Abstract
Overexpression of the Vibrio fischeri sensor kinase RscS induces expression of the syp (symbiosis polysaccharide) gene cluster and promotes biofilm phenotypes such as wrinkled colony morphology, pellicle formation, and surface adherence. RscS is predicted to be a hybrid sensor kinase with a histidine kinase/ATPase (HATPase) domain, a receiver (Rec) domain, and a histidine phosphotransferase (Hpt) domain. Bioinformatic analysis also revealed the following three potential signal detection domains within RscS: two transmembrane helices forming a transmembrane region (TMR), a large periplasmic (PP) domain, and a cytoplasmic PAS domain. In this work, we genetically dissected the contributions of these domains to RscS function. Substitutions within the carboxy-terminal domain supported identification of RscS as a hybrid sensor kinase; disruption of both the HATPase and Rec domains eliminated induction of syp transcription, wrinkled colony morphology, pellicle formation, and surface adherence, while disruption of Hpt resulted in decreased activity. The PAS domain was also critical for RscS activity; substitutions in PAS resulted in a loss of activity. Generation of a cytoplasmic, N-terminal deletion derivative of RscS resulted in a partial loss of activity, suggesting a role for localization to the membrane and/or sequences within the TMR and PP domain. Finally, substitutions within the first transmembrane helix of the TMR and deletions within the PP domain both resulted in increased activity. Thus, RscS integrates both inhibitory and stimulatory signals from the environment to regulate biofilm formation by V. fischeri.
Bacteria commonly employ two-component regulatory systems to sense and respond to changes in their environment (42). These bacterial sensory systems are important during establishment of a symbiotic or pathogenic association; the bacteria detect the presence of a host organism and respond to the various environments encountered during migration through the host to the site of colonization (49). In the marine bioluminescent bacterium Vibrio fischeri, the sensor kinase component of a two-component regulatory pathway, RscS, is required for the bacterium to form a symbiotic association with its host organism, the Hawaiian bobtail squid (Euprymna scolopes) (47). In this symbiosis, colonization is initiated shortly after the juvenile squid hatch; V. fischeri cells form biofilmlike aggregates on the surface of the light organ in mucus secreted by the squid before migrating into the light organ crypts, multiplying to high cell density, and inducing bioluminescence (for reviews, see references 32, 40, 46). Strains in which rscS has been disrupted have a greatly decreased ability to colonize the squid (47) and do not aggregate in the mucus secreted by the squid (50).
RscS has been shown to regulate transcription of the syp (symbiosis polysaccharide) gene cluster (50). The syp cluster is a group of 18 genes that encode proteins important for production of a surface polysaccharide. Most of the genes in this cluster are required for symbiosis, and disruption of these genes results in a severe colonization defect (51). Recently, an increased-activity allele of rscS (rscS1) was identified that greatly increased the transcription of the syp cluster in culture (50). This allele encodes a wild-type protein but has mutations in the transcript that increase protein levels (15a, 50). When present on a multicopy plasmid, rscS1 induces various biofilm phenotypes in culture. These phenotypes include wrinkled colony morphology, adherence to glass surfaces, and pellicle formation. Furthermore, rscS1 dramatically increases the size of the bacterial aggregates on the light organ and confers a dramatic advantage in competitive colonization assays (50).
rscS encodes a large (927-amino-acid) protein predicted to contain multiple domains involved in signal detection and relay. The most conserved domains are a histidine kinase/ATPase (HATPase) domain, a response regulator-like receiver (Rec) domain, and a histidine phosphotransferase (Hpt) domain (Fig. 1), suggesting that after autophosphorylation, the phosphoryl group is relayed within RscS before transfer to a cognate response regulator. One possible role of the phosphorelay of hybrid sensor kinases is to allow multiple regulatory inputs, similar to what occurs in the multiprotein phosphorylation cascade regulating sporulation in Bacillus subtilis. In this phosphorelay, multiple sensor kinases and phosphatases regulate the flow of phosphate and thus signal transduction (35). A second possible role for hybrid sensor kinases may be to act as “rheostats” that modulate the level of the response to the level of the input signal, as has been proposed for the Bordetella hybrid sensor kinase BvgS (8). It is not yet known whether these conserved sequences in RscS are necessary for its activity.
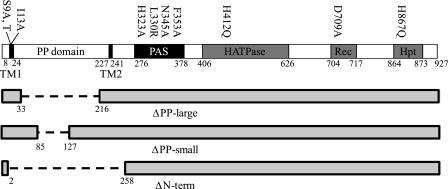
FIG. 1.
Linear map of RscS domains. The locations of amino acid substitutions are indicated above the map, while deletions are shown below the map.
Commonly, the sensor region of a sensor kinase is located in its amino terminus. The sensor regions of sensor kinases are quite diverse, reflecting the wide array of signals detected by these proteins (30). The amino terminus of RscS has three structural features: a transmembrane region (TMR) with two predicted transmembrane (TM) helices (TM1 and TM2) which together define a large, 200-amino-acid periplasmic (PP) domain and a predicted PAS sensor domain located just carboxy terminal of TM2 (16) (Fig. 1). There are examples of sensor kinases that perceive their signal through each of these regions (PP domain, TMR, and PAS domain) (30). The most common class of membrane-bound sensor kinases is the class of sensor kinases thought to receive signals through their PP domains (30). For example, PhoQ in Salmonella detects Mg2+ and antimicrobial peptides through its ∼150-amino-acid periplasmic domain (3, 7, 45). Other examples are the CitA/DcuS-like sensor kinases which detect concentrations of a signal molecule (citrate in the case of CitA) with a PAS-like fold in the PP domain (34, 37). While the large PP domain of RscS exhibits no similarity to PP domains involved in signal detection, this domain is conserved in putative proteins encoded by rscS alleles isolated from other V. fischeri strains (Mark Mandel, personal communication) and is present in a protein encoded by a putative rscS homolog from the coral pathogen Vibrio shilonii AK1 (accession number ZP_01865491). Thus, the PP domain of RscS may play a role in signal detection.
The TMR of membrane-bound sensor proteins has been implicated in sensing cell envelope stress, as well as in transduction of signal from the periplasm to the kinase domain. Many of the sensor kinases thought to perceive signals through their TMR have four or more TM helices (30). The sensor kinase LiaS from B. subtilis and its homologs, however, are predicted to have just two TM helices, like RscS. This sensor kinase responds to the presence of sublethal concentrations of antibiotics active against the cell wall, possibly through interaction of its TMR with the membrane protein LiaF (23). The role of the TMR in signal transduction has been studied most extensively with the bacterial chemoreceptors, in particular the aspartate receptor Tar (30). In these dimeric receptors, the two TM helices of each of the monomers form a four-helix bundle. The orientation of the helices relative to one another changes in response to ligand binding (13). In the case of Tar, ligand binding triggers a pistonlike movement of the signaling helix down toward the cytoplasm. This action regulates autophosphorylation of an associated histidine kinase (12, 33).
The third possible signal detection domain in RscS is its putative PAS domain. PAS domains are found in all kingdoms of life and have a conserved protein fold but limited sequence homology (19). They are important for detection of signals such as changes in light, redox potential, oxygen, and the presence of small ligands, such as ATP (44). Signal detection by PAS domains frequently requires the presence of a bound cofactor (44). Many sensor kinases have cytoplasmic PAS domains implicated in signal detection (30). For example, the O2 sensor FixL from rhizobia possesses a heme-binding PAS domain; the interaction of O2 with the heme cofactor results in inactivation of the kinase activity (17). Escherichia coli ArcB also has a cytoplasmic PAS domain; the interaction of oxidized quinones with this domain is thought to inhibit the kinase activity of this sensor kinase (29).
Despite the importance of the complex regulator RscS in inducing biofilm formation, little is known about the specific sequences essential for RscS activity. In this study, we examined the contribution of each RscS domain to protein activity by generating deletions and site-specific substitutions and evaluating their effects on rscS1-dependent biofilm phenotypes. Our results support the identification of RscS as a hybrid sensor kinase; replacement of predicted phosphorylated residues resulted in a decrease in or loss of biofilm formation. Moreover, our data demonstrate a critical role for PAS in RscS function. Finally, the other predicted sensor domains, TMR and the PP domain, contribute to RscS activity, and the PP domain has an apparently negative effect. These data allowed us to conclude that RscS integrates both positive and negative signals to regulate biofilm formation.
MATERIALS AND METHODS
Strains, plasmids, and media.
Strains used in this study are listed in Table 1. V. fischeri strains constructed in this work were generated by conjugation as previously described (11, 47). A bacterial isolate from E. scolopes, ES114, was the symbiosis-competent parent strain used in this study (4). E. coli strains DH5α (48) and TOP10 (Invitrogen, Carlsbad, CA) were used as hosts for cloning and conjugation. V. fischeri strains were grown in complex medium (LBS [18, 41]) or in HMM (39) containing 0.3% Casamino Acids and 0.2% glucose (HMM-CAA-Glu) (51). Antibiotics were added, as needed, at the following final concentrations: chloramphenicol, 5 μg ml−1; erythromycin, 5 μg ml−1; and tetracycline (Tet), 5 μg ml−1 in LBS and 30 μg ml−1 in HMM. Agar was added to a final concentration of 1.5% for solid media.
TABLE 1.
Plasmids and strains used in this study
| Strain or plasmid | Genotype, characteristics, or construction | Source or reference |
|---|---|---|
| E. coli strains | ||
| TAM1 | mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Active Motif |
| DH5α | endA1 hsdR17(rK− mK+) glnV44 thi-1 recA1 gyrA (Nalr) relA Δ(lacIZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15] | 48 |
| TOP10 F′ | F′ [lacIq Tn10 (Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| V. fischeri strains | ||
| ES114 | Wild type | 4 |
| KV2566 | ES114 att Tn7::PsypA::lacZ | 15a |
| Plasmids | ||
| pJET1 | Commercial cloning vector, Ampr | Fermentas |
| pCR2.1-TOPO | Commercial cloning vector, Ampr Kanr | Invitrogen |
| pKV69 | Mobilizable vector, Cmr Tetr | 47 |
| pKG63 | pKV69 containing rscS1 under control of PlacZ, Cmr Tetr | 15a |
| pKG92 | pKV69 containing rscS1(ΔPP-large), Cmr Tetr | This study |
| pKG94 | pKV69 containing rscS1(ΔPP-small), Cmr Tetr | This study |
| pKG99 | pKV69 containing rscS1(H323A), Cmr Tetr | This study |
| pKG100 | pKV69 containing rscS1(F353A), Cmr Tetr | This study |
| pKG102 | pKV69 containing rscS1(N345A), Cmr Tetr | This study |
| pKG107 | pKV69 containing rscS1(H412Q), Cmr Tetr | This study |
| pKG108 | pKV69 containing rscS1(H867Q), Cmr Tetr | This study |
| pKG115 | pKV69 containing rscS1(ΔN-term), Cmr Tetr | This study |
| pKG116 | pKV69 containing rscS1(L330R), Cmr Tetr | This study |
| pKG123 | pKV69 containing rscS1(S9T), Cmr Tetr | This study |
| pKG124 | pKV69 containing rscS1(S9A), Cmr Tetr | This study |
| pKG125 | pKV69 containing rscS1(I13A), Cmr Tetr | This study |
| pKG127 | pKV69 containing rscS1(D709A), Cmr Tetr | This study |
Molecular techniques.
Plasmids were constructed using standard molecular biology techniques with restriction and modifying enzymes obtained from New England Biolabs (Beverly, MA) or Promega (Madison, WI). Plasmids used or constructed in this study are listed in Table 1. All plasmids used in this study were derived from or are similar to pKG63, which lacks the rscS promoter but bears the rscS1 (50) allele under control of the lacZ promoter (15a). Thus, all rscS alleles described in this work carry the ribosome binding site mutation and the silent mutation at the Leu-25 codon present in rscS1 (50), as well as the described substitutions and deletions.
Site-directed mutagenesis.
The large deletions in the periplasmic domain were generated by divergent PCR. Divergent primers specific for the end points of the desired deletions (Table 2) and Kod HiFi thermostable DNA polymerase (Novagen, Madison, WI) were used to PCR amplify around the appropriate rscS1-containing plasmid. The resulting PCR product was purified (GeneClean; Q-Biogene, Solon, OH), self-ligated, and transformed into TOP10 cells. The H323A, N345A, and F353A substitutions were generated by amplifying the 5′ end of rscS using primer rscS1-RBS-F and the appropriate mutagenic reverse primer and the 3′ end of rscS using the mutagenic forward primer and 9R2 (Table 2). The two fragments were then joined using splicing by overlap extension PCR (21) with rscS1-RBS-F and 9R2. The H867Q substitution was generated using a QuikChange kit (Stratagene, La Jolla, CA). The H412Q substitution was generated using the GeneEditor in vitro site-directed mutagenesis system (Promega, Madison, WI). The remaining substitutions were generated using a Change-It multiple-mutation site-directed mutagenesis kit (USB, Cleveland, OH).
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| ΔPPsmall-F | AAAGAATACACAATCCACACTATTTTT |
| ΔPPsmall-R | ACGACGATTATACAAAAATGAAGC |
| ΔPPlarge-R | ATATTGTTTTACAGGATGGTTCC |
| ΔPPlarge-F | ACCCAATCTGAAACGATAAAAAC |
| H323A-F | GTTAGATAAAATAAAAGCTGCCAATATTATTATCTTGC |
| H323A-R | GCAAGATAATAATATTGGCAGCTTTTATTTTATCTAAC |
| N345A-F | TATCTTTTGAATTAACAGCTCATCATAAAGATGGAAC |
| N345A-R | GTTCCATCTTTATGATGAGCTGTTAATTCAAAAGATA |
| F353A-F | CATAAAGATGGAACTTTAGCCTGGATAGATGTATC |
| F353A-R | GATACATCTATCCAGGCTAAAGTTCCATCTTTATG |
| NewΔNterm-F | GGATCCTTTATAGGAGTTAATGCAATGGAATTAATCCGCTCTGAAAAAGAA |
| D709A-F | TTAATTATCATGGCTAACCATATGCCTG |
| L330R-F | CAATATTATTATCTTGCGTCAATCCTTAGAAACAG |
| S9T-F | TTATTACGTTATACCCTGATCACAATCATT |
| S9A-F | CAATTATTACGTTATGCCCTGATCACAATCATT |
| I13A-F | TATAGCCTTATCACGGCCATTACTTGCACATTA |
| H412Q | GCAACTATGAGCCAGGAGCTGAGGACA |
| H867Q | GATTTAATGCTCAGTCGATAAAAGG |
| H867Q_pur | GATTTAATGCTCAGTCGATAAAAGG |
| Complement_H867Q-pur | CCTTTTATCGACTGAGCATTAAATC |
| rscS1-RBS-F | GGATCCTTTATAGGAGTTAATGCAATGAAA |
| 9R2 | GAGCCATTCCCAATATACC |
Sequencing.
The presence of mutations was confirmed by automated sequencing (Northwestern University Genomics Core Facility, Chicago, IL) of the rscS1 gene on the various plasmids. One plasmid, pKG116 [rscS1(L330R)], contained a silent base change at codon 124. This alteration (GTA to GTG) had a negligible effect on Val codon usage (29% to 17%).
Western analysis.
Cultures were grown in HMM-CAA-Glu with Tet overnight, and then 1-ml samples were removed and pelleted by centrifugation. Cells were resuspended in 50 μl of distilled H2O with a protease inhibitor cocktail for use with bacterial cell extracts (Sigma Aldrich, St. Louis, MO), and a 5-μl aliquot was removed for quantitation by the assay of Lowry et al. (27). After quantitation, cells were diluted as necessary to bring all samples to the same protein concentration, and then 1 volume of 2× sodium dodecyl sulfate dye was added. Cells were lysed by boiling them for 5 min, and then extracts were loaded on an 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. After electrophoretic separation, proteins were transferred to a polyvinylidene difluoride membrane and probed with anti-RscS antibodies as described elsewhere (15a).
Wrinkled colony morphology.
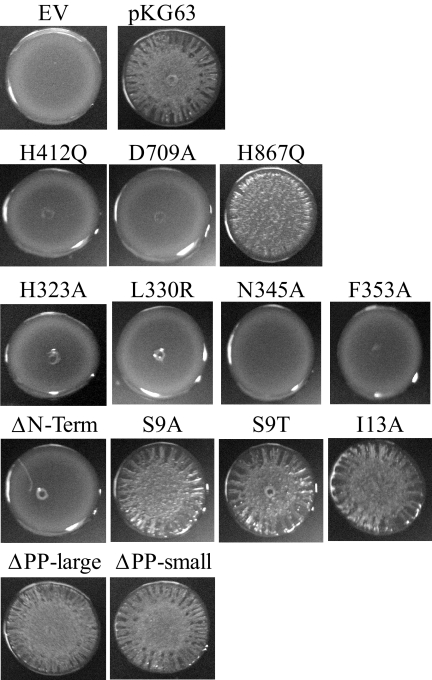
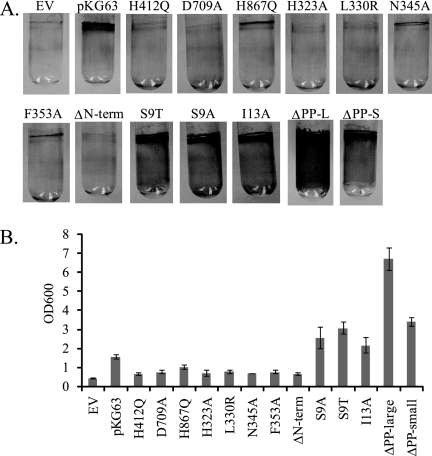
In most experiments, plasmid-bearing strains were streaked onto LBS-Tet plates and incubated at 28°C overnight. To obtain the photographs shown in Fig. 4, the bacteria were grown at 28°C in LBS-Tet for 6 h, and then a 10-μl aliquot was spotted on LBS-Tet plates and incubated at 28°C for 19 h. Spotting produced wrinkled morphology similar to that produced by streaking for single colonies, but it resulted in a uniform size, which allowed more direct comparisons among the different strains.
FIG. 4.
Colony morphology of strains carrying RscS mutants. Bacteria were grown at 28°C in LBS-Tet for 6 h and then spotted on LBS-Tet plates and incubated at 28°C for 19 h. The experiment was performed at least in triplicate; representative results are shown. EV, empty vector.
Surface adherence.
Cultures were grown in HMM-CAA-Glu with Tet overnight and then diluted in 2 ml (final volume) to an optical density at 600 nm (OD600) of 0.1. After 24 h of static incubation in glass tubes at 28°C, cells were stained with 1% (wt/vol) crystal violet for 15 min. Nonadherent cells and excess stain were washed away with several rinses of deionized H2O, and the tubes were allowed to dry overnight. Quantitation was performed by adding 1 g of 1-mm glass beads and 2 ml of 95% ethanol to each tube, vortexing the tube for 10 s, and measuring the OD600 of the ethanol-crystal violet solution. The assay was performed in triplicate for each strain.
Pellicle formation.
Cultures were grown in HMM-CAA-Glu with Tet overnight and then diluted in 3 ml (final volume) to an OD600 of 0.1. After 24 h of static incubation at 28°C, pellicle formation was assessed by drawing a pipette tip across the surface of the well. Formation of a visible film that was disrupted by the pipette tip was scored as positive for pellicle formation (+). Formation of a visible film only after 48 h of incubation was scored as ±. Strains which failed to form a film even after 48 h were scored as negative (−). The assay was performed in triplicate for each strain.
β-Galactosidase activity assays.
Cultures were grown in rich medium (LBS) overnight, and β-galactosidase activity was measured as described previously (31). Total protein concentrations were determined by the method of Lowry et al. (27). The assay was performed in triplicate for each strain.
RESULTS
Putative phosphotransfer residues are important for RscS function.
Sequence alignment predicted that RscS is a hybrid sensor kinase with HATPase, Rec, and Hpt domains (47), but to date, the importance of these regions for RscS activity has not been investigated. To determine if each of these putative domains contributes to RscS activity, we generated substitutions in residues predicted to be the targets of phosphorylation and assessed their effects on rscS1-dependent phenotypes. Specifically, we targeted the predicted autophosphorylated His in the HATPase domain (H412), the Asp site of phosphorylation in the Rec domain (D709), and the His in the Hpt domain (H867) (Fig. 1). Each of the His residues was replaced by Gln (H412Q and H867Q), while D709 was replaced by Ala (D709A).
Because the impact of RscS on biofilm formation has been observed only when an overexpression construct is used, we generated these and all other substitutions in the context of the increased activity allele rscS1 carried on a multicopy plasmid. In each case, transcription of the rscS allele was driven exclusively from the vector-derived lacZ promoter. As controls, we used the unmutagenized rscS1 plasmid pKG63 and vector pKV69. We then moved the resulting plasmids into V. fischeri and assessed RscS function. Specifically, we evaluated the abilities of the mutant alleles to produce protein and induce biofilm phenotypes, including syp transcription, wrinkled colony formation, pellicle formation, and surface attachment.
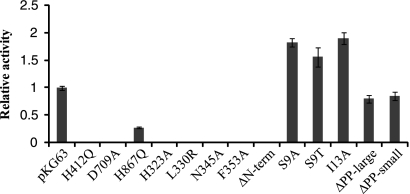
V. fischeri strains carrying plasmids with the H412Q and D709A substitution alleles produced full-length RscS protein, as evaluated by Western blot analysis (Fig. 2), but they exhibited severe defects in inducing biofilm phenotypes. First, these alleles failed to induce syp transcription, as monitored using a reporter strain bearing a chromosomal fusion of the sypA promoter to lacZ (PsypA::lacZ, KV2566 [Table 1]); the level of syp transcription induced by these alleles was 100-fold less than that induced by pKG63 (Fig. 3). Consistent with a loss of syp activation, these substitution alleles also failed to promote wrinkled colony formation under conditions in which pKG63 induces dramatic wrinkling (Fig. 4). Similarly, while pKG63 induced pellicle formation within 24 h, strains carrying the H412Q and D709A substitution alleles failed to form pellicles at the air-liquid interface even after 48 h (Table 3). Finally, these strains also failed to adhere to a glass surface, as visualized by crystal violet staining (Fig. 5A and B).
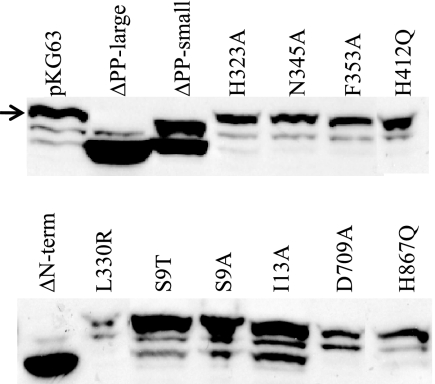
FIG. 2.
Western blot analysis of RscS production by various mutant rscS1 plasmids. The position of full-length RscS is indicated by an arrow. The lower two bands present in the pKG63 lane and the other full-length RscS derivatives represent a cross-reactive species and a putative proteolytic fragment of RscS, respectively (15a). ΔPP-large and ΔN-term roughly comigrate with the proteolytic fragment, while ΔPP-small is approximately the same size as the cross-reactive species.
FIG. 3.
Induction of syp transcription by RscS mutants. The β-galactosidase activity induced in the PsypA::lacZ reporter strain by a plasmid carrying a mutation was divided by the activity induced by pKG63 to derive the relative activity. The error bars represent propagation of the standard deviation.
TABLE 3.
Pellicle formation by RscS mutants
| Allele | Pelliclea |
|---|---|
| Empty vector | − |
| pKG63 | + |
| H412Q | − |
| D709A | − |
| H867Q | ± |
| H323A | − |
| L330R | − |
| N345A | − |
| F353A | − |
| ΔN-term | − |
| S9A | + |
| S9T | + |
| I13A | + |
| ΔPP-large | + |
| ΔPP-small | + |
+, formation of a visible pellicle within 24 h during incubation at 28°C; ±, formation of a visible pellicle within 48 h; −, failure to form a pellicle after 48 h.
FIG. 5.
Surface adherence of strains overexpressing the indicated rscS mutant constructs. (A) Photographs of crystal violet-stained glass tubes, showing variations in surface adherence. The experiment was performed in triplicate, and representative results are shown. (B) Quantitation of crystal violet staining. The error bars indicate the standard deviations. EV, empty vector.
Like the H42Q and D709A substitution alleles, the H867Q allele produced wild-type levels of protein (Fig. 2). However, in contrast to the severe defects observed with the former alleles, H867Q induced moderate biofilm phenotypes. This substitution allele induced syp transcription to a level that was only fourfold less than the control level (Fig. 3). In agreement with this less severe effect on syp transcription, this strain also produced moderate wrinkling (Fig. 4) and formed a pellicle at the air-liquid interface after 48 h (Table 3). Furthermore, the strain carrying the H867Q substitution allele retained some ability to adhere to a glass surface, forming a thin line of crystal violet-stained material at the air-liquid interface (Fig. 5A). These data suggest that the HATPase and Rec domains are required for RscS function, while the Hpt domain is partially dispensable.
RscS possesses a PAS signal detection domain critical for its activity.
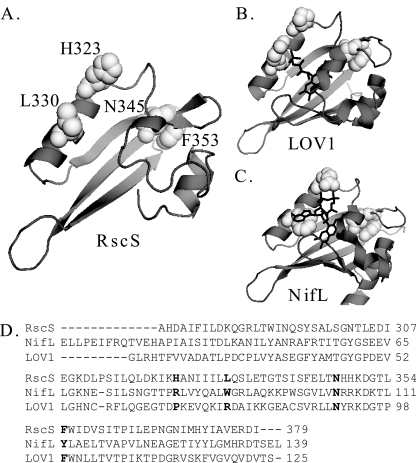
BLAST analysis (1) of the RscS amino acid sequence predicted that there is a PAS domain immediately carboxy terminal to TM2 (data not shown). Since PAS domains are commonly employed to detect environmental signals (44), we predicted that this domain is required to regulate RscS function. To further investigate the function of the putative PAS domain, we first generated a predicted structure. A first-approximation structural model (Fig. 6A) (SwissModel [2]) threaded the RscS PAS sequence onto the LOV1 domain of the Phot1 protein of Chlamydomonas reinhardtii (1N9L [14]), a flavin mononucleotide (FMN)-binding PAS domain. LOV (light, oxygen, or voltage) domains comprise a subset of the PAS domain family and sense blue light through formation of a covalent cysteine-flavin adduct (9, 14). The ability to model the RscS sequence onto a PAS-like structure supported the hypothesis that RscS contains a PAS domain. However, the RscS PAS domain lacks Cys residues, suggesting that it is not a canonical LOV domain. Furthermore, sequence alignment of RscS PAS, LOV1, and the related flavin adenine dinucleotide (FAD)-binding PAS domain of Azotobacter vinelandii NifL (Fig. 6D) revealed that the RscS PAS domain shares homology with NifL, especially in residues predicted to be important for FAD binding (24).
FIG. 6.
RscS PAS domain. (A) RscS PAS domain sequence threaded onto the LOV1 domain of the Phot1 protein of C. reinhardtii (1N9L) (2, 14; http://swissmodel.expasy.org//). Residues subjected to mutagenesis in this study are shown in white spacefill and are labeled. (B) LOV1 domain of the Phot1 protein of C. reinhardtii (1N9L) (14). The FMN cofactor is indicated by black lines. Residues corresponding to the residues mutagenized in RscS are shown in white spacefill. (C) A. vinelandii NifL PAS domain structure (2GJ3) (24). The FAD cofactor is indicated by black lines. Residues corresponding to the residues mutagenized in RscS are shown in white spacefill. (D) Sequence alignment for the putative PAS domain of RscS, the A. vinelandii NifL PAS domain, and the LOV1 domain of the Phot1 protein of C. reinhardtii. Residues targeted for substitution in RscS and the corresponding residues in NifL and LOV1 are indicated by bold type. Crystal structures were visualized using the PyMOL molecular graphics system (http://www.pymol.org).
To determine if the putative PAS domain indeed regulates RscS function, we generated substitutions in PAS domain residues predicted to interact with a flavin cofactor based on the structure prediction and sequence alignment. In both FMN-binding LOV1 and FAD-binding NifL, a conserved Asn residue and an aromatic residue interact with the flavin ring of the respective cofactors (Fig. 6B and C). Replacement of the corresponding residues in RscS (Asn-345 and Phe-353) would thus be predicted to disrupt cofactor binding and RscS function. The NifL PAS domain further interacts with its FAD cofactor through a positive residue (corresponding to His-323 in RscS) and a hydrophobic residue (corresponding to Leu-330 in RscS) (24). Thus, disruption of these residues would be predicted to affect RscS function if the RscS PAS domain binds a FAD cofactor. Therefore, we replaced H323, N345, and F353 with Ala and L330 with Arg.
V. fischeri strains carrying plasmids with the four PAS domain substitutions in rscS produced full-length RscS protein (Fig. 2) but exhibited severe defects in inducing biofilm formation. Each of the four PAS domain substitutions resulted in a profound loss of syp induction, and the transcription levels were approximately 100-fold lower than the level induced by pKG63 (Fig. 3). Consistent with this result, each substitution allele also failed to induce wrinkled colony formation on solid media (Fig. 4) and pellicle formation at the air-liquid interface (Table 3). Strains expressing the H323A and L330R substitution alleles also failed to adhere to a glass surface (Fig. 5A and B), while strains with the N345A and F353A substitution alleles exhibited a moderate ability to adhere to glass (Fig. 5A).
These experiments were performed at 28°C, the optimal growth temperature for V. fischeri. Historically, however, we have observed that incubation at room temperature increases RscS-mediated phenotypes (data not shown), and thus we asked if incubation at a lower temperature would restore activity to our PAS domain substitution alleles. Indeed, this was the case for the N345A and F353A substitutions. When incubated at room temperature, the strains carrying the N345A and F353A alleles both exhibited moderate wrinkling, surface adherence, and pellicle formation and induced syp transcription to levels near pKG63-induced levels (data not shown). However, strains carrying the H323A and L330R substitutions failed to exhibit rscS1-mediated phenotypes regardless of the temperature, and the results were indistinguishable from the results for the vector control (data not shown). Thus, the PAS domain is critical for RscS function, and the severe defect observed for substitutions (H323A and L330R) predicted to disrupt FAD binding but not FMN binding suggests that RscS may bind a FAD cofactor.
A cytoplasmic RscS derivative is partially active.
Some membrane-bound sensor kinases become constitutively active when they are liberated from the membrane (6, 26), while others become nonfunctional (36). To determine what role membrane localization and/or sequences within the TMR and PP domain play in RscS function, we generated a cytoplasmic derivative of RscS lacking the putative TM helices and PP domain but retaining the PAS domain (ΔN-term; amino acids N3 through R257 removed [Fig. 1]) and tested its effect on RscS-dependent culture phenotypes. Western analysis showed that this deletion construct produced protein (Fig. 2). However, the deletion resulted in a loss of RscS function; the truncated protein failed to induce syp transcription (Fig. 3), wrinkled colony formation (Fig. 4), pellicles (Table 3), or surface adherence (Fig. 5A) when the assay was performed at 28°C. In contrast, incubation at a lower temperature restored partial activity, including moderate levels of syp transcription, wrinkled colony formation, thin pellicles, and moderate surface attachment (data not shown). Thus, full RscS activity may require localization to the membrane and/or essential sequences in the amino terminus.
The transmembrane domain is important for RscS function.
The decrease in activity of the N-terminal deletion suggested a positive role for sequences in the PP domain and/or TMR. The TMR may directly detect environmental signals, or it may be important for transduction of the signal across the membrane from the PP domain. To determine the role of the TMR of RscS, we generated substitutions within the first putative TM helix (TM1) and determined their effects on RscS-dependent phenotypes. We focused on TM1 because modeling its sequence onto a helical wheel (data not shown) predicted the presence of a hydrophilic patch along one face of the helix. We predicted that this hydrophilic patch might serve as an interaction surface between the monomers within the RscS dimer or with another membrane protein. If it does, then disrupting this patch would alter these interactions and alter RscS function. To test this hypothesis, we replaced a hydrophilic Ser residue within the patch with a hydrophobic residue (S9A) or with another hydrophilic residue (S9T). Consistent with our hypothesis, strains carrying the S9A substitution allele exhibited an approximately twofold increase in syp transcription (Fig. 3). However, the S9T substitution also caused increased syp transcription, as did replacement of a residue outside the hydrophilic patch (I13A) (Fig. 3). The S9A, S9T, and I13A constructs each also slightly increased surface attachment (Fig. 5A) but induced the formation of wrinkled colonies (Fig. 4) and pellicles (Table 3) to an extent similar to that observed with strains carrying pKG63. Therefore, we propose that the identity of certain residues within TM1, but not their relative hydrophilicity, is important for RscS function.
The periplasmic domain negatively regulates RscS function.
The loss of the N terminus led to a decrease in RscS function, while disruption of TM1 had the opposite effect. To determine what role the PP domain may play, we constructed mutations within this domain, leaving TM1 and TM2 intact. Because the PP domain has no significant homology to other proteins, we had no information to guide mutagenesis; therefore, we elected to examine the function of the PP domain by generating two deletions within it (Fig. 1). The deletion that we designated ΔPP-large removed 183 amino acids (Q33 through I216), leaving an 18-amino-acid linker between the two TM helices, while the deletion that we designated ΔPP-small removed 42 amino acids (amino acids 85 through 127) from the central portion of the domain.
V. fischeri strains carrying plasmids with these rscS deletions produced truncated RscS protein, as detected by Western analysis (Fig. 2). Both the large and small PP deletion constructs induced syp transcription to nearly wild-type levels (Fig. 3), suggesting that the PP domain is dispensable for this phenotype. Consistent with this result, strains carrying these two plasmids also produced wrinkled colonies on solid media (Fig. 4) and thick pellicles at the air-liquid interface (Table 3) to an extent similar to that observed with strains carrying pKG63. These results suggested that the PP domain is not required for RscS function other than as a membrane tether.
However, when we examined attachment, a different story began to emerge. The loss of the PP region of RscS (ΔPP-large) appeared to cause an increase in cell-cell aggregation. This effect was shown by formation of stringy cell aggregates during growth in shaking liquid cultures; these aggregates formed more quickly and to a greater extent in strains containing ΔPP-large than in strains carrying pKG63 (data not shown). Furthermore, strains carrying the ΔPP-large or ΔPP-small deletion alleles exhibited a dramatic increase in adherence to glass (Fig. 5A and B). Thus, these data suggested that removal of the PP domain resulted in increased RscS activity.
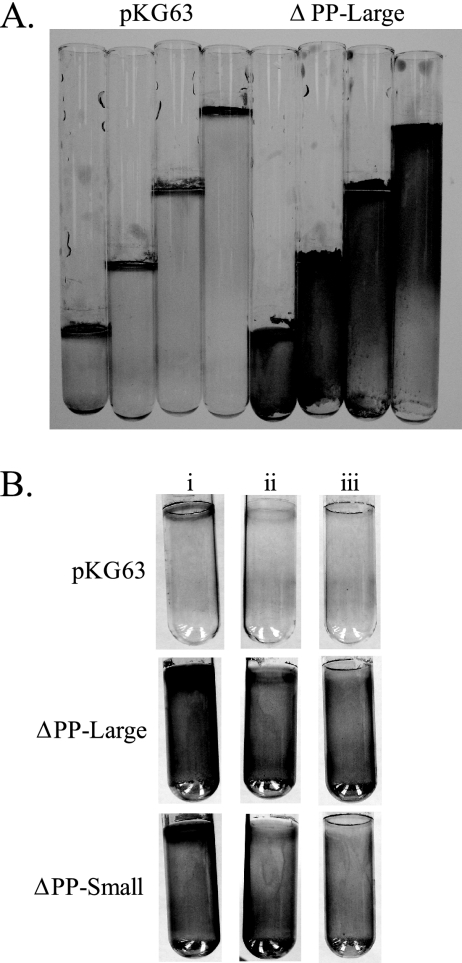
In the assay for surface attachment, the PP domain deletions not only increased the amount of crystal violet-stained material left on the glass surface but also changed the pattern of deposition. The wild-type protein induced surface attachment in a tight ring at the air-liquid interface, while both PP domain deletion alleles induced attachment throughout the culture (Fig. 5A). This effect could be seen quite clearly in surface attachment assays performed with various culture volumes. For pKG63-containing strains, the area of crystal violet staining was limited to the top of the tube, where the air-liquid interface was. In contrast, for strains carrying the ΔPP-large allele of rscS, staining occurred throughout the tube (Fig. 7A).
FIG. 7.
Effect of air limitation on surface attachment. (A) Surface attachment assays performed with increasing volumes of culture. (B) Surface attachment assays performed with (i) normal airflow, (ii) airflow restricted with a cotton plug and Parafilm sealing the top, and (iii) airflow restricted by a mineral oil overlay.
These results suggest that the wild-type protein fails to induce attachment below the air-liquid interface due to a negative signal relayed by the PP domain. For example, cells below the air-liquid interface may be exposed to lower levels of oxygen and thus be in a different redox state. In support of this idea, limiting airflow in the culture tubes using a mineral oil overlay or a cotton plug and Parafilm resulted in loss of the tight ring at the air-liquid interface for both pKG63 and PP domain deletion strains (Fig. 7B). In contrast, attachment below this ring was largely unaffected in cultures expressing the PP domain deletion constructs. The exact signal detected by the PP domain remains to be determined; nonetheless, these data support the conclusion that the PP domain modulates RscS activity under specific environmental conditions.
DISCUSSION
The work described here demonstrates that the V. fischeri symbiosis regulator RscS is a complex protein with multiple inputs for signal integration and relay. In its kinase domain, both the HATPase and Rec domains are required for RscS activity, while the Hpt domain is partially dispensable. In the amino-terminal sensor portion of the protein, disruption of the PP domain and TMR resulted in increased activity, while single amino acid substitutions in the PAS domain eliminated or severely impaired the function.
The identification of RscS as a hybrid sensor kinase suggested that after autophosphorylation the phosphoryl group progresses down the length of RscS through a phosphorelay before ultimately arriving at the target response regulator, as has been shown for the canonical hybrid sensor kinase ArcB (25). However, our results show that disruption of the Hpt domain results in only a partial loss of function. Intriguingly, the putative RscS homolog in V. shilonii has homology to V. fischeri RscS along the length of the protein but lacks a predicted Hpt domain (K. Geszvain, unpublished data). These data raise the possibility that at least some of the phosphoryl transfer along the length of RscS is independent of its Hpt domain. Thus, the phosphorelay may be branched, with some phosphoryl groups being transferred to the Hpt domain on RscS before being transferred to the response regulator and some phosphoryl groups being transferred from Rec to an unknown Hpt domain on a second protein. Such a phosphotransfer pathway is not unprecedented; in the Rcs capsular polysaccharide regulatory pathway of E. coli, the phosphoryl group is transferred from the Rec domain of the hybrid sensor kinase RcsC to the Hpt domain of another sensor kinase, RcsD (28). Alternatively, the phosphoryl group could be transferred directly to the downstream response regulator via H1 (H412), which has been shown to happen in vitro for ArcA/B (15). The syp cluster encodes three two-component regulators: two putative response regulators, SypG and SypE, and a predicted membrane-bound hybrid sensor kinase, SypF. SypG is required for all of the biofilm phenotypes induced by RscS in culture (15a, 22). Furthermore, we have found that SypG is required for biofilm formation induced by RscS(H867Q) and RscS(ΔPP-large) (unpublished data), suggesting that the activity of RscS is ultimately, if not directly, shuttled through SypG. SypE and SypF also play roles in RscS- and/or SypG-dependent phenotypes; however, their roles are complex and poorly understood (9a, 22). Thus, the regulatory consequences of RscS activation appear to be complex, involving interactions with multiple downstream targets.
While substitutions and deletions within the TMR and PP domain resulted in increased RscS activity, deletion of both domains together (ΔN-term allele) resulted in impaired function. Thus, while the TMR and the PP domain may detect or transmit a negative signal, there appears to be a positive role either for the TMR or the PP domain or for membrane localization itself. Possibly, membrane localization is required for activation of RscS through its PAS domain via a signal located predominately there, in much the same way that ArcB is proposed to be negatively regulated by quinones in the membrane (29). However, it is formally possible that the increased activity displayed by the TMR and PP domain mutant alleles is simply due to increased protein levels. This seems unlikely to be the case at least for the PP domain deletion alleles since they do not increase all RscS activities. For example, these alleles greatly increase surface attachment, yet they have little effect on syp transcription. Therefore, our data support a model in which the PP domain negatively regulates RscS function with residues in TM1 involved in transduction of this signal to the cytoplasmic domain, while the PAS domain located at the inner membrane positively regulates RscS activity.
Our results show that the PAS domain is critical for RscS function and support the hypothesis that there is a FAD cofactor bound to the domain. Replacement of H323 and replacement of L330 both resulted in a complete loss of RscS function, while the N345A and F353A substitutions both dramatically reduced activity at higher temperatures. A key difference between the FMN and FAD cofactors is the presence of the adenine base in the FAD cofactor. In NifL, a Trp residue (W87) base stacks with this adenine, stabilizing the interaction of the cofactor with the PAS domain (24). The corresponding residue in the FMN-binding LOV1 is a positively charged Arg (R74) (14), while in RscS it is a Leu residue (L330) (Fig. 6A). Replacement of L330 with Arg resulted in a complete loss of activity, consistent with the hypothesis that this positively charged residue repels binding of the hydrophobic adenine base of a FAD cofactor. The interaction between a positive residue and the phosphate group of FAD is thought to make a major contribution to the stability of FAD binding in flavoproteins (20). In NifL, this residue is R80 (24); the corresponding residue in RscS is H323, while in LOV1 it is P67 (14). Removing this positive residue from RscS (H323A) resulted in a complete loss of activity. Taken together, our data support the hypothesis that a FAD cofactor binds to the RscS PAS domain.
Can our data reveal anything about the nature of the signal(s) detected by RscS? FAD-binding PAS domains commonly act as redox sensors (43). For example, the oxidation state of the FAD cofactor bound in the E. coli aerotaxis sensor Aer is proposed to regulate the response of the protein to oxygen gradients and redox potential (38). Thus, RscS may sense the redox state of the cell through its PAS domain. At the same time, removal of the PP domain resulted in an apparent loss of sensitivity to low O2 concentrations, as shown by increased surface attachment below the air-liquid interface of strains carrying the deletion alleles. This raises the intriguing possibility that a repressive O2/redox state detected by the PP domain and an activating O2/redox state detected by the PAS domain collaborate to limit RscS activity to a narrow window of optimal signal concentration. This is an attractive hypothesis because reactive oxygen species are present in the squid-secreted mucus that V. fischeri cells encounter early in the symbiosis (5, 10). Thus, RscS may detect a change in the redox state of the cell as the bacteria move from the relatively reducing environment of the seawater to the oxidizing environment of the squid.
Acknowledgments
We thank Sean Crosson for his insights into PAS domain structure, Cindy Darnell for technical assistance, Alan Wolfe and members of our lab for critical reading of the manuscript and helpful suggestions, and Mark Mandel for sharing sequence data prior to publication.
This work was supported by NIH grant GM59690 awarded to K.L.V. and by the NIH through Ruth L. Kirschstein National Research Service Award 1 F32 G073523 from the NIGMS awarded to K.G.
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22195-201. [DOI] [PubMed] [Google Scholar]
- 3.Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, H. Le Moual, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122461-472. [DOI] [PubMed] [Google Scholar]
- 4.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 1723701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose, J. L., U. Kim, W. Bartkowski, R. P. Gunsalus, A. M. Overley, N. L. Lyell, K. L. Visick, and E. V. Stabb. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65538-553. [DOI] [PubMed] [Google Scholar]
- 6.Cavicchioli, R., R. C. Chiang, L. V. Kalman, and R. P. Gunsalus. 1996. Role of the periplasmic domain of the Escherichia coli NarX sensor-transmitter protein in nitrate-dependent signal transduction and gene regulation. Mol. Microbiol. 21901-911. [DOI] [PubMed] [Google Scholar]
- 7.Cho, U. S., M. W. Bader, M. F. Amaya, M. E. Daley, R. E. Klevit, S. I. Miller, and W. Xu. 2006. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J. Mol. Biol. 3561193-1206. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. A., and A. M. Jones. 2003. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 11367-373. [DOI] [PubMed] [Google Scholar]
- 9.Crosson, S., and K. Moffat. 2001. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 982995-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Darnell, C. L., E. A. Hussa, and K. L. Visick. 9 May 2008. The putative hybrid sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J. Bacteriol. doi: 10.1128/JB.00197-08. [DOI] [PMC free article] [PubMed]
- 10.Davidson, S. K., T. A. Koropatnick, R. Kossmehl, L. Sycuro, and M. J. McFall-Ngai. 2004. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell. Microbiol. 61139-1151. [DOI] [PubMed] [Google Scholar]
- 11.DeLoney, C. R., T. M. Bartley, and K. L. Visick. 2002. Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J. Bacteriol. 1845121-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draheim, R. R., A. F. Bormans, R. Z. Lai, and M. D. Manson. 2006. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry 4514655-14664. [DOI] [PubMed] [Google Scholar]
- 13.Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz, and M. A. Danielson. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13457-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedorov, R., I. Schlichting, E. Hartmann, T. Domratcheva, M. Fuhrmann, and P. Hegemann. 2003. Crystal structures and molecular mechanism of a light-induced signaling switch: the Phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys. J. 842474-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgellis, D., A. S. Lynch, and E. C. Lin. 1997. In vitro phosphorylation study of the arc two-component signal transduction system of Escherichia coli. J. Bacteriol. 1795429-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Geszvain, K., and K. L. Visick. Multiple factors contribute to keep levels of the symbiosis regulator RscS low. FEMS Microbiol. Lett., in press. [DOI] [PMC free article] [PubMed]
- 16.Geszvain, K., and K. L. Visick. 2006. Roles of bacterial regulators in the symbiosis between Vibrio fischeri and Euprymna scolopes. Prog. Mol. Subcell. Biol. 41:277-290. [DOI] [PubMed] [Google Scholar]
- 17.Gilles-Gonzalez, M. A., G. Gonzalez, and M. F. Perutz. 1995. Kinase activity of oxygen sensor FixL depends on the spin state of its heme iron. Biochemistry 34232-236. [DOI] [PubMed] [Google Scholar]
- 18.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 1766986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hefti, M. H., K. J. Francoijs, S. C. de Vries, R. Dixon, and J. Vervoort. 2004. The PAS fold. A redefinition of the PAS domain based upon structural prediction. Eur. J. Biochem. 2711198-1208. [DOI] [PubMed] [Google Scholar]
- 20.Hefti, M. H., J. Vervoort, and W. J. van Berkel. 2003. Deflavination and reconstitution of flavoproteins. Eur. J. Biochem. 2704227-4242. [DOI] [PubMed] [Google Scholar]
- 21.Horton, R. M. 1997. In vitro recombination and mutagenesis of DNA. SOEing together tailor-made genes. Methods Mol. Biol. 67141-149. [DOI] [PubMed] [Google Scholar]
- 22.Hussa, E. A., C. L. Darnell, and K. L. Visick. 25 April 2008. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J. Bacteriol. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed]
- 23.Jordan, S., A. Junker, J. D. Helmann, and T. Mascher. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 1885153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Key, J., M. Hefti, E. B. Purcell, and K. Moffat. 2007. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism. Biochemistry 463614-3623. [DOI] [PubMed] [Google Scholar]
- 25.Kwon, O., D. Georgellis, and E. C. Lin. 2000. Phosphorelay as the sole physiological route of signal transmission by the Arc two-component system of Escherichia coli. J. Bacteriol. 1823858-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon, O., D. Georgellis, A. S. Lynch, D. Boyd, and E. C. Lin. 2000. The ArcB sensor kinase of Escherichia coli: genetic exploration of the transmembrane region. J. Bacteriol. 1822960-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 28.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59379-405. [DOI] [PubMed] [Google Scholar]
- 29.Malpica, R., G. R. Sandoval, C. Rodriguez, B. Franco, and D. Georgellis. 2006. Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid. Redox Signal. 8781-795. [DOI] [PubMed] [Google Scholar]
- 30.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Nyholm, S. V., and M. J. McFall-Ngai. 2004. The winnowing: establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2632-642. [DOI] [PubMed] [Google Scholar]
- 33.Ottemann, K. M., W. Xiao, Y. K. Shin, and D. E. Koshland, Jr. 1999. A piston model for transmembrane signaling of the aspartate receptor. Science 2851751-1754. [DOI] [PubMed] [Google Scholar]
- 34.Pappalardo, L., I. G. Janausch, V. Vijayan, E. Zientz, J. Junker, W. Peti, M. Zweckstetter, G. Unden, and C. Griesinger. 2003. The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J. Biol. Chem. 27839185-39188. [DOI] [PubMed] [Google Scholar]
- 35.Perego, M., C. Hanstein, K. M. Welsh, T. Djavakhishvili, P. Glaser, and J. A. Hoch. 1994. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell 791047-1055. [DOI] [PubMed] [Google Scholar]
- 36.Piazza, F., P. Tortosa, and D. Dubnau. 1999. Mutational analysis and membrane topology of ComP, a quorum-sensing histidine kinase of Bacillus subtilis controlling competence development. J. Bacteriol. 1814540-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinelt, S., E. Hofmann, T. Gerharz, M. Bott, and D. R. Madden. 2003. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 27839189-39196. [DOI] [PubMed] [Google Scholar]
- 38.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruby, E. G., and K. H. Nealson. 1977. Pyruvate production and excretion by the luminous marine bacteria. Appl. Environ. Microbiol. 34164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stabb, E. V. 2006. The Vibrio fischeri-Euprymna scolopes light organ symbiosis, p. •••-•••. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 41.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, B. L., A. Rebbapragada, and M. S. Johnson. 2001. The FAD-PAS domain as a sensor for behavioral responses in Escherichia coli. Antioxid. Redox Signal. 3867-879. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vescovi, E. G., Y. M. Ayala, E. Di Cera, and E. A. Groisman. 1997. Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+. J. Biol. Chem. 2721440-1443. [DOI] [PubMed] [Google Scholar]
- 46.Visick, K. L., and E. G. Ruby. 2006. Vibrio fischeri and its host: it takes two to tango. Curr. Opin. Microbiol. 9632-638. [DOI] [PubMed] [Google Scholar]
- 47.Visick, K. L., and L. M. Skoufos. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 173469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, J., H. C. Chiang, M. K. Bjursell, and J. I. Gordon. 2004. Message from a human gut symbiont: sensitivity is a prerequisite for sharing. Trends Microbiol. 1221-28. [DOI] [PubMed] [Google Scholar]
- 50.Yip, E. S., K. Geszvain, C. R. DeLoney-Marino, and K. L. Visick. 2006. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 621586-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yip, E. S., B. T. Grublesky, E. A. Hussa, and K. L. Visick. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and σ54-dependent biofilm formation by Vibrio fischeri. Mol. Microbiol. 571485-1498. [DOI] [PubMed] [Google Scholar]