Abstract
DksA is well known for its regulatory role in the transcription of rRNA and genes involved in amino acid synthesis in many bacteria. DksA has also been reported to control expression of virulence genes in pathogenic bacteria. Here, we elucidated the roles of a DksA-like protein (CJJ81176_0160, Cj0125c) in the pathogenesis of Campylobacter jejuni. As in other bacteria, transcription of stable RNA was repressed by the DksA-like protein under stress conditions in C. jejuni. Transcriptomic and proteomic analyses of C. jejuni 81-176 and an isogenic mutant lacking the DksA-like protein showed differential expression of many genes involved in amino acid metabolism, iron-related metabolism, and other metabolic reactions. Also, the C. jejuni DksA-like protein mutant exhibited a decreased ability to invade intestinal cells and induce release of interleukin-8 from intestinal cells. These results suggest that the DksA-like protein plays an important regulatory role in diverse metabolic events and the virulence of C. jejuni.
When exposed to nutrient depletion, many bacteria show global changes in gene expression called the stringent response (15, 38). The effecter molecule of the stringent response is guanosine tetraphosphate (ppGpp). Upon amino acid starvation, the abundance of uncharged tRNA results in ribosome stalling, activating ribosome-associated RelA or SpoT and producing ppGpp (15, 38). ppGpp binds to the β and β′ subunits of the RNA polymerase (RNAP) core enzyme and affects transcription of genes primarily at the stage of transcription initiation during the formation of the open promoter complex (4, 15, 38, 47). In many bacteria, ppGpp-dependent regulation affects bacterial physiology, including the functionality of some sigma factors and the virulence of pathogenic bacteria (15, 18, 58). The best-known types of regulation by ppGpp include negative regulation of rRNA transcription and positive regulation of amino acid biosynthesis, in which ppGpp potentiates direct activation or repression in concert with an RNAP-associated protein designated DksA (6, 7, 20, 48).
DksA was originally identified in Escherichia coli as a multicopy suppressor of the temperature-sensitive phenotype of dnaKJ mutants in 1990 (30). Analysis of bacterial genomes revealed the ubiquitous distribution of dksA open reading frames, which encode highly conserved amino acid sequences (50). DksA is reported to have pleiotropic effects, such as defects in chaperonin function, gene expression, cell division, amino acid biosynthesis, phage sensitivity, and quorum sensing (9, 13, 14, 27, 30, 40, 66). The involvement of DksA in bacterial virulence has also been reported for many pathogenic bacteria, including Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, Shigella flexneri, and enterohemorrhagic E. coli (29, 41, 43, 56, 65). Unlike most transcriptional regulators, DksA does not bind directly to DNA but binds to the secondary channel of RNAP (47, 50). E. coli DksA is composed of a globular domain and a coiled coil with two highly conserved aspartate (Asp) residues at its tip (50). DksA binds directly to RNAP, positioning the Asp residues near the active site to coordinate a ppGpp-bound Mg2+ ion with the Asp residues, thereby stabilizing the ppGpp-RNAP complex (50). In many papers, DksA has been reported to play a role as a cofactor of ppGpp regulation (50). However, Magnusson and colleagues recently demonstrated that DksA regulates gene expression without ppGpp, suggesting that DksA may have another mechanism of gene regulation which is independent of ppGpp (39).
Campylobacter jejuni is a microaerophilic, spiral-shaped, gram-negative bacterium belonging to the epsilon group of the Proteobacteria. C. jejuni is a major causative agent of human bacterial gastroenteritis throughout the world (17). C. jejuni is a commensal gastrointestinal bacterium in birds, and consumption of undercooked poultry meat may result in campylobacteriosis ranging from mild diarrhea to bloody enteritis (61). Campylobacter infection is also associated with Guillain-Barré syndrome, a form of neuromuscular paralysis (57). However, how C. jejuni causes campylobacteriosis or how C. jejuni can survive in hostile environments is not clearly understood. Factors known to be associated with the virulence of C. jejuni include motility, colonization, adhesion to intestinal cells, invasion of intestinal cells, N-linked protein glycosylation, and production of cytolethal distending toxin (33, 60, 64). Like many other bacteria, C. jejuni mounts a SpoT (Cj1272c)-mediated stringent response upon nutrient deprivation (18). A C. jejuni mutant with spoT deleted showed defects in aerotolerance, rifampin resistance, stationary-phase survival, and adherence to, invasion of, and survival in epithelial intestinal cells (18). The stringent response mediated by SpoT in C. jejuni is considered to be important for the survival and virulence of this bacterium, because C. jejuni lacks the stationary-phase sigma factor rpoS (46), which is essential for stress and stationary-phase responses in many gram-negative bacteria (26). Since C. jejuni is able to use ppGpp as its stringent signal, DksA is expected to play a regulatory role in concert with ppGpp and to affect the physiology and virulence of C. jejuni. To examine this possibility, we characterized a DksA-like protein in C. jejuni. It was found that the DksA-like protein in C. jejuni modulates gene expression and affects the virulence, as well as the physiology, of this bacterium.
MATERIALS AND METHODS
Bacterial strains, plasmids, cell lines, and growth conditions.
C. jejuni 81-176 (11), a pathogenic strain, and derivatives of this strain were used in this study. E. coli DH5α was used as a cloning host. C. jejuni 81-176 and an isogenic mutant lacking the DksA-like protein (FMB1104) were cultured using Mueller-Hinton (MH) broth or agar at 37°C under microaerobic conditions generated with MART (Anoxomat). Mannitol-free morpholinepropanesulfonic acid (MOPS)-MGS medium (50 mM MOPS [pH 7.4], 2 mM MgSO4, 0.25 mM CaCl2, 19 mM glutamic acid, 0.004 mM biotin) was used as a minimal medium for C. jejuni to induce the stringent response (18). A human intestinal epithelial cell line, INT-407, was used for the invasion assay and was grown in minimal essential medium supplemented with 10% fetal bovine serum (MEM-FBS) containing penicillin (100 U/ml) and streptomycin (50 μg/ml) in a humidified atmosphere containing 5% CO2 and 95% air.
DNA manipulations.
All DNA manipulations, including cloning, transformation into E. coli, isolation of DNA from agarose gels, PCRs, and DNA sequencing, were performed by using standard techniques (55) and the manufacturers’ instructions. The oligonucleotide primers used in PCRs are described elsewhere (http://plaza.snu.ac.kr./∼fmb/main/pu-01.htm). DNA sequencing was performed using a BigDye terminator cycle sequencing kit (Applied Biosystems) and an ABI Prism 3700 DNA analyzer (Perkin-Elmer) at the National Instrumentation Center for Environmental Management (Seoul, Korea).
Construction of a mutant with cat inserted in CJJ81176_0160.
C. jejuni 81-176 was used as a parental strain. The chloramphenicol resistance cassette (cat) was amplified from pRY112 (69) with Vent polymerase (New England Biolabs) using primers catF and catR. To construct the mutant with a mutation in the DksA-like protein (CJJ81176_0160), a DNA fragment containing CJJ81176_0160 and a flanking region was amplified using primers Cj0125cF1 and Cj0125cR1, digested with PstI and EcoRI, and then ligated into pUC19 digested with the same enzymes. The resultant plasmid was digested with SspI, and then the cat cassette was inserted into the SspI site. The orientation of the cat cassette was confirmed by sequencing. The plasmid with the cat cassette inserted in the same orientation as CJJ81176_0160 was designated pFMB1104, which was electroporated into C. jejuni 81-176. The mutant with cat inserted into CJJ81176_0160 was screened by growing it on MH agar plates containing chloramphenicol (12.5 μg/ml). The mutant was confirmed by PCR and designated C. jejuni FMB1104.
RNA manipulation.
For reverse transcription (RT)-PCR, total RNA was isolated from C. jejuni cultures using an RNeasy mini kit (Qiagen). After DNase treatment of the total RNA solution using a Turbo DNA-free kit (Ambion), cDNA was synthesized using an Omniscript RT kit (Qiagen) and random hexamers (Invitrogen) according to the manufacturers’ instructions. A negative control for RT-PCR was prepared by omitting reverse transcriptase at the cDNA synthesis step. For RNA quantification, total RNA was prepared from 5 ml of each C. jejuni culture using Trizol reagent (Invitrogen) according to the manufacturer's instructions. We changed the RNA purification method to obtain total RNA, because the other RNA preparation method using columns resulted in partial removal of rRNA. Then the RNA concentration was determined by measuring the optical densities at 260 and 280 nm of the RNA solution using GeneQuant pro (Amersham Pharmacia Biotech).
Proteomic analyses. (i) Protein preparation and two-dimensional gel electrophoresis (2DGE).
The wild type and the mutant were grown to mid-log phase in 50 ml MH broth in Erlenmeyer flasks (500 ml) at 37°C with rotation at 130 rpm for 6 h. Protein was prepared from the wild type and the mutant as described by Yoon et al. (72). Protein concentrations were determined by the Bradford assay (12). For first-dimension separation, 400 or 900 μg of protein from each sample was separated by isoelectric focusing using 13-cm IPG strips (GE Healthcare). For the second-dimension separation, proteins were separated based on molecular weight by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 10 or 12% acrylamide gels. Gels were visualized by colloidal Coomassie blue staining (Invitrogen).
(ii) Image analysis and protein identification.
Quantitative data were obtained from multiple gel runs using ImageMaster 2D Platinum version 6.0 (GE Healthcare). A twofold difference in expression was used as a threshold during image analysis to detect proteins that were differentially expressed. Protein spots showing different levels were excised from the stained gels, which was followed by in-gel digestion using the method of Jensen et al. (28). Peptide mass fingerprinting was done with a Voyager-DETM STR biospectrometry workstation (PerSeptive Biosystems, Framingham, MA), and proteins were identified by peptide mass fingerprinting with MASCOT (http://www.matrixscience.com) supplemented with the option for Campylobacter in the NCBI database.
Transcriptomic analyses. (i) Preparation of total RNA for a DNA microarray.
Total RNA was extracted from six independent cultures of the wild type and the mutant, which were cultured under the same conditions that were used for 2DGE. C. jejuni strains were inoculated into MH broth to an optical density at 600 nm of 0.05. Bacterial cultures were grown at 37°C with shaking at 130 rpm for 6 h. Total RNA was isolated using RNAprotect bacterial reagent (Qiagen) and an RNeasy kit (Qiagen) according to the manufacturer's protocol with optional on-column DNase treatment with RNase-free DNase (Qiagen). The RNA concentration was determined with a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE).
(ii) cDNA synthesis and microarray hybridization.
cDNA was synthesized using aminoallyl deoxynucleoside triphosphates with the ChipShot indirect cDNA labeling system (Promega) according to the manufacturer's instructions. After purification, aminoallyl-labeled cDNA was fluorescently labeled with a CyDye postlabeling reactive pack (Amersham Bioscience, Piscataway, NJ). After purification, the fluorescent labeling of the cDNA was quantified, and equal amounts of labeled cDNA of the wild type and the mutant were mixed and dried in a speed vacuum concentrator. After resuspension with hybridization buffer provided by the slide manufacturer, the mixture was denatured by heating at 95°C for 3 min. The hybridization mixture was loaded onto a microarray slide (OciChip; Ocimum Biosolutions, Indianapolis, IN) and hybridized in a shaking incubator at 42°C for 24 h. The slides were washed using the manufacturer's recommendations and dried by centrifugation. In total, six slides were hybridized using RNA extracted from six independent cultures. For three slides, the wild-type strain was labeled with Cy3 and the mutant strain was labeled with Cy5. For the remaining three slides, the dye-sample combination was reversed to control for differences in dye labeling of the cDNA samples. Further control for dye bias was achieved by adjusting the volume of cDNA hybridized based on the actual concentration of Cy dye incorporated as measured by a NanoDrop ND-1000 (NanoDrop Technologies).
(iii) Image acquisition and analysis.
The data analysis was performed essentially as described by Madsen et al. (37). Using a ScanArray Express laser scanner (Applied Biosystems Inc., Foster City, CA), each dye channel of each slide was scanned at 10-μm resolution with three predetermined laser power settings (high, medium, and low) to prevent saturation of high signals while ensuring adequate sensitivity for genes with low signals. ImaGene software (BioDiscovery, El Segundo, CA) was used to measure the spot-specific mean signal strength and the spot-specific background median of each spot. Using the R statistical package (version 2.0.1; The R Foundation for Statistical Computing), the spot-specific background median was subtracted from the spot-specific signal mean to obtain a background-subtracted value for each spot. The natural logarithm of each background-subtracted value was obtained, and median centering was performed as previously described (37). The median of the individual spot signals for each gene obtained at different power settings was determined and used for all further calculations. Data obtained from the two dye channels were subjected to locally weighted scatterplot smoother (LOWESS) normalization (68) to balance the fluorescence intensities of the two dyes (Cy3 and Cy5) used to label cDNA. This normalization process was used to rescale the fluorescence intensity of each transcript, preventing the dye bias problem (16, 35).
(iv) Statistical analysis.
The normalized data generated for individual probes of each array by dye combination were subjected to an independent mixed linear model using the SAS statistical software package (67). The model included fixed effects for treatment (wild type versus mutant) and dye (Cy3 versus Cy5) in addition to the random effect for slides. A t test was performed using the null hypothesis that there was no difference in the signal intensities of the treatment groups. The resulting P values for each of the probes were sorted in ascending order and converted to a Q value by the method of Storey and Tibshirani (59). Q values are used to estimate a false discovery rate for a subset of gene targets. Complete results of these experiments are available at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE9866.
Quantitative real-time RT-PCR (qRT-PCR).
Total RNA was isolated from C. jejuni cultures using an RNeasy mini kit (Qiagen). After DNase treatment of the total RNA solution with a Turbo DNA-free kit (Ambion), cDNA was synthesized using an Omniscript RT kit (Qiagen) and random hexamers (Invitrogen). Quantification of cDNA was carried out using IQ SYBR green PCR Supermix (Bio-Rad), and real-time amplification of PCR products was analyzed using the iCycler optical module (Bio-Rad). The amplification program consisted of one cycle of 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 30 s. The mRNA expression levels for target genes were normalized to the 16S rRNA gene (rrsA) expression level. The relative amount of cDNA was calculated using the 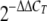 method (36). The primers used for real-time PCR are described elsewhere (http://plaza.snu.ac.kr./∼fmb/main/pu-01.htm).
method (36). The primers used for real-time PCR are described elsewhere (http://plaza.snu.ac.kr./∼fmb/main/pu-01.htm).
Invasion assay.
Approximately 1 × 105 cells of INT-407 were seeded in each well of a 24-well plate and grown to semiconfluence in antibiotic-free MEM-FBS for 16 h. Then each strain of C. jejuni in exponential phase was added at a multiplicity of infection of 100. The plates were centrifuged at 200 × g for 5 min to maximize bacterium-host cell contact and were incubated for 4 h at 37°C in the presence of 5% CO2. After three washes with phosphate-buffered saline, the monolayers were incubated in MEM-FBS containing gentamicin (100 μg/ml) for 2 h to kill extracellular bacteria. After three additional washes with phosphate-buffered saline, infected epithelial cells were lysed with 1% Triton X-100 for 5 min. Intracellular bacteria were enumerated by plating serial dilutions on MH agar plates.
Interleukin-8 secretion assay.
As in the invasion assay, approximately 1 × 105 INT-407 cells were infected with C. jejuni at a multiplicity of infection of 100. The plates were centrifuged at 200 × g for 5 min to maximize bacterium-host cell contact and incubated for 4 h at 37°C in the presence of 5% CO2. The culture medium was harvested and centrifuged at 15,000 × g for 10 min to pellet the residual bacteria. The supernatant was collected and stored at −80°C until analysis. The secreted interleukin-8 (IL-8) levels were determined using a BD OptEIA human IL-8 enzyme-linked immunosorbent assay II kit (BD Biosciences).
Ferrous iron uptake assay.
The wild type and the mutant were grown to mid-log phase under microaerobic conditions in MEMα (Invitrogen) at 37°C with rotation at 130 rpm for 6 h. The uptake of ferrous iron into C. jejuni was measured using 55FeCl3 (Perkin-Elmer) as described by Naikare et al. (42). The activity, expressed in cpm, was determined using a Wallac 1400 liquid scintillation counter (Turku, Finland), and the values were converted to picomoles of 55Fe using a previously determined standard curve.
Other computer programs.
Sequence manipulation and sequence alignment were performed with DNASTAR software (DNASTAR Inc.). Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) was used to design primers used in qRT-PCR. GraphPad Prism 4 (GraphPad Inc.) was used to plot the charts.
RESULTS AND DISCUSSION
Identification of a DksA homolog in C. jejuni.
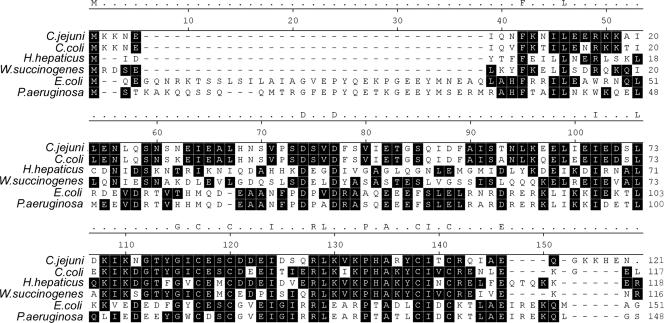
Analysis of the genomic sequence revealed that Campylobacter possesses DksA-like proteins, which are designated CJJ81176_0160 and Cj0125c in C. jejuni 81-176 and C. jejuni NCTC 11168 (46), respectively. All the C. jejuni strains whose genome sequences were available by March 2008 also possess DksA-like proteins (data not shown). The DksA-like protein is composed of 120 amino acids with a calculated molecular mass of 13.5 kDa. A BLASTP search showed that CJJ81176_160 (Cj0125c) was similar to E. coli DksA (26% identity and 53% similarity). CJJ81176_160 showed high levels of identity (ca. 63 to 89%) to the DksA-like proteins of bacteria belonging to the epsilon group of the Proteobacteria, such as other species of Campylobacter, Helicobacter hepaticus, and Wolinella succinogenes (2). However, Helicobacter pylori does not have a DksA homolog (1, 61). The DksA-like protein of C. jejuni contains conserved sequences of DksA proteins (Fig. 1).
FIG. 1.
Amino acid sequence alignment of DksA homologs. The DksA-like protein of C. jejuni (CJJ81176_0160, YP_999849.1) was aligned with DksA homologs of Campylobacter coli RM2228 (ZP_00367836.1), H. hepaticus ATCC 51449 (NP_860767.1), W. succinogenes DSM 1740 (NP_906418.1) E. coli K-12 (NP_414687.1), and P. aeruginosa PAO1 (NP_253411.1). A black background indicates dominant amino acid residues in the aligned sequences. Two Asp residues that are known to be critical for fixing the position of the ppGpp bound to Mg2+ (54) are indicated by asterisks. Four Cys residues that are required for a canonical C4 Zn finger motif (54) are indicated by plus signs.
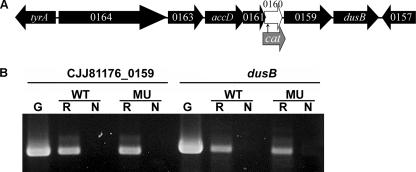
In C. jejuni 81-176, CJJ81176_0160 is located in a cluster containing seven genes, CJJ81176_0164 to dusB (Fig. 2A). There are four genes upstream of CJJ81176_0160 and two genes downstream. Among the six flanking genes, only accD (CJJ81176_0162) and dusB (CJJ81176_0158) are annotated, as genes encoding acetyl-coenzyme A carboxylase and tRNA dihydrouridine synthase B. The other genes encode proteins with unknown functions. RT-PCR generated an RT-PCR product spanning two adjacent open reading frames with all seven genes in the cluster (data not shown), suggesting that CJJ81176_0160 belongs to an operon composed of CJJ81176_0164 to dusB (CJJ81176_0158).
FIG. 2.
Operon structure of CJJ81176_0160 and flanking genes. (A) Genomic structure of CJJ81176_0160 and flanking genes and site of nonpolar insertion of a cat cassette. (B) RT-PCR results obtained with mRNA isolated from the wild type (WT) and the mutant (FMB1104) (MU), confirming the transcription of CJJ81176_0159 and dusB, which are downstream of CJJ81176_0160. G, PCR performed with genomic DNA of the wild type; R, PCR performed with reverse transcriptase and C. jejuni mRNA; N, PCR performed without reverse transcriptase but with C. jejuni mRNA (negative control).
To define the functions of the DksA-like protein (CJJ81176_160), we disrupted CJJ81176_0160 in C. jejuni 81-176. To minimize the polar effect of a cat insertion, the cat gene was cloned in the same orientation as CJJ81176_0160, which was confirmed by sequencing. RT-PCR demonstrated that the transcription of CJJ81176_0159 and dusB (CJJ81176_0158) was not affected (Fig. 2B). Also, qRT-PCR revealed that the levels of expression of CJJ81176_0161, CJJ81176_0159, and CJJ81176_0158 in C. jejuni FMB1104 are approximately 102, 74, and 82% of those in wild-type C. jejuni, respectively (data were not shown). Therefore, the insertion in CJJ81176_0160 did not have a polar effect on the downstream genes. In addition, the mutant did not show growth defects in rich medium (MH medium) compared with the wild type in 24 h (data not shown).
The best-known regulatory mechanism of DksA is regulation of the transcription of stable RNA, such as rRNA in E. coli (47, 53). In concert with ppGpp, DksA regulates rRNA transcription negatively by reducing the open complex lifetime and enhancing the effect of ppGpp (47). The effect of DksA on rRNA transcription has also been demonstrated in other bacteria, such as P. aeruginosa and S. flexneri (51, 56). Cessation of stable RNA synthesis results in a net decrease in the total RNA, because more than 90% of the total RNA is stable RNA. Total RNA was prepared from C. jejuni 81-176 and FMB1104 grown to the mid-log phase, and then the cultures were either maintained in rich medium (MH broth) or shifted to minimal medium (MOPS-MGS medium). The nutrient downshift from rich medium to minimal medium was reported previously to induce stringent responses by producing ppGpp (18). Viability (CFU) and RNA levels were monitored for 3 h under microaerobic conditions. As expected, neither strain of C. jejuni grew in the minimal medium, but there was no loss of viability for 3 h (data not shown). The total amount of RNA in the mutant (282.1 ± 85.9 fg/CFU) was similar to that in the wild type (244.1 ± 58.2 fg/CFU) in rich medium (P > 0.05). Following the nutrient downshift, the total amount of RNA was greatly decreased in the wild type (158.2 ± 33.7 fg/CFU), whereas the total amount of RNA was only slightly reduced in the mutant (241.4 ± 22.9 fg/CFU), which resulted in a 1.4-fold-higher level of total RNA in the mutant (P < 0.05). These results support the hypothesis that the DksA-like protein functions as a negative regulator of stable RNA transcription under stringent conditions, which is consistent with the findings for other bacteria (47, 51).
Mutation of the DksA-like protein affected the expression of multiple genes in C. jejuni.
The DksA-like protein was expected to be a transcriptional regulator in C. jejuni, as it is in other bacteria. Transcriptomic profiles of the wild type (C. jejuni 81-176) and the isogenic mutant lacking the DksA-like protein were compared by using a DNA microarray. Since a microarray for C. jejuni 81-176 was not commercially available, we used a commercial oligomer chip (OciChip; Ocimum Biosolutions, Indianapolis, IN) that was designed based on the genome sequence of C. jejuni NCTC 11168. Thus, the genetic differences between C. jejuni NCTC 11168 and C. jejuni 81-176 had to be taken into consideration when the results were interpreted, because the 38 genes unique to the C. jejuni 81-176 chromosome and the genes of two plasmids in C. jejuni 81-176, pVir and pTet, were not present in the microarray slide (5, 10, 24, 44). A total, of 51 of 1,633 genes on the array were determined to be differentially regulated by more than 1.5-fold (P < 0.05) based on the results of six microarray slides. Twenty-one genes were down-regulated, and 30 genes were up-regulated in the mutant. These genes are described elsewhere (http://plaza.snu.ac.kr./∼fmb/main/pu-01.htm).
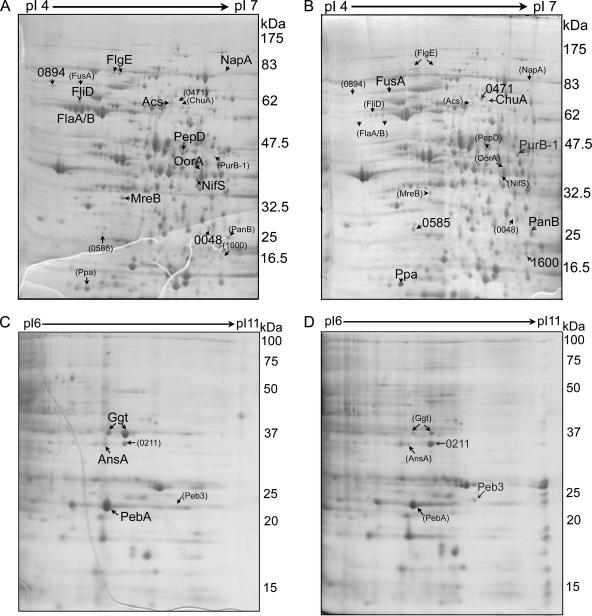
Profiles of protein expression for the wild type and the mutant were compared by using 2DGE. As shown in Fig. 3, many protein spots that showed differences in expression between the wild type and the mutant were detected, and protein spots with more-than-twofold differences were selected and identified by matrix-assisted laser desorption ionization-time of flight analyses after digestion with trypsin. Fourteen proteins were down-regulated in the mutant, whereas nine proteins were up-regulated in the mutant. The identified proteins are indicated in Fig. 3 and are described elsewhere (http://plaza.snu.ac.kr./∼fmb/main/pu-01.htm). The proteins showing differential expression in the wild type and the mutant identified by 2DGE did not agree with the results obtained from the transcriptome analyses. Correlations between DNA microarray and proteomic results are known to be highly variable, and inconsistency in findings between the two methods has been reported in many studies (16, 21). The discrepancy between the transcriptomic and proteomic data is ascribed to the intrinsic differences between these two methods and the biological differences between the transcription and translation processes, such as posttranscriptional regulation, posttranslational modification, and proteolysis (16, 21). Despite the discrepancy, genes or proteins involved in amino acid metabolism and iron-related metabolism were determined to be expressed differently in the wild type and the mutant by both methods. These genes and proteins are categorized and discussed below.
FIG. 3.
2DGE images of whole-cell lysates from the wild type (A and C) and the mutant lacking the DksA-like protein (B and D). Proteins were separated by 2DGE using two ranges, pH 4 to 7 (A and B) and pH 6 to 11 (C and D). Protein spots with increased expression (more than twofold greater) in one gel are indicated by large letters, while the corresponding spots in the other gel are indicated by small letters and numbers in parentheses. A full list of the identified proteins is given elsewhere.
The DksA-like protein is involved in amino acid metabolism in C. jejuni.
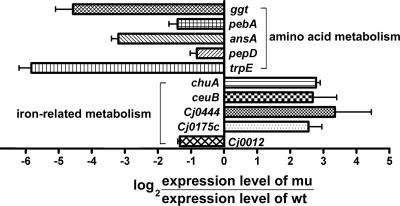
The expression of several genes involved in amino acid synthesis and transport was affected by mutation of the DksA-like protein in C. jejuni, which is consistent with findings for other bacteria (43, 52). The results of 2DGE demonstrated that there were reductions in the expression of Ggt (γ-glutamyltransferase [GGT]), PebA (periplasmic ABC transporter of amino acids), AnsA (cytoplasmic l-asparaginase), PepD (aminoacyl-histidine dipeptidase), and CJJ81176_0265 (Cj0240c, cysteine desulfurase). Also, the DNA microarray results revealed that the transcriptional levels of the trpABDEF (tryptophan synthesis) operon, pepA (leucyl amino transferase), and Cj1258 (CJJ81176_1274, possible phosphotyrosine protein phosphatase) were reduced in the mutant. The down-regulation of these genes was confirmed by real-time PCR (Fig. 4).
FIG. 4.
Confirmation of expression levels by qRT-PCR. The transcription levels of genes that exhibited differential expression in the wild type (wt) and the mutant lacking the DksA-like protein (mu) as determined by transcriptomic or proteomic analyses were confirmed by qRT-PCR. Negative values indicate down-regulation in the mutant, while positive values indicate up-regulation in the mutant.
Some of these genes have previously been reported to be involved in bacterial virulence. The GGT (EC.2.3.2.2) catalyzes transpeptidation and hydrolysis of the γ-glutamyl group of glutathione and related compounds and hence is known to be involved in glutathione metabolism in eukaryotes and prokaryotes (74). GGT is not present in C. jejuni NCTC 11168 but is present in C. jejuni 81-176 (24). Barnes et al. recently reported that the ggt gene is present in approximately 19.4% of C. jejuni isolates (8). Inactivation of ggt in C. jejuni reduced colonization in mice (24) and chickens (8). C. jejuni GGT also influenced the invasiveness of this bacterium and is involved in apoptosis of colon epithelial cells (8). Mutation of the DksA-like protein in C. jejuni also reduced the expression of PebA (or Peb1A as annotated in C. jejuni NCTC 11168). PebA is reported to be involved in amino acid transport as a component of an ABC transporter of aspartate and glutamate (34). PebA was originally identified as cell-binding factor 1, and inactivation of this protein in C. jejuni reduces interactions with epithelial cells and intestinal colonization of mice (49). Although other genes have not been functionally characterized in terms of amino acid metabolism, the multiple changes caused by mutation of the DksA-like protein strongly suggest that the DksA-like protein influences amino acid utilization and metabolism in C. jejuni.
The DksA-like protein is involved in iron-related metabolism in C. jejuni.
Like other bacteria, Campylobacter has an absolute requirement for iron as a nutrient. However, iron also participates in the formation of reactive oxygen species, and bacteria must maintain intracellular iron homeostasis to cope with oxidative stress (62). To achieve successful iron homeostasis, many bacteria have a Fur (ferric uptake regulator) system regulating ferric ion transport proteins (3). Fur with its corepressor Fe2+ binds to a consensus sequence in the promoters of Fur-regulated genes. Thus, the transcription of Fur-regulated genes is repressed when the Fe2+ concentration is high. C. jejuni possesses several mechanisms to obtain iron from the environment, although there is diversity in the iron uptake system among C. jejuni strains (24). C. jejuni 81-176 contains genes encoding FeoAB for ferrous iron uptake, ChuABCD for hemin/hemoglobin uptake, CeuBCDE for enterochelin transport, a putative iron uptake transport system (Cfbp, CJJ81176_0209 to CJJ81176_0211), and a TonB-dependent siderophore receptor (CJJ81176_0471) (24). Expression of all of these genes except feoAB is dependent on Fur (25, 45, 54). In C. jejuni, there is another Fur-like regulator, PerR (63). PerR is known to form a PerR-Fe2+ complex like Fur and to act as a repressor of genes involved in oxidative stress responses (63).
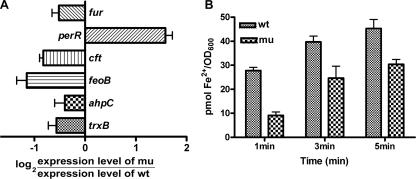
Transcriptomic and proteomic analyses showed that mutation of the DksA-like protein in C. jejuni caused changes in the expression levels of several genes involved in iron-related metabolism. Proteomic analysis showed that the expression of iron transporter proteins, including ChuA, CJJ81176_0211 (Cj0175c, ferric iron ABC transporter periplasmic iron-binding protein), and CJJ81176_0471 (Cj0444, putative TonB-dependent iron uptake protein) was higher in the mutant than in the wild type. ChuZ (heme oxygenase, CJJ81176_1600 or Cj0163c) also was up-regulated in the mutant and was reported to be a Fur regulon (54). Transcriptomic analysis also demonstrated that there was up-regulation of genes associated with iron uptake (Cj0175c and Cj0173c) and chuZ and down-regulation of Cj0012c (rbr) encoding a nonheme iron protein, which was reported to be activated upon iron addition (25, 45). Changes in the transcriptional levels of these iron-related genes were also confirmed by qRT-PCR (Fig. 4 and 5A). The expression levels of other proteins that may be important for iron-related metabolism were also investigated by qRT-PCR (Fig. 5A). The expression of fur was reduced approximately 50% in the mutant compared to the wild type. Interestingly, the expression of perR was increased in C. jejuni FMB1104, unlike the expression of fur (Fig. 5A). The expression levels of perR-regulated genes, such as ahpC and trxB, were reduced, which is consistent with the finding that perR expression was increased by the mutation (Fig. 5A).
FIG. 5.
Involvement of the DksA-like protein in iron-related metabolism of C. jejuni. (A) Comparison of transcription levels in the wild type (wt) and the mutant lacking the DksA-like protein (mu) for iron metabolism-related genes other than the Fur regulons. (B) Ferrous iron uptake was reduced in the mutant compared with the wild type. OD600, optical density at 600 nm.
The expression levels of a ferrous iron uptake gene (feoB) and the gene encoding ferritin (cft), an iron storage protein, were also down-regulated in the mutant. The down-regulation of feoB transcription in the mutant was verified by a ferrous iron uptake assay, in which less ferrous iron was taken up by the mutant with the DksA-like mutant protein than by the wild type (Fig. 5B). Based on all the findings, we concluded that mutation of the DksA-like protein causes down-regulation of feoAB, which results in a reduction in ferrous iron uptake and a concomitant decrease in the concentration of the Fur-Fe2+ complex. Therefore, Fur regulons are up-regulated in the mutant.
There has been only one report concerning the relationship between DksA and iron-related metabolism, and this report indicated that the transcription of fur in S. flexneri was reduced twofold in the absence of dksA (56). The relationship between iron-related metabolism and DksA in other bacteria has not been reported. Also, there have been no reports about ppGpp-dependent iron metabolism, except for one report for Geobacter sulfurreducens, which showed that genome-wide expression profiling of a ppGpp-negative mutant (relGsu deletion mutant) of G. sulfurreducens identified genes that possess Fur-regulated motifs (32). The ppGpp-negative strain of C. jejuni (the spoT mutant) did not show any changes in iron-related metabolism in C. jejuni (18). Thus, the DksA regulation of iron-related genes is likely mediated in a ppGpp-independent manner. However, it is not certain whether the regulation of iron-related metabolism by the DksA-like protein in C. jejuni results directly from the regulation by the DksA-like protein or indirectly from other changes in the mutant, such as changes in amino acid metabolism.
Other metabolic activities affected by the DksA-like protein.
Other genes exhibiting differential expression in the wild type and the mutant included genes involved in flagellar synthesis, N-linked protein glycosylation, energy metabolism, and small-molecule metabolism.
Proteomic analysis of mutations in the DksA-like protein also demonstrated that some flagellar genes were down-regulated in the mutant. These genes encode flagellar hook protein (FlgE), flagellar capping protein (FliD), flagellin family protein (CJJ81176_0894, FlaD), and flagellin (CJJ81176_1338 and CJJ81176_1339, FlaAB); however, the results of the DNA microarray analysis did not show any noticeable changes in flagellar genes. Also, some of the genes, such as wlaGH (pglAC), which are involved in N-linked protein glycosylation (60), showed up-regulation in C. jejuni FMB1104. Of the 30 putative glycoproteins (73), 5 (Cj0175c, PEB3, Cj0420, FusA, and Ppa) were up-regulated in C. jejuni FMB1104 according to the results of 2DGE and microarray analyses.
Some genes involved in energy metabolism showed differential expression in the wild type and C. jejuni FMB1104. In the mutant, putative cytochrome c genes (Cj0037c and Cj1163), napA (periplasmic nitrate reductase), and oorA (2-oxoglutarate ferredoxin oxidoreductase) were down-regulated, whereas the nuoD operon (NADH dehydrogenase) was up-regulated. Genes that are involved in small-molecule metabolism were also influenced by the mutation in the DksA-like protein. In the mutant, acs (acetyl-coenzyme A synthetase) was down-regulated, whereas panB (3-methyl-2-oxobutanoate hydroxymethyltransferase), ppa (inorganic pyrophosphatase), and purB (adenylosuccinate lyase) were up-regulated.
Invasion and IL-8 secretion were impaired by mutation of the DksA-like protein.
C. jejuni 81-176 is known to be a highly invasive strain among campylobacters. Human intestinal INT-407 cells were infected with C. jejuni 81-176 or the isogenic mutant lacking the DksA-like protein, and the total numbers of bacteria that invaded the INT-407 cells were determined. For the wild type the invasion level was 0.22% ± 0.03% of the input bacteria, while for the mutant the invasion level was 0.0018% ± 0.0006% of the input bacteria, which represents a 120-fold reduction in invasiveness. The differences were statistically significant (P < 0.001). Previous studies of the DksA protein of S. flexneri showed that DksA was involved in the intercellular spread of this bacterium but not in invasion (41).
The invasion of epithelial cells by C. jejuni is associated with its virulence in an animal model (71). Many factors have been reported to affect the invasiveness of C. jejuni. Motility is one factor associated with the invasion of host cells by C. jejuni (31, 70). Other factors affecting the invasiveness of C. jejuni include FlaC, Cia proteins, FspA, and Cj0977, and all of these proteins except Cj0977 require a flagellar export apparatus for secretion (19, 31, 52, 58). This is associated with the finding that mutation in the DksA-like protein of C. jejuni resulted in down-regulation of some flagellar genes at the protein level. In addition, the spoT gene of C. jejuni was up-regulated during bacterial infection of the INT-407 cells, and the spoT mutant of C. jejuni was defective in invasion and intracellular survival (18), suggesting that the stringent response of C. jejuni is associated with the invasiveness of this bacterium. Based on all the findings, the defect in invasiveness caused by mutation of the DksA-like protein in C. jejuni is likely associated with the stringent response.
Campylobacter enterocolitis is typically associated with a local acute inflammatory response, inducing inflammatory cytokine responses. IL-8 is a proinflammatory cytokine and a mediator of localized inflammatory responses. It has been reported that the invasiveness of C. jejuni is associated with IL-8 secretion (22). To determine if a mutation in the DksA-like protein influences IL-8 secretion, INT-407 cells were infected with C. jejuni 81-176 and FMB1104, and IL-8 secretion into the culture medium was determined. The level of IL-8 secretion induced by the wild type was 83.54 ± 7.61 pg/ml after 4 h of infection, while the level of IL-8 secretion induced by the mutant was 54.88 ± 3.91 pg/ml, which is approximately 65% of the wild-type value. The difference in IL-8 secretion was statistically significant (P < 0.05). The reduced secretion of IL-8 was likely due to the significant decrease in the invasiveness of the mutant. However, the reduction was modest considering the remarkable invasion defect, probably because the transcription of cytolethal distending toxin genes, another factor that affects IL-8 secretion (23), was not influenced by mutation of the DksA-like protein (data not shown).
Concluding remarks.
The DksA-like protein of C. jejuni was involved in expression of genes for amino acid metabolism, iron-related metabolism, and other metabolic activities. In addition, mutation of the DksA-like protein in C. jejuni resulted in reductions in invasiveness and IL-8 secretion. The results of this study suggest that the DksA-like protein plays a regulatory role in C. jejuni. The mechanism of these changes was not explained; this mechanism may be dependent on or independent of the stringent response and warrants further investigation.
Acknowledgments
We thank M. Blaser for providing C. jejuni 81-176 and P. Guerry for providing pRY112.
This work was supported by the Korea Research Foundation (grant KRF-2006-005-J04702) and by a National Research Initiative competitive grant (2007-35201-18278) from the USDA Cooperative State Research, Education, and Extension Service. J. Yun was a recipient of a graduate fellowship provided by the Ministry of Education through the Brain Korea 21 Project.
Footnotes
Published ahead of print on 2 May 2008.
REFERENCES
- 1.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397176-180. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27215-237. [DOI] [PubMed] [Google Scholar]
- 4.Artsimovitch, I., V. Patlan, S.-I. Sekine, M. N. Vassylyeva, T. Hosaka, K. Ochi, S. Yokoyama, and D. G. Vassylyev. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117299-310. [DOI] [PubMed] [Google Scholar]
- 5.Bacon, D. J., R. A. Alm, L. Hu, T. E. Hickey, C. P. Ewing, R. A. Batchelor, T. J. Trust, and P. Guerry. 2002. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176. Infect. Immun. 706242-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of PNAP mutants and competition for RNAP. J. Mol. Biol. 305689-702. [DOI] [PubMed] [Google Scholar]
- 7.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305673-688. [DOI] [PubMed] [Google Scholar]
- 8.Barnes, I. H. A., M. C. Bagnall, D. D. Browning, S. A. Thompson, G. Manning, and D. G. Newell. 2007. γ-Glutamyl transpeptidase has a role in the persistent colonization of the avian gut by Campylobacter jejuni. Microb. Pathog. 43198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass, S., Q. Gu, and A. Christen. 1996. Multicopy suppressors of prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated R1pA. J. Bacteriol. 1781154-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batchelor, R. A., B. M. Pearson, L. M. Friis, P. Guerry, and J. M. Wells. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 1503507-3517. [DOI] [PubMed] [Google Scholar]
- 11.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobater jejuni infection in humans. J. Infect. Dis. 157472-479. [DOI] [PubMed] [Google Scholar]
- 12.Bollag, D. M., and S. J. Edelstein. 1991. Protein methods. Wiley-Liss, Inc., New York, NY.
- 13.Branny, P., J. P. Pearson, E. C. Pesci, T. Kohler, B. H. Iglewski, and C. Van Delden. 2001. Inhibition of quorum sensing by a Pseudomonas aeruginosa dksA homologue. J. Bacteriol. 1831531-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown, L., D. Gentry, T. Elliott, and M. Cashel. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 1844455-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichi coli and Salmonella: cellular and molecular biology, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 16.Cook, S. A., and A. Rosenzweig. 2002. DNA microarrays: implications for cardiovascular medicine. Circ. Res. 91559-564. [DOI] [PubMed] [Google Scholar]
- 17.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, DC.
- 18.Gaynor, E. C., D. H. Wells, J. K. MacKichan, and S. Falkow. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 568-27. [DOI] [PubMed] [Google Scholar]
- 19.Goon, S., C. P. Ewing, M. Lorenzo, D. Pattarini, G. Majam, and P. Guerry. 2006. A σ28-regulated nonflagella gene contributes to virulence of Campylobacter jejuni 81-176. Infect. Immun. 74769-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haugen, S. P., M. B. Berkmen, W. Ross, T. Gaal, C. Ward, and R. L. Gourse. 2006. rRNA promoter regulation by nonoptimal binding of σ region 1.2: an additional recognition element for RNA polymerase. Cell 1251069-1082. [DOI] [PubMed] [Google Scholar]
- 21.Hegde, P. S., I. R. White, and C. Debouck. 2003. Interplay of transcriptomics and proteomics. Curr. Opin. Biotechnol. 14647-651. [DOI] [PubMed] [Google Scholar]
- 22.Hickey, T. E., S. Baqar, A. L. Bourgeois, C. P. Ewing, and P. Guerry. 1999. Campylobacter jejuni-stimulated secretion of interleukin-8 by INT407 cells. Infect. Immun. 6788-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hickey, T. E., A. L. McVeigh, D. A. Scott, R. E. Michielutti, A. Bixby, S. A. Carroll, A. L. Bourgeois, and P. Guerry. 2000. Campylobacter jejuni cytolethal distending toxin mediates release of interleukin-8 from intestinal epithelial cells. Infect. Immun. 686535-6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofreuter, D., J. Tsai, R. O. Watson, V. Novik, B. Altman, M. Benitez, C. Clark, C. Perbost, T. Jarvie, L. Du, and J. E. Galan. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 744694-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes, K., F. Mulholland, B. M. Pearson, C. Pin, J. McNicholl-Kennedy, J. M. Ketley, and J. M. Wells. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151243-257. [DOI] [PubMed] [Google Scholar]
- 26.Ishihama, A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 38499-518. [DOI] [PubMed] [Google Scholar]
- 27.Ishii, Y., H. Yamada, T. Yamashino, K. Ohashi, E. Katoh, H. Shindo, T. Yamazaki, and T. Mizuno. 2000. Deletion of the yhhP gene results in filamentous cell morphology in Escherichia coli. Biosci. Biotechnol. Biochem. 64799-807. [DOI] [PubMed] [Google Scholar]
- 28.Jensen, O. N., M. Wilm, A. Shevchenko, and M. Mann. 1999. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol. Biol. 112513-530. [DOI] [PubMed] [Google Scholar]
- 29.Jude, F., T. Kohler, P. Branny, K. Perron, M. P. Mayer, R. Comte, and C. van Delden. 2003. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J. Bacteriol. 1853558-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang, P. J., and E. A. Craig. 1990. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 1722055-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konkel, M. E., J. D. Klena, V. Rivera-Amill, M. R. Monteville, D. Biswas, B. Raphael, and J. Mickelson. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 1863296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krushkal, J., B. Yan, L. N. DiDonato, M. Puljic, K. P. Nevin, T. L. Woodard, R. M. Adkins, B. A. Methe, and D. R. Lovley. 2007. Genome-wide expression profiling in Geobacter sulfurreducens: identification of Fur and RpoS transcription regulatory sites in a relGsu mutant. Funct. Integr. Genomics 7229-255. [DOI] [PubMed] [Google Scholar]
- 33.Lara-Tejero, M., and J. E. Galan. 2000. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290354-357. [DOI] [PubMed] [Google Scholar]
- 34.Leon-Kempis Mdel, R., E. Guccione, F. Mulholland, M. P. Williamson, and D. J. Kelly. 2006. The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 601262-1275. [DOI] [PubMed] [Google Scholar]
- 35.Leung, Y. F., and D. Cavalieri. 2003. Fundamentals of cDNA microarray data analysis. Trends Genet. 19649-659. [DOI] [PubMed] [Google Scholar]
-
36.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real time quantitative PCR and the
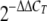 method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
method. Methods 25402-408. [DOI] [PubMed] [Google Scholar] - 37.Madsen, M. L., D. Nettleton, E. L. Thacker, R. Edwards, and F. C. Minion. 2006. Transcriptional profiling of Mycoplasma hyopneumoniae during heat shock using microarrays. Infect. Immun. 74160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnusson, L. U., A. Farewell, and T. Nystrom. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13236-242. [DOI] [PubMed] [Google Scholar]
- 39.Magnusson, L. U., B. Gummesson, P. Joksimovic, A. Farewell, and T. Nystrom. 2007. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol. 1895193-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mallik, P., B. J. Paul, S. T. Rutherford, R. L. Gourse, and R. Osuna. 2006. DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J. Bacteriol. 1885775-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mogull, S. A., L. J. Runyen-Janecky, M. Hong, and S. M. Payne. 2001. dksA is required for intercellular spread of Shigella flexneri via an RpoS-independent mechanism. Infect. Immun. 695742-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naikare, H., K. Palyada, R. Panciera, D. Marlow, and A. Stintzi. 2006. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 745433-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakanishi, N., H. Abe, Y. Ogura, T. Hayashi, K. Tashiro, S. Kuhara, N. Sugimoto, and T. Tobe. 2006. ppGpp with DksA controls gene expression in locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 61194-205. [DOI] [PubMed] [Google Scholar]
- 44.Okuda, J., M. Fukumoto, Y. Takeda, and M. Nishibuchi. 1997. Examination of diarrheagenicity of cytolethal distending toxin: suckling mouse response to the products of the cdtABC genes of Shigella dysenteriae. Infect. Immun. 65428-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 1864714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403665-668. [DOI] [PubMed] [Google Scholar]
- 47.Paul, B. J., M. M. Barker, W. Ross, D. A. Schneider, C. Webb, J. W. Foster, and R. L. Gourse. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118311-322. [DOI] [PubMed] [Google Scholar]
- 48.Paul, B. J., M. B. Berkmen, and R. L. Gourse. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. USA 1027823-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pei, Z., C. Burucoa, B. Grignon, S. Baqar, X.-Z. Huang, D. J. Kopecko, A. L. Bourgeois, J.-L. Fauchere, and M. J. Blaser. 1998. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66938-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perederina, A., V. Svetlov, M. N. Vassylyeva, T. H. Tahirov, S. Yokoyama, I. Artsimonitch, and D. G. Vassylyev. 2004. Regulation through the secondary channel-structural framework for ppGpp-DksA synergism during transcription. Cell 118297-309. [DOI] [PubMed] [Google Scholar]
- 51.Perron, K., R. Comte, and C. van Delden. 2005. DksA represses ribosomal gene transcription in Pseudomonas aeruginosa by interacting with RNA polymerase on ribosomal promoters. Mol. Microbiol. 561087-1102. [DOI] [PubMed] [Google Scholar]
- 52.Poly, F., C. Ewing, S. Goon, T. E. Hickey, D. Rockabrand, G. Majam, L. Lee, J. Phan, N. J. Savarino, and P. Guerry. 2007. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect. Immun. 753859-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potrykus, K., D. Vinella, H. Murphy, A. Szalewska-Palasz, R. D'Ari, and M. Cashel. 2006. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J. Biol. Chem. 28115238-15248. [DOI] [PubMed] [Google Scholar]
- 54.Ridley, K. A., J. D. Rock, Y. Li, and J. M. Ketley. 2006. Heme utilization in Campylobacter jejuni. J. Bacteriol. 1887862-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook, J., and D. W. Russel. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 56.Sharma, A. K., and S. M. Payne. 2006. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol. Microbiol. 62469-479. [DOI] [PubMed] [Google Scholar]
- 57.Snelling, W. J., M. Matsuda, J. E. Moore, and J. S. G. Dooley. 2005. Campylobacter jejuni. Lett. Appl. Microbiol. 41297-302. [DOI] [PubMed] [Google Scholar]
- 58.Song, M., H.-J. Kim, E. Y. Kim, M. Shin, H. C. Lee, Y. Hong, J. H. Rhee, H. Yoon, S. Ryu, S. Lim, and H. E. Choy. 2004. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J. Biol. Chem. 27934183-34190. [DOI] [PubMed] [Google Scholar]
- 59.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 1009440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3225-237. [DOI] [PubMed] [Google Scholar]
- 61.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388539-547. [DOI] [PubMed] [Google Scholar]
- 62.van Vliet, A. H., J. M. Ketley, S. F. Park, and C. W. Penn. 2002. The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26173-186. [DOI] [PubMed] [Google Scholar]
- 63.van Vliet, A. H. M., M.-L. A. Baillon, C. W. Penn, and J. M. Ketley. 1999. Campylobacter jejuni contains two Fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 1816371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wassenaar, T. M., and M. J. Blaser. 1999. Pathophysiology of Campylobacter jejuni infections of humans. Microbes Infect. 11023-1033. [DOI] [PubMed] [Google Scholar]
- 65.Webb, C., M. Moreno, M. Wilmes-Riesenberg, R. Curtiss III, and J. W. Foster. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 34112-123. [DOI] [PubMed] [Google Scholar]
- 66.Wei, P., and C. R. Stewart. 1995. Genes that protect against the host-killing activity of the E3 protein of Bacillus subtilis bacteriophage SPO1. J. Bacteriol. 1772933-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comp. Biol. 8625-637. [DOI] [PubMed] [Google Scholar]
- 68.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method for addressing single and multiple slide systematic variation. Nucleic Acids Res. 30e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130127-130. [DOI] [PubMed] [Google Scholar]
- 70.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14883-893. [DOI] [PubMed] [Google Scholar]
- 71.Yao, R., D. H. Burr, and P. Guerry. 1997. CheY-mediated modulation of Campylobacter jejuni virulence. Mol. Microbiol. 231021-1031. [DOI] [PubMed] [Google Scholar]
- 72.Yoon, H., S. Lim, S. Heu, S. Choi, and S. Ryu. 2003. Proteome analysis of Salmonella enterica serovar Typhimurium fis mutant. FEMS Microbiol. Lett. 226391-396. [DOI] [PubMed] [Google Scholar]
- 73.Young, N. M., J.-R. Brisson, J. Kelly, D. C. Watson, L. Tessier, P. H. Lanthier, H. C. Jarrell, N. Cadotte, F. St. Michael, E. Aberg, and C. M. Szymanski. 2002. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 27742530-42539. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, H., H. J. Forman, and J. Choi. 2005. γ-Glutamyl transpeptidase in glutahione biosynthesis. Methods Enzymol. 401468-483. [DOI] [PubMed] [Google Scholar]