Abstract
Mutants of Salmonella enterica lacking apbC have nutritional and biochemical properties indicative of defects in [Fe-S] cluster metabolism. Here we show that apbC is required for S. enterica to use tricarballylate as a carbon and energy source. Tricarballylate catabolism requires three gene products, TcuA, TcuB, and TcuC. Of relevance to this work is the TcuB protein, which has two [4Fe-4S] clusters required for function, making it a logical target for the apbC effect. TcuB activity was 100-fold lower in an apbC mutant than in the isogenic apbC+ strain. Genetic data show that derepression of the iscRSUA-hscAB-fdx-orf3 operon or overexpression of iscU from a plasmid compensates for the lack of ApbC during growth on tricarballylate. The studies described herein provide evidence that the scaffold protein IscU has a functional overlap with ApbC and that ApbC function is involved in the synthesis of active TcuB.
Three systems for iron-sulfur ([Fe-S]) cluster biosynthesis have been identified. The first system is encoded by the nif (nitrogen fixation) operon in Azotobacter vinelandii and is required for the biosynthesis of nitrogenase (reviewed in reference 15). The second system is encoded by the iscSUA-hscAB-fdx-orf3 (iron sulfur cluster) operon of Azotobacter vinelandii. In Escherichia coli, the iscSUA-hscAB-fdx-orf3 operon encodes housekeeping [Fe-S] cluster biosynthetic functions (20, 35, 55) and is regulated by the IscR repressor (49). A third system for the biosynthesis/repair of the [Fe-S] cluster has been described for E. coli. In this bacterium, the sufABCDSE (sulfur utilization factor) operon is induced during times of limited Fe availability and oxidative stress (20, 28, 35, 37, 44, 54, 63). As in E. coli, the genome of Salmonella enterica carries both the isc and suf operons, and cellular viability requires the presence of one of the two (36).
The [Fe-S] cluster biosynthetic systems mentioned above have two general functional components. The cysteine desulfurase enzymes NifS, SufS, and IscS catalyze the removal of inorganic sulfide from l-cysteine (23, 33, 34, 43, 62), while the scaffolding proteins NifU, IscU, IscA, and SufA appear to bind and transfer labile [Fe-S] clusters to apoproteins (12, 27, 41, 56). Additional components can be specific to each system.
It was recently shown that cluster transfer from IscU to apoferredoxin is stimulated by the addition of HscA, HscB, and Mg·ATP (10). These data emphasized a role for ATP-hydrolyzing proteins, e.g., HscA and SufC, in the process of cluster maturation (10, 38, 50).
Work with Salmonella enterica serovar Typhimurium LT2 identified several loci outside the above-mentioned operons that impact [Fe-S] cluster metabolism. These loci include apbC, apbE, rseC, and yggX, all of which encode proteins with no characterized biochemical function (3, 4, 18, 45). Initially isolated as conditional thiamine auxotrophs, strains with lesions in these loci displayed phenotypic behavior similar to that of strains lacking isc operon functions (51-53). The apbC locus was the most common location of conditional thiamine auxotrophs identified in general screens (45). ApbC is a 40-kDa cytoplasmic protein that contains two conserved C-terminal cysteine residues separated by two amino acids (CXXC) and a Walker A box used for ATP binding and hydrolysis (25, 51).
Strains with lesions in apbC were independently isolated as mutants unable to use tricarballylate as the sole carbon and energy source for growth (A. R. Horswill and J. C. Escalante-Semerena, unpublished data). The tricarballylate catabolic genes tcuRABC (tricarballylate utilization) have been previously described (31), and a mechanism for this catabolism has been proposed (29). In this model, TcuC transports tricarballylate across the inner membrane, where it is oxidized by the flavoprotein TcuA to cis-aconitate, which can then enter the Krebs cycle (29). During growth on tricarballylate, the recycling of the reduced flavin adenine dinucleotide of TcuA is achieved by TcuB, a membrane-bound protein that contains two [4Fe-4S] clusters and heme (30).
The demonstration that TcuB contains iron-sulfur clusters, in combination with work on ApbC and homologs, led to the hypothesis that the [Fe-S] clusters of TcuB were compromised in an apbC strain, preventing growth on tricarballylate. Consistent with this hypothesis, we show herein that TcuB activity is 100-fold lower in a strain lacking ApbC. The data further show that derepression of the isc operon or overexpression of iscU compensated for the lack of ApbC during growth of an apbC strain on tricarballylate.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
All strains used in this study are derived from S. enterica serovar Typhimurium LT2, and their genotypes are given in Table 1. MudJ refers to the MudI1734 insertion element (9), and Tn10d(Tc) refers to the transposition-defective mini-Tn10 described by Way et al. (59). No-carbon essential (NCE) medium of Berkowitz et al. (5) was made with Milli-Q filtered water and supplemented with 1 mM MgSO4 and trace minerals (1, 11, 58). Glucose and tricarballylate were added to NCE medium at 11 mM and 20 mM, respectively. Difco nutrient broth (8 g/liter) with NaCl (5 g/liter) or lysogenic broth (6, 7) was used as rich medium. Difco BiTek agar was added (15 g/liter) for solid medium. When present in the medium, supplements were provided at the following final concentrations: thiamine, 10 nM or 100 nM; adenine, 0.4 mM; and nicotinic acid, 20 μM. When needed, antibiotics were added to the following concentrations in rich and minimal medium, respectively: tetracycline, 20 and 10 μg/ml; kanamycin, 50 and 125 μg/ml; chloramphenicol, 20 and 4 μg/ml; ampicillin, 50 and 15 μg/ml; and gentamicin, 6 and 6 μg/ml. All chemicals were purchased from Sigma-Aldrich.
TABLE 1.
Strains and plasmidsa
| Strain or plasmidb | Descriptionc | Source |
|---|---|---|
| Strains | ||
| S. enterica | ||
| TR6583 | metE205 ara-9 | J. Roth |
| DM10310 | ara-9 (wild type) | |
| DM9678 | apbE42::Tn10d(tet) | |
| DM9682 | rseC::kan | |
| DM9684 | cyaY::cat | |
| DM10780 | ara-9 apbC55::Tn10d(tet) rseC::kan | |
| DM10779 | ara-9 apbC::MudJ (kan) apbE42::Tn10d(tet) | |
| DM10296 | ara-9 yggX::Gm | |
| DM10300 | ara-9 apbC55::Tn10d(tet) | |
| DM10302 | ara-9 apbC55:Tn10d(tet) yggX::Gm | |
| DM10696 | ara-9 iscR2::MudJ (kan) | |
| DM10697 | ara-9 apbC55:Tn10d(tet) iscR2::MudJ (kan) | |
| DM10325 | ara-9 ΔiscRSUA::cat | |
| DM10681 | ara-9 apbC55::Tn10d(tet) Δisc5::cat | |
| DM10326 | ara-9 sufS::cat | |
| DM10682 | ara-9 apbC55::Tn10d(tet) sufS::cat | |
| DM10667 | ara-9 iscA1::MudJ (kan) | |
| DM10683 | ara-9 apbC55::Tn10d(tet) iscA1::MudJ (kan) | |
| DM10604 | ara-9 apbC::MudJ (kan) | |
| DM10608 | ara-9 apbC::MudJ (kan) iscR11 | |
| JE10432 | ara-9 iscR::kan | J. C. Escalante-Semerena |
| DM10698 | ara-9 ΔiscR11 | |
| JE10435 | ara-9 apbC55::Tn10d(tet) ΔiscR11 | J. C. Escalante-Semerena |
| DM10685 | ara-9 apbC55::Tn10d(tet) yggX::Gm iscR11 | |
| DM10474 | ara-9 apbC55::Tn10d(tet) stm2545::Tn10d(cat) iscR6 | |
| DM10476 | ara-9 apbC55::Tn10d(tet) stm2545::Tn10d(cat) iscR7 | |
| DM10480 | ara-9 apbC55::Tn10d(tet) stm2545::Tn10d(cat) iscR8 | |
| DM10482 | ara-9 apbC55::Tn10d(tet) stm2545::Tn10d(cat) iscR9 | |
| DM10484 | ara-9 apbC55::Tn10d(tet) stm2545::Tn10d(cat) iscR10 | |
| DM10771 | ara-9 staC::Tn10d(tet) stm2545::Tn10d(cat) yggX::Gm | |
| DM10772 | ara-9 apbC::MudJ (kan) staC::Tn10d(tet) stm2545::Tn10d(cat) yggX::Gm | |
| DM10770 | ara-9 apbC::MudJ (kan) stm2545::Tn10d(Cm) yggX::Gm iscR7 | |
| DM10769 | ara-9 staC::Tn10d(tet+) stm2545::Tn10d(cat+) yggX::Gm iscR7 | |
| E. coli | ||
| C43(λDE3) | F−ompT gal hsdSB(rB− mB−) dcm lon | J. C. Escalante-Semerena |
| JE10465 | F−ompT gal hsdSB(rB− mB−) dcm lon apbC::kan+ | J. C. Escalante-Semerena |
| DH5α/F′ | F′/endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA (Nar1r) relA1 Δ(lacZYA-argF)U169 deoR[80dlac Δ(lacZ)M15] | New England Biolabs |
| Plasmids (vectors) | ||
| pSU19 (pSU19) | None | 2 |
| pIscR1 (pSU19) | iscR (S. enterica) | 52 |
| pIscS1 (pSU19) | iscS (S. enterica) | 52 |
| pIscA1 (pSU19) | iscA (S. enterica) | 52 |
| pIscA-orf3 (pSU19) | iscA-hscB-hscA-fdx-orf3 (S. enterica) | 52 |
| pIscU1 (pSU19) | iscU (S. enterica) | |
| pTCU21 (pBAD30) | tcuABC (S. enterica) | 31 |
| pCTH-ApbC (pET20b) | apbC (S. enterica) | 51 |
| pDB1282 (pAra13) | iscSUA-hscB-hscA-fdx-orf3 (A. vinelandii) | Dennis Dean |
| pHscB-orf3 (pSU19) | hscB-hscA-fdx-orf3 (S. enterica) | |
| pGSO164 (pBAD) | sufABCDSE (E. coli) | 43 |
| pIsc2 (pFZY1) | iscR operator | 26d |
| pTCU55 (pET15b) | tcuB | 30 |
| pApbC (pSU19) | apbC |
Unless indicated otherwise in the description, all strains are S. enterica serovar Typhimurium LT2 constructed for this study.
All S. enterica strains constructed for this study are derivatives of TR6583 that were transduced to metE+. The TR6583 strain was from the Escalante-Semerena laboratory.
Genotypes are given for strains, and inserts (hosts) are given for plasmids. The following alleles have been previously described: apbC55::Tn10d (46), cyaY::Cm (57), rseC::Kan (3, 53), apbE42::Tn10d(Tc) (4), yggX::Gm (19), iscR2::MudJ (52), Δisc5::Cm (53), and iscA2::MudJ (52). Tc, tetracycline; Cm, chloramphenicol; Gm, gentamicin; Kan, kanamycin.
Source also includes Lewis et al. (submitted).
Genetic methods. (i) Mutant isolation.
Nine independent cultures of DM10300 (apbC) were grown to full density in nutrient broth medium. One hundred microliters of each culture was spread onto individual minimal tricarballylate thiamine plates. Colonies spontaneously arose after 48 h of incubation at 37°C. One colony derived from each culture was saved.
(ii) Isolation of linked insertions.
The methods for transduction and the purification of transductants have been previously described (13, 47, 48).
Transposons [Tn10d(cat)] (14) genetically linked to the suppressor mutations were isolated by standard genetic techniques (24). In each case, mutant strains were reconstructed and verified phenotypically prior to characterization. The relevant insertions were mapped by sequencing using a PCR-based protocol (University of Wisconsin Biotechnology DNA Sequence Facility) (8, 60).
(iii) Phenotypic analysis.
Nutritional requirements were assessed on solid medium and by the quantification of growth in liquid medium using either 5-ml cultures in 25-ml shake tubes or 200-μl cultures in a 96-well plate. Protocols for each have been previously described (3, 4). The starting A650 was routinely between 0.03 and 0.08, with a final A650 between 0.5 and 1.1. Each culture had at least three replicates. Growth on solid medium was scored after replica printing to the relevant medium and after incubation at 37°C for 48 to 60 h.
Molecular biology.
Restriction enzymes and DNA ligase were purchased from Promega, and Pfu DNA polymerase was purchased from Stratagene. The iscU and the hscA-orf3 genes were amplified from wild-type S. enterica by using genomic DNA as the template. The primers were as follows: for iscU, the forward primer was 5′-CCGAAGCTTATGGCTTACAGCGAAAAAG-3′ and the reverse primer was 5′-CGGGGATCCTTATTTCGCTTCGCGTTTG-3′; for hscA-orf3, the forward primer was 5′-GGGCAAGCTTTGGATTACTTCACCCTCTT-3′ and the reverse primer was 5′-CCTCGGATCCTTACTCTGCTTCATCCAACC-3′.
The PCR product of iscU was purified and digested with BamHI and HindIII. The resulting products were purified and ligated into similarly digested pSU19 (2), creating pIscU. The PCR product containing hscA-orf3 was blunt end ligated into HincII-digested pSU19, resulting in pHscA-orf3. Plasmids were moved into the appropriate strains via electroporation (42), and their identities were confirmed by restriction digestion and/or sequencing. The plasmids used are given in Table 1.
Enzyme assays. (i) β-Galactosidase.
β-Galactosidase assays were performed according to the method of Winston et al. (61).
(ii) TcuB activity.
His6-TcuB was overproduced from plasmid pTCU55 as previously described (29). Cells from either strain JE6664 [C43(λDE3)] or strain JE10465 [C43(λDE3) apbC::kan] containing pTCU55 were grown at 18°C on Terrific broth to an optical density at 600 nm of ∼0.4 and then induced with 300 μM isopropyl-β-d-thiogalactopyranoside (IPTG) overnight. His6-TcuB-enriched extracts were obtained and assayed as previously described (29). Briefly, 200-μl reaction mixtures contained 2-(N-morpholino)ethanesulfonic acid (MES) (100 mM, pH 6.5, at 30°C), dithiothreitol (1 mM), 1 μg TcuA, and 20 μg His6-TcuB-enriched extract. Reaction mixtures were incubated at 30°C for 10 min following the addition of tricarballylate (10 mM). Reactions were stopped by the addition of 40-μl samples to 60 μl of 166.7 mM H2SO4. Fifty microliters of each sample was used to quantify cis-aconitate production using a high-performance liquid chromatography protocol (29).
RESULTS
ApbC, but not Isc or Suf protein, is required for growth on tricarballylate.
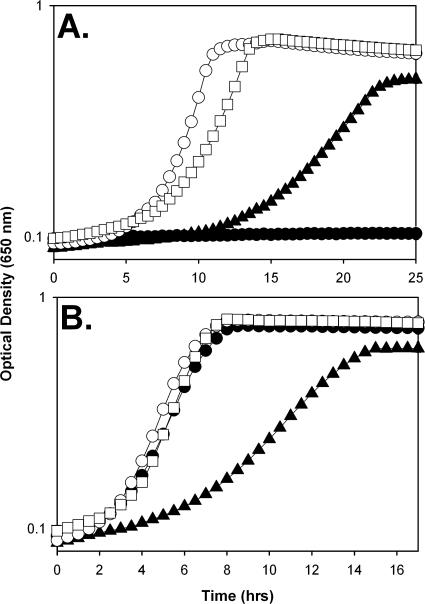
An apbC mutant does not grow on tricarballylate as a carbon and energy source, although it is proficient for growth on glucose (Fig. 1) and other carbon sources (e.g., succinate and gluconate) (data not shown). The expression of the tcuABC operon in trans from a nonnative promoter did not restore growth on tricarballylate, consistent with a posttranscriptional effect. A strain carrying a deletion of the iscRSUA-hscAB-fdx-orf3 operon or a polar insertion in sufS grew well on tricarballylate (Fig. 1; Table 2). The growth of the ΔiscRSUA-hscAB-fdx-orf3 mutant was reduced compared to that of the wild type on both tricarballylate and glucose, consistent with a previously reported global defect (52). The growth of strains lacking other genes involved in [Fe-S] cluster metabolism was also assessed on tricarballylate; no defect was found for mutants lacking apbE, rseC, cyaY, or yggX (data not shown). These growth data suggested a specific role for ApbC function during tricarballylate catabolism.
FIG. 1.
apbC mutants fail to grow on tricarballylate. Strains were grown at 37°C in NCE medium supplemented with thiamine and nicotinic acid and with a sole carbon and energy source. Growth of strains DM10310 (wild type) (○), DM10300 (apbC) (•), DM10325 (iscSUA-hscAB-fdx-orf3) (▴), and DM10667 (iscA-hscAB-fdx-orf3) (□) was monitored on tricarballylate (A) and glucose (B).
TABLE 2.
An apbC mutant is unable to grow on tricarballylate medium
| Strain | Relevant genotypeb | Doubling time (h) on medium containinga:
|
Ratio of doubling timesd | |
|---|---|---|---|---|
| Tricarballylatec | Glucosec | |||
| DM10310 | Wild type | 1.7 ± 0.1 | 2.1 ± 0.1 | 0.81 |
| DM10667 | iscA-hscAB-fdx-orf3 | 1.9 ± 0.1 | 2.1 ± 0.1 | 0.90 |
| DM10325 | iscSUA-hscAB-fdx-orf3 | 4.5 ± 0.2 | 4.3 ± 0.1 | 1.1 |
| DM10326 | sufS | 1.8 ± 0.1 | 2.2 ± 0.0 | 0.82 |
| DM10300 | apbC | NG | 2.1 ± 0.0 | |
Doubling times were calculated using the formula μ = ln(X/Xo)/T, where μ is the growth rate, X and Xo are optical density measurements at 650 nm, T is the time between the absorbance readings X and Xo, and the doubling time (g) was (ln 2)/μ (39). Values are averages of three independent cultures. NG, no growth.
Relevant genotypes indicate the genes that are defective due to the presence of a polar mutation or deletion.
The defined medium included the indicated carbon source and was supplemented with thiamine and nicotinic acid.
Ratio of the doubling time on tricarballylate to the doubling time on glucose.
TcuB is less active in apbC mutants.
To determine if ApbC function was involved in the synthesis of active TcuB, we measured the activity of this protein in vitro in a strain lacking ApbC. E. coli strain C43(λDE3) and an apbC mutant derivative of that strain were used to overproduce TcuB from plasmid pTCU55 (30). The apbC mutant extract produced 100-fold less cis-aconitate than that of the wild-type strain (220 ± 20 and 25,300 ± 700 pmol cis-aconitate produced after 10 min, respectively). In contrast, the activities of aconitase, succinate dehydrogenase, and the non-[Fe-S] cluster protein malate dehydrogenase were indistinguishable in the apbC mutant and wild-type extracts (data not shown).
Conditional growth of an apbC mutant.
An apbC strain did not grow in liquid medium with tricarballylate as a carbon source but grew on solid tricarballylate medium after ∼48 h. This observation suggested a functionally redundant system working at low efficiency. Insertions in yggX, iscA, or iscR eliminated residual growth of the apbC mutants on tricarballylate medium, while apbC strains with insertions in sufS, apbE, or rseC retained growth. None of these above-mentioned loci were required for growth on tricarballylate in an otherwise wild-type strain.
Suppressor analysis provides insight into ApbC function.
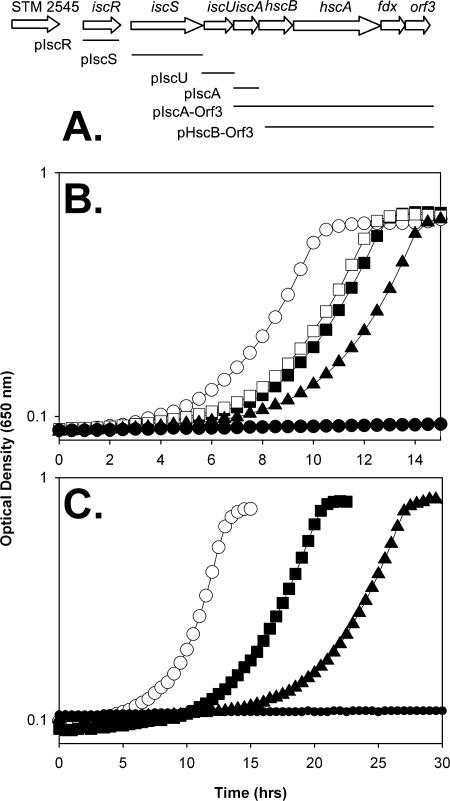
Nine independent spontaneous mutations that allowed the growth of strain DM10300 (apbC) on tricarballylate were identified. Genetic analysis determined that a Tn10d(cat) insertion in open reading frame STM2545 was ∼85% cotransducible by phage P22 with five of the nine suppressor mutations, placing the mutations near the isc operon (Fig. 2A). Sequence analysis determined that each of the five mutations was in iscR, encoding the repressor of the isc operon (49). The suppressor mutations resulted in variant IscR proteins with the following amino acid changes: L109Q (iscR6), S38F (iscR7), Q94Z (stop) (iscR8), Y41S (iscR9), and G64A (iscR10). The four substitutions were located in the predicted helix-turn-helix DNA binding domain of IscR (49) and were expected to generate inactive proteins and result in the constitutive expression of the isc operon.
FIG. 2.
The overexpression of isc genes allows growth of an apbC mutant on tricarballylate. Strains were grown at 37°C in NCE medium supplemented with thiamine and nicotinic acid and with tricarballylate as a carbon and energy source. (A) A schematic shows the genetic organization of the S. enterica isc operon. The borders of inserts used to generate plasmids are diagrammed below the operon. (B) Growth of strains DM10310 (wild type) (○), JE10435 (iscR11 apbC) (▪), DM10698 (iscR11) (□), DM10474 (apbC iscR7) (▴), and DM10300 (apbC) (•) on tricarballylate NCE medium. (C) Growth of strains DM10310 (wild type) with pSU19 (○), DM10300 (apbC) with pSU19 (•), DM10300 (apbC) with pIscU (▴), and DM10474 (apbC iscR7) with pSU19 (▪) on tricarballylate NCE medium.
Derepression of the isc operon restores the growth of an apbC mutant on tricarballylate.
Three results confirmed that the suppressor mutations disrupted IscR function and that the resulting derepression of the isc operon was sufficient to allow an apbC mutant to grow on tricarballylate. First, the introduction of an in-frame deletion of iscR (iscRΔ11) restored the growth of an apbC mutant on tricarballylate (Fig. 2B). Second, the expression of the wild-type allele of iscR in trans eliminated growth on tricarballylate of strain JE10435 (apbC iscRΔ11) and strains containing the other iscR alleles (data not shown).
Third, transcription was monitored using a lacZ reporter under the control of the iscR promoter (piscR-lacZ transcriptional fusion [plasmid pIsc2]) (J. A. Lewis, J. M. Boyd, D. M. Downs, and J. C. Escalante-Semerena, submitted for publication). Data given in Table 3 show that the chromosomal iscR alleles increased the expression of the reporter as efficiently as the chromosomal deletion of iscR. Together, these results confirmed that the IscR variants encoded by mutant iscR alleles failed to repress iscRSUA-hscAB-fdx-orf3 transcription and that derepression of the iscRSUA-hscAB-fdx operon was sufficient to restore growth on tricarballylate in an apbC strain.
TABLE 3.
Inactivation of IscR derepresses the isc operona
| Strain | Relevant genotype | β-Galactosidase activity (Miller units) |
|---|---|---|
| DM10310 | Wild type | 70 ± 4 |
| DM10300 | apbC | 70 ± 2 |
| JE10432 | iscR::Kan | 390 ± 72 |
| DM10474 | apbC iscR6 | 270 ± 28 |
| DM10476 | apbC iscR7 | 430 ± 47 |
| DM10480 | apbC iscR8 | 350 ± 31 |
| DM10482 | apbC iscR9 | 340 ± 28 |
| DM10484 | apbC iscR10 | 290 ± 22 |
All strains contained a plasmid that carried the isc reporter-lacZ fusion as described in the text. Strains were grown in LB-ampicillin medium to the mid-exponential phase of cell growth. β-Galactosidase assays were performed as described in Materials and Methods.
IscU has functional redundancy with ApbC.
The above-mentioned results strongly suggested that one or more proteins encoded by the isc operon had a functional overlap with ApbC. Plasmids encoding one or more isc genes (Fig. 2A) were introduced into strain DM10300 (apbC), and growth on tricarballylate was assessed. Of the plasmids tested, only pIscU affected the growth of the apbC mutant strain on tricarballylate (Fig. 2C). Several points can be taken from the data in Fig. 2C. First, pIscU restored the growth of the apbC mutant compared to the growth of the same strain with the vector-only control. The uniformly increased lag did not alter the conclusion that the growth of an apbC mutant on tricarballylate was allowed by either the derepression of the isc operon or the overexpression of iscU. Each of the strains retained the pattern of growth upon reinoculation, indicating that the growth was not due to a mutant overpopulating the culture. The doubling times for the strains were 1.5 ± 0.0, 1.7 ± 0.0, and 2.3 ± 0.1 h for the wild type with pSU19, the apbC iscR7 mutant with pSU19, and the apbC mutant with pIscU, respectively.
IscU is not sufficient to compensate for ApbC.
Plasmid pIscU failed to restore growth to an apbC mutant that was also defective in the isc operon (data not shown). This result was particularly significant for the apbC iscA genetic background, since the iscA mutation alone did not affect growth on tricarballylate (Table 2). These data suggested that the ability of IscU overproduction to allow the growth of an apbC mutant on tricarballylate required iscA and/or at least one gene downstream in the operon.
DISCUSSION
This study was initiated to understand the inability of S. enterica apbC mutants to grow with tricarballylate as a carbon and energy source. Previous studies implicating ApbC in [Fe-S] cluster metabolism (51, 53) and the report that TcuB contained [Fe-S] clusters (30) generated a hypothesis for the growth defect. The result that strains lacking apbC displayed 100-fold less TcuB activity than the wild type supported this hypothesis and has provided an additional system that can be exploited to dissect the role of ApbC in Salmonella. The physiological studies described herein provide data that IscU has a functional overlap with ApbC and, furthermore, that ApbC has a specific role in tricarballylate utilization that distinguishes it from the general [Fe-S] cluster biosynthesis systems encoded by the isc and suf operons.
Mutants lacking either the complete isc operon or the suf operon had no growth defect specific to tricarballylate medium. Thus, the lack of growth on tricarballylate was the first defect described for an apbC mutant that was not shared by strains lacking the major [Fe-S] cluster biosynthetic system isc. Recently, ErpA, a protein essential for isoprenoid biosynthesis in E. coli, was shown to specifically transfer [Fe-S] clusters to IspG (32). Similarly, Iba57 was shown to be essential for mitochondrial aconitase maturation in yeast (16). The results with ErpA and Iba57 were parallel to those with ApbC, since in all cases neither the isc system nor the suf system expressed at physiological levels could functionally replace these proteins in isoprenoid biosynthesis, aconitase maturation, or tricarballylate catabolism, respectively.
Null alleles of iscR restored the growth of the apbC mutant on tricarballylate and suggested that a gene(s) in the isc operon had a functional overlap with apbC. This result was confirmed in a study by Lewis et al. that showed that growth of an apbC mutant on tricarballylate could be restored by physiological conditions that increased the expression of the isc operon (Lewis et al., submitted). When provided in trans, iscU restored growth on tricarballylate to an apbC mutant. The expression of iscU in trans failed to restore growth on tricarballylate to an apbC isc double mutant, showing that IscU required at least one of the iscA, hscAB, or orf3 genes to restore growth. These data were reminiscent of studies on the functional redundancy of the U-type scaffolds in Azotobacter vinelandii. Johnson and coworkers found that target specificity distinguished the isc and nif systems when these operons were expressed at chromosomal levels, which was altered if the relevant genes were overexpressed (21, 22). Transcriptional studies found no effect of IscR on apbC expression or vice versa (reference 17 and data not shown).
Eukaryotic ApbC homologues Npb35 and Cfd1 can independently bind [Fe-S] clusters and rapidly and efficiently transfer these clusters to the Leu1 apoenzyme (40). Recent data show that ApbC can similarly transfer an [Fe-S] cluster to Leu1 in vitro (J. M. Boyd, A. J. Pierik, D. J. Aguilar-Netz, R. Lill, and D. M. Downs, submitted for publication). The data presented herein suggest that the TcuB protein provides a physiologically relevant system to further explore the biochemical function of ApbC and to address its specificity in vitro.
Acknowledgments
This work was supported by competitive grants GM47296 (D.M.D.) and GM62203 (J.C.E.-S.) and a Kirschstein postdoctoral training grant (GM079938-02) (J.M.B.) from the National Institutes of Health. Funds were also provided from a 21st Century Scientists Scholars Award from the J. M. McDonnell fund to D.M.D.
The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 10275-78. [DOI] [PubMed] [Google Scholar]
- 3.Beck, B. J., L. E. Connolly, A. De Las Peñas, and D. M. Downs. 1997. Evidence that rseC, a gene in the rpoE cluster, has a role in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 1796504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck, B. J., and D. M. Downs. 1998. The apbE gene encodes a lipoprotein involved in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 180885-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz, D., J. M. Hushon, H. J. Whitfield, Jr., J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caetano-Anolles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 385-94. [DOI] [PubMed] [Google Scholar]
- 9.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. J. Bacteriol. 158488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandramouli, K., and M. K. Johnson. 2006. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry 4511087-11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 12.Dos Santos, P. C., A. D. Smith, J. Frazzon, V. L. Cash, M. K. Johnson, and D. R. Dean. 2004. Iron-sulfur cluster assembly: NifU-directed activation of the nitrogenase Fe protein. J. Biol. Chem. 27919705-19711. [DOI] [PubMed] [Google Scholar]
- 13.Downs, D. M., and L. Petersen. 1994. apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J. Bacteriol. 1764858-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, T., and J. R. Roth. 1988. Characterization of Tn10d-Cam: a transposition-defective Tn10 specifying chloramphenicol resistance. Mol. Gen. Genet. 213332-338. [DOI] [PubMed] [Google Scholar]
- 15.Frazzon, J., and D. R. Dean. 2003. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol. 7166-173. [DOI] [PubMed] [Google Scholar]
- 16.Gelling, C., I. W. Dawes, N. Richhardt, R. Lill, and U. Mühlenhoff. 2008. Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol. Cell. Biol. 281851-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giel, J. L., D. Rodionov, M. Liu, F. R. Blattner, and P. J. Kiley. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 601058-1075. [DOI] [PubMed] [Google Scholar]
- 18.Gralnick, J., and D. Downs. 2001. Protection from superoxide damage associated with an increased level of the YggX protein in Salmonella enterica. Proc. Natl. Acad. Sci. USA 988030-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gralnick, J. A., and D. M. Downs. 2003. The YggX protein of Salmonella enterica is involved in Fe(II) trafficking and minimizes the DNA damage caused by hydroxyl radicals: residue CYS-7 is essential for YggX function. J. Biol. Chem. 27820708-20715. [DOI] [PubMed] [Google Scholar]
- 20.Hantke, K. 2002. Members of the Fur protein family regulate iron and zinc transport in E. coli and characteristics of the Fur-regulated fhuF protein. J. Mol. Microbiol. Biotechnol. 4217-222. [PubMed] [Google Scholar]
- 21.Johnson, D. C., P. C. Dos Santos, and D. R. Dean. 2005. NifU and NifS are required for the maturation of nitrogenase and cannot replace the function of isc-gene products in Azotobacter vinelandii. Biochem. Soc. Trans. 3390-93. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. C., M.-C. Unciuleac, and D. R. Dean. 2006. Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii. J. Bacteriol. 1887551-7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kambampati, R., and C. T. Lauhon. 1999. IscS is a sulfurtransferase for the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. Biochemistry 3816561-16568. [DOI] [PubMed] [Google Scholar]
- 24.Kleckner, N., J. Roth, and D. Botstein. 1977. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J. Mol. Biol. 116125-159. [DOI] [PubMed] [Google Scholar]
- 25.Koonin, E. V. 1993. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 2291165-1174. [DOI] [PubMed] [Google Scholar]
- 26.Koop, A. H., M. E. Hartley, and S. Bourgeois. 1987. A low-copy-number vector utilizing beta-galactosidase for the analysis of gene control elements. Gene 52245-256. [DOI] [PubMed] [Google Scholar]
- 27.Krebs, C., J. N. Agar, A. D. Smith, J. Frazzon, D. R. Dean, B. H. Huynh, and M. K. Johnson. 2001. IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry 4014069-14080. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J. H., W. S. Yeo, and J. H. Roe. 2004. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 511745-1755. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, J. A., and J. C. Escalante-Semerena. 2006. The FAD-dependent tricarballylate dehydrogenase (TcuA) enzyme of Salmonella enterica converts tricarballylate into cis-aconitate. J. Bacteriol. 1885479-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis, J. A., and J. C. Escalante-Semerena. 2007. Tricarballylate catabolism in Salmonella enterica. The TcuB protein uses 4Fe-4S clusters and heme to transfer electrons from FADH2 in the tricarballylate dehydrogenase (TcuA) enzyme to electron acceptors in the cell membrane. Biochemistry 469107-9115. [DOI] [PubMed] [Google Scholar]
- 31.Lewis, J. A., A. R. Horswill, B. E. Schwem, and J. C. Escalante-Semerena. 2004. The tricarballylate utilization (tcuRABC) genes of Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 1861629-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loiseau, L., C. Gerez, M. Bekker, S. Ollagnier-de Choudens, B. Py, Y. Sanakis, J. Teixeira de Mattos, M. Fontecave, and F. Barras. 2007. ErpA, an iron-sulfur (Fe-S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 10413626-13631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loiseau, L., S. Ollagnier-de-Choudens, L. Nachin, M. Fontecave, and F. Barras. 2003. Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J. Biol. Chem. 27838352-38359. [DOI] [PubMed] [Google Scholar]
- 34.Loiseau, L., S. Ollagnier-de Choudens, D. Lascoux, E. Forest, M. Fontecave, and F. Barras. 2005. Analysis of the heteromeric CsdA-CsdE cysteine desulfurase, assisting Fe-S cluster biogenesis in Escherichia coli. J. Biol. Chem. 28026760-26769. [DOI] [PubMed] [Google Scholar]
- 35.Massé, E., C. K. Vanderpool, and S. Gottesman. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 1876962-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413852-856. [DOI] [PubMed] [Google Scholar]
- 37.Nachin, L., M. El Hassouni, L. Loiseau, D. Expert, and F. Barras. 2001. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 39960-972. [DOI] [PubMed] [Google Scholar]
- 38.Nachin, L., L. Loiseau, D. Expert, and F. Barras. 2003. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell. Sinauer Associates, Inc., Sunderland, MA.
- 40.Netz, D. J., A. J. Pierik, M. Stumpfig, U. Muhlenhoff, and R. Lill. 2007. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3278-286. [DOI] [PubMed] [Google Scholar]
- 41.Ollagnier-de Choudens, S., L. Nachin, Y. Sanakis, L. Loiseau, F. Barras, and M. Fontecave. 2003. SufA from Erwinia chrysanthemi. Characterization of a scaffold protein required for iron-sulfur cluster assembly. J. Biol. Chem. 27817993-18001. [DOI] [PubMed] [Google Scholar]
- 42.O'Toole, G. A., M. R. Rondon, and J. C. Escalante-Semerena. 1993. Analysis of mutants of Salmonella typhimurium defective in the synthesis of the nucleotide loop of cobalamin. J. Bacteriol. 1753317-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Outten, F. W., M. J. Wood, F. M. Munoz, and G. Storz. 2003. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J. Biol. Chem. 27845713-45719. [DOI] [PubMed] [Google Scholar]
- 44.Patzer, S. I., and K. Hantke. 1999. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 1813307-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petersen, L., and D. M. Downs. 1996. Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J. Bacteriol. 1785676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen, L., J. Enos-Berlage, and D. M. Downs. 1996. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics 14337-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts, G. P. 1978. Isolation and characterization of informational suppressors in Salmonella typhimurium. Ph.D. thesis. University of California, Berkeley.
- 48.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 11975-88. [DOI] [PubMed] [Google Scholar]
- 49.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 9814895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silberg, J. J., and L. E. Vickery. 2000. Kinetic characterization of the ATPase cycle of the molecular chaperone Hsc66 from Escherichia coli. J. Biol. Chem. 2757779-7786. [DOI] [PubMed] [Google Scholar]
- 51.Skovran, E., and D. M. Downs. 2003. Lack of the ApbC or ApbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar Typhimurium. J. Bacteriol. 18598-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skovran, E., and D. M. Downs. 2000. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. J. Bacteriol. 1823896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skovran, E., C. T. Lauhon, and D. M. Downs. 2004. Lack of YggX results in chronic oxidative stress and uncovers subtle defects in Fe-S cluster metabolism in Salmonella enterica. J. Bacteriol. 1867626-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi, Y., and U. Tokumoto. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in Archaea and plastids. J. Biol. Chem. 27728380-28383. [DOI] [PubMed] [Google Scholar]
- 55.Tokumoto, U., and Y. Takahashi. 2001. Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron-sulfur proteins. J. Biochem. 13063-71. [DOI] [PubMed] [Google Scholar]
- 56.Unciuleac, M. C., K. Chandramouli, S. Naik, S. Mayer, B. H. Huynh, M. K. Johnson, and D. R. Dean. 2007. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry 466812-6821. [DOI] [PubMed] [Google Scholar]
- 57.Vivas, E., E. Skovran, and D. M. Downs. 2006. Salmonella enterica strains lacking the frataxin homolog CyaY show defects in Fe-S cluster metabolism in vivo. J. Bacteriol. 1881175-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 21897-106. [PubMed] [Google Scholar]
- 59.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32369-379. [DOI] [PubMed] [Google Scholar]
- 60.Webb, E., K. Claas, and D. Downs. 1998. thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J. Biol. Chem. 2738946-8950. [DOI] [PubMed] [Google Scholar]
- 61.Winston, F., D. Botstein, and J. H. Miller. 1979. Characterization of amber and ochre suppressors in Salmonella typhimurium. J. Bacteriol. 137433-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng, L., V. L. Cash, D. H. Flint, and D. R. Dean. 1998. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 27313264-13272. [DOI] [PubMed] [Google Scholar]
- 63.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 1834562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]