Abstract
Sequence-specific transactivation by p53 is essential to its role as a tumor suppressor. A modified tetracycline-inducible system was established to search for transcripts that were activated soon after p53 induction. Among 9,954 unique transcripts identified by serial analysis of gene expression, 34 were increased more than 10-fold; 31 of these had not previously been known to be regulated by p53. The transcription patterns of these genes, as well as previously described p53-regulated genes, were evaluated and classified in a panel of widely studied colorectal cancer cell lines. “Class I” genes were uniformly induced by p53 in all cell lines; “class II” genes were induced in a subset of the lines; and “class III” genes were not induced in any of the lines. These genes were also distinguished by the timing of their induction, their induction by clinically relevant chemotherapeutic agents, the absolute requirement for p53 in this induction, and their inducibility by p73, a p53 homolog. The results revealed substantial heterogeneity in the transcriptional responses to p53, even in cells derived from a single epithelial cell type, and pave the way to a deeper understanding of p53 tumor suppressor action.
The fact that p53 function is impaired in the majority of human cancers has stimulated efforts to understand the function of this gene in normal and neoplastic states. A large number of functions have been attributed to p53, including cell-cycle checkpoints, apoptosis, angiogenesis, and genetic stability (1–7). In cells containing wild-type (wt) p53 genes, the p53 protein is induced by a variety of stimuli, including chemotherapeutic agents, oxidative stress, hypoxia, nucleotide depletion, and oncogene expression (6, 7).
Though several models for the biochemical basis of p53 action have been proposed, the most well studied involves its ability to bind to specific genomic sequences and activate transcription of adjacent genes (7, 8). The DNA-binding domain has been defined thoroughly through biochemical studies, and its interaction with DNA has been illuminated through x-ray crystallography (9). The active p53 polypeptide exists in tetrameric form, binding to four palindromic copies of its consensus sequence (5′-PuPuPuGA/T-3′). The carboxyl terminus of p53 contains the amino acids required for tetramerization, whereas the amino terminus contains a potent transcriptional activation domain that is essential for its ability to regulate downstream genes (10–12). Tumor-derived mutants are almost always defective in sequence-specific transactivation (SST), providing compelling evidence for the hypothesis that SST is essential for p53 tumor suppression.
Investigators have described several genes that are apparently controlled by p53 and have postulated that these p53-regulated genes are responsible for mediating the various effects of p53 (13, 14). These studies have employed different cell types, often from different species, and have used a large and varied number of agents to elevate p53 expression. It has therefore been difficult to put these transcriptional targets into perspective or to interpret and compare the induction patterns observed.
In an effort to understand the transcriptional responses to p53 better, we sought to characterize the genes induced by p53 systematically in a panel of cell lines derived from the same epithelial cell type (neoplastic cells of the human colon). We identified a number of potential transcriptional targets and found marked heterogeneity in the extent, timing, and p53 dependence of both previously identified and unidentified targets. This heterogeneity suggests that, even in tumor cells derived from the same stem cell type, the response to p53 expression varies considerably. These results provide insights into the basis for cell-type-specific responses to p53 and their associated pleiotropic biologic effects.
Materials and Methods
Cell Culture.
Derivation and growth of the lines used in this study have been previously described (15, 16). Cells were treated with chemotherapeutic agents for 24 hr at concentrations of 0.2 μg/ml (adriamycin) and 50 μg/ml (5-fluorouracil; 5-FU). Transfections were carried out with either Lipofectamine (Life Technologies, Grand Island, NY) or Fugene 6 (Roche Molecular Biochemicals) according to the manufacturers’ instructions.
Inducible Lines.
A two-step procedure was used to establish a tetracycline (tet)-off system in which the “off” mode was achieved by maintaining cells in the presence of 20 ng/ml doxycycline. In brief, DLD-1 cells were transfected with tTA-IRES-Neo (tTA, tet activator; IRES, internal ribosome entry site; Neo, G418 resistance gene) and selected with 800 μg/ml of G418 (Geneticin, Life Technologies). Single clones were obtained by limiting dilution and tested by transient transfection with a reporter plasmid, pBI-GL (CLONTECH), which expressed luciferase and β-galactosidase in a tTA-dependent fashion. One clone, DLDtet14, was chosen to generate inducible cell lines of interest.
tTA-IRES-Neo was generated by cloning PCR-amplified tTA cDNA from pUHD15-1 (17) into the XbaI site of plasmid pMk10-59 (18). The tet-responsive reporter plasmid pBI-EGFP was purchased from CLONTECH and modified by the inclusion of a polylinker to create pBI-MCS-EGFP. Tet-responsive p53 expression constructs were made by cloning restriction fragments of full-length cDNA of wt p53 and p53R175H into pBI-MCS-EGFP. Tet-responsive p73 expression constructs were made in a similar fashion by using pCDNA3-HA-p73α and pCDNA3-HA-p73α292 (HA, hemagglutinin) as the sources of cDNA (19, 20). These p53 and p73 constructs were linearized and cotransfected into DLDtet14 cells with pTK-hygro (CLONTECH) at ≈5 to 1 molar ratio. Single colonies were obtained by limiting dilution with 400 μg/ml G418 and 250 μg/ml hygromycin B (Calbiochem) in the presence of 20 ng/ml doxycycline for 2–3 weeks. Clones that had low background green fluorescent protein (GFP) and homogeneous GFP induction were selected and analyzed for the expression of p53 by Western blot analysis.
Serial Analysis of Gene Expression (SAGE).
SAGE was performed essentially as described (21). In cases in which tags could not be assigned to a Unigene cluster, a PCR-based technique was used to amplify the sequences specific to a given tag (modified from ref. 22; detailed protocol available from the authors on request). Comparisons between libraries were carried out by using sage analysis software 3.04 beta, and differentially expressed tags were considered significant when P < 0.001 and the fold of induction was above 10.
p53 Adenoviruses and Infections.
High-titer adenoviruses expressing both GFP and either the wt or the R175H mutant form of p53 were generated through the AdEasy system (23) and used to infect cells for 4 hr. Cells were washed with normal growth medium once and allowed to recover for 16 hr before total RNA and protein was prepared and analyzed as described (21, 22).
Western Blots and Other Assays.
The p53, p21, and HA-tagged p73 proteins were detected by mAbs 1801, EA10 (Oncogene Science), and 3F10 (Roche Molecular Biochemicals), respectively. The β-catenin protein was detected by using the mAb C19220 from Transduction Laboratories (Lexington, KY). Luciferase assays and β-galactosidase assays were carried out by using Luciferase Assay Reagents (Promega) and AURORATM Gal Xe (ICN) according to the manufacturers’ instructions. To determine the fraction of apoptotic cells, cells were trypsinized and pelleted at 400 g. Resuspended cells were stained with the DNA-binding dye H33258 and evaluated by fluorescence microscopy and flow cytometry (24).
Results
Colon Cancer Cell Lines with Inducible p53.
Although tTA-based vectors have been used to establish inducible expression of a variety of genes in many systems, we were unable to establish p53-inducible clones with tTA-based (or other inducible) vectors in colon cancer cell lines (17). This failure was due to the “leakiness” of standard inducible systems and the silencing of tTA that often occurred during long-term culture of clones that initially seemed promising (25).
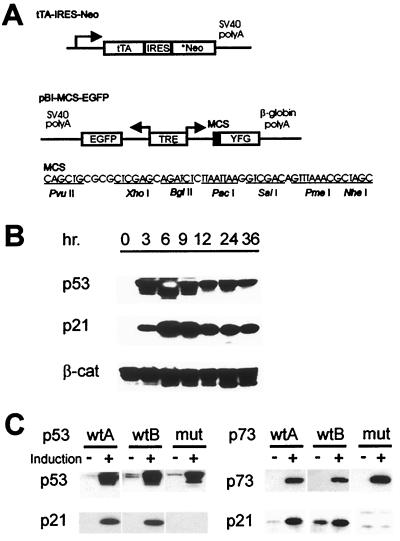
To overcome these problems, a different tTA expression vector was generated (Fig. 1A). The vector incorporated a bicistronic transcription unit with the tet-activator separated from the G418 resistance gene (Neo) by an improved IRES, ensuring the constitutive expression of tTA in geneticin-resistant clones (18, 26). A bidirectional tTA-responsive promoter regulates expression of GFP from one side and of p53 from the other (Fig. 1A). We established this system in DLD-1 cells that have endogenous mutant p53 alleles (27). Two clones expressing wt p53 (wtA and wtB) were selected for further analysis and compared with an analogous clone (mut) expressing a common tumor-derived mutant form of p53 (R175H).
Figure 1.
Modified tet-inducible expression system. (A) Diagram of the modified tet-off system constructs. *Neo denotes a optimized Neo-resistance gene (18). (B) Induction of p53 in p53 wtA cells. Cells were induced by removing doxycycline for the indicated times and assayed by Western blot analysis for p53 and p21 expression. β-Catenin served as the loading control. (C) Inducible expression of p53 and p73 resulted in induction of p21, but mutant forms of p53 and p73 did not. A second clone expressing mutant p53 and p73 yielded results identical to those shown.
Effects of p53 Expression.
The p53 protein in this system was induced rapidly after removal of doxycycline (examples in Fig. 1B). The induction of wt p53, but not mutant p53, stimulated a relatively rapid induction of p21, a prototypical p53 target gene, as well as a marked apoptotic response (Figs. 1C and 2 A and B). As expected from its induction of apoptosis, prolonged expression of p53 virtually abolished the cells’ ability to form colonies, whereas mutant p53 had no effect (data will be available at www.sagenet.org/findings.htm).
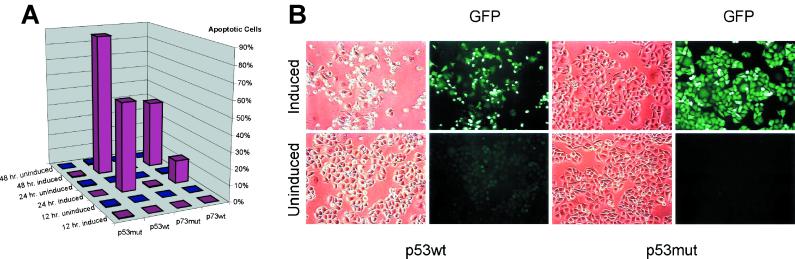
Figure 2.
Effects of p53 expression. (A) Mutant or wt p53- and p73-inducible clones were assayed for nuclear morphology by fluorescence microscopy at the indicated time points. Red bars represent p53- or p73-induced cells, and blue bars represent uninduced cells. At least 200 cells were counted for each determination. (B) Morphology after p53 induction. The same fields are shown as viewed under phase contrast or fluorescence microscopy. The four panels at Left were p53 wtA cells, and the four panels at Right were p53 mut cells grown in induction (Induced) or normal medium (Uninduced) for 24 hr.
This system also provided a way to determine the “commitment time” in these cells, i.e., the minimal exposure to p53 required to commit the cells irreversibly to death. Exponentially growing cells were placed in induction medium (without doxycycline) for varying time periods, and then doxycycline was added back to the growth medium. A large fraction of cells were already committed to apoptosis as early as 9 hr after p53 induction, long before any morphological characteristics of apoptosis could be observed (data will be available at www.sagenet.org/findings.htm.).
Identification of Previously Unidentified p53-Regulated Genes.
The above results stimulated us to use SAGE to analyze the transcripts present at 9 hr after p53 induction. SAGE involves the generation of short sequence tags derived from the 3′ ends of transcripts through a series of enzymatic manipulations. The relative abundance of individual tags in SAGE “libraries” provides a direct measure of the abundance of the corresponding transcripts. A SAGE library from p53 wtA cells was compared with an analogous library from p53mut cells both generated from cells 9 hr after doxycycline withdrawal. A total of 56,182 and 57,893 tags were obtained from the two libraries, respectively. Collation of the tags revealed that they represented 9,954 unique transcripts (see Materials and Methods). The great majority of these genes were expressed at equivalent levels in cells expressing either mutant or wt p53. Only 34 tags (0.3% of the total) were elevated by more than 10-fold in wtA cells compared with mut cells, whereas 12 tags were expressed at 10-fold lower levels in the wtA cells compared with the mut cells.
In light of much evidence indicating the importance of p53 in transcriptional activation (see the introduction), we focused on the 34 tags expressed at higher levels after wt p53 induction. We were able to assign 27 of these tags to known genes or to expressed sequence tags (ESTs) through a combination of database searching and PCR-based experimental techniques. Only three of these transcripts had previously been identified as p53-regulated. A summary of the 34 tags and their levels (tag numbers) is presented in Table 1; transcripts that had not been described previously in the literature (available only as EST entries or not at all) were designated “PETs” (for p53 early transcripts).
Table 1.
PETs identified by SAGE
| Gene | Tag sequence | p53 wt, no. | p53 mut, no. | Unigene | Description |
|---|---|---|---|---|---|
| PET-1* | CATGAGCTAAGTTT | 25 | 0 | 19447 | ESTs |
| PET-2* | CATGCCTGACCAGG | 20 | 0 | 150378 | ESTs |
| PET-3* | CATGGTGCTGGTGC | 17 | 0 | 9613 | ESTs |
| PET-4* | CATGCCTGGACGCT | 13 | 0 | ESTs | |
| PET-5* | CATGGGAGAATTTT | 13 | 0 | 115274 | ESTs, highly similar to hedgehog (Homo sapiens) |
| PET-6* | CATGTCCTGTAAAG | 13 | 0 | 74034 | H. sapiens clone 24651 mRNA sequence |
| PET-7* | CATGTACCCAGGGC | 12 | 0 | 101395 | ESTs |
| PET-8* | CATGGCCCTGACCT | 22 | 1 | 97871 | ESTs |
| PET-9* | CATGGTGCTGCTCC | 11 | 0 | 7936 | ESTs |
| PET-10† | CATGCCGTCTTTCC | 12 | 0 | No match | |
| PET-11* | CATGGCGAGCTGGC | 12 | 0 | 146344 | ESTs |
| PET-12† | CATGACAACTTAAA | 11 | 0 | No match | |
| PET-13* | CATGCACACACCAG | 11 | 0 | 22824 | ESTs |
| PET-14† | CATGGGCCGGGCAC | 11 | 0 | No match | |
| PET-15† | CATGTGCGCTGGCC | 11 | 0 | No match | |
| PET-16† | CATGCCTTCCAGCT | 15 | 1 | No match | |
| PET-17† | CATGTTTTGCTTTT | 15 | 1 | No match | |
| PET-18† | CATGGCAGTGGGCT | 21 | 2 | No match | |
| ET2* | CATGCCTCAGCCAG | 22 | 0 | 1407 | Endothelin 2 |
| Cav* | CATGTGGATCAACC | 20 | 0 | 119333 | Caveolin, caveolae protein, 22 kDa |
| Htk* | CATGTCTGTTTCCA | 16 | 0 | 155227 | Human tyrosine kinase (HTK) |
| IPK* | CATGGAGGTCCTTC | 14 | 0 | 6453 | Human inositol 1,3,4-trisphosphate 5/6-kinase |
| RTP* | CATGGGACTTTCCT | 22 | 1 | 75789 | Human mRNA for RTP |
| B61* | CATGTGTACATTCT | 11 | 0 | 1624 | Ephrin-related ligand precursor I |
| DR5‡ | CATGAAATTGTTGG | 20 | 1 | 120932 | Human DR5/killer |
| Mat8* | CATGGCAGGGCCTC | 19 | 1 | 92323 | H. sapiens mRNA for MAT8 protein |
| Gpc* | CATGGCTTGTTCTC | 17 | 1 | 2699 | Glypican 1 |
| Jun-B* | CATGACCCACGTCA | 16 | 1 | 89792 | Jun B protooncogene |
| Tsp‡ | CATGGCTCGACTCAG | 32 | 2 | 87409 | Thrombospondin 1 |
| Tap1* | CATGGTGCAGGCTC | 16 | 1 | 158164 | Antigen peptide transporter |
| HO1* | CATGCGTGGGTGGG | 31 | 2 | 75967 | Heme oxygenase (decycling) 1 |
| ATDC* | CATGTTGCATATCA | 15 | 1 | 82237 | AT group D-associated protein |
| EVPL* | CATGGTGATGGGCT | 14 | 1 | 25482 | Envoplakin |
| p21‡ | CATGTGTCCTGGTT | 153 | 11 | 74984 | cdk-inhibitor |
PETs identified through SAGE analysis; 34 highly induced tags (14 bp) are shown. Columns 3 and 4 indicate the numbers of each tag in two libraries.
Transcripts that matched Unigene entries but had not been known to be regulated by p53.
† Previously unidentified transcripts that had no matches to Unigene entries.
‡ Previously described p53-regulated genes.
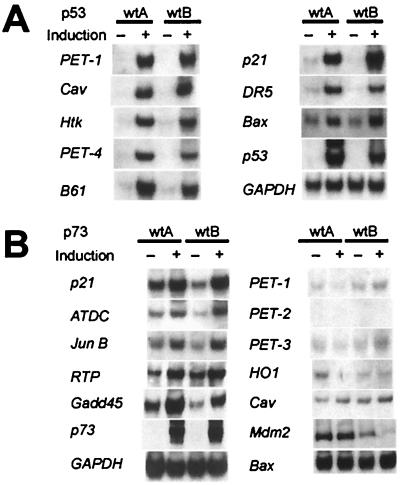
To confirm the SAGE data, Northern blot analyses were performed on all 27 assigned transcripts. In each of the 27 cases, the expression of the transcripts was substantially induced in both of the p53 wt-inducible clones and in neither of the p53 mut-inducible clones (see examples in Fig. 3A). Using similar Northern blots, we also examined the expression of nine genes that did not register as highly induced in the SAGE analysis but had been reported previously to be transcriptionally regulated by p53 (14). Of these nine, three (cyclin G1, Fas, and cathepsin D) were not found to be induced at the early time point studied, whereas six were indeed induced but at lower levels than the 27 transcripts noted above (see examples in Fig. 3 A and B and complete list in Table 2). Examination of the SAGE data showed that these six genes (Bax, IGBP3, PIG3, 14-3-3σ, MDM2, and Gadd45) were each induced by p53 at 9 hr, at levels between 2-fold and 10-fold.
Figure 3.
Confirmation of SAGE results by Northern blot analysis. Representative Northern blots are shown with RNA prepared from cells induced (“+”) for 9 hr or uninduced (“−”). (A) RNA from two independent p53-inducible clones was analyzed to confirm induction of transcripts by p53. No induction of these transcripts was observed in RNA prepared from cells with inducible mutant p53 genes (data not shown). (B) RNA from two independent p73-inducible clones was used to assay induction of transcripts by p73.
Table 2.
Classification of p53-regulated genes
| Gene | Class | Time | Adr. Ind. | 5-FU Ind. | p53-dep. | p73-Ind. |
|---|---|---|---|---|---|---|
| Cav | I | E | 1 | 1 | − | − |
| DR5 | I | E | 1 | 2 | − | − |
| Gpc | I | E | 1 | 1 | + | − |
| 14-3-3σ* | I | E | 2 | 2 | − | + |
| p21 | I | E | 2 | 2 | + | + |
| HO1 | I | I | 2 | 1 | − | − |
| Htk | I | I | 1 | 1 | − | − |
| Bax* | I | I | 2 | 2 | − | − |
| PIG3* | I | I | 2 | 2 | + | − |
| Jun-B | II | E | 0 | 0 | − | + |
| Mdm2* | II | E | 2 | 2 | + | − |
| PET-3 | II | E | 2 | ND | − | − |
| PET-7 | II | E | 2 | ND | − | − |
| ATDC | II | E | 0 | ND | N/A | + |
| B61 | II | I | 2 | 2 | − | − |
| PET-1 | II | I | 1 | 0 | − | − |
| PET-8 | II | I | 1 | 1 | − | − |
| PET-4 | II | I | 1 | 2 | − | − |
| Gadd45* | II | L | 1 | 0 | − | + |
| RTP | II | I | 1 | 1 | − | + |
| IGBP3* | II | I | 1 | 0 | N/A | − |
| ET2 | II | I | 0 | 0 | − | − |
| Mat8 | II | I | 2 | 0 | + | − |
| PET-9 | II | I | 1 | ND | − | − |
| Fas* | II | N/A | 1 | 0 | − | − |
| PET-11 | II | ND | 1 | ND | − | − |
| Cln G1* | III | N/A | 0 | 0 | N/A | − |
| Tsp | III | E | 1 | 1 | − | − |
| PET-2 | III | I | 0 | ND | N/A | − |
| PET-5 | III | I | 0 | ND | N/A | − |
| PET-6 | III | I | 0 | ND | N/A | − |
| IPK | III | I | 1 | ND | − | − |
| CatD* | III | N/A | 0 | 0 | N/A | ND |
| PET-13 | III | ND | 1 | ND | − | ND |
| Tap1 | ND | I | ND | ND | ND | − |
| EVPL | ND | I | ND | ND | ND | − |
Class: class I, induced by Ad-p53 wt in all five colorectal cancer cell lines studied; class II, induced in only a subset of these lines; class III, induced in none of the lines; N/A, not applicable; ND, not determined. Time: E, early (detectably induced within 3 hr of doxycycline withdrawal); I, intermediate (detectably induced after 6 hr); L, late (detectably induced only after 12 hr). Adriamycin (Adr.) or 5-FU induced (ind.): 2, induced in two wt p53 cell lines; 1, induced in one cell line; 0, induced in neither cell line. p53 dependence (dep.) was determined only for transcripts induced by adriamycin or 5-FU in HCT116 cells. “+” denotes that induction was reduced substantially or absent in p53 KO cells, and “−” indicates that comparable induction was observed in the p53 KO cells. p73 induced; “+” denotes that transcript was induced in two p73 wt clones, and “−” indicates that induction was not observed in both clones.
Genes not identified herein but shown to be p53-regulated in other studies. All other genes were identified through the SAGE analysis reported in this paper.
The tTA system depicted in Fig. 1A was also used to obtain inducible expression of the p73 gene (19, 20). The induction of wt, but not mutant, p73 stimulated apoptosis of DLD-1 cells, though not quite as dramatically as p53 (Fig. 2A). Only 6 of the 36 p53-induced transcripts described above were found to be induced significantly in the p73 wt clones (Table 2 and Fig. 3B). These data establish that the genes regulated by p53 and p73 are considerably different, even when similarly expressed through the identical promoter in the same cells.
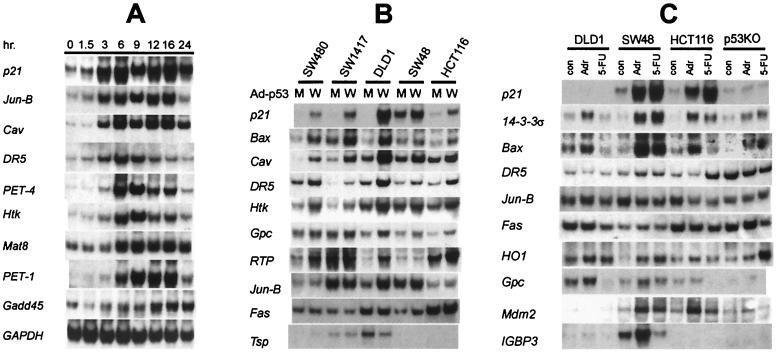
Northern blot analysis was also used to determine the earliest times at which induction of these genes could be detected (Fig. 4A; complete listing in Table 2). Increased levels of 11 genes could be detected as early as 3 hr after doxycycline withdrawal (e.g., p21, caveolin, DR5, and Jun-B). Of the other 20 genes, most were expressed by 6 hr after doxycycline withdrawal (e.g., PET-1 and PET-4), whereas one (Gadd45) was not induced until 12 hr after removal of doxycycline.
Figure 4.
Heterogeneity in the induction of p53-responsive genes. (A) Time course of induction. The p21, Jun-B, Caveolin, and DR-5 genes were detectably induced by 3 hr after doxycycline withdrawal, whereas Gadd45 was induced 12 hr later. RNA was prepared from wtA cells at the indicated time points. (B) Induction by p53 adenoviruses. Class I genes included p21, Bax, Caveolin (Cav), DR5, Htk, and Gpc, whereas RTP, Jun-B, and Fas were class II genes, and thrombospondin (Tsp) was a class III gene. “W” and “M” refer to the adenoviruses expressing wt p53 and mutant p53, respectively. (C) Induction by chemotherapeutic agents. RNA from the indicated colorectal cancer cells lines treated with adriamycin (Adr) or 5-FU for 24 hr was analyzed. RNA from untreated cells was used as control (con). The expression of p21 was strongly induced by both drugs in both cell lines expressing wt p53 but was not induced in DLD-1 cells containing endogenous mutant p53 or in p53 KO cells. In contrast, Bax was induced by adriamycin but not by 5-FU. The induction of Bax by adriamycin was considerably stronger in the two lines with wt p53 than in DLD-1 or p53 KO cells. Another pattern was observed with Fas, in which little induction was observed in any of the lines studied.
Uniformity of Expression of p53-Activated Genes in Colorectal Cancer Cells.
Replication-deficient adenoviruses containing a GFP gene plus either a mutant or a wt p53 gene were generated by using AdEasy vectors (23) and used to infect five different colorectal cancer cell lines. The five lines were chosen on the basis of infectability with adenoviruses and were representative of those with endogenous wt p53 genes (HCT116 and SW48) or endogenous mutant p53 genes (DLD-1, SW480, and SW1417). All five cell lines were infected to similar extents, based on GFP fluorescence and p53 protein expression assessed by Western blots (data not shown).
The transcripts from 34 genes, including the 25 genes revealed by SAGE and the 9 genes identified through other methods, were assessed in these lines (Table 2). The expression of these transcripts and their inducibility by p53 varied widely among the lines (Fig. 4B and Table 2). Three classes of transcripts were identified. Class I contained 9 genes, which were induced in all five lines (e.g., p21 and caveolin); class II contained 17 genes, which were induced in a subset of the lines (e.g., RTP and Jun-B); and class III contained 8 genes, which were not induced in any of the lines by adenovirus infection (e.g., thrombospondin). Cell lines that expressed one class II gene were no more likely to express a different class II gene than another cell line. However, every gene induced by p53 adenovirus infection of DLD-1 cells was also induced in the tTA system, suggesting that the activation of downstream genes by p53 was largely determined by the cell type rather than the mode by which exogenous p53 was introduced.
Expression of p53-Activated Genes After Treatment with Chemotherapeutic Agents.
The two systems described above (tTA and adenovirus infection) employ exogenous p53 expressed at levels likely to be higher than those ever encountered in vivo. To assess transcriptional induction of these genes at more physiologic levels of p53, we treated cells with two agents known to induce endogenous p53: adriamycin, a prototypical DNA damaging agent, and 5-FU, an antimetabolite that is the mainstay of adjuvant therapy for colorectal cancer in the clinic. Expression of each of the 34 genes in the panel was examined in response to these drugs in four colorectal cancer cell lines (Table 2). Two of these lines (SW48 and HCT116) had wt p53 genes, one had a mutant p53 gene (DLD-1), and one had its endogenous wt p53 gene disrupted by homologous recombination (p53 KO; ref. 16). The majority of the 36 tested genes were found to be induced by adriamycin in at least one of the cell lines with wt p53 (Fig. 4C and Table 2). Interestingly, however, only six genes were found to be induced by both adriamycin and 5-FU, though identical levels of p53 resulted from treatment with both drugs (28). For example, Bax was induced by adriamycin in HCT116 cells but not by 5-FU, even though 5-FU treatment stimulated extensive apoptosis in this line (Table 2 and Fig. 4C). Thus, in addition to cell-type dependency, there was a clear dependence on the nature of the induction signal. To our surprise, analysis of the p53 KO line revealed that p53 was not required for induction in most cases. Only 3 of the 36 genes studied (p21, Mdm2, and PIG3) were induced to considerably higher levels in parental cells than in the p53 KO cells after treatment with these two drugs.
Discussion
Initial descriptions of oncogene and tumor suppressor gene action concentrated on relatively simple linear pathways. As more has become known about signal transduction, it has become clear that important cellular pathways are much more complex than originally believed (29). These complexities involve downstream and upstream branching as well as positive and negative feedback controls.
Viewed in the above context, the relatively large number of genes that we found to be induced by p53 was not surprising. The heterogeneity in induction, however, was unexpected. Only a small number of genes (class I) were induced in all of the five lines studied, even by very high levels of p53 expression, despite the fact that the lines were derived from the same precursor cell type. The responses to drugs that induce p53 at more physiologic levels were even more heterogeneous. These results suggest that most of the genes transcriptionally activated by p53 are codependent on other transcription factors. The presence or absence of such factors is in turn likely to depend on other genetic alterations accumulated during the long history (decades) of neoplastic cells before their removal from patients. This heterogeneity helps explain the diverse patterns of expression observed in published studies of p53 effectors. Based on the results reported here, one would expect even less uniformity in the levels of expression of these genes in different cell types from different species.
Several other features of potential p53-regulated genes were notable. First, the induction of most of the genes was not strictly p53-dependent, because similar levels of induction were observed in isogenic controls differing only in the targeted deletion of the p53 gene (Fig. 4C). That p53 is not required for these genes’ induction should not be interpreted to mean that p53 is not important for their induction. These genes likely respond to several transducers of the same initiating signal. Reactive oxygen species (ROS), for example, induce p53 and p21 independently. With p53 present, p21 is induced to greater levels than in its absence, but p21 is still induced by ROS in the absence of p53 (30). Second, more genes were induced by high exogenous levels of p53 than by the lower endogenous levels of p53 stimulated by treatment with chemotherapeutic agents. There were no exceptions to this pattern, in that all genes induced by chemotherapeutic agents were also induced by the p53 adenovirus. These differences likely reflect differences in the affinity of various promoters for p53, such that some are responsive only to high levels of p53 (31).
In summary, p53 activates the expression of many genes whose effects, in aggregate, determine important components of neoplastic cell behavior. The particular genes induced likely depend on the cell’s normal tissue derivation as well as on the genetic alterations that have occurred during the tumorigenic process. It is unlikely that mutation of any single gene could recapitulate all the effects of p53, and such recapitulation should not be expected in experimental studies. Recognition of these facts, which likely apply to other tumor suppressor genes, should prove useful for designing and interpreting future studies on downstream effectors.
Acknowledgments
We thank Akiko Tanaka for providing IRES construct pMK10-59, Manfred Gossen for tTA expression plasmid pUHD15-1, and William G. Kaelin for p73 expression plasmids. Under an agreement between Calbiochem and Johns Hopkins University, K.W.K and B.V. are entitled to a share of the sales royalty for the anti-p21 antibodies received by the University from Calbiochem. The terms of these arrangements are being managed by the University in accordance with its conflict of interest policies. This work was supported by the Clayton Fund and by National Institutes of Health Grants CA57345 and CA62924.
Abbreviations
- wt
wild-type
- tet
tetracycline
- 5-FU
5-fluorouracil
- GFP
green fluorescent protein
- SAGE
serial analysis of gene expression
- HA
hemagglutinin
- tTA
tet activator
- IRES
internal ribosome entry site
- EST
expressed sequence tag
- PET
p53 early transcript
References
- 1.Bates S, Vousden K H. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oren M. Semin Cancer Biol. 1994;5:221–227. [PubMed] [Google Scholar]
- 3.Dameron K M, Volpert O V, Tainsky M A, Bouck N. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 4.Cross S M, Sanchez C A, Morgan C A, Schimke M K, Ramel S, Idzerda R L, Raskind W H, Reid B J. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 5.Lanni J S, Jacks T. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 7.Prives C, Hall P A. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 8.Vogelstein B, Kinzler K W. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 9.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 10.Raycroft L, Wu H Y, Lozano G. Science. 1990;249:1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields S, Jang S K. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 12.Jeffrey P D, Gorina S, Pavletich N P. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 13.Gottlieb T M, Oren M. Semin Cancer Biol. 1998;8:359–368. doi: 10.1006/scbi.1998.0098. [DOI] [PubMed] [Google Scholar]
- 14.El-Deiry W S. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 15.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 16.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 17.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi M, Yamauchi Y, Tanaka A, Shimamura S. Biotechniques. 1996;21:398–402. doi: 10.2144/96213bm12. [DOI] [PubMed] [Google Scholar]
- 19.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J C, Valent A, Minty A, Chalon P, Lelias J M, Dumont X, et al. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 20.Jost C A, Marin M C, Kaelin W G., Jr Nature (London) 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 22.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature (London) 1997;389:300–304. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 23.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waldman T, Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 25.Lengauer C, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaminski A, Howell M T, Jackson R J. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata D, Peinado M A, Ionov Y, Malkhosyan S, Perucho M. Nat Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- 28.Bunz F, Hwang P M, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler K W, Vogelstein B. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pawson T, Saxton T M. Cell. 1999;97:675–678. doi: 10.1016/s0092-8674(00)80779-5. [DOI] [PubMed] [Google Scholar]
- 30.Russo T, Zambrano N, Esposito F, Ammendola R, Cimino F, Fiscella M, Jackman J, O’Connor P M, Anderson C W, Appella E. J Biol Chem. 1995;270:29386–29391. doi: 10.1074/jbc.270.49.29386. [DOI] [PubMed] [Google Scholar]
- 31.Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. EMBO J. 1998;17:4668–4679. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]