% CO2) in brain heart infusion broth (BHI; Difco, Sparks, MD) or on BHI agar plates. Filter-sterilized glucose was supplemented when indicated from a freshly prepared stock solution (20%). Escherichia coli DH5α cells were grown aerobically at 37°C in Luria-Bertani medium (Difco). When required, antibiotics were supplemented at the following concentrations: spectinomycin at 500 μg ml−1 for S. sanguinis and S. gordonii and at 100 μg ml−1 for E. coli, kanamycin at 300 μg ml−1 for S. sanguinis and S. gordonii and at 25 μg ml−1 for E. coli, and ampicillin 100 μg ml−1 for E. coli.
DNA manipulations.
Standard recombinant DNA manipulations were used. Restriction enzymes and DNA ligase were obtained from New England Biolabs (Beverly, MA) and used as specified by the manufacturer. PCR products were cloned into the pGEM-T kit from Promega (Madison, WI). All plasmids were extracted and purified from E. coli with a Qiagen Miniprep kit (Valencia, CA). DNA extracted from agarose gels (1%) was purified with a Qiagen QIAquick gel extraction kit. PCR was performed with a Mastercycler thermocycler (Eppendorf, Westbury, NY) according to the manufacturer's protocol. GoTaq-DNA polymerase was obtained from Promega, and Phusion high-fidelity DNA polymerase was obtained from New England Biolabs. Primer sequences (Table 1) were designed using sequence data obtained from the Los Alamos National Laboratory Oral Pathogens Sequence Database (http://www.oralgen.lanl.gov) and synthesized by Integrated DNA Technologies (Coralville, IA).
TABLE 1.
Primers used in this study
| Primer | Function | Nucleotide sequence (5′ to 3′) |
|---|---|---|
| poxCF | pox complementation | CGGGTACCTAGTTTCTTCGAAAGAACACAGC |
| poxCR | pox complementation | ACGCGTCGACTTATTTAATTGCGCGTGATTGC |
| poxKOF | pox inactivation | GGATCCCATGGGGCGTAGATACCATC |
| poxKOR | pox inactivation | AAGCTTACCACCAAATCCAGCGTAGA |
| nlmA RT F | Real-time PCR nlmA | TATCGGGTGGAGAAGCAGTC |
| nlmA RT R | Real-time PCR nlmA | TCCCCAAGTGCCTACACAAT |
| mutA RT F | Real-time PCR mutA | CTATTGTGGCAAGCAACGAC |
| mutA RT R | Real-time PCR mutA | AAACTAGGATTTTTCACCCCTGT |
| comE RT F | Real-time PCR comE | AGCCCATAAGCTCTGCCTTT |
| comE RT R | Real-time PCR comE | AGCGATGGCACTGAAAAAGT |
| comC RT F | Real-time PCR comC | GAGATTATATGGGCGGAAGC |
| comC RT R | Real-time PCR comC | TTTCCCAAAGCTTGTGTAAAA |
| 16S RT F S.m | Real-time PCR 16S rRNA | GATAATTGATTGAAAGATGCAAGC |
| 16S RT R S.m | Real-time PCR 16S rRNA | ATTCCCTACTGCTGCCTCCC |
| 16S strep F | DNA release quantification | AAGCAACGCGAAGAACCTTA |
| 16S strep R | DNA release quantification | GTCTCGCTAGAGTGCCCAAC |
Construction of Pox− mutants and complemented strains.
For the insertional inactivation of the spxB gene, which encodes pyruvate oxidase (Pox), an internal DNA fragment of the spxB gene from S. sanguinis was amplified by PCR using the primers poxKOF and poxKOR (Table 1), which incorporated restriction sites for BamHI and HindIII. The PCR fragments were cloned into the vector pGEM-T. After digestion with BamHI/HindIII, the fragment was ligated into compatible sites on the suicide vector pFW5, which contains a spectinomycin resistance marker (gene aad9) (39), to generate pFW5-pox. The resulting constructs were confirmed by restriction analysis before integration into the chromosome of S. sanguinis and S. gordonii via single-crossover homologous recombination using a transformation protocol described previously (52). Antibiotic-resistant isolates were tested by PCR to confirm the genotypes. Representative clones were named JKH2 (SK36 Pox−) and JKH1 (DL1 Pox−) and were frozen for storage. To complement the Pox− mutants, the spxB gene, including the promoter, was PCR amplified from S. sanguinis chromosomal DNA by using Phusion high-fidelity DNA polymerase and the primers poxCF and poxCR. These primers incorporated restriction sites for KpnI and SalI. The PCR fragment was sequentially digested with KpnI and SalI and ligated into the streptococcal shuttle plasmid pDL276 (9). The resulting plasmid, pDL276-pox, was transformed into JKH2 and JKH1. Transformants were then tested for the ability to inhibit growth of S. mutans.
RNA isolation, cDNA synthesis, and real-time PCR.
S. mutans was inoculated on BHI plates with a sterile cotton tip. The plates were incubated anaerobically or aerobically, and the cells were subsequently scraped from the plate after 7 h of growth. RNA was isolated as described previously (31), except that the DNase I (Promega) treatment was extended to 5 h. RNA concentration was determined spectrophotometrically. RNA (10 μg) was reverse transcribed into cDNA with random hexamer primers (Promega) as described previously (55) using M-MLV reverse transcriptase (Promega) according to the manufacturer's instructions. The expression of the mutacin I and IV genes, mutA and nlmA, and the com genes, comC and comE, was measured. Real-time PCR was performed using the FullVelocity Sybr Green QPCR master mix (Stratagene, Cedar Creek, TX) according to the manufacturer's instructions. Quantification was performed by using an Mx3000P real-time PCR system (Stratagene) for amplification and fluorescent detection. Total cDNA abundance between test samples was normalized by using the 16S rRNA gene as a housekeeping gene control.
H2O2 concentration.
The concentration of H2O2 was determined according to a protocol described previously (33). Briefly, bacteria were grown in glucose-free tryptic soy broth (TSB; Difco) supplemented with 1% glucose when indicated. Samples (1 ml) were taken at the times indicated, centrifuged (16,000 × g) for 5 min, and transferred (0.2 ml) to a new incubation tube. A reaction solution (0.8 ml of 10 mM phosphate buffer [pH 7.4] with 0.16 mM o-dianisidine [ICN, Aurora, OH], 1.2 μg of horseradish peroxidase (Pierce, Rockford, IL) ml−1, 0.02% Triton X-100) was added, and the reaction mixture was incubated at 37°C for 20 min. The absorbance at 570 nm was determined, and the concentration was calculated from a standard curve prepared for each experiment from a 30% H2O2 stock solution (Sigma, St. Louis, MO).
Preparation of S. sanguinis and S. gordonii conditioned media for S. mutans growth curves.
S. sanguinis or S. gordonii were grown overnight in BHI media (containing 0.2% glucose in original formula) and inoculated into 40 ml of BHI in tubes to an A600 of 0.025. When indicated, 30 μg of catalase (Sigma) was added to the culture. The cells were then incubated aerobically on a rocking platform at 37°C until the A600 reached 0.6 (ca. 107 bacteria ml−1). The cultures were harvested by centrifugation at 1,700 × g, and 20 ml of the supernatant (conditioned medium) was removed and filter sterilized. The sterile conditioned medium was supplemented with 0.25% glucose, S. mutans was inoculated to an A600 of 0.025, and the growth of static cultures was monitored by measuring the absorbance at the indicated time points.
DNA isolation from culture supernatants.
To determine the amount of DNA released during growth in coculture, S. sanguinis, S. gordonii, and the isogenic Pox− mutants were separately grown overnight in BHI (with 500 μg of spectinomycin ml−1 for the mutants). Next, separate tubes for S. sanguinis wild-type plus S. gordonii wild-type and for JKH1 plus JKH2 were inoculated in 5 ml of BHI medium (no antibiotic added) as a 1-to-1 mixed culture. The cells were incubated overnight anaerobically as static cultures or aerobically on a rocking platform. At harvest, the A600 and CFU were determined by serial dilution and plating. Cells were removed by centrifugation for 2 min at 16,000 × g, 1 ml of the supernatant was transferred to a new tube, and 0.5 ml of phenol was added and vortex mixed for 30 s. To precipitate DNA, the mixture was centrifuged for 5 min at 16,000 × g, and 0.8 ml of the aqueous solution was removed and mixed with 80 μl of sodium acetate (3 M, pH 5.2) and 500 μl of 100% 2-propanol. The solution was then centrifuged for 10 min at 16,000 × g, the liquid was decanted, and the precipitated sample was air dried and suspended in 25 μl of deionized H2O. Using universal primers for 16S rRNA genes of S. sanguinis and S. gordonii (16S strep F and 16S strep R), the relative quantity of DNA was estimated to a standard curve of dilutions (1:1, 1:5, 1:25, 1:125, and 1:625) of chromosomal DNA of S. sanguinis. DNA from the standard dilutions and samples from S. sanguinis and S. gordonii were amplified by real-time PCR, and the relative quantity of DNA in the streptococcal samples was calculated in comparison to the cycle threshold (CT) values of the DNA dilutions as described previously (17). Real-time PCR was performed by using the FullVelocity Sybr Green QPCR Master Mix (Stratagene) according to the manufacturer's instructions. The experiment was repeated three times on different days.
Competition assays on solid medium and in liquid medium.
To assess competitive growth, a protocol described previously was used with modifications (18). Briefly, 8 μl of an overnight culture of each species in BHI medium was inoculated onto a BHI agar plate (supplemented with 1% glucose when indicated) as the pioneer colonizer. After incubation overnight (16 h) under the specified conditions, 8 μl of the competing species was inoculated next to the pioneer colonizer such that the colonies almost touched each other. The plate was incubated overnight. Growth inhibition was assessed by the presence of a proximal zone of inhibition at the intersection with the pioneer colony. For competition assays in liquid media, cells were grown in BHI medium overnight, and either S. sanguinis or S. gordonii (15 μl of each) and S. mutans (15 μl) was inoculated simultaneously into 1 ml of fresh BHI (supplemented with 1% glucose when indicated) in a Costar 24-well cell culture cluster (Corning, Inc., Corning, NY) for biofilm growth. The cells were incubated overnight in static culture in the presence of 5% CO2. Cells were dispersed by vigorous pipetting, serially diluted (10−5 to 10−7), and plated on BHI agar plates in duplicates, and the CFU counts were determined.
Statistical analysis.
Statistical analysis of data was performed with the QuickCalcs online calculators (http://www.graphpad.com/quickcalcs/index.cfm) using the t test software to compare the means of two groups. The data were considered significantly different if the two-tailed P value was ≤0.05.
RESULTS
Influence of oxygen availability on interspecies competition.
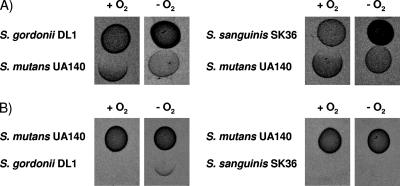
To test the hypothesis that competitiveness increases under aerobic conditions which favor H2O2 production, we analyzed the inhibitory potential of oral streptococci when grown with (aerobic: 5% CO2) or without (anaerobic: 5% CO2, 5% H2, 90% N2) oxygen. Under aerobic growth conditions, the first inoculated competing species inhibited the growth of the opposing strain (Fig. 1). During anaerobic growth, however, S. gordonii and S. sanguinis no longer inhibited S. mutans (Fig. 1A). S. mutans inhibited S. gordonii and S. sanguinis during anaerobic growth (Fig. 1B).
FIG. 1.
Inhibition ability of S. sanguinis, S. gordonii, and S. mutans when grown with or without oxygen. (A) S. gordonii or S. sanguinis was inoculated first and grown for 16 h at 37°C with (+O2) or without oxygen (−O2), S. mutans was then inoculated next to the pioneer colonizer, and the plates incubated overnight. (B) S. mutans was inoculated first and grown for 16 h at 37°C with (+O2) or without oxygen (−O2), and then S. gordonii or S. sanguinis was inoculated next to the pioneer colonizer and the plates were incubated overnight.
Pox-mediated inhibition of S. mutans under aerobic growth conditions.
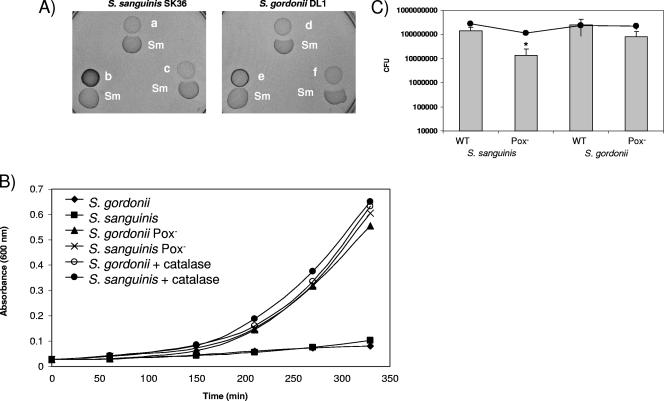
During aerobic growth, the Pox (pyruvate oxidase) converts pyruvate into acetyl phosphate, carbon dioxide, and H2O2. The Pox-expressing wild-type S. sanguinis (Fig. 2A; a) and S. gordonii (Fig. 2A;d) inhibited S. mutans. S. mutans was not inhibited by the tested Pox− mutants of S. sanguinis (Fig. 2Ab) and S. gordonii (Fig. 2Ae). Complementation of the spxB mutation restored the growth-inhibiting ability of both Pox− mutants (Fig. 2Ac and f), indicating that under aerobic conditions competitive growth inhibition caused by H2O2 production is Pox dependent.
FIG. 2.
Inhibition ability and competitiveness of wild-type and Pox− mutant of S. sanguinis and S. gordonii. (A) S. sanguinis or S. gordonii or their derivatives were inoculated first and incubated aerobically at 37°C. S. mutans was inoculated 16 h later, and the plates were incubated overnight. a, SK36; b, JKH2 Pox−; c, JKH2/pDL276-pox; d, DL1; e, JKH1 Pox−; f, JKH1/pDL276-pox. (B) Growth of S. mutans in sterile conditioned medium from S. sanguinis or S. gordonii cultures. The conditioned medium was prepared from exponentially growing S. sanguinis or S. gordonii cells or their derivatives, filter sterilized, supplemented with 0.25% glucose, and immediately inoculated with S. mutans at a titer of 107 bacteria ml−1. Cells were incubated aerobically at 37°C, and 1 ml was removed to determine the A600 at the indicated time points. The data presented are representative of two independent experiments with similar results. (C) Competitiveness of S. sanguinis or S. gordonii wild-type and Pox− mutants in submerged dual-species biofilms with S. mutans. Dual-species biofilms were grown in BHI. After overnight growth, the cells were dispersed by vigorous pipetting, serially diluted, and plated. The CFU values for each strain and the standard deviations were calculated from three independent experiments performed on different days. *, P ≤ 0.05. Gray bars, S. sanguinis or S. gordonii; black line, S. mutans.
Growth inhibition of S. mutans during planktonic growth in conditioned medium.
To confirm that the inhibition of S. mutans was caused by H2O2, S. mutans was grown in filter-sterilized conditioned medium from aerobically grown wild-type cells in the presence or absence of catalase or from the Pox− mutants of S. sanguinis and S. gordonii. The conditioned medium from wild-type cells supported slow growth, with approximately two doublings of S. mutans in 5.5 h (Fig. 2B). In contrast, S. mutans grown in conditioned medium from the Pox− mutants or from wild-type cells grown in the presence of catalase showed growth after a lag phase, with approximately 4.5 doublings in 5.5 h.
Fitness of wild-type streptococci and Pox− mutants in dual-species biofilms.
Growth exclusion of the tested streptococci was most evident when the growth of a pioneer colonizer preceded a potentially competing species. To test whether H2O2 production gives S. sanguinis and S. gordonii a competitive advantage during growth, we determined the simultaneous growth of each species in dual-species biofilms with S. mutans (Fig. 2C). The wild-type species and Pox− mutants grew with similar generation times (data not shown), with the Pox− mutants yielding slightly higher final cell densities. When inoculated simultaneously at a 1:1 ratio with S. mutans, the Pox− mutants of S. sanguinis and S. gordonii showed less growth than their wild-type counterparts. For S. sanguinis, this difference was significant (P ≤ 0.05).
Influence of glucose on the competitiveness of S. sanguinis and S. gordonii.
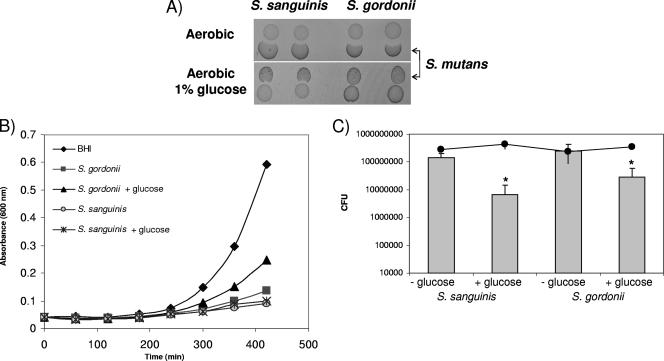
To learn whether glucose might repress Pox as described for Lactobacillus plantarum (25) and influence the fitness of S. gordonii and S. sanguinis in competition with S. mutans, BHI was supplemented with 1% glucose. In the presence of 1% glucose, S. gordonii was unable to inhibit growth of S. mutans; S. sanguinis activity against S. mutans was slightly reduced (Fig. 3A). Conversely, the addition of glucose to S. mutans did not influence the ability to inhibit S. sanguinis and S. gordonii (data not shown). To confirm these results, conditioned media from S. sanguinis and S. gordonii cultures grown with or without 1% glucose were compared for inhibiting activity against S. mutans. In the presence of glucose, S. gordonii conditioned medium inhibited S. mutans growth less than in the absence of glucose (Fig. 3B). Conditioned medium from S. sanguinis also inhibited growth of S. mutans, but glucose did not reverse the inhibition (Fig. 3B). Since changes in the generation time could explain the appearance of inhibition, growth was monitored for all strains with or without the addition of 1% glucose in liquid growth medium. S. mutans, S. sanguinis, and S. gordonii showed similar generation times with or without glucose, but the glucose-supplemented cultures yielded higher densities (data not shown). Interestingly, when tested as dual-species biofilms as described above, the addition of 1% glucose to the growth medium had an effect similar to that of the Pox− mutation on the outcome. S. sanguinis and S. gordonii grew significantly less in the presence of 1% glucose than in unsupplemented cultures, whereas S. mutans was unaffected (Fig. 3C).
FIG. 3.
Influence of glucose on the competitiveness of S. sanguinis or S. gordonii. (A) S. mutans growth inhibition assay in the presence or absence of 1% glucose. (B) Growth of S. mutans in sterile conditioned medium from S. sanguinis or S. gordonii cultures grown with or without 1% added glucose. The conditioned medium was prepared from cells growing to the mid log growth phase and immediately inoculated with S. mutans. Cells were incubated aerobically at 37°C, and 1 ml removed to determine the A600 at the indicated time points. The data presented are representative of two independent experiments with similar results. (C) Competitiveness of S. sanguinis or S. gordonii in submerged dual-species biofilms with S. mutans. Dual-species biofilms were grown in BHI plus 1% glucose when indicated. After overnight growth, the cells were dispersed by vigorous pipetting, serially diluted, and plated. CFU values and standard deviations were calculated from three independent experiments performed on different days. *, P ≤ 0.05. Gray bars, S. sanguinis or S. gordonii; black line, S. mutans.
Differential H2O2 production by S. sanguinis and S. gordonii.
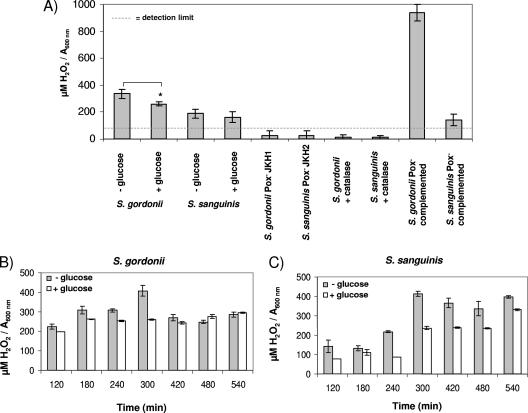
S. gordonii grown in media with 1% glucose showed reduced ability to inhibit S. mutans. To determine whether the presence of 1% glucose reduces generation of H2O2, H2O2 production by S. sanguinis and S. gordonii was determined. In TSB growth medium, H2O2 production by S. gordonii was reduced more than S. sanguinis by the addition of 1% glucose (S. gordonii, P ≤ 0.05; Fig. 4A). The Pox− mutants and cells grown with catalase did not produce detectable amounts of H2O2. H2O2 production was restored in S. sanguinis and S. gordonii by complementation of Pox (Fig. 4A). During growth in the presence of 1% glucose S. gordonii and S. sanguinis H2O2 production peaked during late log transition into stationary phase (after 300 min), followed by a 30% reduction in stationary phase for S. gordonii (Fig. 4B) and a slight reduction for S. sanguinis (Fig. 4C). In the presence of 1% glucose, S. sanguinis H2O2 production was maximal in stationary phase (Fig. 4C). H2O2 production by both species was generally higher in the absence of glucose (Fig. 4A to C).
FIG. 4.
Production of H2O2 during growth of S. sanguinis or S. gordonii. (A) SK36 or DL1 wild type with or without 1% glucose or catalase (30 μg), Pox− mutants JKH1 and JKH2, and complemented strains grown for 3 h until mid-exponential phase in TSB medium. (B) Time course of DL1 H2O2 production with or without glucose. (C) Time course of SK36 H2O2 production with or without glucose. Hydrogen peroxide concentration was adjusted to the absorbance at 600 nm. Gray bars, TSB; white bars, TSB plus 1% glucose. *, P ≤ 0.05 (n = 3).
Differential expression of S. mutans mutacin IV gene.
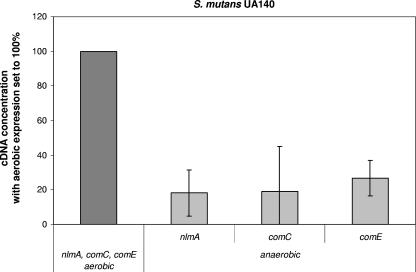
The expression of the genes for mutacin I and IV, the two characterized mutacins of S. mutans UA140 (40), were compared when cells were grown aerobically or anaerobically. Expression of mutA, the structural gene for mutacin I, was similar under aerobic and anaerobic conditions (data not shown). In aerobic conditions, the expression of nlmA, the structural gene for mutacin IV, was at least fivefold higher (Fig. 5). The expression of nlmA is controlled by the ComDE two-component system in response to the competence-stimulating peptide CSP (17, 19). To learn whether oxygen-dependent regulation of nlmA is controlled by ComDE, the expression of comC and comE was measured in aerobic and anaerobic cultures. In aerobic conditions, comC and comE were also expressed at about fivefold higher levels than under anaerobic conditions (Fig. 5).
FIG. 5.
Differential expression of nlmA, comC, and comE from S. mutans during aerobic and anaerobic growth. Gene expression was compared from cells growing on BHI agar plates for 7 h with or without oxygen. The values are given as relative cDNA abundance, with the expression under aerobic conditions set to 100%. Transcript levels were measured by real-time PCR using the 16S RNA as a housekeeping control. cDNA abundance was normalized against the 16S cDNA. The data presented represent the means and standard deviations of two (comC and comE) or three (nlmA) independent experiments performed on different days.
Aerobic growth triggers increased DNA release from mixed S. sanguinis and S. gordonii cultures.
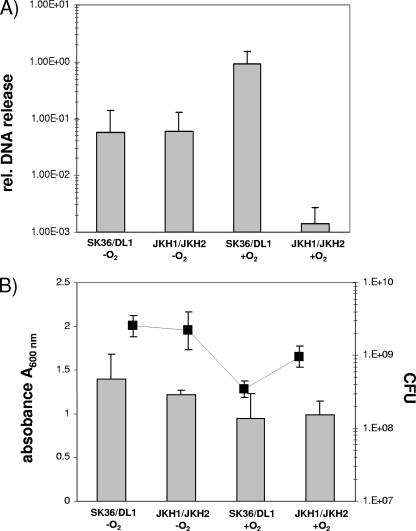
Mutacin IV can lyse closely related streptococci, and the released DNA can be taken up by competent S. mutans cells (17). We next determined whether higher expression of the competence system under aerobic growth conditions correlates with DNA release from mixed cultures of S. sanguinis and S. gordonii. After overnight growth, the mixed anaerobic cultures of S. sanguinis and S. gordonii wild-type cells and the Pox− mutants released similar amounts of DNA (Fig. 6A) and reached comparable cell densities and CFU counts (Fig. 6B). Under aerobic conditions, however, the DNA released from the mixed wild-type cultures increased more than 10-fold (Fig. 6A), although the final cell density was less than under anaerobic conditions (Fig. 6B). Under aerobic conditions the release of DNA appeared to depend upon the expression of a functional Pox gene, since mixed cultures with the Pox− mutants released less DNA than the other mixed culture conditions tested (Fig. 6A). DNA release from aerobically grown wild-type cultures was reduced by the addition of catalase to the levels of the Pox− mutants (data not shown). Catalase addition also leads to higher wild-type cell densities, a finding consistent with a role for H2O2 in DNA release (data not shown).
FIG. 6.
Quantification of DNA release and cell densities from mixed cultures of S. sanguinis and S. gordonii during growth under aerobic and anaerobic conditions. (A) Real-time PCR quantification of DNA release from mixed cultures of wild-type SK36/DL1 and Pox− mutants JKH1/JKH2 after overnight incubation with or without oxygen using universal primers for 16S RNA genes of S. gordonii and S. sanguinis (16S strep F and 16S strep R). The DNA release in each individual sample is reported relative to a serial dilution of chromosomal DNA from S. sanguinis (see Materials and Methods for details) by comparison of the CT values for the standard curve and the samples. (B) CFU counts and A600 values of the overnight cultures prior to DNA quantification. The data presented represent the means and standard deviations of three independent experiments performed on different days. Gray bars, CFU; black line, absorbance (A600).
DISCUSSION
The oral biofilm is a biological system of exceptionally high complexity. This complexity is largely reflected by the broad diversity of microbial species, the spatial and temporal arrangement of the individual members, and the high cell density. In addition, the oral environment is a habitat with constant changes in conditions, e.g., nutrient limitation, saliva flow, and oxygen availability. Members of the oral biofilm have evolved to cope with change and compete with other oral bacteria for this ecological niche. For example, adhesion through specific proteins enables S. gordonii to successfully compete with S. sanguinis during initial biofilm formation (31), and Streptococcus oligofermentans can use the lactic acid produced by S. mutans to increase its production of H2O2 to inhibit growth of the competitor (49). Hydrogen peroxide production by streptococci is well known to inhibit the growth of other bacterial species and contribute to the antagonism between S. mutans and S. sanguinis in oral biofilms (18, 43).
Oral biofilm formation and H2O2 production are influenced by oxygen (1, 2, 6). Oxygen in the oral biofilm allows for respiration of plaque bacteria and H2O2 production (26). H2O2 is produced by Pox as a by-product of aerobic metabolism (7). Using genomic analysis, the genes encoding the Pox of S. sanguinis (54) and S. gordonii (51) showed 95% homology when compared at the DNA level (data not shown). In agreement with its inability to produce H2O2 (1), the S. mutans genome contains no ortholog of a Pox gene (4). The production of H2O2 during aerobic respiration is considered an aggressive by-product that may eliminate competitors. H2O2 is strongly suggested to be a competitive factor in oral biofilms based upon in vitro studies (18) and our results demonstrating the Pox-dependent inhibition of S. mutans growth under aerobic conditions. Interspecies inhibition mediated by H2O2 has also been reported for Streptococcus pneumoniae (43) and Lactobacillus paracasei (32) toward Staphylococcus aureus.
The production of H2O2 by S. sanguinis and S. gordonii is likely to have other important consequences to the biofilm community. Under aerobic conditions, H2O2 production by mixed wild-type S. sanguinis/S. gordonii cultures caused release of 10-fold more DNA than cells grown under anaerobic conditions. Pox− mutant mixed cultures grown under aerobic conditions also released DNA, but less DNA than when H2O2 was produced. In aerobic conditions, wild-type cocultures yielded fewer viable cells correlating with the increased DNA release. Consistent with our data, spontaneous cell death and lysis of the bacterial pathogen S. pneumoniae has recently been shown to be H2O2 dependent (42). Indeed, H2O2 induced an apoptosis-like response, resulting in the loss of DNA content from H2O2-damaged cells (42). Besides triggering the lysis of competitors during initial colonization, H2O2-dependent cell lysis might confer an alternative competitive advantage. The biofilm matrix consists of proteins, carbohydrates, water, lipids, and DNA. The addition of DNase to S. mutans or Staphylococcus epidermidis cultures reduces the ability to form a biofilm (38, 41), suggesting that DNA stabilizes the biofilm. Moreover, the availability of DNA in the biofilm increases the chance for competence-dependent DNA uptake and genetic exchange, since competence increases when S. mutans grows in a biofilm (23). Hence, H2O2 is likely to regulate interspecies interactions of oral streptococci in the oral biofilm in several ways that were previously unknown.
In S. sanguinis and S. gordonii the promoter and gene sequences of the spxB genes share high homology. In the presence of glucose, which is a major nutritional carbon source whose concentration increases up to 1,000-fold during food intake (1), only S. sanguinis was able to inhibit S. mutans. During planktonic growth, both species produced less H2O2 in the presence of 1% glucose. To explain regulation in response to glucose, a putative cre box was identified in the promoter regions of the spxB genes of S. gordonii and S. sanguinis (data not shown). The cre box is the binding site for the major regulator of carbon catabolite repression in gram-positive bacteria, CcpA (13). In L. plantarum, CcpA represses the Pox gene poxB by binding to the experimentally confirmed cre box in the poxB promoter (25). However, a transcript for poxB could be measured in cells grown in the presence of 2% glucose. Transcription increased markedly when glucose was exhausted (25). In the results presented here, S. sanguinis was able to inhibit S. mutans in the presence of 1% glucose. CcpA may regulate differently in S. gordonii, allowing for more complete repression of the spxB gene. Differences in Pox half-life and the H2O2-degrading abilities of S. sanguinis and S. gordonii might also contribute to this process. Interestingly, the Pox-complemented strain of S. gordonii produced more H2O2 and slightly greater inhibition ability than the wild type (Fig. 2Ad and f and Fig. 4A), suggesting the presence of a potential regulator, which is titrated in the complemented strain by the presence of several spxB promoter sequences on the low-copy shuttle plasmid. Further work is needed to verify that the spxB genes in S. gordonii and S. sanguinis are in fact repressed by the binding of CcpA to the putative cre boxes and that the transcription is indeed controlled differently.
The glucose-mediated repression of H2O2 production shown in vitro might have a larger impact on the in vivo oral biofilm community. S. mutans, S. sanguinis, and S. gordonii all utilize glucose and possess multiple transport systems for this substrate (50). When BHI medium was supplemented with glucose, the growth rates of the three species were similar (data not shown). The reduced production of H2O2, however, could impair S. gordonii and S. sanguinis inhibition of S. mutans in vivo. S. mutans would acquire a competitive advantage, because the production of mutacins is not influenced by glucose. In fact, our biofilm experiments with mixed S. mutans and S. gordonii/S. sanguinis wild types grown simultaneously with or without glucose showed that S. gordonii and S. sanguinis CFU counts were significantly reduced in the presence of glucose in vitro.
The repression of Pox by glucose could have additional consequences for the metabolism of S. sanguinis and S. gordonii beyond those identified in the present study. The product of Pox enzymatic activity is acetyl-phosphate. Acetyl-phosphate is a potential phosphoryl donor for intracellular processes (14) and has been shown to be a global signal during biofilm development of E. coli. Mutants in the generation of acetyl-phosphate were able to form biofilms, but the architecture and physiological properties of the biofilm were changed (53). Similarly, S. pneumoniae SpxB− yields significantly less acetyl-phosphate (37) and is impaired in its ability to adhere to human endothelial cells (48). Adhesion ability is restored, however, when acetate is supplied in the growth medium and acetyl-phosphate can be generated by acetate kinase (48). Since inactivation of the Pox influences production of H2O2 and acetyl-phosphate, the role of acetyl-phosphate in biofilm development and interspecies competition requires further investigation.
Surprisingly, the competence system of S. mutans appears to be optimized for aerobic conditions, given that heterologous DNA is present in aerobic cultures of S. gordonii and S. sanguinis. Consistent with our data, S. mutans transformability is higher under aerobic conditions (38). Furthermore, S. pneumoniae controls the expression of the competence operon comCDE in an oxygen-dependent process via the two-component signal transduction system CiaRH (10). The expression of the competence genes in S. mutans is coordinated with the expression of a group of bacteriocins and bacteriocin-like genes (19); accordingly, the expression of the bacteriocin mutacin IV was elevated under aerobic conditions in our studies. During the preparation of the present study, Ahn et al. (3) reported the oxygen-dependent upregulation of the mutacin IV gene and the competence system for S. mutans using DNA microarrays, further supporting our results. Mutacin IV was shown to be involved in the lysis and release of plasmid DNA from S. gordonii (17). Interspecies interference by S. mutans appears to be equally effective in aerobic and anaerobic growth conditions and in the presence of glucose, based upon inhibition of the competitors S. gordonii and S. sanguinis. This preliminary result might partially explain the ability of S. mutans to increase in numbers and promote caries development when the diet is rich in glucose, a cariogenic carbohydrate (30), and the environment is anaerobic, even when no sucrose is available.
In conclusion, we have demonstrated that the environmental factor oxygen and the nutritional compound glucose substantially influence the interspecies competitiveness of the tested streptococci in our in vitro assays. The presented experiments were performed in liquid cultures and as plate assays, and the main outcomes were comparable between both experimental conditions. It is noteworthy that the oxygen availability is reduced in static liquid cultures due to rapid consumption. The experimental outcome could therefore as well be influenced by a specific biofilm phenotype. In future experiments, other ecological and environmental factors need to be incorporated into our model to understand the interplay between the members of the oral biofilm. The greatest challenge will be to confirm the basis of interspecies competition in vivo.
Acknowledgments
This study was supported by NIH grants 1K99DE018400-01 to J.K. and R01 DE08590 to M.C.H.
We thank C. F. Schachtele (University of Minnesota, Minneapolis) for helpful comments on the manuscript.
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Abbe, K., J. Carlsson, S. Takahashi-Abbe, and T. Yamada. 1991. Oxygen and the sugar metabolism in oral streptococci. Proc. Finn. Dent. Soc. 87477-487. [PubMed] [Google Scholar]
- 2.Ahn, S. J., and R. A. Burne. 2007. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J. Bacteriol. 1896293-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, S. J., Z. T. Wen, and R. A. Burne. 2007. Effects of oxygen on virulence traits of Streptococcus mutans. J. Bacteriol. 1898519-8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 401001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradshaw, D. J., P. D. Marsh, C. Allison, and K. M. Schilling. 1996. Effect of oxygen, inoculum composition, and flow rate on development of mixed-culture oral biofilms. Microbiology 142(Pt. 3)623-629. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson, J., M. B. Edlund, and S. K. Lundmark. 1987. Characteristics of a hydrogen peroxide-forming pyruvate oxidase from Streptococcus sanguis. Oral Microbiol. Immunol. 215-20. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49711-745. [DOI] [PubMed] [Google Scholar]
- 9.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 571194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echenique, J. R., S. Chapuy-Regaud, and M. C. Trombe. 2000. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol. Microbiol. 36688-696. [DOI] [PubMed] [Google Scholar]
- 11.Evaldson, G., A. Heimdahl, L. Kager, and C. E. Nord. 1982. The normal human anaerobic microflora. Scand. J. Infect. Dis. Suppl. 359-15. [PubMed] [Google Scholar]
- 12.Hammill, H. A. 1989. Normal vaginal flora in relation to vaginitis. Obstet. Gynecol. Clin. N. Am. 16329-336. [PubMed] [Google Scholar]
- 13.Hueck, C. J., and W. Hillen. 1995. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol. Microbiol. 15395-401. [DOI] [PubMed] [Google Scholar]
- 14.Klein, A. H., A. Shulla, S. A. Reimann, D. H. Keating, and A. J. Wolfe. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 1895574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolenbrander, P. E., R. J. Palmer, Jr., A. H. Rickard, N. S. Jakubovics, N. I. Chalmers, and P. I. Diaz. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 4247-79. [DOI] [PubMed] [Google Scholar]
- 16.Kolstad, R. A. 1976. Strain typing of oral streptococci by the use of bacterial antagonism. J. Dent. Res. 55A154-A165. [DOI] [PubMed] [Google Scholar]
- 17.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighboring species. Mol. Microbiol. 57392-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 1877193-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreth, J., J. Merritt, L. Zhu, W. Shi, and F. Qi. 2006. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol. Lett. 26511-17. [DOI] [PubMed] [Google Scholar]
- 20.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 9614547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuboniwa, M., G. D. Tribble, C. E. James, A. O. Kilic, L. Tao, M. C. Herzberg, S. Shizukuishi, and R. J. Lamont. 2006. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol. Microbiol. 60121-139. [DOI] [PubMed] [Google Scholar]
- 22.Kuramitsu, H. K., X. He, R. Lux, M. H. Anderson, and W. Shi. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71653-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorquet, F., P. Goffin, L. Muscariello, J. B. Baudry, V. Ladero, M. Sacco, M. Kleerebezem, and P. Hols. 2004. Characterization and functional analysis of the poxB gene, which encodes pyruvate oxidase in Lactobacillus plantarum. J. Bacteriol. 1863749-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquis, R. E. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J. Ind. Microbiol. 15198-207. [DOI] [PubMed] [Google Scholar]
- 27.Marsh, P. D. 1999. Microbiologic aspects of dental plaque and dental caries. Dent. Clin. N. Am. 43599-614. [PubMed] [Google Scholar]
- 28.Marteau, P., F. Daniel, P. Seksik, and R. Jian. 2004. Inflammatory bowel disease: what is new? Endoscopy 36130-136. [DOI] [PubMed] [Google Scholar]
- 29.Mikx, F. H., J. S. van der Hoeven, A. J. Plasschaert, and K. G. Konig. 1976. Establishment and symbiosis of Actinomyces viscosus, Streptococcus sanguis and Streptococcus mutans in germ-free Osborne-Mendel rats. Caries Res. 10123-132. [DOI] [PubMed] [Google Scholar]
- 30.Moynihan, P. J. 1998. Update on the nomenclature of carbohydrates and their dental effects. J. Dent. 26209-218. [DOI] [PubMed] [Google Scholar]
- 31.Nobbs, A. H., Y. Zhang, A. Khammanivong, and M. C. Herzberg. 2007. Streptococcus gordonii Hsa environmentally constrains competitive binding by Streptococcus sanguinis to saliva-coated hydroxyapatite. J. Bacteriol. 1893106-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ocana, V. S., A. A. de Ruiz Holgado, and M. E. Nader-Macias. 1999. Growth inhibition of Staphylococcus aureus by H2O2-producing Lactobacillus paracasei subsp. paracasei isolated from the human vagina. FEMS Immunol. Med. Microbiol. 2387-92. [DOI] [PubMed] [Google Scholar]
- 33.Ocana, V. S., A. A. Pesce de Ruiz Holgado, and M. E. Nader-Macias. 1999. Selection of vaginal H2O2-generating Lactobacillus species for probiotic use. Curr. Microbiol. 38279-284. [DOI] [PubMed] [Google Scholar]
- 34.Pakula, R., and W. Walczak. 1963. On the nature of competence of transformable streptococci. J. Gen. Microbiol. 31125-133. [DOI] [PubMed] [Google Scholar]
- 35.Palmer, R. J., Jr., P. I. Diaz, and P. E. Kolenbrander. 2006. Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J. Bacteriol. 1884117-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 1833770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pericone, C. D., S. Park, J. A. Imlay, and J. N. Weiser. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J. Bacteriol. 1856815-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen, F. C., L. Tao, and A. A. Scheie. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 1874392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lutticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177137-147. [DOI] [PubMed] [Google Scholar]
- 40.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 6715-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin, Z., Y. Ou, L. Yang, Y. Zhu, T. Tolker-Nielsen, S. Molin, and D. Qu. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 1532083-2092. [DOI] [PubMed] [Google Scholar]
- 42.Regev-Yochay, G., K. Trzcinski, C. M. Thompson, M. Lipsitch, and R. Malley. 2007. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J. Bacteriol. 1896532-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regev-Yochay, G., K. Trzcinski, C. M. Thompson, R. Malley, and M. Lipsitch. 2006. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J. Bacteriol. 1884996-5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rickard, A. H., P. Gilbert, N. J. High, P. E. Kolenbrander, and P. S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 1194-100. [DOI] [PubMed] [Google Scholar]
- 45.Rickard, A. H., R. J. Palmer, Jr., D. S. Blehert, S. R. Campagna, M. F. Semmelhack, P. G. Egland, B. L. Bassler, and P. E. Kolenbrander. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 601446-1456. [DOI] [PubMed] [Google Scholar]
- 46.Sbordone, L., and C. Bortolaia. 2003. Oral microbial biofilms and plaque-related diseases: microbial communities and their role in the shift from oral health to disease. Clin. Oral Investig. 7181-188. [DOI] [PubMed] [Google Scholar]
- 47.Schachtele, C. F., A. Nobbs, Y. Zhang, M. Costalonga, and M. C. Herzberg. 2007. Oral streptococci: commensals and opportunistic pathogens. Horizon Scientific Press, Norfolk, VA.
- 48.Spellerberg, B., D. R. Cundell, J. Sandros, B. J. Pearce, I. Idanpaan-Heikkila, C. Rosenow, and H. R. Masure. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19803-813. [DOI] [PubMed] [Google Scholar]
- 49.Tong, H., W. Chen, J. Merritt, F. Qi, W. Shi, and X. Dong. 2007. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol. Microbiol. 63872-880. [DOI] [PubMed] [Google Scholar]
- 50.Vadeboncoeur, C., and M. Pelletier. 1997. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 19187-207. [DOI] [PubMed] [Google Scholar]
- 51.Vickerman, M. M., S. Iobst, A. M. Jesionowski, and S. R. Gill. 2007. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J. Bacteriol. 1897799-7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, B. Y., B. Chi, and H. K. Kuramitsu. 2002. Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol. Immunol. 17108-112. [DOI] [PubMed] [Google Scholar]
- 53.Wolfe, A. J., D. E. Chang, J. D. Walker, J. E. Seitz-Partridge, M. D. Vidaurri, C. F. Lange, B. M. Pruss, M. C. Henk, J. C. Larkin, and T. Conway. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48977-988. [DOI] [PubMed] [Google Scholar]
- 54.Xu, P., J. M. Alves, T. Kitten, A. Brown, Z. Chen, L. S. Ozaki, P. Manque, X. Ge, M. G. Serrano, D. Puiu, S. Hendricks, Y. Wang, M. D. Chaplin, D. Akan, S. Paik, D. L. Peterson, F. L. Macrina, and G. A. Buck. 2007. Genome of the opportunistic pathogen Streptococcus sanguinis. J. Bacteriol. 1893166-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Y., Y. Lei, A. Khammanivong, and M. C. Herzberg. 2004. Identification of a novel two-component system in Streptococcus gordonii V288 involved in biofilm formation. Infect. Immun. 723489-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]