Abstract
Despite the fact that heliobacteria are the only phototrophic representatives of the bacterial phylum Firmicutes, genomic analyses of these organisms have yet to be reported. Here we describe the complete sequence and analysis of the genome of Heliobacterium modesticaldum, a thermophilic species belonging to this unique group of phototrophs. The genome is a single 3.1-Mb circular chromosome containing 3,138 open reading frames. As suspected from physiological studies of heliobacteria that have failed to show photoautotrophic growth, genes encoding enzymes for known autotrophic pathways in other phototrophic organisms, including ribulose bisphosphate carboxylase (Calvin cycle), citrate lyase (reverse citric acid cycle), and malyl coenzyme A lyase (3-hydroxypropionate pathway), are not present in the H. modesticaldum genome. Thus, heliobacteria appear to be the only known anaerobic anoxygenic phototrophs that are not capable of autotrophy. Although for some cellular activities, such as nitrogen fixation, there is a full complement of genes in H. modesticaldum, other processes, including carbon metabolism and endosporulation, are more genetically streamlined than they are in most other low-G+C gram-positive bacteria. Moreover, several genes encoding photosynthetic functions in phototrophic purple bacteria are not present in the heliobacteria. In contrast to the nutritional flexibility of many anoxygenic phototrophs, the complete genome sequence of H. modesticaldum reveals an organism with a notable degree of metabolic specialization and genomic reduction.
Heliobacteria are photosynthetic bacteria that uniquely employ bacteriochlorophyll (Bchl) g as the major antenna pigment and primary electron donor within a type I reaction center (RC) (3, 55). Bchl g is related to chlorophyll (Chl) a but has an ethylidine functional group at the C-81 position and is esterified with farnesol rather than phytol (33). An oxidized form of Chl a, 81-hydroxy-Chl a, is the primary electron acceptor from the RC special pair (3, 56). Unlike other anoxygenic phototrophic bacteria, heliobacteria have no Bchl-containing internal membranes or structures, such as lamellae (purple bacteria) or chlorosomes (green bacteria). In the heliobacteria, photosynthetic pigments are confined to RCs in the cytoplasmic membrane (16, 37). Carotenoids are also unusual in heliobacteria in that they consist of C30 pigments rather than the C40 derivatives present in other phototrophs (53). The dominant carotenoid in nonalkaliphilic heliobacteria is 4,4′-diaponeurosporene (52).
In addition to their unique photosynthetic properties, heliobacteria can be distinguished from all other anaerobic anoxygenic phototrophs in at least three major ways. In terms of carbon metabolism, heliobacteria are obligately heterotrophic. Growth occurs either photoheterotrophically (anoxic, with light) on a limited range of organic substrates or by fermentation of pyruvate in the dark (22, 34). By contrast, autotrophic growth, the hallmark of photosynthetic organisms, has not been observed with cultures of any heliobacterium species (34). Heliobacteria are phylogenetically unique, as they are the only phototrophic organisms that group within the bacterial phylum Firmicutes using 16S rRNA gene sequence analyses (8, 32). Finally, heliobacteria are unique among all phototrophs in that they produce endospores (24), a key property of nonphototrophic Firmicutes, such as Bacillus and Clostridium.
A complete genome sequence analysis of Heliobacterium modesticaldum, the first heliobacterial genome to be sequenced, reveals an organism with a full complement of nitrogen fixation genes but only a limited capacity for carbon metabolism and no apparent mechanism for autotrophic growth. Many genes linked to endosporulation in Bacillus subtilis were not found in H. modesticaldum, which may have relevance for the ambiguous sporulation patterns observed in heliobacterial cultures (24). These results describe H. modesticaldum strain Ice1, the type strain of this species isolated from Icelandic hot spring volcanic soils (23).
MATERIALS AND METHODS
Genome sequencing.
H. modesticaldum strain Ice1T genomic DNA was fragmented randomly by kinetic shearing, and two shotgun libraries were constructed: a small insert library in plasmid pOTWI3 (using 3- to 4-kb size fractions) and a large insert fosmid library in pEpiFOS-5 (with insert sizes ranging from 28 to 47 kb), which was used as a scaffold. The relative amounts of sequence coverage obtained from the small and large insert libraries were approximately 11× and 2×, respectively. The whole genome sequence was established from 51,795 end sequences derived from these libraries using dye terminator chemistry with Applied Biosystems 3730xl automated sequencers. The sequence was assembled with the program ARACHNE (5) and finished as described previously (51).
Annotation.
Initial automated annotation of the genome was performed with the TIGR/JCVI annotation engine (www.tigr.org/AnnotationEngine), and it was processed by The Institute for Genome Research's prokaryotic annotation pipeline using gene finding with Glimmer, SignalP predictions, and BLAST extend-repraze (BER), HMM, and TMHMM searches. Automatic annotations were created using AutoAnnotate. Manatee (manatee.sourceforge.net) was used to manually review and confirm the annotation of all genes. Pseudogenes contained one or more mutations that could ablate expression; each inactivating mutation was subsequently checked against the original sequencing data. A circular genome map was created using the program CGView (50).
Phylogenetic analyses.
Sequence identity analyses were performed using the Basic Local Alignment Search Tool (BLAST) (2). Gene sequences were aligned and phylogenetic trees were constructed using MEGA, version 3.1 (28). Bootstrap values were determined using 500 replicates. Sequences used in phylogenetic analyses were obtained from NCBI genome databases (http://www.ncbi.nlm.nih.gov/).
Nucleotide sequence accession number.
The complete and annotated sequence of the genome of H. modesticaldum strain Ice1T has been deposited in the DDBJ/EMBL/GenBank database under accession number CP000930.
RESULTS AND DISCUSSION
Genome properties.
H. modesticaldum strain Ice1T has a single 3,075,407-bp circular chromosome containing 3,138 open reading frames (ORFs), eight pseudogenes, and no plasmids (Table 1). The genomic G+C content of H. modesticaldum, 56.0%, is at the upper end of the range for heliobacteria (33) (Table 1 and Fig. 1). The total protein-encoding content of the chromosome is 87%, and the average gene length is 882 nucleotides. Eight rRNA operons are randomly dispersed throughout the chromosome. Each operon contains one copy each of 5S, 16S, and 23S rRNA genes, giving a total of 24 rRNA genes. This number is high but consistent with the numbers for other endospore-forming Firmicutes (25). A summary of the number and percentage of genes in each primary category is shown in Table 2.
TABLE 1.
Features of the H. modesticaldum strain Ice1T (= ATCC 51547T) genome
| Characteristic | Value |
|---|---|
| Chromosome size (bp) | 3,075,407 |
| G+C content (%) | 56.0 |
| % Genome coding | 87 |
| Total no. of ORFs | 3,138 |
| Avg ORF length (bp) | 882 |
| % ATG initiation codons | 62.1 |
| % GTG initiation codons | 19.1 |
| % TTG initiation codons | 18.8 |
| No. of rRNAs (no. of genes/no. of operons) | 24/8 |
| No. of tRNAs | 104 |
| No. of structural RNAs | 1 |
| No. of tmRNA | 1 |
| % Conserved hypothetical proteins | 11.1 |
| % Hypothetical proteins | 23.8 |
| No. of transposases | 70 |
| No. of pseudogenes | 8 |
FIG. 1.
Circular genome map of the 3.1-Mb H. modesticaldum chromosome. The rings indicate (from outside to inside) all the genes and insertion elements, color coded by functional category (rings 1 and 2), the deviation from the average G+C content (ring 3), and the GC skew (ring 4). The approximate location of the origin of replication is at the beginning of the dnaA gene. The colors indicate the following: turquoise, small-molecule biosynthesis; yellow, central or intermediary metabolism; orange, energy metabolism; red, signal transduction; light blue, DNA metabolism; blue, transcription; purple, protein synthesis/fate; dark green, surface-associated features; gray, miscellaneous features; pink, phage and insertion elements; light green, unknown function; dark gray, conserved hypothetical proteins; black, hypothetical proteins; brown, pseudogenes.
TABLE 2.
Characterization of selected gene categories of the H. modesticaldum strain Ice1T (=ATCC 51547T) genome
| Category | No. of genes | % of genome content |
|---|---|---|
| Energy and central intermediary metabolism | 389 | 13.0 |
| Amino acid biosynthesis | 113 | 3.8 |
| Transport | 157 | 5.2 |
| Cofactor and prosthetic group biosynthesis | 152 | 5.1 |
| DNA metabolism | 135 | 4.5 |
| Transcription | 44 | 1.5 |
| Protein synthesis, modification, and degradation | 247 | 8.2 |
| Regulatory functions and signal transduction | 177 | 5.9 |
| Cellular processes (cell division, motility, sporulation, etc.) | 273 | 9.1 |
| Fatty acid and phospholipid metabolism | 45 | 1.5 |
| Phage/insertion elements | 123 | 4.1 |
| Surface features | 145 | 4.8 |
An unusual feature of the H. modesticaldum genome is the prevalence of gene strand bias, where large numbers of genes cluster predominately on one strand or the other throughout the chromosome (Fig. 1). One strand of the chromosome contains approximately two-thirds of the protein coding sequences (CDSs) (Fig. 1, outer ring), while the other strand contains the other one-third. This degree of strand asymmetry of coding sequences is unusual in bacteria and, to our knowledge, is equaled only by the close phylogenetic but not phototrophic relative Desulfitobacterium hafniense Y51 among completely sequenced prokaryotes having a circular chromosome (40).
CDSs encoding conserved hypothetical proteins make up approximately 11% of the total CDSs, and an additional 1.7% of the ORFs contain domain regions having sequence identity to known motifs. The genome contains 70 ORFs that show significant sequence identity to genes that encode transposases. Although this number is high compared to the numbers for many other completely sequenced Firmicutes, it is comparable to the 71 (2 confirmed and 69 putative) transposase genes found in D. hafniense Y51 (40) and the 60 transposase genes of Desulfotomaculum reducens MI-1 (U.S. DOE Joint Genome Institute, NCBI genome database); these two strains are nonphototrophic gram-positive bacteria that are close phylogenetic relatives of Heliobacterium spp. (48).
Most of the 20 common aminoacyl-tRNAs can be synthesized directly in H. modesticaldum; the only exception is asparaginyl-tRNA, which cannot be synthesized due to the apparent absence of a gene encoding asparaginyl-tRNA synthetase. An alternative pathway that could allow asparaginyl-tRNA synthesis involves aspartyl/glutamyl-tRNA amidotransferase, a heterotrimeric enzyme that can catalyze the conversion of aspartyl-tRNA to asparaginyl-tRNA in Pseudomonas aeruginosa (1). Although this function is more commonly observed in Archaea (12), genes predicted to encode all three subunits of aspartyl-tRNA amidotransferase (gatABC) are present in H. modesticaldum. These genes may have functions similar to those of P. aeruginosa genes for synthesis of a full complement of aminoacyl-tRNAs. In addition, the H. modesticaldum genome contains the selABCD genes, which allow the synthesis and incorporation of selenocysteine into proteins.
Carbon metabolism.
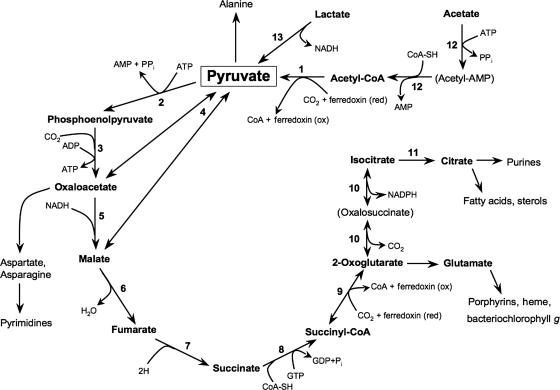
Genomic analyses of H. modesticaldum supported the restricted capacity for organic carbon photoassimilation previously observed in physiological studies (23). Although photoheterotrophic growth using fatty acids, such as butyrate and propionate, has been observed for some species of heliobacteria (4, 6, 9, 43), H. modesticaldum is limited to photoassimilation of pyruvate, lactate, or acetate (23). Growth of Heliobacterium gestii, but not growth of H. modesticaldum, is supported by ethanol plus CO2 (43). Although three genes predicted to encode alcohol dehydrogenase are present in H. modesticaldum, a lack of peripheral enzymes, such as NAD-dependent aldehyde dehydrogenase, may preclude the catabolism of alcohols. Glucose and fructose utilization has also been reported in H. gestii (33). However, despite the presence of genes predicted to encode a four-subunit monosaccharide-transporting ATPase (rbsABCD) in the genome, sugars did not support growth of H. modesticaldum (23). Genes corresponding to complete glycolytic and nonoxidative pentose phosphate pathways were identified (data not shown), so it remains unclear why monosaccharides are unsuitable growth substrates for H. modesticaldum.
Pyruvate supports both photoheterotrophic and fermentative (dark anoxic) growth of heliobacteria (33). Consistent with biochemical studies carried out by Pickett et al. (44), no gene encoding pyruvate dehydrogenase was found in the H. modesticaldum genome. However, the oxidation of pyruvate to acetyl coenzyme A (acetyl-CoA) can alternatively be catalyzed by a predicted pyruvate:ferredoxin oxidoreductase (HM1_0807). This enzyme showed 65 to 70% amino acid sequence identity to the pyruvate:ferredoxin oxidoreductase of closely related, nonphototrophic Firmicutes and 83% sequence identity to the pyruvate:ferredoxin oxidoreductase of Heliobacillus mobilis. During photoheterotrophic growth, the oxidation of lactate to pyruvate (and the concomitant reduction of NAD+ to NADH) is catalyzed by a putative lactate dehydrogenase (HM1_2756) (Fig. 2). Acetate is presumably photometabolized through an acetyl adenylate intermediate to acetyl-CoA via an AMP-forming acetyl-CoA synthetase (HM1_0951) (Fig. 2). A gene predicted to encode acetate kinase (HM1_2157), which phosphorylates acetate to acetyl-P, was also found in the genome.
FIG. 2.
Putative pathway of carbon metabolism in H. modesticaldum (adapted from reference 44). A partial reverse citric acid cycle with CO2 incorporation via PEP carboxykinase is shown. The enzymes involved in the reduction of acetate to pyruvate putatively function in an oxidative direction during chemotrophic (dark) growth on pyruvate. The oxidation of pyruvate to acetyl-CoA is likely accompanied by hydrogen evolution via an [FeFe] hydrogenase. The numbers indicate the following enzymes: 1, pyruvate:ferredoxin oxidoreductase; 2, pyruvate-phosphate dikinase; 3, PEP carboxykinase; 4, oxaloacetate decarboxylase; 5, malate dehydrogenase; 6, fumarase; 7, fumarate reductase; 8, succinyl-CoA synthetase; 9, 2-oxoglutarate:ferredoxin oxidoreductase; 10, NADP-dependent isocitrate dehydrogenase; 11, aconitate hydratase; 12, AMP-forming acetyl-CoA synthetase; 13, lactate dehydrogenase.
The mechanism of pyruvate fermentation in H. modesticaldum is complicated by the lack of a gene encoding phosphotransacetylase, which converts acetyl-CoA to acetyl-phosphate. Either a highly unusual phosphotransacetylase is present, or there is an alternative mechanism of pyruvate fermentation in H. modesticaldum. One possibility for this is reverse activity of enzymes involved in the photoassimilation of acetate (reactions 1 and 12 in Fig. 2). In this scenario, CO2 and reduced ferredoxin are products of the oxidation of pyruvate to acetyl-CoA. The production of ATP would occur with the conversion of acetyl-adenylate (from acetyl-CoA) to acetate via acetyl-CoA synthetase (Fig. 2), an enzyme known to have reversible activity (57).
Genes encoding a putative [FeFe] hydrogenase that could oxidize ferredoxin with the production of H2 were identified in H. modesticaldum and are shown schematically in Fig. 3A. These genes, nuoEF (HM1_1028) and nuoG (hydA; HM1_1029), showed levels of sequence identity of 79 and 81%, respectively, to genes of Heliobacillus mobilis. The arrangement of nuoE and nuoF varies among Firmicutes containing an [FeFe] hydrogenase. In both H. modesticaldum and Heliobacillus mobilis, these genes are fused to produce a single transcript, but in some other Firmicutes, such as Pelotomaculum thermopropionicum SI, the genes are separate (30) (Fig. 3A). The fusion of nuoE and nuoF does not occur exclusively in heliobacteria, however, since these genes are also fused in Symbiobacterium thermophilum, an uncultivated commensal bacterium phylogenetically related to Firmicutes (30).
FIG. 3.
Schematic representation of hydrogenase and photosynthesis gene clusters in H. modesticaldum. Arrows represent individual genes and indicate the direction of transcription. (A) Putative [FeFe] hydrogenase genes in H. modesticaldum and the close phylogenetic relative P. thermopropionicum SI. As in Heliobacillus mobilis, nuoE and nuoF are fused in H. modesticaldum, suggesting that this feature may be universal in heliobacteria. Colors indicate the following: gold, NADH dehydrogenase subunits; orange, [FeFe] hydrogenase structural genes. (B) Uptake [NiFe] hydrogenase genes in related Firmicutes. The genes are located in a single operon in H. modesticaldum, whereas they are dispersed in different regions of the D. hafniense Y51 chromosome. Colors indicate the following: blue, [NiFe] hydrogenase structural genes; purple, hydrogenase expression/formation; red, hydrogenase assembly/maturation. (C) Photosynthesis gene clusters from H. modesticaldum and the purple bacterium R. capsulatus. Shared genes are outlined with bold lines. Lines indicate gene synteny, as follows: black lines, single gene rearrangements; red lines, inverted genes; blue lines, inverted genes with a gene insertion. Dashed boxes indicate R. capsulatus photosynthesis genes absent from H. modesticaldum. The colors of the arrows indicate the following: green, Bchl biosynthesis (bch); orange, carotenoid biosynthesis (crt); pink, proteobacterial reaction centers (puf) and light harvesting complexes (puh); olive, heliobacterial reaction center (psh); teal, regulatory proteins; light green, electron transport (pet); red, cofactor biosynthesis; purple, cell division and sporulation; light blue, nitrogen fixation; gray, transcription; light gray, other nonphotosynthetic genes; white, uncharacterized genes.
No mechanism for autotrophic growth has been identified in any heliobacterial species, in contrast to all other anaerobic anoxygenic phototrophs (18). Consistent with these physiological observations, genes encoding essential enzymes of all known autotrophic pathways were not found in the H. modesticaldum genome, including genes encoding ribulose 1,5-bisphosphate carboxylase and phosphoribulokinase (Calvin cycle), citrate lyase (reverse citric acid cycle), carbon monoxide dehydrogenase (reductive acetyl-CoA pathway), and malyl-CoA lyase (3-hydroxypropionate pathway). Moreover, there is no evidence for a gene encoding 4-hydroxybutyryl-CoA dehydratase, a key enzyme of the recently described 3-hydroxypropionate/4-hydroxybutyrate pathway of carbon fixation (7, 54). The absence of citrate lyase may be the key factor distinguishing the carbon metabolism of heliobacteria from that of green sulfur bacteria, autotrophic phototrophs that fix CO2 via the reverse citric acid cycle. In addition, no gene encoding citrate synthase was found in the genome. Thus, H. modesticaldum appears to have an incomplete citric acid cycle, which is typical of organisms using this cycle for biosynthesis.
A putative anapleurotic carbon fixation pathway for heliobacteria, in which CO2 assimilation occurs through phosphoenolpyruvate (PEP) carboxylase activity, was proposed by Pickett et al. (44) and recently examined again by Heinnickel and Golbeck (18). A similar mechanism was proposed as a means of mixotrophic growth in the aerobic phototrophic proteobacterium Roseobacter denitrificans (51). The enzymes needed to carry out these transformations have been identified in the H. modesticaldum genome, and thus a small amount of nonautotrophic CO2 assimilation likely takes place. A summary of the proposed pathways of carbon metabolism in H. modesticaldum is shown in Fig. 2.
Nitrogen fixation.
H. modesticaldum is an active dinitrogen fixer, and this species and the green sulfur bacterium Chlorobaculum tepidum are the only cultured anoxygenic phototrophs that are capable of N2 fixation at temperatures above 50°C (23, 31). The organization of the nif regulon in H. modesticaldum is identical to that in Heliobacterium chlorum (15). The nif regulon consists of 11 genes: nifI1, nifI2, nifH, nifD, nifK, nifE, nifN, nifX, fdxB, nifB, and nifV. In a phylogenetic analysis using a concatenated alignment of the nitrogenase structural genes nifHDK, H. modesticaldum grouped within a clade containing both a closely related species belonging to the Firmicutes (D. hafniense) and a distantly related deltaproteobacterium (Geobacter sulfurreducens) (Fig. 4). The high level of similarity of heliobacterial nifHDK genes to the genes of Geobacter was also noted in phylogenetic analyses using nif genes from H. chlorum (15) and suggests that either these genes had a common ancestor or there was a lateral transfer event.
FIG. 4.
Phylogenetic tree showing the relationship of concatenated bchXYZ, bchLNB, and nifHDK genes from H. modesticaldum to those of other organisms containing these genes. Although not present in H. modesticaldum, alternative nitrogenase genes, anfHDK and vnfHDK, are also included to balance the tree. Organisms containing more than one set of genes used in the comparison appear multiple times in the tree. The following organisms were included in the analysis: Anabaena siamensis strain TISTR8012, Azospirillum brasilense, Azotobacter vinelandii, Bradyrhizobium sp. strain BTAi1, Bradyrhizobium sp. strain ORS278, Chlorobaculum tepidum, Chloroflexus aurantiacus, Clostridium acetobutylicum, Clostridium pasteurianum, Clostridium kluyveri, Desulfitobacterium hafniense strain Y51, Geobacter sulfurreducens, Heliobacterium modesticaldum, Jannaschia sp. strain CCS1, Klebsiella pneumoniae, Methanothermobacter thermoautotrophicus, Methanococcus maripaludis, Methanosarcina acetivorans strain C2A, Nostoc sp. strain PCC7120, Prosthecochloris aestuarii, Rhodobacter capsulatus, Rhodopseudomonas palustris, Roseiflexus castenholzii, Roseobacter denitrificans, Synechococcus elongatus strain PCC6301, and Synechocystis sp. strain PCC6803.
Although some heliobacteria contain an alternative iron (Fe)-only group III type nitrogenase (21, 29), H. modesticaldum contains a molybdenum-dependent, group I nitrogenase (45). The products of nifI1 and nifI2, which encode proteins belonging to the PII signal transduction family, are located between the nifH and nifD products in group II and III nitrogenases, but they are not present in most group I nitrogenases (15, 45). The presence of nifI1 and nifI2 upstream of nifH in H. modesticaldum, as well as in other species of Heliobacterium and in D. hafniense Y51, suggests that the nitrogenase of these organisms may be an evolutionary intermediate between group I and group II/III nitrogenases (14, 15).
The H. modesticaldum genome contains an ORF (HM1_0869) with 90 and 74% sequence identity to the recently described orf1 in H. gestii and H. chlorum, respectively (14). The orf1 product shares some amino acid sequence identity (∼27%) with the hutP product, a positive regulator of the histidine utilization (hut) operon in B. subtilis. Similar to the regulatory mechanism for histidine utilization by HutP, orf1, situated upstream of the nif regulon, may be involved in ammonia switch-off regulation of nif gene expression in heliobacteria (14, 31). The activation of the orf1 product in the absence of sufficient levels of fixed nitrogen may allow transcription to continue through a terminator-like structure in the intergenic region of orf1 and nifI1, resulting in expression of the nif structural genes (14). In H. modesticaldum the terminator-like structure shows 94 and 83% sequence identity to the terminator regions of H. gestii and H. chlorum, respectively. This regulatory mechanism is distinct from that of most diazotrophs, which employ the nifA gene product, which is not present in heliobacteria (10, 14), as a transcriptional activator (36).
An operon containing genes involved in the assembly of an uptake [NiFe] hydrogenase was also found in the genome of H. modesticaldum. In contrast to the [FeFe] hydrogenase involved in hydrogen production during pyruvate fermentation, the uptake [NiFe] hydrogenase is likely associated with nitrogenase, catalyzing the oxidation of hydrogen produced in the following nitrogen-fixing reaction: 8H+ + 8e− + N2 → 2NH3 + H2 (46). A total of 10 genes are present in the uptake [NiFe] hydrogenase operon (Fig. 3B). Six genes (hypABCDEF) encode assembly/maturation proteins, and hypA (HM1_1482) encodes the Ni insertion protein. Three genes encode hydrogenase structural proteins; hupS, hupL, and hupC encode the small, large, and b-type cytochrome subunits, respectively. One gene, hupD (HM1_1481), encodes an accessory formation protein. Genes encoding an uptake [NiFe] hydrogenase, including three copies each of hupC, hupL, and hupS, are also present in D. hafniense Y51. In contrast to the single gene cluster present in H. modesticaldum, however, these genes occur in separate operons found in different regions of the D. hafniense chromosome (Fig. 3B).
Endospore formation.
Endosporulation appears to be universal among heliobacteria (24), and the ability of endospores to survive pasteurization (heating to 80°C for 15 min) can be exploited for selective enrichment of these organisms (33, 49). H. modesticaldum contains genes encoding all five sigma factors deemed essential for sporulation in B. subtilis (σH, σE, σF, σG, and σK). However, in a genome-wide comparison of the two organisms, several sporulation-specific genes were not found in H. modesticaldum.
An ortholog of the spo0M gene, which is involved in the regulation of endospore formation in B. subtilis (17), was not identified in H. modesticaldum. Although this gene is not essential for viability of B. subtilis, inactivation of it significantly hinders initiation of endosporulation (17). The absence of this gene in H. modesticaldum is curious since it is present in many species of Bacillus, as well as in sporulating organisms more closely related to heliobacteria, such as D. reducens and P. thermopropionicum. Likewise, no homolog of the spoIIB gene, which is involved in septum formation during stage II of sporulation in B. subtilis, could be found in the H. modesticaldum genome. Although genetic studies in which the B. subtilis spoIIB gene was altered resulted in only minor impairment of sporulation (35), the apparent absence of both spoIIB and spo0M in heliobacteria could adversely affect the ability of these organisms to sporulate. Consistent with this hypothesis is the fact that sporulation of pure cultures of heliobacteria is rarely observed (24).
None of the 20 known cot genes in B. subtilis, whose products form the inner and outer spore coat and ostensibly function in a protective capacity (39), showed significant sequence identity to genes in the H. modesticaldum genome. Although it is not known whether the lack of cot genes is universal among heliobacteria, heliobacterial endospores do not appear to be compromised in terms of heat resistance, as evidenced by the presence of viable cells in pasteurized cultures (49).
Pigment biosynthesis and photosynthetic proteins.
Bchl g is one of two Chl derivatives (along with Bchl b) that contain an ethylidene substituent on ring B. The biosynthetic pathway of Bchl g (or Bchl b) has not been established yet. The genome data for H. modesticaldum revealed no gene predicted to encode a divinyl reductase; however, this cannot be correlated to Bchl b-containing organisms since, to date, the genomes of none of these organisms have been sequenced. Homologues of neither bciA nor slr1923, genes recently shown to encode C-8 divinyl reductases in C. tepidum (11) and Synechocystis sp. strain PCC6803 (20), respectively, were identified in the H. modesticaldum genome. Thus, it is possible that heliobacteria contain a third, unknown type of divinyl reductase, as has been proposed for species of Roseiflexus, a filamentous anoxygenic phototroph that also lacks bciA and slr1923 (20).
However, a proposed alternative pathway of Bchl g synthesis that circumvents the absence of divinyl reductase and incorporates the somewhat unexpected presence of the chlorophyllide reductase enzyme complex (BchX, BchY, and BchZ) is shown in Fig. 5. We propose that in this pathway, the C-7-C-8 double bond is reduced by chlorophyllide reductase, and the ethylidene group is then formed by isomerization of the C-81 vinyl substituent. Although the gene that encodes the enzyme responsible for this isomerase activity has not been identified, this mechanism seems plausible, as it would require the reduction of only one double bond on ring B and is completely consistent with the gene presence/absence patterns described above.
FIG. 5.
Proposed pathway of later steps in Bchl g biosynthesis in H. modesticaldum. Divinyl protochlorophyllide a is reduced to 8-vinyl chlorophyllide a by the activity of the bchLNB gene products. This is followed by the reduction of the C-7—C-8 double bond via the bchXYZ gene products, which yields C-8 vinyl bacteriochlorophyllide a. Bacteriochlorophyllide g is produced by the isomerization of the 81-vinyl group to an ethylidene group. Bchl synthetase (BchG) then catalyzes the addition of a farnesyl group, which yields the completed Bchl g.
Farnesol, the esterifying alcohol of Bchl g, is synthesized in a complete nonmevalonate pathway, in which pyruvate and glyceraldehyde 3-phosphate are condensed to yield isopentenyl-pyrophosphate and dimethylallyl-pyrophosphate through a cascade of enzymatic reactions (13). Farnesyl-pyrophosphate is produced from these precursors through a geranyl-pyrophosphate intermediate (13).
The gene encoding Bchl synthetase (bchG) in H. modesticaldum (HM1_0692) has a high level of sequence identity (74.7%) to bchG from Heliobacillus mobilis (see Fig. S1 in the supplemental material). As it is in Heliobacillus mobilis (58), bchG is situated between bchJ (HM1_0693), a gene recently shown to not encode divinyl reductase in green sulfur bacteria (11), and pshA (HM1_0690) in H. modesticaldum (Fig. 3C). PshA is the type I RC core polypeptide homologous to PscA and PsaA of other phototrophs with type I RCs (see Fig. S2 in the supplemental material). These genes are members of a large photosynthesis gene cluster (PGC) that also contains genes encoding all four cytochrome bc complex subunits (PetA, PetB, PetC, and PetD), the 18-kDa cytochrome c553 (PetJ), a carotenoid biosynthesis protein (CrtN), and several proteins involved in vitamin biosynthesis (58) (Fig. 3C). The bchXYZ genes are located on the opposite strand in an adjacent operon (Fig. 3C). The phylogenetic relationship of concatenated amino acid sequences from protochlorophyllide reductase (bchLNB) and chlorophyllide reductase (bchXYZ) genes from H. modesticaldum and other organisms is shown in Fig. 4. The distinctly separate lineages of the H. modesticaldum bchXYZ and bchLNB genes and the genes of other anoxygenic phototrophs are evident.
The grouping of photosynthetic genes into superoperonal clusters is a trait found only in heliobacteria and purple bacteria. However, as these organisms are phylogenetically distinct phototrophs, it is significant that the PGC of purple bacteria shows little gene synteny with that of heliobacteria. A comparison of the PGCs of Rhodobacter capsulatus and H. modesticaldum shows that most of the photosynthesis genes in R. capsulatus are not present in H. modesticaldum (Fig. 3C). It is also notable that while nearly all of the genes in the PGC of R. capsulatus encode proteins with photosynthetic functions, the products of more than one-half of the genes in the PGC of H. modesticaldum appear to be involved in nonphotosynthetic processes (Fig. 3C).
The relative simplicity of heliobacterial photosynthesis is suggested first by the absence of three Bchl biosynthesis genes in H. modesticaldum that are present in R. capsulatus (bchCPF). Conversely, R. capsulatus contains all 13 bch genes present in H. modesticaldum (Fig. 3C). Perhaps even more indicative of the simplicity of the heliobacterial photosynthetic apparatus is the lack of a heteromeric RC, such as that encoded by the pufLM genes in purple bacteria, and the absence of additional light-harvesting complexes, which are encoded by the pufAB and pucAB genes in purple bacteria. It is therefore difficult to envision a close evolutionary connection between the PGCs of heliobacteria and purple bacteria due to extensive gene rearrangements and divergent transcription. Instead, the high degree of genetic disparity suggests that these superoperon clusters developed convergently rather than having a common origin. The possible functional significance of this is not apparent.
Electron transfer pathways.
Genes predicted to encode all 14 subunits of NADH:quinone oxidoreductase (nuoA to nuoN) were identified in the H. modesticaldum genome. Presumably, electrons from NADH are transferred to menaquinone through this complex and donated to the cytochrome bc complex, which reduces the Rieske [2Fe-2S] subunit (PetC) and cytochrome bL (18) (Fig. 6). Electrons from the cytochrome bc complex are transferred to the RC primary electron donor, P798, via cytochrome c553 (42). A complete ATP synthase, having all eight subunits encoded in a single conserved operon, catalyzes ATP synthesis. Phylogenetic and physiological similarities between photosystem I and the heliobacterial RC, as well as kinetic studies conducted with Heliobacillus mobilis (27), suggest that proton motive force generation through cyclic electron flow occurs in H. modesticaldum, but this has not been confirmed.
FIG. 6.
Diagram showing a putative pathway of electron transfer based on genetic components present in H. modesticaldum. Cyclic electron transfer has not been confirmed in heliobacteria. In addition, the reduction of NAD+ by cytoplasmic ferredoxin has not been confirmed, as a gene encoding FNR was not identified in the genome. Despite this, genes encoding all 14 subunits of NADH:quinone oxidoreductase (nuoA to nuoN) were putatively identified.
Electron transfer within the RC of heliobacteria is not well understood. It is known that rapid transfer occurs from the excited state of the primary electron donor, P798, to the primary acceptor, 81-OH-Chl a (A0), but the existence of a secondary quinone acceptor (A1) has not been confirmed (41). PshA exists as a homodimer containing two cysteine residues per subunit that are believed to bind to an FX-like [4Fe-4S] cluster (26, 38, 41). In addition to pshA, a pshB gene (HM1_1462), which encodes an RC-associated [4Fe-4S]-binding ferredoxin, was confirmed to be present in the H. modesticaldum genome. A recent study showed that PshB binds the terminal Fe/S dicluster electron acceptors, FA and FB (19). Electrons are likely transferred from the Fe/S clusters to a cytoplasmic ferredoxin, which can supply reducing equivalents for cellular processes, such as carbon assimilation and nitrogen fixation (Fig. 6).
No clear homolog of a gene encoding ferredoxin:NADP+ reductase (FNR) is present in the genome. A well-conserved thioredoxin reductase encoded by a gene in the H. modesticaldum genome (HM1_0984) showed 30 and 28% sequence identity to the FNR of Bacillus thuringiensis and C. tepidum, respectively. The gene encoding FNR in C. tepidum was originally annotated as a putative thioredoxin reductase gene, but the product has since been shown to have FNR activity (47). However, the low levels of sequence identity of these enzymes to HM1_0984, as well as the equivalog-level support of the ORF as a thioredoxin reductase gene, suggest that the enzyme is not FNR. Therefore, the mechanism of NADP+ reduction in H. modesticaldum remains unknown.
Our analysis of the Heliobacterium genome provides the first glimpse of the genetic capacity of the unique anoxygenic phototrophs belonging to this genus. Further genomic analyses, especially analyses of species of heliobacteria that inhabit agricultural soils (such as Heliophilum fasciatum) or alkaline soils (such as Heliorestis species), should further unravel the unusual patterns of carbon metabolism, photosynthetic energy conversion, and sporulation found here.
Supplementary Material
Acknowledgments
This work was supported by the U.S. National Science Foundation Phototrophic Prokaryotes Sequencing Project (grant 0412824). W.D.S. was funded by a Japanese Society for Promotion of Science Postdoctoral Fellowship for Foreign Researchers (grant P07141). M.C. was funded by Australian Research Council Discovery Project grant DP0665169. Partial support for the participation of P.C.C. and L.E.K. in this research was provided by a grant to Washington University from the Howard Hughes Medical Institute through the Undergraduate Biological Sciences Education Program. D.O.J. was partially supported by National Science Foundation grant MCB0237567.
We thank Aaron Collins for helpful discussions and assistance with graphics.
Footnotes
Published ahead of print on 25 April 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akochy, P.-M., D. Bernard, P. H. Roy, and J. Lapointe. 2004. Direct glutaminyl-tRNA biosynthesis and indirect asparaginyl-tRNA biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186767-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amesz, J. 1995. The antenna-reaction center complex of heliobacteria, p. 687-697. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 4.Asao, M., D. O. Jung, L. A. Achenbach, and M. T. Madigan. 2006. Heliorestis convoluta sp. nov., a coiled, alkaliphilic heliobacterium from the Wadi El Natroun, Egypt. Extremophiles 10403-410. [DOI] [PubMed] [Google Scholar]
- 5.Batzoglou, S., D. B. Jaffe, K. Stanley, J. Butler, S. Gnerre, E. Mauceli, B. Berger, J. P. Mesirov, and E. S. Lander. 2002. ARACHNE: a whole-genome shotgun assembler. Genome Res. 12177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beer-Romero, P., and H. Gest. 1987. Heliobacillus mobilis, a peritrichously flagellated anoxyphototroph containing bacteriochlorophyll g. FEMS Microbiol. Lett. 41109-114. [Google Scholar]
- 7.Berg, I. A., D. Kockelkorn, W. Buckel, and G. Fuchs. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 3181782-1786. [DOI] [PubMed] [Google Scholar]
- 8.Bryantseva, I. A., V. M. Gorlenko, E. I. Kompantseva, L. A. Achenbach, and M. T. Madigan. 1999. Heliorestis daurensis, gen. nov. sp. nov., an alkaliphilic rod-to-coiled-shaped phototrophic heliobacterium from a Siberian soda lake. Arch. Microbiol. 172167-174. [DOI] [PubMed] [Google Scholar]
- 9.Bryantseva, I. A., V. M. Gorlenko, E. I. Kompantseva, T. P. Tourova, B. B. Kuznetsov, and G. A. Osipov. 2000. Alkaliphilic heliobacterium Heliorestis baculata sp. nov. and emended description of the genus Heliorestis. Arch. Microbiol. 174283-291. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J. S. 2004. Nitrogen fixation in the clostridia, p. 53-64. In W. Klipp, B. Masepohl, J. R. Gallon, and W. E. Newton (ed.), Genetics and regulation of nitrogen fixation in free-living bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 11.Chew, A. G. M., and D. A. Bryant. 2007. Characterization of a plant-like protochlorophyllide a divinyl reductase in green sulfur bacteria. J. Biol. Chem. 2822967-2975. [DOI] [PubMed] [Google Scholar]
- 12.Curnow, A. W., D. L. Tumbula, J. T. Pelaschier, B. Min, and D. Söll. 1998. Glutamyl-tRNAGln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl. Acad. Sci. USA 9512838-12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubey, V. S., R. Bhalla, and R. Luthra. 2003. An overview of the non-mevalonate pathway for terpenoid biosynthesis in plants. J. Biosci. 28637-646. [DOI] [PubMed] [Google Scholar]
- 14.Enkh-Amgalan, J., H. Kawasaki, H. Oh-oka, and T. Seki. 2006. Cloning and characterization of a novel gene involved in nitrogen fixation in Heliobacterium chlorum: a possible regulatory gene. Arch. Microbiol. 186327-337. [DOI] [PubMed] [Google Scholar]
- 15.Enkh-Amgalan, J., H. Kawasaki, and T. Seki. 2006. Molecular evolution of the nif gene cluster carrying nifI1 and nifI2 genes in the Gram-positive phototrophic bacterium Heliobacterium chlorum. Int. J. Syst. Evol. Microbiol. 5665-74. [DOI] [PubMed] [Google Scholar]
- 16.Fuller, R. C., S. G. Sprague, H. Gest, and R. E. Blankenship. 1985. A unique photosynthetic reaction center from Heliobacterium chlorum. FEBS Lett. 182345-349. [Google Scholar]
- 17.Han, W.-D., S. Kawamoto, Y. Hosoya, M. Fujita, Y. Sadaie, K. Suzuki, Y. Ohashi, F. Kawamura, and K. Ochi. 1998. A novel sporulation-control gene (spo0M) of Bacillus subtilis with a σH-regulated promoter. Gene 21731-40. [DOI] [PubMed] [Google Scholar]
- 18.Heinnickel, M., and J. H. Golbeck. 2007. Heliobacterial photosynthesis. Photosynth. Res. 9235-53. [DOI] [PubMed] [Google Scholar]
- 19.Heinnickel, M., G. Shen, and J. H. Golbeck. 2007. Identification and characterization of PshB, the dicluster ferredoxin that harbors the terminal electron acceptors FA and FB in Heliobacterium modesticaldum. Biochemistry 462530-2536. [DOI] [PubMed] [Google Scholar]
- 20.Ito, H., M. Yokono, R. Tanaka, and A. Tanaka. 2008. Identification of a novel vinyl reductase gene essential for the biosynthesis of monovinyl chlorophyll in Synechocystis sp. PCC6803. J. Biol. Chem. 2839002-9011. [DOI] [PubMed] [Google Scholar]
- 21.Kimble, L. K., and M. T. Madigan. 1992. Evidence for an alternative nitrogenase in Heliobacterium gestii. FEMS Microbiol. Lett. 100255-260. [Google Scholar]
- 22.Kimble, L. K., A. K. Stevenson, and M. T. Madigan. 1994. Chemotrophic growth of heliobacteria in darkness. FEMS Microbiol. Lett. 11551-55. [DOI] [PubMed] [Google Scholar]
- 23.Kimble, L. K., L. Mandelco, C. R. Woese, and M. T. Madigan. 1995. Heliobacterium modesticaldum, sp. nov., a thermophilic heliobacterium of hot springs and volcanic soils. Arch. Microbiol. 163259-267. [Google Scholar]
- 24.Kimble-Long, L. K., and M. T. Madigan. 2001. Molecular evidence that the capacity for endosporulation is universal among phototrophic heliobacteria. FEMS Microbiol. Lett. 199191-195. [DOI] [PubMed] [Google Scholar]
- 25.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 661328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleinherenbrink, F. A. M., H. C. Chiou, R. LoBrutto, and R. E. Blankenship. 1994. Spectroscopic evidence for the presence of an iron-sulfur center similar to FX of photosystem I in Heliobacillus mobilis. Photosynth. Res. 41115-123. [DOI] [PubMed] [Google Scholar]
- 27.Kramer, D. M., B. Schoepp, U. Liebl, and W. Nitschke. 1997. Cyclic electron transfer in Heliobacillus mobilis involving a menaquinol-oxidizing cytochrome bc complex and an RCI-type reaction center. Biochemistry 364203-4211. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 29.Loveless, T. M., and P. E. Bishop. 1999. Identification of genes unique to Mo-independent nitrogenase systems in diverse diazotrophs. Can. J. Microbiol. 45312-317. [PubMed] [Google Scholar]
- 30.Ludwig, M., R. Schulz-Friedrich, and J. Appel. 2006. Occurrence of hydrogenases in cyanobacteria and anoxygenic photosynthetic bacteria: implications for the phylogenetic origin of cyanobacterial and algal hydrogenases. J. Mol. Evol. 63758-768. [DOI] [PubMed] [Google Scholar]
- 31.Madigan, M. T. 1995. Microbiology of nitrogen fixation by anoxygenic photosynthetic bacteria, p. 915-928. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 32.Madigan, M. T. 2001. Heliobacteriaceae, p. 625-630. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, NY. [Google Scholar]
- 33.Madigan, M. T. 2006. The family Heliobacteriaceae, p. 951-964. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 4th ed. Springer, New York, NY.
- 34.Madigan, M. T., and J. G. Ormerod. 1995. Taxonomy, physiology, and ecology of heliobacteria, p. 17-30. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 35.Margolis, P. S., A. Driks, and R. Losick. 1993. Sporulation gene spoIIB from Bacillus subtilis. J. Bacteriol. 175528-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merrick, M. J. 2004. Regulation of nitrogen fixation in free-living diazotrophs. p. 197-223. In W. Klipp, B. Masepohl, J. R. Gallon, and W. E. Newton (ed.), Genetics and regulation of nitrogen fixation in free-living bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 37.Miller, K. R., J. S. Jacob, U. Smith, S. Kolaczkowski, and M. K. Bowman. 1986. Heliobacterium chlorum: cell organization and structure. Arch. Microbiol. 146111-114. [DOI] [PubMed] [Google Scholar]
- 38.Miyamoto, R., M. Iwaki, H. Mino, J. Harada, S. Itoh, and H. Oh-oka. 2006. ESR signal of the iron-sulfur center FX and its function in the homodimeric reaction center of Heliobacterium modesticaldum. Biochemistry 456306-6316. [DOI] [PubMed] [Google Scholar]
- 39.Naclerio, G., L. Baccigalupi, R. Zilhao, M. De Felice, and E. Ricca. 1996. Bacillus subtilis spore coat assembly requires cotH gene expression. J. Bacteriol. 1784375-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nonaka, H., G. Keresztes, Y. Shinoda, Y. Ikenaga, M. Abe, K. Naito, K. Inatomi, K. Furukawa, M. Inui, and H. Yukawa. 2006. Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 1882262-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh-oka, H. 2007. Type 1 reaction center of photosynthetic heliobacteria. Photochem. Photobiol. 83177-186. [DOI] [PubMed] [Google Scholar]
- 42.Oh-oka, H., M. Iwaki, and S. Itoh. 2002. Electron donation from membrane-bound cytochrome c to the photosynthetic reaction center in whole cells and isolated membranes of Heliobacterium gestii. Photosynth. Res. 71137-147. [DOI] [PubMed] [Google Scholar]
- 43.Ormerod, J. G., L. K. Kimble, T. Nesbakken, Y. A. Torgersen, C. R. Woese, and M. T. Madigan. 1996. Heliophilum fasciatum gen. nov. sp. nov. and Heliobacterium gestii sp. nov.: endospore-forming heliobacteria from rice field soils. Arch. Microbiol. 165226-234. [DOI] [PubMed] [Google Scholar]
- 44.Pickett, M. W., M. P. Williamson, and D. J. Kelly. 1994. An enzyme and 13C-NMR study of carbon metabolism in heliobacteria. Photosynth. Res. 4175-88. [DOI] [PubMed] [Google Scholar]
- 45.Raymond, J., J. L. Siefert, C. R. Staples, and R. E. Blankenship. 2004. The natural history of nitrogen fixation. Mol. Biol. Evol. 21541-554. [DOI] [PubMed] [Google Scholar]
- 46.Rey, F. E., Y. Oda, and C. S. Harwood. 2006. Regulation of uptake hydrogenase and effects of hydrogen utilization on gene expression in Rhodopseudomonas palustris. J. Bacteriol. 1886143-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo, D., and H. Sakurai. 2002. Purification and characterization of ferredoxin-NAD(P)+ reductase from the green sulfur bacterium Chlorobium tepidum. Biochim. Biophys. Acta 1597123-132. [DOI] [PubMed] [Google Scholar]
- 48.Stackebrandt, E., C. Sproer, F. A. Rainey, J. Burghardt, O. Pauker, and H. Hippe. 1997. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Intl. J. Syst. Bacteriol. 471134-1139. [DOI] [PubMed] [Google Scholar]
- 49.Stevenson, A. K., L. K. Kimble, C. R. Woese, and M. T. Madigan. 1997. Characterization of new phototrophic heliobacteria and their habitats. Photosynth. Res. 531-12. [Google Scholar]
- 50.Stothard, P., and D. S. Wishart. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21537-539. [DOI] [PubMed] [Google Scholar]
- 51.Swingley, W. D., S. Sadekar, S. D. Mastrian, H. J. Matthies, J. Hao, H. Ramos, C. R. Acharya, A. L. Conrad, H. L. Taylor, L. C. Dejesa, M. K. Shah, M. E. O'Huallachain, M. T. Lince, R. E. Blankenship, J. T. Beatty, and J. W. Touchman. 2007. The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism. J. Bacteriol. 189683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takaichi, S., K. Inoue, M. Akaike, M. Kobayashi, H. Oh-oka, and M. T. Madigan. 1997. The major carotenoid in all known species of heliobacteria is the C30 carotenoid 4,4′-diaponeurosporene, not neurosporene. Arch. Microbiol. 168277-281. [DOI] [PubMed] [Google Scholar]
- 53.Takaichi, S. 1999. Carotenoids and carotenogenesis in anoxygenic photosynthetic bacteria, p. 39-69. In H. A. Frank, R. J. Cogdell, A. Young, and G. Britton (ed.), The photochemistry of carotenoids: applications in biology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 54.Thauer, R. K. 2007. A fifth pathway of carbon fixation. Science 3181732-1733. [DOI] [PubMed] [Google Scholar]
- 55.Trost, J. T., and R. E. Blankenship. 1989. Isolation of a photoactive photosynthetic reaction center-core antenna complex from Heliobacillus mobilis. Biochemistry 289898-9904. [DOI] [PubMed] [Google Scholar]
- 56.van de Meent, E. J., M. Kobayashi, C. Erkelens, P. A. van Veelen, J. Amesz, and T. Watanabe. 1991. Identification of 81-hydroxychlorophyll a as a functional reaction center pigment in heliobacteria. Biochim. Biophys. Acta 1058356-362. [Google Scholar]
- 57.Webster, L. T. 1967. Studies of the acetyl coenzyme A synthetase reaction. J. Biol. Chem. 2421232-1240. [PubMed] [Google Scholar]
- 58.Xiong, J., K. Inoue, and C. E. Bauer. 1998. Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis. Proc. Natl. Acad. Sci. USA 9514851-14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.