Abstract
We demonstrate that in Listeria monocytogenes, temperature-responsive transcriptional control of flagellar genes does not rely on the phosphorylation of the conserved phosphorylation site (D55) in the receiver domain of response regulator DegU. Furthermore, proper control of DegU-regulated genes involved in ethanol tolerance and virulence is independent of receiver phosphorylation.
In Listeria monocytogenes, a facultative intracellular bacterium, the expression of flagellum-based motility is regulated in response to the growth temperature (7, 13), with the permissive temperature being 30°C and below. Temperature-dependent transcriptional regulation of flagellar genes in L. monocytogenes relies on three regulatory proteins: the repressor protein MogR, the response regulator DegU, and GmaR, an antirepressor of MogR (3, 4, 9, 10, 13). The MogR protein was shown to bind directly to multiple TTTT-N5-AAAA recognition sites within the promoter regions of the motility genes, thus preventing their transcription at the nonpermissive temperature (3, 9). Response regulator DegU is required to relieve MogR-mediated repression by enabling expression of GmaR at low temperatures. In vitro DNA-binding experiments demonstrated that GmaR is able to both interfere with the binding of MogR to flagellar gene promoters by protein-protein interaction and disrupt preformed repressor-DNA complexes (10). Apparently, DegU is also involved in the regulation of flagellin expression on the posttranscriptional level, since compared to the wild type, a mogR- and degU-negative mutant of L. monocytogenes produced reduced amounts of the FlaA protein, albeit increased transcription of flaA (9). Since transcription of degU is not responsive to temperature and the DegU protein could be detected at elevated and low temperatures (9, 13), it is expected that the activity state of the transcriptional activator DegU is modulated in response to temperature. Usually the activity of a response regulator is controlled by receiver phosphorylation via a cognate histidine kinase which autophosphorylates in response to the appropriate environmental stimulus (6). However, DegU of L. monocytogenes is an orphan response regulator which apparently lacks a cognate histidine kinase (2, 12). Since interaction of a noncognate histidine kinase with DegU is conceivable, we analyzed the contribution of receiver phosphorylation to the transcriptional control of DegU-regulated target genes by complementing a nonmotile in-frame degU deletion mutant of L. monocytogenes EGD (12) with a mutated degU allele carrying a substitution of the putative phosphate-accepting aspartic acid residue (D55) for asparagine. D55 in L. monocytogenes DegU corresponds to D56 in the orthologous response regulator protein of Bacillus subtilis, which was shown to be the target of phosphorylation by the cognate histidine kinase DegS (1). In fact, recombinant His6-tagged DegU of L. monocytogenes was phosphorylated by DegS of B. subtilis in vitro, while phosphoryl group transfer to the mutated response regulator protein DegU(D55N) was not observed (data not shown).
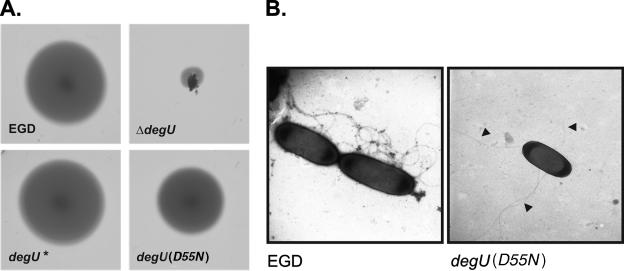
The mutated degU allele was integrated into the chromosome of the L. monocytogenes ΔdegU strain by homologous recombination via DNA fragments flanking the degU gene by using a two-step pLSV1-based integration-excision procedure described earlier (12). To construct the degU(D55N) allele with 5′ and 3′ flanking sequences, recombinant PCR was performed by amplification of a 428-bp fragment by using primer pair degU-u5/degU(D55N)1 and a 729-bp fragment by using primer pair degU(D55N)2/degU-d3 and by annealing of the purified PCR products, followed by PCR-based filling-in reactions. After digestion with EcoRI and BamHI, the recombinant PCR fragment was ligated into the mutagenesis plasmid pLSV1 (14), yielding knockout plasmid pLSV-degU(D55N). A control construct, pLSV-degU, was obtained by PCR amplification of the wild-type degU gene and its flanking sequences with primer pair degU-u5/degU-d3 and by ligation of the resulting 1,155-bp EcoRI-BamHI fragment into pLSV1 vector DNA. Electrotransformation of the L. monocytogenes ΔdegU strain with pLSV-degU(D55N) and pLSV-degU, respectively, followed by appropriate selection procedures, yielded the L. monocytogenes degU(D55N) and L. monocytogenes ΔdegU* strains. These strains were tested for swimming motility by stabbing the bacteria into semisolid brain heart infusion (BHI) agar (0.25% agar; Difco) and by measurement of the diameters of the swimming halos which were formed around the inoculation point after 48 h of incubation at 24°C. In three independent experiments, the diameters of at least 12 swimming halos were determined, and the diameters obtained were normalized to the mean diameter of halos formed by the wild-type strain EGD. In contrast to the completely nonmotile L. monocytogenes ΔdegU parent strain, the ΔdegU* strain exhibited swimming motility which was not significantly different from that of wild-type L. monocytogenes EGD (Student's t test; P > 0.05). Swimming motility was also observed for the L. monocytogenes degU(D55N) strain; however, the swimming halos produced by this mutant showed 31%-reduced diameters compared to those of L. monocytogenes EGD (P < 0.001) (Fig. 1A). In agreement with this result, in electron microscopical images, fewer flagella per cell were observed, on average, in cases of the L. monocytogenes degU(D55N) strain than in cases of the wild-type strain EGD (Fig. 1B).
FIG. 1.
(A) Swimming motility of the L. monocytogenes EGD, ΔdegU, ΔdegU*, and degU(D55N) strains. Bacteria from single colonies of the respective strains were stabbed into BHI soft agar, and the plates were incubated for 48 h at 24°C. Swimming halos obtained in a representative experiment are shown. The diameter of halos produced by the L. monocytogenes ΔdegU* and degU(D55N) strains averaged 106% ± 18% and 69% ± 8%, respectively, of the mean diameter of the wild-type strain EGD. (B) Electron micrographs of the L. monocytogenes EGD and degU(D55N) strains. Bacteria were grown without shaking in BHI broth at 24°C and were then examined under a transmission electron microscope (TEM 100; Zeiss) after negative staining with 0.5% uranyl acetate for 1 min. The number of flagella of 50 bacteria were counted per strain. On average, L. monocytogenes EGD exhibited seven flagella per cell, while 4.5 flagella were counted in the case of the degU(D55N) strain. The flagella are indicated by arrowheads. Original magnification, ×20,000.
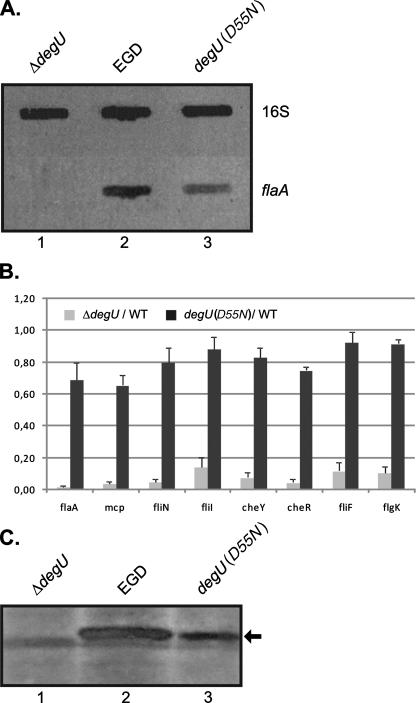
In order to analyze the transcript amounts of flagellum-specific genes, RNA was prepared from the L. monocytogenes EGD, ΔdegU, and degU(D55N) strains grown at 24°C in BHI broth to an optical density of 1.0 at 600 nm (13). RNA slot blot analysis was performed according to the protocol of Pflock et al. (8) by using a flaA-specific hybridization probe that was PCR amplified with primer pair flaA-S5/flaA-S3. While no flaA-specific transcript could be detected in the L. monocytogenes ΔdegU strain, the complemented degU(D55N) strain produced flaA-specific mRNA; however, the amount of transcript was reduced to about 50% of that of the wild-type strain L. monocytogenes EGD (Fig. 2A). In addition, transcription of the flagellar genes flaA (lmo0690), flgK (lmo0705), fliF (lmo0713), fliN (lmo0675), and fliI (lmo0716) and the chemotaxis genes cheY (lmo0691), cheR (lmo0683), and mcp (lmo0723) was monitored by quantitative real-time PCR (qRT-PCR), which was performed essentially as described by Mertins et al. (5). In accordance with previous results (13), transcription of the flagellar and chemotaxis genes was almost abolished in the L. monocytogenes ΔdegU strain, while the amounts of the flaA-, flgK-, fliF-, fliN-, fliI-, cheY-, cheR-, and mcp-specific transcripts were found to be modestly reduced in the L. monocytogenes degU(D55N) strain (Fig. 2B; Table 1). Transcription was affected most prominently in cases of mcp (65% of wild-type efficiency), cheR (75% of wild-type efficiency), and flaA (75% of wild-type efficiency). Transcription of the motility genes in the L. monocytogenes EGD and degU(D55N) strains grown at 37°C was also assessed by qRT-PCR, and equal amounts of transcript were observed (data not shown). Moreover, supernatant proteins were isolated from L. monocytogenes cultures grown at 24°C, as described previously (13), and were subjected to immunoblot analysis with a polyclonal rabbit antiserum (H-AB; Denka Seiken UK Ltd.) directed against the flagella of L. monocytogenes serotype 1/2a. In accordance with reduced transcription of flaA (Fig. 2A and B), the amount of flagellin expressed in the L. monocytogenes degU(D55N) strain was decreased to about 55% of the expression level of FlaA in wild-type strain EGD (Fig. 2C). As observed previously (9, 13), no FlaA protein could be detected in the L. monocytogenes ΔdegU strain (Fig. 2C). These results demonstrate that the phosphorylation of DegU at the conserved receiver phosphorylation site is not a prerequisite for its function as a temperature-responsive activator of expression of the antirepressor protein GmaR. However, reduced transcription of flaA in the L. monocytogenes degU(D55N) strain indicates that the DegU(D55N) protein is less efficacious as a transcriptional activator, which might be due to a conformational distortion caused by the receiver mutation.
FIG. 2.
Expression analysis of motility genes in the L. monocytogenes EGD, ΔdegU, and degU(D55N) strains. (A) Slot blot hybridization carried out with equal amounts of RNA extracted from the L. monocytogenes EGD (lane 2), ΔdegU (lane 1), and degU(D55N) (lane 3) strains. Hybridization was performed with flaA- and 16S rRNA-specific probes on RNA from three independent preparations. The 16S rRNA-specific hybridization probe was PCR amplified with primer pair 16S-S5/16S-S3. The results of a representative experiment are shown. (B) The relative changes in transcription of flaA, fliN, fliI, fliF, flgK, cheY, cheR, and mcp in the L. monocytogenes ΔdegU and degU(D55N) mutant strains, compared to the wild-type strain EGD (WT), were assessed by qRT-PCR. The ratios of the transcript amount detected in the respective mutant to that of the wild type (ratio, mutant/WT) are depicted. The indicated ratios represent the means of results of three qRT-PCR experiments performed in duplicate with cDNA which was reverse transcribed from independent RNA preparations extracted from bacteria grown at 24°C. The relative transcription of the genes under study was normalized to that of the housekeeping gene rpoB, as described by Mertins et al. (5). The primer pairs generating the respective amplicons are listed in Table 1. Error bars indicate the standard deviations from the means. (C) Western blot analysis was performed with equal amounts of supernatant proteins prepared from liquid cultures of the L. monocytogenes EGD (lane 2), ΔdegU (lane 1), and degU(D55N) (lane 3) strains grown at 24°C using a polyclonal rabbit antiserum directed against flagella of L. monocytogenes serotype 1/2a. FlaA is indicated by an arrow. The results of one of three independent experiments are shown.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ to 3′)a | Restriction recognition site | Strandb | Positionc |
|---|---|---|---|---|
| degU(D55N)1 | AGTTGGCATATTAATgTtCATTAAAAC | + | 2678814-2678842 | |
| degU(D55N)2 | GATATTGTTTTAATGaAcATTAATATGCC | − | 2678814-2678842 | |
| degU-u5 | taatatggatccAATTACATTTTATAGGCATATAGG | BamHI | − | 2679202-2679225 |
| degU-d3 | GTGAATGAATTCTCCTGGGGC | EcoRI | + | 2678064-2678084 |
| 16S-S5 | GGAAACTGGAAGACTGGAGT | + | 238114-238134 | |
| 16S-S3 | GCTGATCCACGATTAGTAGCG | − | 238817-238838 | |
| flaA-S5 | TTAGATGCAGCAAGCAAAAAC | + | 725070-725091 | |
| flaA-S3 | AGTTGCGATGGATTGATTGTT | − | 725589-725610 | |
| flaA-A5 | CGTGAACAATCAATCCATCG | + | 725585-725605 | |
| flaA-A3 | ACATTTGCGGTGTTTGGTTT | − | 725717-725737 | |
| fliF-A5 | TGCAAGAAAAAGTTGGCACA | + | 744264-744284 | |
| fliF-A3 | TTCCTGCAGCGGTTCCTTTT | − | 744264-744284 | |
| fliI-A5 | CCAAAAAGGGAAAATCCACA | + | 747278-747298 | |
| fliI-A3 | GCTTTGATTAAACGGGAGCA | − | 747418-747438 | |
| fliN-A5 | GGCAGTACTTGCGGGAATTT | + | 711162-711182 | |
| fliN-A3 | CAACACCTTTTCCCCGTCT | − | 711303-711322 | |
| flgK-A5 | CTGGCAAATGGATCCTGAAT | + | 739350-739370 | |
| flgK-A3 | GTGGCAACCTCGGTAATGAT | − | 739490-739510 | |
| cheY-A5 | CGGAAATGGATGGCTTAGAA | + | 726203-726223 | |
| cheY-A3 | CGCTTGGAAAGGTTTTACGA | − | 726335-726355 | |
| cheR-A5 | ACGCAAATGAAACGTCGAAT | + | 718565-718585 | |
| cheR-A3 | ATTGCGATTACGGAAAAACG | − | 718701-718721 | |
| mcp-A5 | AGTGGCGACCTATCATACCG | + | 754186-754206 | |
| mcp-A3 | ACGTTGTCGCTCGAAGATTT | − | 754318-754338 | |
| rpoB-A5 | CACCCTGAAGCTCCATTTGT | + | 274959-274979 | |
| rpoB-A3 | ACACGACGAACCCAGATTTC | − | 275070-275090 |
Sequences in uppercase letters are derived from the genome sequences of L. monocytogenes EGD-e (2). Sequences introduced for cloning purposes are shown in lowercase letters, and restriction recognition sequences are underlined.
+, positive; −, negative.
Nucleotide positions refer to the genome sequence of L. monocytogenes EGD-e (2).
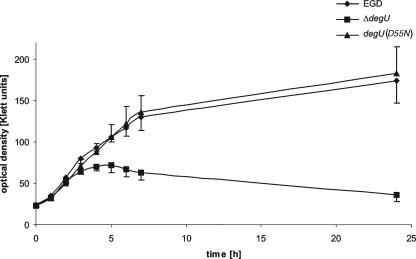
In addition to the lack of flagellar gene expression, the L. monocytogenes ΔdegU strain exhibited increased sensitivity toward ethanol and reduced virulence in mice after intravenous infection (12). Attenuation of virulence cannot be attributed to the nonmotile phenotype of the degU-deficient mutant, since the virulence of L. monocytogenes 10403S, an aflagellate mutant lacking flaA, was not affected when the bacteria were administered orogastrically or intravenously to mice (11). The DegU-regulated genes responsible for both mutant phenotypes have not been identified yet. The L. monocytogenes EGD, ΔdegU, and degU(D55N) strains were grown at 37°C in BHI broth supplemented with 5% ethanol. While the L. monocytogenes ΔdegU strain is unable to grow in the presence of 5% ethanol (12), no significant difference was observed in growth of wild-type and L. monocytogenes degU(D55N) mutant bacteria, suggesting that the D55N substitution in the response regulator DegU does not affect the expression of genes involved in ethanol tolerance (Fig. 3). To investigate whether receiver phosphorylation of DegU is a prerequisite for the proper regulation of virulence-relevant genes, BALB/c mice were intravenously infected with the L. monocytogenes degU(D55N) strain according to standard protocols (12). The experiment was performed three times, independently, with groups of five animals. At day 3 of infection, no significant differences in the bacterial loads in the livers and spleens of mice infected with the L. monocytogenes degU(D55N) strain were observed compared to the loads in the respective organs of animals to which the wild-type strain L. monocytogenes EGD had been administered. As a control, the degU gene was PCR amplified from chromosomal DNA of the L. monocytogenes degU(D55N) strain reisolated from mice, and the presence of the D55N mutation was confirmed by sequence analysis. Therefore, we reason that the D55N mutation in the receiver sequence of DegU does not interfere with the expression control of virulence-relevant genes.
FIG. 3.
Growth of the L. monocytogenes EGD, ΔdegU, and degU(D55N) strains at 37°C in BHI broth in the presence of 5% ethanol. Overnight cultures of the respective strains were diluted 1:50 in BHI broth supplemented with 5% ethanol. The optical densities (in Klett units) were determined at the indicated time points. The growth curve represents the means of results of four independent experiments. Error bars indicate the standard deviations from the means.
In conclusion, we have shown that phosphorylation of response regulator DegU at the conserved phosphorylation site is not crucial for the control of target genes whose differential expression is responsible for the complex phenotype of the L. monocytogenes strain lacking DegU. The mechanism of temperature-responsive, DegU-dependent regulation of GmaR expression, which might require an additional regulatory protein acting in concert with DegU, remains to be elucidated.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (BE 1543/5-1) and ERA-NET PathoGenoMics, project SPATELIS (0313939C).
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Dahl, M. K., T. Msadek, F. Kunst, and G. Rapoport. 1991. Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase. J. Bacteriol. 1732539-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chétouani, E. Couvé, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K.-D. Entian, H. Fsihi, F. Garcia-Del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L.-M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. Mata Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J.-C. Pérez-Dias, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J.-A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294849-852. [DOI] [PubMed] [Google Scholar]
- 3.Gründling, A., L. S. Burrack, H. G. Bouwer, and D. E. Higgins. 2004. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. USA 10112318-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knudsen, G. M., J. E. Olsen, and L. Dons. 2004. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol. Lett. 240171-179. [DOI] [PubMed] [Google Scholar]
- 5.Mertins, S., B. Joseph, M. Goetz, R. Ecke, G. Seidel, M. Sprehe, W. Hillen, W. Goebel, and S. Müller-Altrock. 2007. Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J. Bacteriol. 189473-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 2671-112. [DOI] [PubMed] [Google Scholar]
- 7.Peel, M., W. Donachie, and A. Shaw. 1988. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and Western blotting. J. Gen. Microbiol. 1342171-2178. [DOI] [PubMed] [Google Scholar]
- 8.Pflock, M., N. Finsterer, B. Joseph, H. Mollenkopf, T. F. Meyer, and D. Beier. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 1883449-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen, A., and D. E. Higgins. 2006. The MogR transcriptional repressor regulates non-hierarchal expression of flagellar motility genes and virulence in Listeria monocytogenes. PLoS Pathog. 2e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen, A., H. D. Kamp, A. Gründling, and D. E. Higgins. 2006. A bifunctional O-GlcNAc transferase governs flagellar motility trough anti-repression. Genes Dev. 203283-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Way, S. S., L. J. Thompson, J. E. Lopes, A. M. Hajjar, T. R. Kollmann, N. E. Freitag, and C. B. Wilson. 2004. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell. Microbiol. 6235-242. [DOI] [PubMed] [Google Scholar]
- 12.Williams, T., S. Bauer, D. Beier, and M. Kuhn. 2005. Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect. Immun. 733152-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams, T., B. Joseph, D. Beier, W. Goebel, and M. Kuhn. 2005b. Response regulator DegU of Listeria monocytogenes regulates the expression flagella-specific genes. FEMS Microbiol. Lett. 252287-298. [DOI] [PubMed] [Google Scholar]
- 14.Wuenscher, M. D., S. Köhler, W. Goebel, and T. Chakraborty. 1991. Gene disruption by plasmid integration in Listeria monocytogenes: insertional inactivation of the listeriolysin determinant lisA. Mol. Gen. Genet. 228177-182. [DOI] [PubMed] [Google Scholar]