Abstract
Proteins belonging to the Omp85 family are involved in the assembly of β-barrel outer membrane proteins or in the translocation of proteins across the outer membrane in bacteria, mitochondria, and chloroplasts. The cell envelope of the thermophilic bacterium Thermus thermophilus HB27 is multilayered, including an outer membrane that is not well characterized. Neither the precise lipid composition nor much about integral membrane proteins is known. The genome of HB27 encodes one Omp85-like protein, Omp85Tt, representing an ancestral type of this family. We overexpressed Omp85Tt in T. thermophilus and purified it from the native outer membranes. In the presence of detergent, purified Omp85Tt existed mainly as a monomer, composed of two stable protease-resistant modules. Circular dichroism spectroscopy indicated predominantly β-sheet secondary structure. Electron microscopy of negatively stained lipid-embedded Omp85Tt revealed ring-like structures with a central cavity of ∼1.5 nm in diameter. Single-channel conductance recordings indicated that Omp85Tt forms ion channels with two different conducting states, characterized by conductances of ∼0.4 nS and ∼0.65 nS, respectively.
The thermophilic bacterium Thermus thermophilus, with a growth temperature ranging from 45° to 85°C, belongs to one of the oldest branches of bacterial evolution and forms a phylum together with the genus Deinococcus. It most likely represents an evolutionary intermediate between today's gram-positive and -negative bacteria (18). The genome sequences of two T. thermophilus strains, HB8 and HB27, are available (20). The chromosomes are highly conserved with an identity of 94%, but variations are found, predominantly in cell envelope structures (6). The cell envelope of T. thermophilus is multilayered, comprising, as first identified in strain HB8, a thin murein layer (39), an outer membrane (OM) (7), an S-layer (8), and an outermost layer of amorphous material (9). The lipid composition of the OM is not precisely known. However, recently two glycolipids and one phosphoglycolipid were identified in whole-cell membranes of T. thermophilus strain Samu-SA1, reflecting adaptation to heat (27). Almost no information is available about integral OM proteins (OMPs). In black lipid bilayer experiments done using OM preparations from HB8, one 185-kDa protein was identified as a putative porin that formed unusually large pores (31). For HB27, Rumszauer et al. described OMPs involved in pilus biogenesis (42). In addition, a recent phylogenetic sequence analysis of the Omp85 protein family revealed that the genome of HB8 codes for one Omp85-like protein (4).
Proteins of the Omp85 family are integral OMPs found in all gram-negative bacteria sequenced to date as well as in mitochondria and chloroplasts (46). In respect to the putative function, the Omp85 family of proteins can be grouped in two classes. One class is proposed to be involved in insertion of β-barrel proteins into the OM and the other in the transport of proteins across the OM (46). Examples of Omp85-like proteins involved in the translocation of proteins are Toc75 from chloroplasts (22), nOmp85 (formerly alr2269) from the cyanobacterium Nostoc sp. strain PCC7120 (4, 13), and the integral membrane components, called TpsB, of the two-partner secretion system in gram-negative bacteria. TpsB proteins secrete large, mostly β-helical proteins called TpsA proteins through the OM. A well-studied TpsB protein is FhaC of Bordetella pertussis, which secretes a filamentous hemagglutinin (23). A well-characterized eukaryotic Omp85-like protein suggested to be involved in the insertion of β-barrel proteins into the mitochondrial OM is Tob55 from Saccharomyces cerevisiae (37), also called Sam50 (25). Omp85 of Neisseria meningitidis was the first example shown to function in the biogenesis of OMPs in gram-negative bacteria (51). Later, the same properties were found for the Escherichia coli Omp85 homologue YaeT (53). Both, Omp85Nm and YaeT are essential for cell viability, and depletion resulted in reduced amounts of OMPs in E. coli (53) or in the accumulation of unfolded OMPs in N. meningitidis (51). Apart from the TpsB proteins, Omp85 proteins are often part of multiprotein complexes (46). Neither the mechanism of protein secretion nor the mechanism of insertion of β-barrel OMPs by Omp85 family proteins in the absence of an obvious energy source is understood in detail.
Based on the amino acid sequence and secondary structure predictions, the Omp85 family is currently defined by the presence of two modules: an N-terminal periplasmic module containing one to five polypeptide transport-associated (POTRA) domains and a C-terminal transmembrane domain composed of 16 predicted β-strands (15). The POTRA domains are proposed to mediate protein-protein interaction, while the membrane-integrated C terminus forms a pore (44). Recently the structure of FhaC, a TpsB-type Omp85 family protein, was solved (11), showing two POTRA domains and 16 antiparallel β-strands. There is no complete structure of an Omp85 family protein involved in the assembly of β-barrel proteins, but the structure of the four N-terminal POTRA domains of YaeT has been solved (24).
In this paper we report the characterization of an Omp85 family protein from T. thermophilus HB27. We purified Omp85Tt in native form from T. thermophilus and demonstrated that it exists primarily as a monomer in detergent-containing solution. Omp85Tt reconstituted into liposomes appeared as ring-like structures in the electron microscope (EM) and showed pore activity in lipid bilayer experiments.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
T. thermophilus strains HB27 (DSM7039; obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) and HB27::nar (35) as well as E. coli strain Mach1-T1 (Invitrogen) were used in this study. T. thermophilus was grown at 70°C in medium containing 8g/liter Trypticase, 4g/liter yeast extract, and 3 g/liter NaCl dissolved in distilled water (pH 7,5) (34). For transformation of T. thermophilus HB27::nar, Ca2+- and Mg2+-rich medium containing 8 g/liter tryptone, 4 g/liter yeast extract, and 3 g/liter NaCl dissolved in Evian mineral water (pH 7.7) was used (5). Expression of proteins from the nar promoter of pMKE2 in T. thermophilus HB27::nar was performed as previously described (34). E. coli was grown in LB medium at 37°C. Kanamycin was used at 25 μg/ml for T. thermophilus and E. coli.
Plasmid construction.
DNA work was carried out using standard procedures (43). Chromosomal DNA from T. thermophilus was prepared as described recently for E. coli (16). Phusion DNA polymerase (Finnzymes) was used for PCRs.
To obtain a plasmid overexpressing Omp85Tt in T. thermophilus, TTC0193 encoding Omp85Tt was amplified by PCR from chromosomal DNA of HB27 using the primers 5′ACCATGGAGCGGCTTCTCGCCCTGGGC3′ (NcoI site underlined) and 5′CTCGAGTGCGGCCGCTTAGAACATGGGCCCGATGCG3′ (NotI site underlined). The PCR product was cloned into TOPO vector pCR4 (Invitrogen), cut out by digestion with NcoI and NotI, and ligated into NcoI/NotI-digested pMKE2 (34). The insert was sequenced (GATC). Due to the introduction of the restriction site NcoI, the resulting plasmid, pMKE2-Omp85Tt, expresses Omp85Tt with a modified signal sequence (MERLLALGLLALTALA instead of MKRLLALGLLALTALA).
Protein analysis.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (26) and stained with Coomassie brilliant blue R250 (Serva). The protein concentration was measured by either the bicinchoninic acid method (49) or the method of Lowry (29).
Blue native-PAGE (BN-PAGE) was performed as described recently (45, 52).
To identify the protein, purified Omp85Tt was subjected to mass spectrometry analysis (Proteome Factory, Berlin).
Far-UV circular dichroism (CD) spectra were recorded at room temperature on a Jasco 715 CD spectrometer (Jasco, Tokyo, Japan) using a 0.5-mm cuvette. Three scans were acquired from 190 to 250 nm, with a response time of 8 s, a bandwidth of 1 nm, and a scan speed of 20 nm/min. The background spectrum of the detergent solution in the absence of Omp85Tt was subtracted. The recorded CD spectra were normalized to obtain the mean residue molar ellipticity, [θ](λ) in degrees cm2 dmol−1, from the recorded ellipticity, θ(λ), where [θ](λ) = 100 × θ(λ)/(c × n × l) and c is the concentration of Omp85Tt in mol/liter, l is the path length of the cuvette in cm, and n is the number of amino acid residues (806) in Omp85Tt.
Dynamic light scattering (DLS) was performed using a DynaProE (Protein Solutions) instrument. All samples were measured at 20°C after filtering (filter pore size, 0.02 μm). Control experiments with buffers containing detergent concentrations above the critical micellar concentration (0.05% β-dodecylmaltoside [DDM] [Merck], 0.05% Cymal-6 [Anatrace], and 0.5% n-octyltetraoxyethylene [C8E4; Bachem]) showed monodisperse distributions of detergent micelles. The concentration of Omp85Tt in the different detergent-containing buffers was 3 mg/ml. DLS data were analyzed with Dynamics version 6 software (Protein Solutions). The molecular mass was calculated from the average hydrodynamic radius, assuming a globular shape of the measured molecules.
Proteinase digestion was performed with purified Omp85Tt samples. Five micrograms of Omp85Tt was treated with either 5 μg of trypsin or proteinase K for 5 h at 56°C. The reaction was stopped by the addition of 1 mM phenylmethylsulfonyl fluoride, and fragments were blotted onto a polyvinylidene difluoride membrane (Pall). The membrane was stained with Coomassie blue G, and the two fragments were subjected to five steps of Edman degradation (Proteome Factory, Berlin, Germany).
Cell fractionation.
Cell pellets of T. thermophilus were resuspended in 50 mM Tris (pH 8) and lysed by passing the suspensions through a French pressure cell. Unbroken cells were removed by centrifugation at 5,000 × g for 10 min, and cell envelopes were obtained as a pellet after centrifugation of the supernatant at 100,000 × g for 1 h. Cell envelopes were washed once with 50 mM Tris (pH 8) under the same conditions, resuspended in 50 mM Tris (pH 8), and loaded onto a step gradient of 30%, 40%, 50%, 55%, and 65% sucrose as previously reported (31). The gradient was centrifuged at 110,000 × g for 17 h at 20°C. Inner membrane fractions appeared as yellow and OM vesicles as white bands.
Purification of native Omp85Tt.
The OM fraction was isolated from the sucrose gradient (see “cell fractionation” above). The extract was diluted threefold with 50 mM Tris (pH 8), and OM vesicles were recovered after centrifugation at 100,000 × g for 1 h at 20°C. Proteins were solubilized in 20 mM Tris (pH 8.5) containing 0.5% Cymal-6 (Anagrade) for 1 h at 37°C, followed by centrifugation at 100,000 × g for 1 h. The supernatant was loaded on a MonoQ HR5/5 column (Amersham). After extensive washing with 20 mM Tris (pH 8.5) containing 0.5% Cymal-6, bound proteins were eluted with a linear gradient of 0 to 1 M NaCl in 20 mM Tris (pH 8.5)-0.05% Cymal-6. Fractions containing Omp85Tt were further purified by gel filtration chromatography (HiLoad 26/60 Superdex 200 prep grade; Amersham) using 20 mM Tris (pH 8.5)-150 mM NaCl-0.05% Cymal-6. If necessary, the detergent was exchanged by reloading Omp85Tt onto the MonoQ HR5/5 column after dialysis against 20 mM Tris (pH 8.5).
STEM.
A Vacuum Generators (East Grinstead) HB-5 scanning transmission EM (STEM) interfaced to a modular computer system (Tietz Video and Image Processing Systems) was used. The Omp85Tt sample in phosphate-buffered saline (PBS) containing 0.02% DDM was diluted fivefold in the same buffer immediately before the mass measurements. Aliquots of 5 μl were adsorbed to glow-discharged thin-carbon films spanning perforated carbon layers supported by 200-mesh-per-inch, gold-plated copper grids (STEM grids). The grids were blotted, washed on 6 droplets of quartz-bidistilled water, plunge frozen, and freeze-dried in the microscope. Digital dark-field images were recorded at doses of between 443 and 1,027 electrons/nm2 (pixel size, 0.905 nm; accelerating voltage, 80 kV; nominal magnification, × 200,000). Beam-induced mass loss was experimentally assessed and corrected for as detailed previously (36). Tobacco mosaic virus particles (kindly provided by R. Diaz Avalos) were used for absolute mass calibration. Mass analysis was achieved with the IMPSYS program package as described earlier (36). In the final step, mass values were binned into a histogram and described by multiple Gauss curves. The overall experimental uncertainty of the results was estimated from the corresponding standard error (standard deviation/√n) and the ≈5% uncertainty in the calibration of the instrument.
Reconstitution of Omp85Tt into lipid vesicles and negative-stain transmission EM (TEM).
To achieve reconstitution, Omp85Tt (0.1 mg/ml) in 20 mM Tris (pH 8.5)-150 mM NaCl-0.05% Cymal-6 was mixed with either E. coli lipids or dimyristoylphosphatidylcholine (DMPC) (Avanti Polar Lipids) at a lipid-to-protein ratio (LPR) of 0.2 and incubated at 4°C for 2 days in the presence of Bio-Beads SM2 (Bio-Rad) to remove the detergent (40). E. coli lipids in 20 mM Tris (pH 8.5)-150 mM NaCl-0.05% Cymal-6 were similarly incubated and served as a control.
For the EM, 5-μl aliquots of the reconstituted proteoliposomes or the pure liposomes were adsorbed to glow-discharged STEM grids, washed on several droplets of water, and stained on 2 drops of 2% uranyl acetate or 2% phosphotungstic acid. Images were recorded on Eastman Kodak Co. SO-163 sheet film with a Hitachi H-7000 TEM operated at 100 kV or a Philips CM-100 EM operated at 80 kV. The electron micrographs were digitized using a Heidelberg Primescan D 7100 at 4 Å/pixel at the specimen level. Particles were manually selected for single-particle analysis and averaged using the EMAN software (30).
FFEM.
To obtain the low degree of protein reconstitution required for freeze fracture electron microscopy (FFEM), Omp85Tt (0.05 mg/ml) in 20 mM Tris (pH 8.5)-150 mM NaCl-0.35% C8E4 was mixed with E. coli lipids at an LPR of 40 and a final detergent concentration of 0.68% and incubated at 4°C overnight in the presence of Bio-Beads SM2. E. coli lipids at 1.4 mg/ml in the same buffer were similarly incubated and served as a control.
The proteoliposomes or pure liposomes were centrifuged at 200,000 × g in a TL-100 ultracentrifuge (Beckman) for 20 min at 4°C. The pellets were cryoprotected with glycerol (30%, vol/vol). A small drop (3 μl) of the sample was put on top of a copper holder and plunge-frozen in liquid propane cooled by liquid nitrogen. The frozen samples were placed in a BAF 300 freeze fracture device (Balzers AG, Liechtenstein) and fractured at −125°C in vacuo at ≤5 × 10−7 torr using a liquid nitrogen-cooled razor blade. The fractured samples were unidirectionally metal shadowed with platinum-carbon at an elevation angle of 45°C to yield an average metal film thickness of 1 to 1.5 nm and were subsequently reinforced with carbon deposited by 5 to 7 s of thermionic evaporation from a pointed carbon rod. After thawing, the replicas were floated onto a 2% aqueous solution of SDS, washed with distilled water, and picked up on 200-mesh-per-inch gold-plated copper microscopy grids. They were observed in a Philips CM-100 EM operated at 80 kV. Images were recorded at a nominal magnification of ×21,000 on Eastman Kodak Co. SO-163 sheet film. The electron micrographs were digitized using a Heidelberg Primescan D 7100 at 9.5 Å/pixel at the specimen level. The negative images (in which platinum looks white and the shadows are black) were in general much more informative and have been used in Fig. 4. The ImageJ software (1) was employed to measure particle dimensions; only particles with complete shadows were used.
FIG. 4.
FFEM of reconstituted Omp85Tt. (A) Proteoliposome formed from Omp85Tt and E. coli lipids. It is peppered with shadowed particles, revealing the distribution of the Omp85Tt protein on the lipidic surface. The individual particles have an average diameter of 9 nm, compatible with the insertion of monomeric Omp85Tt in the lipid bilayer. (B) Control liposomes formed by pure E. coli lipids. Note the orange-skin appearance of their surface and the absence of larger shadowed particles. Bar, 100 nm.
Single-channel conductance across black lipid bilayers.
The single-channel conductance across diphytanoylphosphatidylcholine (diPhPC) bilayers was recorded as described previously (38, 48), but with Tris buffer (10 mM Tris, 5 mM CaCl2 [pH 7.4] containing 1 M KCl) in each of the two 6-ml compartments of the Teflon cell. A potential of 20 mV was applied to the front electrode. One to 3 μl of Omp85Tt (0.1 g/liter) in 20 mM Tris (pH 8.5), 100 mM NaCl, and 0.05% Cymal-6 was added to the cis compartment while the current across the membrane was recorded. Control experiments with Cymal-6 detergent alone did not cause any conductance changes. The conductance (G) was calculated from the recorded current.
Bioinformatic analysis.
Domains of Omp85Tt were identified using the SMART database (47). The beta-barrel domain was predicted using PRED-TMBB (http://bioinformatics.biol.uoa.gr/PRED-TMBB/input.jsp). BacMap (50) was used to search the T. thermophilus HB27 genome.
RESULTS
Omp85Tt can be overexpressed in T. thermophilus and purified in native form from OM preparations.
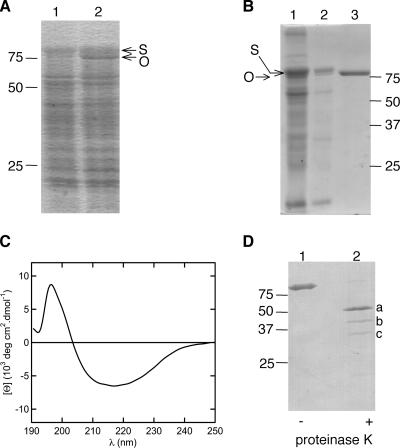
In the genome sequence of T. thermophilus HB27 (20), one open reading frame, TTC0193, encodes an Omp85 family protein, which we have named Omp85Tt. Attempts to inactivate TTC0193 by insertion of a thermostable bleomycin (5) or kanamycin (32) cassette failed, suggesting that Omp85Tt might be essential. It was not possible to overexpress Omp85Tt in native form in E. coli (data not shown), and therefore we tested plasmid-mediated overexpression in T. thermophilus. An inducible expression system based on the presence of nitrate and absence of oxygen was recently described (34). Using this system, Omp85Tt containing a slightly modified signal sequence could be overexpressed in T. thermophilus (Fig. 1A). Subsequent cell fractionation studies, including sucrose gradient centrifugation, showed that Omp85Tt was localized together with the S-layer protein in the OM fraction (Fig. 1B, lane 2). A multistep purification protocol was established to purify Omp85Tt from the OM and resulted in a preparation of highly enriched Omp85Tt (Fig. 1B, lane 3). Solubilization of Omp85Tt from the OM was best when the detergent Cymal-6 was used. Cymal-6 was also used during purification but was often changed afterwards to DDM or C8E4 without causing any aggregation.
FIG. 1.
Expression, purification, secondary structure, and stability of Omp85Tt. (A) Expression of Omp85Tt in HB27::nar from plasmid pMKE2. After overnight induction, whole-cell extracts were separated by SDS-PAGE and stained with Coomassie blue. Lane 1, HB27::nar pMKE2. Lane 2, HB27::nar pMKE2-Omp85Tt. The size markers are indicated on the left. The S-layer protein (S) and the Omp85Tt (O) are indicated by arrows. (B) The OM (lane 2) was separated from the whole membrane (lane 1) by sucrose gradient centrifugation, and solubilization and purification resulted in highly enriched Omp85Tt (lane 3). (C) CD spectrum of Omp85Tt in PBS with 0.05% DDM, indicating that the predominant secondary structure is the β-sheet. The spectrum was normalized by the concentration of Omp85Tt and by the number of amino acid residues in Omp85Tt to obtain the mean residue molar ellipticity. (D) Module architecture of Omp85Tt. Omp85Tt in 20 mM Tris (pH 8.5)-150 mM NaCl-0.05% Cymal-6 was digested for 5 h at 56°C with proteinase K, and fragments were identified by Edman degradation. A Coomassie blue-stained polyvinylidene difluoride membrane is shown. Lane 1, purified Omp85Tt without treatment. Lane 2, proteinase K-digested Omp85Tt. The bands indicated correspond to the N-terminal module of Omp85Tt (a), the C-terminal domain of Omp85Tt (b), and proteinase K (c). The other, less predominant, bands were not determined.
We examined the secondary structure of Omp85Tt by CD spectroscopy (Fig. 1C). The far-UV CD spectrum of Omp85Tt in PBS with 0.05% DDM was characterized by a broad minimum at ∼216.5 nm and a zero-ellipticity crossover at 203 nm, indicating that Omp85Tt has predominantly β-sheet secondary structure. Secondary structure analysis with the CDSSTR deconvolution algorithm (12) suggested ∼55% β-sheet, ∼16% α-helix, and 29% random-coil secondary structure in Omp85Tt. This is consistent with the prediction of a periplasmic N-terminal module containing α-helices and β-sheets and a C-terminal β-sheet-rich domain.
Mature Omp85Tt (88.3 kDa) is predicted to have five POTRA domains within the predicted 449-amino-acid (aa) N terminus (49.5 kDa) and 16 transmembrane β-strands within the predicted 357-aa C terminus forming one β-barrel (38.8 kDa). This two-module model was verified by proteinase digestion (Fig. 1D). Omp85Tt was resistant to trypsin (data not shown), but treatment with proteinase K for 5 h at 56°C resulted in two stable fragments. The fragments were sequenced by Edman degradation. One fragment (Fig. 1D, lane 2, a) started with APLER, corresponding to the predicted N terminus of the mature protein, and the other (Fig. 1D, lane 2, b) started with AFKET, a sequence 6 aa downstream of the predicted β-barrel domain.
Purified Omp85Tt is primarily monomeric in the presence of detergents.
Several proteins of the Omp85 family are reported to form oligomers in solution (37, 41). The oligomeric state of purified Omp85Tt was investigated by different techniques. DLS revealed that purified Omp85Tt is monodisperse in buffer, independent of the detergent used. Using this technique, the calculated masses of Omp85Tt-detergent complexes were 119 kDa in the presence of Cymal-6, 146 kDa in the presence of DDM, and 114 kDa in the presence of C8E4. The micellar masses of the different detergents in the absence of Omp85Tt were found to be 40 kDa for Cymal-6, 57 kDa for DDM, and 43 kDa for C8E4. Taking the micellar masses of the different detergents into account, these data suggest that Omp85Tt (88.3 kDa, calculated molecular mass from the sequence) is a monomer in detergent micelles.
Analysis of purified Omp85Tt by BN-PAGE confirmed that the monomeric form is predominant (Fig. 2A). Omp85Tt in buffer containing Cymal-6 migrated mainly at an apparent mass of approximately 160 kDa, corresponding most likely to the monomer, since Coomassie blue G dye contributes to the molecular masses of membrane proteins in BN gels (21). The two additional, less prominent bands at higher molecular mass were also identified as Omp85Tt in the second-dimension SDS-PAGE (Fig. 2A). The higher-molecular-mass forms probably represent dimers and trimers of Omp85Tt. Similar results were obtained when the detergent was changed to DDM or C8E4 (data not shown), indicating that the formation of oligomers is independent of the detergent.
FIG. 2.
Purified Omp85Tt is primarily a monomer. (A) Two-dimensional BN/SDS-PAGE analysis of purified Omp85Tt in Cymal-6-containing buffer. In the upper part the 4 to 24% first-dimension BN-PAGE is shown. A second identical lane was cut off and subjected to SDS-PAGE in the second dimension. Molecular weight markers from the first and second dimensions are indicated. (B) STEM mass analysis of purified Omp85Tt in buffer containing DDM. The histogram shows the mass distributions for the purified Omp85Tt. The Gaussian is at 166 ± 40 kDa (n = 891) for the monomer, 265 ± 45 kDa (n = 239) for the dimer, 356 ± 53 kDa (n = 122) for the trimer, and 483 ± 39 kDa (n = 49) for the tetramer.
The mass of Omp85Tt in the presence of DDM was also determined by STEM. Evaluation of the images yielded the histogram shown in Fig. 2B. Considering the micellar mass of the associated DDM detergent (57 kDa), the main peak at 166 ± 40 kDa indicated that around 69% of the 1,301 Omp85Tt (molecular mass from sequence, 88.3 kDa) particles investigated were monomeric. In addition, the peaks at higher mass suggested that 18% of the particles were dimers, 9% trimers, and 4% tetramers.
Projection structure of Omp85Tt reconstituted into liposomes.
Omp85Tt solubilized in 20 mM Tris (pH 8.5)-150 mM NaCl-0.05% Cymal-6 was reconstituted into liposomes formed by DMPC or E. coli lipids, negatively stained either with 2% uranyl acetate or 2% phosphotungstic acid, and examined in a TEM. In all cases the incorporated protein was visible as rings on the liposome surface (Fig. 3). The insets in Fig. 3A show the average structures of such top views. Omp85Tt had two different diameters, approximately 5.1 nm (Fig. 3A, upper inset) and 5.9 nm (Fig. 3A, lower inset), suggesting that the protein had inserted into the bilayer in both the “up” and “down” directions. The central stain-filled cavities indicated the presence of an ∼1.5-nm-wide pore. Omp85Tt could also be discerned at the edges of some vesicles and protruded ∼4 to 5 nm from the lipid bilayer (Fig. 3B and C). An uncharged lipid vesicle is shown in the inset of Fig. 3B for comparison.
FIG. 3.
Negative-stain TEM images of Omp85Tt reconstituted into lipid vesicles. (A) Omp85Tt reconstituted into DMPC liposomes and negatively stained with 2% uranyl acetate. The reconstituted protein is visible as rings on the liposome surface (arrows) The insets show averages of the two different types of Omp85Tt ring seen; the diameter of the upper averages approximately 5.1 nm, and that of the lower averages approximately 5.9 nm. (B) Omp85Tt reconstituted into DMPC liposomes and negatively stained with 2% phosphotungstic acid. Omp85Tt protrudes roughly 4 to 5 nm from the membrane, giving the edge of the liposomes a rather lacy appearance (arrows). The inset shows liposomes prepared in the absence of protein. (C) Omp85Tt reconstituted into E. coli liposomes and negatively stained with 2% uranyl acetate. End views (black arrow) and side views (white arrow) of the protein can be distinguished. Bars, 50 nm (insets in panel A, 5 nm).
Both to confirm this result and to demonstrate reconstitution when starting from another detergent system, FFEM was employed to examine a reconstitution made using Omp85Tt in 20 mM Tris (pH 8.5)-150 mM NaCl-0.35% C8E4 and E. coli lipids. A very high LPR of 40 was chosen for this experiment to ensure that individual protein entities would be visible on the liposome surface. A typical proteoliposome is shown in Fig. 4A. The reconstituted proteins appear as particles peppered across the liposome surface. Uncharged liposomes prepared as a control exhibited the expected “orange-skin” surface but lacked these larger particles (Fig. 4B). The shadowing/replication step required by this microscopy technique makes their apparent diameter, 9 ± 1 nm (n = 31), considerably larger than that measured by negative-stain microscopy, but the value is still compatible with the presence of Omp85Tt monomers.
Omp85Tt forms channels in lipid bilayers.
To date, all OMPs of the Omp85 family have been found to form ion channels, with single-channel conductances ranging from 0.4 nS for YaeT from E. coli to 2.1 nS for nOmp85 from Nostoc (4, 13, 33, 37, 41). We tested pore formation by Omp85Tt in planar lipid bilayers. Purified native Omp85Tt in 20 mM Tris (pH 8.5) containing 100 mM NaCl and 0.05% Cymal-6 was reconstituted into black lipid membranes of diPhPC. Conductance measurements revealed the opening and closing of pores (Fig. 5A and B). Out of 640 recorded channel conductance events, ∼46% displayed a conductance change ΔG2 of ∼0.4 nS at 20 mV in 1 M KCl (Fig. 5C). In addition, 9 to 10% of the channel events displayed a change in conductance ΔG1 of ∼0.65 nS (Fig. 5C). The conductance recordings indicated that the large conductance change ΔG1 of ∼0.65 nS always preceded the small conductance change, leading to a total conductance change ΔG of ∼1.1 nS (Fig. 5A and B).
FIG. 5.
Single-channel conductance recordings indicate that Omp85Tt forms ion channels in black lipid films. (A) Stepwise increase of the conductance (G) recorded as a function of time (t), showing that Omp85Tt exhibits two channel states. (B) Expanded time window of panel A, indicating frequent channel openings with a conductance ΔG2 of ∼0.4 nS. These openings required a preceding conductance change ΔG1 of ∼0.65 nS. (C) Histogram of Omp85Tt channel conductance events, indicating that about half of all channel events show this conductance ΔG of ∼0.4 nS. Conductance events with ΔG1 of ∼0.65 were far less frequent. A total of 640 channel events were recorded and evaluated.
DISCUSSION
In the present work, we have analyzed the first protein of the Omp85 family that originates from a thermophilic bacterium, Omp85Tt. We were able to overexpress and purify Omp85Tt as a native, stable protein from the OM of T. thermophilus. Omp85Tt showed a high content of β-sheet secondary structure, similar to most OMPs of gram-negative bacteria, indicating that it forms a transmembrane β-barrel. The composition of the secondary structure (∼55% β-sheet, ∼16% α-helix, and 29% random coil) closely resembles that of FhaC (∼49% β-sheet and ∼9.1% α-helix) (11). The significant amount of α-helix secondary structure detected in Omp85Tt agrees well with the predicted presence of five POTRA domains within the periplasmic N terminus. The structures of several POTRA domains were recently solved (11, 24). Although the primary sequence similarity between different POTRA domains is very low, their structures are similar and comprise a three-stranded β-sheet and two α-helices. POTRA domains are known to mediate protein-protein interactions, and the presence of at least one is essential for the function of proteins of the Omp85 family (3, 11, 19). They are reported to interact with unfolded substrates (11, 19) and to mediate homo-oligomerization (e.g., nOmp85 trimerizes via the POTRA domains [13]) and hetero-oligomerization (e.g., YaeT interacts with the four lipoproteins NlpB, SmpA, YfiO, and YfgL via the POTRA domains [24]). Incorporated into liposomes, Omp85Tt was found to protrude ∼4 to 5 nm from the membrane, giving a rough idea of the size of the N terminus with its five predicted POTRA domains.
Three different techniques, DLS, BN-PAGE, and STEM, indicated that native Omp85Tt is present mainly as a monomer in detergent solutions. On reconstitution into lipid bilayers, Omp85Tt also appeared as single particles scattered across liposomes by FFEM (E. coli lipids) or as monomeric, ring-like particles with a central cavity by negative-stain TEM (DMPC and E. coli lipids). In the presence of detergent, higher-molecular-mass forms of Omp85Tt were observed by BN-PAGE analysis and STEM, suggesting that Omp85Tt has the capability to form homo-oligomers. Compared to those obtained by the other techniques, the STEM data suggested a relatively high content of oligomeric particles (around 31%). However, STEM is a single-molecule rather than a bulk technique, and not every particle imaged could be measured due to the close proximity of neighbors, which would have influenced the statistics. Some purified Omp85 family proteins have been reported to exist as multimers. In native form the TpsB-type Omp85 protein HMW1B was found to be a dimer (28). The cyanobacterial Omp85 protein nOmp85 was reported to exist as homotrimers when reconstituted into artificial membranes (4). Purified, refolded YaeT was suggested to form tetramers (41). In contrast, FhaC was crystallized as a monomer, suggesting that it is also predominantly monomeric in solution (11). Omp85 family proteins are thus able to oligomerize, but there are clearly differences in the degree. At present it is not understood whether the protein-trafficking activity of Omp85 family proteins in vivo depends on this property. Further, oligomerization might also be dependent on the association of other proteins to form one active complex.
Omp85Tt showed channel activity when incorporated into planar lipid bilayers. The most frequent change in conductance at 1 M KCl found for Omp85Tt (ΔG2, ∼0.4 nS) is similar to that published for Omp85 proteins involved in the insertion of β-barrel proteins, e.g., 0.4 nS (4) or 0.5 nS (41) for the YaeT of E. coli and 0.56 nS for the Sam50 of Drosophila melanogaster mitochondria (4). Larger channel conductances have been observed for Omp85 proteins proposed to be involved in the translocation of proteins, e.g., 2.1 nS for the nOmp85 of the cynaobacterium Nostoc, 2 nS for the Toc75 of Pisum sativum chloroplasts (4), and 1.2 nS for FhaC of Bordetella pertussis (33).
The channel conductance of Omp85Tt always displayed a two-step increase across the lipid bilayer. The first conductance increase (ΔG1, ∼0.65 nS) was about twice as large as the second (ΔG2, ∼0.4 nS). Two different interpretations would explain this behavior. Omp85Tt could insert as a monomer and first partially open into a substate before it opens fully. The conductance of the fully open state would then correspond to ∼1.1 nS. This behavior might indicate gating, as was assumed for nOmp85 from Nostoc. Interestingly, the β-barrel domain of nOmp85 itself did not show any subconductance steps, but these were induced when the N-terminal module of nOmp85 was added (13). This suggests that the POTRA domains might be involved in gating of the pore formed by the β-barrel domain, at least in some Omp85 family members. Alternatively, Omp85Tt might not be active as a monomer in this assay. Rather, dimers or trimers might have to be formed in the diPhPC bilayer. Our results would then indicate that one Omp85Tt channel is mostly open, while another opens and closes more frequently. Channel activity of an oligomeric form was suggested for refolded YaeT. It was found that every subunit of the active YaeT tetramer forms a pore that can open and close independently (41).
A first indication that the channel-forming C termini of Omp85 family proteins might be dynamic came from work on FhaC (11). While the conductivity of 1.2 nS measured for FhaC suggests a channel of 8 to 10 Å in diameter (33), the structure of FhaC revealed a narrow channel that is just 3 Å wide because it is occluded by a loop and an α-helix (11). This channel would be too narrow to allow the transport of a protein, and indeed the available evidence suggests that the loop is flexible and opens the channel by moving outwards when the substrate filamentous hemagglutinin is present (17). Omp85Tt reconstituted into liposomes showed a stain-filled cavity, implying a pore with a maximum diameter of approximately 15 Å. We propose that the pore of Omp85Tt is also largely occluded, as observed for FhaC. In agreement with this idea, sequence analysis of FhaC and Omp85Tt showed that while the helix occluding the channel of FhaC is not conserved in Omp85Tt, the loop is (data not shown).
Our preliminary data suggest that Omp85Tt is essential. Since we found that Omp85Tt fractionates with the OM/S-layer envelope, this would further suggest that the OM is essential. So far it is known only that the S-layer protein is not essential, although it attaches the OM to the secondary cell wall polymer, a layer covalently bound to the peptidoglycan (14). An S-layer mutant looses its shape and produces instead so-called multicellular bodies, spherical shells formed by the OM that contain a large number of cells (10).
The function of Omp85Tt is not known. In gram-negative bacteria, genes encoding Omp85 family proteins are often flanked by genes indicating their function. In T. thermophilus the DNA region surrounding TTC0193 (omp85Tt) does not allow such speculation. Bioinformatics did not reveal homologues of other proteins involved in the biogenesis of OMPs in gram-negative bacteria (data not shown). Only the Sec system and the Omp85 are present, but neither the cognate chaperones nor the cognate lipoproteins (2) seem to be conserved. The presence of only one Omp85 family protein in T. thermophilus suggests that Omp85Tt might be involved in the insertion of β-barrel proteins. In line with this, the C-terminal end of Omp85Tt (IHFRIGPMF) shows the typical OMP signature sequence known for gram-negative bacteria (41), with phenylalanine at the ultimate position and hydrophobic residues at positions 5, 7, and 9 (underlined) from the C terminus. The C-terminal signature sequence was recently found to interact with YaeT (Omp85) of E. coli (41). Therefore, it is tempting to speculate that Omp85Tt needs itself in order to integrate into the OM. Using bioinformatics, we have identified two more potential β-barrel OM proteins harboring this C-terminal signature sequence (unpublished results). In further studies we will characterize these putative β-barrel proteins and evaluate them as potential substrates for Omp85Tt.
Acknowledgments
We are grateful to Hans-Jürgen Apell (University of Konstanz) for help with the conductance recordings and for useful discussions. We also thank Andreas Engel (University of Basel) for his support.
The work in Konstanz was funded by the Deutsche Forschungsgemeinschaft (grant SFB/TR 11). CD experiments and single-channel recordings performed by G.J.P. were supported by grant TP B3 to J.H.K. The microscopy in Basel was supported by Swiss National Foundation grant 501221 and by the Maurice E. Müller Foundation of Switzerland.
Footnotes
Published ahead of print on 2 May 2008.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 1136-42. [Google Scholar]
- 2.Bos, M. P., V. Robert, and J. Tommassen. 2007. Biogenesis of the Gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61191-214. [DOI] [PubMed] [Google Scholar]
- 3.Bos, M. P., V. Robert, and J. Tommassen. 2007. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 81149-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bredemeier, R., T. Schlegel, F. Ertel, A. Vojta, L. Borissenko, M. T. Bohnsack, M. Groll, A. von Haeseler, and E. Schleiff. 2007. Functional and phylogenetic properties of the pore-forming beta-barrel transporters of the Omp85 family. J. Biol. Chem. 2821882-1890. [DOI] [PubMed] [Google Scholar]
- 5.Brouns, S. J., H. Wu, J. Akerboom, A. P. Turnbull, W. M. de Vos, and J. van der Oost. 2005. Engineering a selectable marker for hyperthermophiles. J. Biol. Chem. 28011422-11431. [DOI] [PubMed] [Google Scholar]
- 6.Bruggemann, H., and C. Chen. 2006. Comparative genomics of Thermus thermophilus: plasticity of the megaplasmid and its contribution to a thermophilic lifestyle. J. Biotechnol. 124654-661. [DOI] [PubMed] [Google Scholar]
- 7.Castan, P., M. A. de Pedro, C. Risco, C. Valles, L. A. Fernandez, H. Schwarz, and J. Berenguer. 2001. Multiple regulatory mechanisms act on the 5′ untranslated region of the S-layer gene from Thermus thermophilus HB8. J. Bacteriol. 1831491-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caston, J. R., J. Berenguer, M. A. de Pedro, and J. L. Carrascosa. 1993. S-layer protein from Thermus thermophilus HB8 assembles into porin-like structures. Mol. Microbiol. 965-75. [DOI] [PubMed] [Google Scholar]
- 9.Caston, J. R., J. L. Carrascosa, M. A. de Pedro, and J. Berenguer. 1988. Identification of a crystalline surface layer on the cell envelope of the thermophilic eubacterium Thermus thermophilus. FEMS Microbiol. Lett. 51225-230. [Google Scholar]
- 10.Cava, F., M. A. de Pedro, H. Schwarz, A. Henne, and J. Berenguer. 2004. Binding to pyruvylated compounds as an ancestral mechanism to anchor the outer envelope in primitive bacteria. Mol. Microbiol. 52677-690. [DOI] [PubMed] [Google Scholar]
- 11.Clantin, B., A. S. Delattre, P. Rucktooa, N. Saint, A. C. Meli, C. Locht, F. Jacob-Dubuisson, and V. Villeret. 2007. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317957-961. [DOI] [PubMed] [Google Scholar]
- 12.Compton, L. A., and W. C. Johnson, Jr. 1986. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 155155-167. [DOI] [PubMed] [Google Scholar]
- 13.Ertel, F., O. Mirus, R. Bredemeier, S. Moslavac, T. Becker, and E. Schleiff. 2005. The evolutionarily related beta-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. J. Biol. Chem. 28028281-28289. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Herrero, L. A., G. Olabarria, J. R. Caston, I. Lasa, and J. Berenguer. 1995. Horizontal transference of S-layer genes within Thermus thermophilus. J. Bacteriol. 1775460-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentle, I. E., L. Burri, and T. Lithgow. 2005. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 581216-1225. [DOI] [PubMed] [Google Scholar]
- 16.Grimberg, J., S. Maguire, and L. Belluscio. 1989. A simple method for the preparation of plasmid and chromosomal E. coli DNA. Nucleic Acids Res. 178893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guedin, S., E. Willery, J. Tommassen, E. Fort, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2000. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 27530202-30210. [DOI] [PubMed] [Google Scholar]
- 18.Gupta, R. S. 2000. The natural evolutionary relationships among prokaryotes. Crit. Rev. Microbiol. 26111-131. [DOI] [PubMed] [Google Scholar]
- 19.Habib, S. J., T. Waizenegger, A. Niewienda, S. A. Paschen, W. Neupert, and D. Rapaport. 2007. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial beta-barrel proteins. J. Cell Biol. 17677-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henne, A., H. Bruggemann, C. Raasch, A. Wiezer, T. Hartsch, H. Liesegang, A. Johann, T. Lienard, O. Gohl, R. Martinez-Arias, C. Jacobi, V. Starkuviene, S. Schlenczeck, S. Dencker, R. Huber, H. P. Klenk, W. Kramer, R. Merkl, G. Gottschalk, and H. J. Fritz. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22547-553. [DOI] [PubMed] [Google Scholar]
- 21.Heuberger, E. H., L. M. Veenhoff, R. H. Duurkens, R. H. Friesen, and B. Poolman. 2002. Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J. Mol. Biol. 317591-600. [DOI] [PubMed] [Google Scholar]
- 22.Hinnah, S. C., K. Hill, R. Wagner, T. Schlicher, and J. Soll. 1997. Reconstitution of a chloroplast protein import channel. EMBO J. 167351-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40306-313. [DOI] [PubMed] [Google Scholar]
- 24.Kim, S., J. C. Malinverni, P. Sliz, T. J. Silhavy, S. C. Harrison, and D. Kahne. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317961-964. [DOI] [PubMed] [Google Scholar]
- 25.Kozjak, V., N. Wiedemann, D. Milenkovic, C. Lohaus, H. E. Meyer, B. Guiard, C. Meisinger, and N. Pfanner. 2003. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 27848520-48523. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 27.Leone, S., A. Molinaro, B. Lindner, I. Romano, B. Nicolaus, M. Parrilli, R. Lanzetta, and O. Holst. 2006. The structures of glycolipids isolated from the highly thermophilic bacterium Thermus thermophilus Samu-SA1. Glycobiology 16766-775. [DOI] [PubMed] [Google Scholar]
- 28.Li, H., S. Grass, T. Wang, T. Liu, and J. W. St. Geme III. 2007. Structure of the Haemophilus influenzae HMW1B translocator protein: evidence for a twin pore. J. Bacteriol. 1897497-7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 30.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 12882-97. [DOI] [PubMed] [Google Scholar]
- 31.Maier, E., G. Polleichtner, B. Boeck, R. Schinzel, and R. Benz. 2001. Identification of the outer membrane porin of Thermus thermophilus HB8: the channel-forming complex has an unusually high molecular mass and an extremely large single-channel conductance. J. Bacteriol. 183800-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mather, M. W., and J. A. Fee. 1992. Development of plasmid cloning vectors for Thermus thermophilus HB8: expression of a heterologous, plasmid-borne kanamycin nucleotidyltransferase gene. Appl. Environ. Microbiol. 58421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meli, A. C., H. Hodak, B. Clantin, C. Locht, G. Molle, F. Jacob-Dubuisson, and N. Saint. 2006. Channel properties of TpsB transporter FhaC point to two functional domains with a C-terminal protein-conducting pore. J. Biol. Chem. 281158-166. [DOI] [PubMed] [Google Scholar]
- 34.Moreno, R., A. Haro, A. Castellanos, and J. Berenguer. 2005. High-level overproduction of His-tagged Tth DNA polymerase in Thermus thermophilus. Appl. Environ. Microbiol. 71591-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno, R., O. Zafra, F. Cava, and J. Berenguer. 2003. Development of a gene expression vector for Thermus thermophilus based on the promoter of the respiratory nitrate reductase. Plasmid 492-8. [DOI] [PubMed] [Google Scholar]
- 36.Müller, S. A., K. Goldie, R. Burki, R. Haring, and A. Engel. 1992. Factors influencing the precision of quantitative scanning transmission electron microscopy. Ultramicroscopy 46317-334. [Google Scholar]
- 37.Paschen, S. A., T. Waizenegger, T. Stan, M. Preuss, M. Cyrklaff, K. Hell, D. Rapaport, and W. Neupert. 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426862-866. [DOI] [PubMed] [Google Scholar]
- 38.Pocanschi, C. L., H. J. Apell, P. Puntervoll, B. Hogh, H. B. Jensen, W. Welte, and J. H. Kleinschmidt. 2006. The major outer membrane protein of Fusobacterium nucleatum (FomA) folds and inserts into lipid bilayers via parallel folding pathways. J. Mol. Biol. 355548-561. [DOI] [PubMed] [Google Scholar]
- 39.Quintela, J. C., E. Pittenauer, G. Allmaier, V. Aran, and M. A. de Pedro. 1995. Structure of peptidoglycan from Thermus thermophilus HB8. J. Bacteriol. 1774947-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rigaud, J. L., G. Mosser, J. J. Lacapere, A. Olofsson, D. Levy, and J. L. Ranck. 1997. Bio-Beads: an efficient strategy for two-dimensional crystallization of membrane proteins. J. Struct. Biol. 118226-235. [DOI] [PubMed] [Google Scholar]
- 41.Robert, V., E. B. Volokhina, F. Senf, M. P. Bos, P. Van Gelder, and J. Tommassen. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rumszauer, J., C. Schwarzenlander, and B. Averhoff. 2006. Identification, subcellular localization and functional interactions of PilMNOWQ and PilA4 involved in transformation competency and pilus biogenesis in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 2733261-3272. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 44.Sanchez-Pulido, L., D. Devos, S. Genevrois, M. Vicente, and A. Valencia. 2003. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci. 28523-526. [DOI] [PubMed] [Google Scholar]
- 45.Schagger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199223-231. [DOI] [PubMed] [Google Scholar]
- 46.Schleiff, E., and J. Soll. 2005. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 61023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 955857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shanmugavadivu, B., H. J. Apell, T. Meins, K. Zeth, and J. H. Kleinschmidt. 2007. Correct folding of the beta-barrel of the human membrane protein VDAC requires a lipid bilayer. J. Mol. Biol. 36866-78. [DOI] [PubMed] [Google Scholar]
- 49.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 15076-85. [DOI] [PubMed] [Google Scholar]
- 50.Stothard, P., G. Van Domselaar, S. Shrivastava, A. Guo, B. O'Neill, J. Cruz, M. Ellison, and D. S. Wishart. 2005. BacMap: an interactive picture atlas of annotated bacterial genomes. Nucleic Acids Res. 33D317-D320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299262-265. [DOI] [PubMed] [Google Scholar]
- 52.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue native PAGE. Nat. Protoc. 1418-428. [DOI] [PubMed] [Google Scholar]
- 53.Wu, T., J. Malinverni, N. Ruiz, S. Kim, T. J. Silhavy, and D. Kahne. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121235-245. [DOI] [PubMed] [Google Scholar]