Abstract
Lipoprotein signal peptidase (lsp) is responsible for cleaving the signal peptide sequence of lipoproteins in gram-positive bacteria. Investigation of the role of Lsp in Streptococcus uberis, a common cause of bovine mastitis, was undertaken using the lipoprotein MtuA (a protein essential for virulence) as a marker. The S. uberis lsp mutant phenotype displayed novel lipoprotein processing. Not only was full-length (uncleaved) MtuA detected by Western blotting, but during late log phase, a lower-molecular-weight derivative of MtuA was evident. Similar analysis of an S. uberis double mutant containing insertions disrupting both lsp and eep (a homologue of the Enterococcus faecalis “enhanced expression of pheromone” gene) indicated a role for eep in cleavage of lipoproteins in the absence of Lsp. Such a function may indicate a role for eep in maintenance of secretion pathways during disruption of normal lipoprotein processing.
Streptococcus uberis is a major cause of bovine mastitis worldwide. This pathogen is transmitted to the udder largely from environmental sources, and the incidence of intramammary infection with S. uberis is not well controlled by hygienic milking practice (19). The “manganese transporter uberis” protein (MtuA) of S. uberis has been shown to be essential for this organism to grow in milk and for infection/disease in lactating dairy cattle (36). MtuA, a predicted lipoprotein, contains a lipobox motif within a typical lipoprotein signal peptide (36) and has been shown by enzyme-linked immunosorbent assay, immunoblotting, and electron microscopy (23, 24) to locate to the cell membrane.
Most secreted proteins are produced with a signal peptide at the amino terminus which is removed by a signal peptidase during the secretion process (40, 45). Lipoproteins typically contain a specialized signal peptide containing a lipobox motif in the carboxyl region of the signal peptide (37, 40). This motif is believed to target the lipoprotein to the correct posttranslational processing pathway (47). The consensus motif of the lipobox is LxxC, in which the cysteine residue is the invariable target for lipidation by lipoprotein diacylglyceryl transferase (Lgt). Lipidation at this residue serves to anchor the lipoprotein to the membrane (37, 44). Lipidation has been considered to be a prerequisite for the action of lipoprotein signal peptidase (Lsp), which removes the signal peptide and leaves the cysteine of the lipobox as the new amino-terminal residue of the mature lipoprotein (44).
Lsp was first identified in Escherichia coli (20, 33, 43, 49) and subsequently in gram-positive bacteria, where it was first identified in Staphylococcus aureus (50). Lsp has since been found to be a ubiquitous bacterial enzyme and has been studied in several bacterial species, including Mycobacterium tuberculosis (34), Listeria monocytogenes (32), Enterobacter aerogenes (22), Streptococcus suis (10), and Streptococcus pneumoniae (28).
The role of Lsp in Bacillus subtilis has been studied in detail, where it was shown not to be essential for cell viability, although it was required for growth at higher temperatures (41). In a strain lacking Lsp, a lipoprotein responsible for protein folding, PrsA, was not processed to its mature form. Around 50% of the PrsA was visualized at a size comparable with the full-length protein (pre-PrsA), while the remainder was shown to have been processed to a form with a lower molecular weight but which appeared to be greater in size than the mature form of the protein. This observation implied that an alternative pathway existed for processing lipoproteins in B. subtilis. Both forms of the PrsA protein were correctly localized to the membrane, and it was concluded by Tjalsma et al. (41) that both retained their biological function.
Lsp enzymes are predicted to have four transmembrane-spanning regions (31), and five regions of conserved sequence have been identified by comparison of 18 sequences of Lsp (42). Point mutagenesis has been used to define individual residues important for the activity of Lsp. These are all predicted to be localized close to the external surface of the cytoplasmic membrane. Asp-14 was shown to be important for the structural stability of the enzyme, while five other residues were directly involved in the catalysis of the signal peptide removal. Two strictly conserved aspartic acid residues were essential for the activity of this enzyme, suggesting that Lsp belongs to a group of enzymes known as aspartic peptidases. These data were reinforced by the observation that Lsp was inhibited by pepstatin, a known inhibitor of aspartic peptidases (12). Observations by Tjalsma et al. (42) suggested that Lsp belonged to a novel class of aspartic peptidases that evolved exclusively in eubacteria in which Asp-102 and Asp-129 (in that from B. subtilis) were shown to act as a catalytic dyad. Other highly conserved residues have been implicated in stabilizing the active site and/or recognizing the diacylglyceryl-modified cysteine residue in the lipobox of preproteins (42).
Interestingly, a second peptidase has been described in Enterococcus faecalis that has been implicated in the processing of signal peptides of some lipoproteins (3). Enhanced expression of pheromone (Eep) is a predicted metallopeptidase and has been shown to cleave the signal peptides of lipoproteins to yield octapeptides that act as bacterial pheromones, inducing conjugation between different strains of E. faecalis (2, 3, 9).
The antibiotic globomycin has been invaluable in studying the activity and function of Lsp enzymes. This antibiotic is a potent, reversible, noncompetitive inhibitor of Lsp (21). It functions by binding to the enzyme and in doing so prevents cleavage of signal peptides from target lipoproteins (11).
In this communication, the role of Lsp in S. uberis was described by comparative analysis of the wild-type organism with isogenic mutants lacking Lsp.
MATERIALS AND METHODS
Identification of lipoprotein sequences by pattern searching.
Lipoproteins from the genome of S. uberis strain 0140J (http://www.sanger.ac.uk/Projects/S_uberis/) were found using a slight modification of the G+Lpp published by Sutcliffe and Harrington (37). The pattern [MV]-X(0,13)-[RK]-, (6,20)-[LIVMFESTAG]-[LVIAMF]-[IVMSTAFGC]-[AG]-C (I. C. Sutcliffe, personal communication) was entered in the pattern search option of the PEDANT website (15, 16), and the proteins were cross referenced with the open reading frames (ORFs) predicted by the Sanger Centre annotation (M. Holden, personal communication). Candidate proteins were checked to determine that the pattern was located at the N terminus of the protein and that a signal peptide sequence could be predicted using SignalP (4). Putative functions for each of the predicted lipoproteins were ascribed using the BLASTp algorithm (1) (http://www.ncbi.nlm.nih.gov/BLAST/).
Bacterial growth conditions and oligonucleotide primers.
S. uberis was routinely grown on Todd-Hewitt agar or sheep blood-esculin agar plates and in Todd-Hewitt broth (THB; Oxoid, Basingstoke, United Kingdom) at 37°C as a standing culture. Uncured pGh9+::ISS1 mutants were grown in the same medium with the addition of 1 μg/ml erythromycin at 37°C. Oligonucleotide primers P432 (GCTCTTCGGATTTTCGGTATC) and P571 (AATATCTTCAGCTTCATAATCC) were used to amplify the lsp locus, and primers eep fwd (TCAGTTTGGAGACTTAGTGG) and eep rev (TTTCATTTATCTGTATTTCTCC) were used to amplify the eep locus of S. uberis. Oligonucleotide primers P358 (CATTTTCCACGAATAGAAGGACTGTC) and P247 (GCTCTTCGGATTTTCGGTATC) were used to screen pGh9+::ISS1 mutant banks, as described below, and a digoxigenin-labeled ISS1 probe was used for Southern blotting as described previously (48).
Growth of S. uberis in the presence of globomycin.
A stock of globomycin (kindly supplied by Shunichi Miyakoshi, Sankyo Company Ltd., Japan) was prepared at a concentration of 10 mg/ml in ethanol and stored at −20°C. Bacteria were grown overnight in THB, prior to dilution in fresh THB to an optical density at 550 nm of 0.01. Cultures were grown at 37°C to the start of exponential phase (optical density at 550 nm of 0.1) before the addition of globomycin to 100 μg/ml. Bacteria were harvested at appropriate time points by centrifugation (12,000 × g, 5 min, 4°C). Whole-cell lysates and trichloroacetic acid supernatant precipitates were prepared.
Preparation of chromosomal DNA from S. uberis.
Chromosomal DNA from 3 ml of culture was prepared by using a variation of the method of Hill and Leigh (18). Bacteria were harvested by centrifugation (10,000 × g, 5 min), and the cell pellet was washed with 500 μl of 10 mM Tris-Cl-5 mM EDTA (pH 7.8). Bacterial cell walls were disrupted by resuspending bacteria in 375 μl of 10 mM Tris-Cl-5 mM EDTA (pH 7.8) containing 30 U/ml mutanolysin (Sigma) and 10 mg/ml lysozyme (Sigma). The bacterial suspension was incubated at 37°C for 30 min. Total cell lysis was achieved by addition of 20 μl of a sodium dodecyl sulfate solution (20% [wt/vol] in 50 mM Tris-Cl, 20 mM EDTA, pH 7.8) and proteinase K (Sigma) at a final concentration of 150 μg/ml and incubation at 37°C for 1 h. Cell wall material was precipitated by the addition of 200 μl of saturated NaCl solution and subsequent centrifugation (13,000 × g, 15 min, 4°C). The cleared supernatant was transferred to a clean tube, and contaminating proteins were removed by phenol-chloroform extraction. The DNA was precipitated by addition of 2 volumes of absolute ethanol. DNA pellets were washed with 70% aqueous ethanol and air dried prior to resuspension in Tris-EDTA buffer containing 20 μg/ml of RNase A (Sigma).
Southern blotting to determine location and distribution of ISS1 insertion.
Southern blot analysis was performed upon genomic DNA digested with EcoRI or HindIII. Hybridization of the digoxigenin-dUTP (Roche Diagnostics Ltd.)-labeled probe was carried out at 65°C overnight. Hybridizing fragments were visualized with the chemiluminescent substrate nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (Roche Diagnostics Ltd.) as instructed by the manufacturer.
Isolation of mutants from an S. uberis strain 0140J pGhost9+::ISS1 mutant bank.
PCR amplification was carried out on DNA from pools of 96 mutants representing a single microtiter tray from an S. uberis strain 0140J pGh9+::ISS1 mutant bank, comprised of 8,800 individual clones (39). A screen for ISS1 insertions at a specific locus was carried out by PCR using paired ISS1 and gene-specific primers as first described by Taylor et al. (39). Amplification was carried out using 2 U Taq polymerase (New England Biolabs). Each pooled DNA sample was screened twice using a gene-specific primer (listed above) with either P247 or P358 to enable detection of chromosomally inserted ISS1 in either orientation at the relevant locus. Clones from individual wells were cultured to obtain single colonies and cured as previously described (48); the presence of the ISS1 insertion was confirmed by Southern blotting, described above.
Sequence analysis to define the ISS1 insertion point (48) was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, United Kingdom), according to the manufacturer's instructions. Sequence determination was carried out by L. Richardson (University of Oxford, United Kingdom).
Random mutagenesis in lsp mutant S. uberis.
The procedures described previously by Maguin et al. (30) and Smith et al. (36) were used to generate a bank of random mutants within the lsp mutant of S. uberis by using the pGh9+::ISS1 vector. Isolation of a mutant carrying an insertion within eep was conducted as described above for isolation of mutant strains carrying single lesions.
Preparation of cleared whole-cell lysates.
Bacteria were harvested from cultures (2 ml to 100 ml) at required time points by centrifugation (12,000 × g, 5 min), and the pellets were washed three times in an equal volume of phosphate-buffered saline (PBS). Washed cells were suspended in 100 to 500 μl of PBS and mixed with 170- to 180-μm-diameter glass beads (0.5 g; Braun Biotech International). Bacteria were disrupted by rapid agitation (twice for 40 s each) by using a Cell Homogenizer-MSK instrument (Braun Biotech International). During disruption, samples were cooled with liquid CO2. Following disruption, samples were kept on ice for 1 h in the presence of 1% (vol/vol) Triton X-100 (BDH). Cell debris, unbroken cells, and beads were removed by centrifugation (12,000 × g, 5 min), and the supernatant was stored at −20°C.
Subcellular fractionation of S. uberis.
The following fractionation procedures were used sequentially on 100-ml cultures. All buffers contained 1× Complete EDTA-free protease inhibitors (Roche Diagnostics Ltd., United Kingdom).
Preparation of capsule fraction.
Bacteria were harvested by centrifugation (11,500 × g, 10 min). The supernatant was discarded, and the pelleted bacteria were resuspended and washed three times in 10 ml ice-cold PBS (0.01 M phosphate buffer, 0.0027 M potassium chloride, and 0.137 M sodium chloride, pH 7.4, at 25°C) with a final resuspension in 250 μl PBS containing hyaluronidase (100 U ml−1; Sigma). Bacteria were incubated for 2 h at 37°C, and cells were collected by centrifugation (9,500 × g, 5 min). The supernatant fraction (capsular extract) was removed and stored at −20°C.
Preparation of cell wall fraction.
Cells harvested from the capsule preparation procedure were resuspended and washed three times in 1 ml ice-cold PBS before being resuspended in 250 μl PBS containing 40% (wt/vol) sucrose and 170 U mutanolysin (Sigma). Samples were incubated for 2 h at 37°C, and intact bacterial cells were harvested by centrifugation (400 × g, 15 min, 4°C) while the supernatant fraction (cell wall extract) was removed and stored at −20°C.
Preparation of cell content fraction.
The pellet from the cell wall preparation (see above) was resuspended and washed three times in 1 ml ice-cold PBS containing 40% (wt/vol) sucrose with a final resuspension in 250 μl membrane buffer (100 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 100 mM NaCl). The cell protoplasts were freeze-thawed (−80°C to +37°C) three times to promote cell lysis. Residual intact cells and debris were removed by centrifugation (9,500 × g, 5 min, 4°C), and the supernatant fraction (cell contents) was harvested and stored at −20°C.
Preparation of membrane fraction.
The pellet remaining from the cell content procedure was resuspended and washed three times in 1 ml cold PBS, with a final resuspension in 250 μl membrane buffer containing Triton X-100 (1%, vol/vol) prior to incubation at room temperature (20°C) for 1 h to solubilize the membrane components. Insoluble debris was removed by centrifugation (9,500 × g, 5 min, 4°C), and the supernatant fraction (membrane extract) was harvested and stored at −20°C.
Quantitation of protein within cell fractions.
The protein content of cell fractions was determined using the bicinchoninic acid protein assay kit (Perbio) as directed by the manufacturer. Protein concentrations were calculated from mean values of triplicate readings for each sample by using a standard curve prepared using bovine serum albumen.
Detection of MtuA within cell fractions prepared from S. uberis.
The proteins present in cell fractions (1 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12.5% gels or 10% Bis-Tris gels (Invitrogen) and detected by Western blotting using MtuA antiserum (24) at a concentration of 1:2,500 and a secondary goat anti-rabbit-horseradish peroxidase conjugate (Sigma) at a concentration of 1:2,500.
RESULTS
Bioinformatic analysis of the complete genome of S. uberis strain 0140J for genes encoding putative lipoproteins.
Thirty-one ORFs were predicted to encode lipoproteins, using the G+Lpp search pattern; each contained a signal peptide sequence that was predicted by SignalP (4) (see Table S1 in the supplemental material). A pattern search was also performed using the Prosite pattern PS00013 for bacterial lipoproteins which identified a further 55 proteins; however, all of these sequences were considered false positives due to incorrect/unsuitable positioning of the lipobox motif and/or the absence of a signal peptide sequence as predicted by SignalP. Where possible, a putative function was ascribed to the ORF identified by using BLAST-p (1) (see Table S1 in the supplemental material). Many of the proteins identified from S. uberis displayed homology to known lipoproteins such as ABC transporters and the chaperone protein PrsA. The amino acid sequence of the previously studied MtuA protein of S. uberis was identified as containing a signal peptide typical of lipoproteins (24, 36).
Processing of the essential lipoprotein MtuA was affected by the antibiotic globomycin.
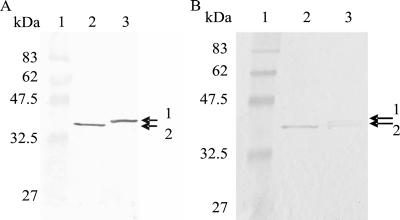
Protein samples prepared from cultures grown to the onset of stationary phase in the presence of globomycin yielded a single MtuA protein band approximately 2 kDa larger than that detected from equivalent cultures lacking the antibiotic (Fig. 1A). Analysis of samples produced from cultures incubated for 24 h (approximately 14 h after the onset of stationary phase) in the presence of globomycin revealed two proteins reactive to the MtuA antibody (Fig. 1B). The lower-molecular-weight protein (indicated by arrow 2 in Fig. 1B) corresponded in size to that detected in the absence of globomycin while the larger protein (indicated by arrow 1 in Fig. 1B) corresponded in size to that detected previously in the presence of globomycin after 14 h of incubation (Fig. 1A). MtuA was not detectable in the supernatant of wild-type S. uberis in the presence or absence of globomycin (data not shown).
FIG. 1.
Western blot analysis of wild-type S. uberis grown in the presence and absence of the Lsp inhibitor globomycin. Globomycin (100 μg/ml) was added where appropriate to bacterial cultures at the start of exponential phase. Whole-cell lysates were prepared from bacteria cultured in the presence (lanes 3) and absence (lanes 2) of globomycin grown to the start of stationary phase (A) or for 24 h (B). Markers are shown with molecular masses (lanes 1). Arrows 1 and 2, higher- and lower-molecular-mass forms of MtuA, respectively, recognized by specific antiserum.
The lsp locus in the S. uberis genome of strain 0140J.
An ORF (SUB0729) showing homology to lsp from B. subtilis (accession no. Q45479), S. aureus (accession no. P31024), E. coli (accession no. AAA24092), and L. monocytogenes (accession no. CAC99922) was identified within the genome of S. uberis strain 0140J (http://www.sanger.ac.uk/Projects/S_uberis). Lsp of S. uberis was found to be 148 amino acids in length. The amino acid homology and conservation between the S. uberis and B. subtilis Lsp proteins were 40% and 61%, respectively. Analysis of the sequence using the transmembrane helix prediction algorithm TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) (27) revealed the presence of the four transmembrane domains present in other Lsp proteins (see Fig. S1 in the supplemental material). The lsp locus is conserved in other genomes of streptococci including S. suis and S. pneumoniae (10, 28). In S. uberis the gene was flanked by a putative transcriptional regulator, lysR (SUB0728), and a putative ribosomal large subunit, pseudouridine synthase, rluD (SUB0730) (see Fig. S2 in the supplemental material).
Isolation of lsp mutants from an S. uberis pGh9+::ISS1 random mutant bank.
A mutant carrying an insertion in lsp (lsp::ISS1or lsp mutant) was identified in a bank of approximately 8,800 random insertion mutants by PCR screening. DNA sequence analysis placed the ISS1 insertion at 132 bp downstream of the start codon of the lsp coding sequence (see Fig. S2 in the supplemental material). Excision of the pGh9+ vector from the S. uberis lsp mutant and subsequent PCR amplification across the lsp locus in the cured mutant revealed an increase in the size of the amplicon of approximately 800 bp, consistent with the presence of the residual ISS1 element within the lsp gene. The presence of only a single insertion sequence element was confirmed by Southern blotting (data not shown).
The lipoprotein MtuA localized to the membrane in the wild-type and lsp mutant strains.
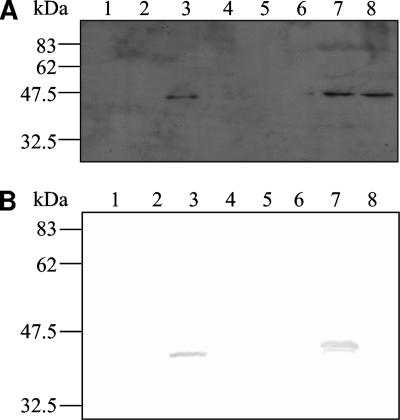
The putative lipoprotein MtuA was visualized by Western blotting with anti-MtuA (24) within cell fractions prepared from the wild-type and lsp mutant strains (Fig. 2). Fractions from cells grown to mid-exponential phase (Fig. 2A) revealed MtuA in the membrane-enriched fraction of both strains and additionally the cell content fraction in the lsp mutant strain. A single band corresponding to a molecular mass of approximately 37 kDa was detected in the wild type in agreement with previous data (23). However, in the lsp mutant a single band, approximately 2 kDa larger (39 kDa), was detected. The size of this band corresponds to the larger-molecular-mass band detected after the wild type had been treated with globomycin (Fig. 1). Intriguingly, analysis of samples produced from bacteria grown for 24 h revealed an additional immunoreactive band in the lsp mutant. This protein was slightly larger than the mature form seen in the wild type but smaller than that previously detected in the lsp mutant (Fig. 2B). This suggested that the alternatively cleaved form of MtuA from the lsp mutant detected at 24 h was likely to be shorter than that detected in mid-exponential cultures of the lsp mutant but longer than that detected in the wild-type strain.
FIG. 2.
Western blot showing that MtuA in the lsp mutant localizes to the membrane. (A) Cell fractions prepared from bacteria grown to mid-exponential phase. Lanes 1 to 4, wild-type capsule, cell wall, membrane, and cell contents, respectively; lanes 5 to 8, lsp mutant capsule, cell wall, membrane, and cell contents, respectively. (B) Cell fractions prepared from bacteria grown to 24 h. Lanes 1 to 4, wild-type capsule, cell wall, membrane, and cell contents, respectively; lanes 5 to 8, lsp mutant capsule, cell wall, membrane, and cell contents, respectively. Molecular masses of markers (not shown) are displayed (kDa).
A homologue of the gene encoding the E. faecalis protein Eep (enhanced expression of pheromone) is present within the genome of S. uberis strain 0140J.
The metallopeptidase Eep from Enterococcus faecalis has been shown to cleave the signal peptides of certain lipoproteins with the consequent release of short peptides that act as aggregation pheromones (3, 8, 14). This includes the peptide cAD1 (2), which is derived from the eight amino-terminal amino acids preceding the cysteine residue of the lipobox, the predicted cleavage point for Lsp (9).
A homologue of eep (accession number AAD47948) was found within the genome of S. uberis strain 0140J (ORF SUB0254) by using the BLAST algorithm (1); the identity between the predicted amino acid sequence of S. uberis SUB0254 and that of E. faecalis Eep was 304/427 (71%) (see Fig. S3 in the supplemental material). The HEXXH zinc binding signature motif, characteristic of M50A peptidases such as Eep (26), was also present in SUB0254. Eep of E. faecalis was predicted to be a membrane protein and, by use of the MEMSAT prediction software (25), was predicted to have four regions that span the membrane. An equivalent number of domains were found in SUB0254.
Identification of eep mutants within the S. uberis 0140J and S. uberis 0140J lsp mutant banks.
Genotypic PCR screening of the 0140J pGh9+::ISS1 mutant bank (39) permitted isolation of an eep::ISS1 mutant (eep mutant) with an insertion located 362 bp downstream of the start codon of SUB0254 (see Fig. S4 in the supplemental material). Similarly, screening of an lsp mutant pGh9+::ISS1 mutant bank permitted isolation of a double mutant carrying an insertion 34 bp downstream of the start codon of SUB0254 (see Fig. S4 in the supplemental material). The growth of the eep and lsp/eep mutants in THB and in skimmed milk was comparable to that of the wild type (data not shown).
MtuA is not cleaved in the lsp/eep mutant.
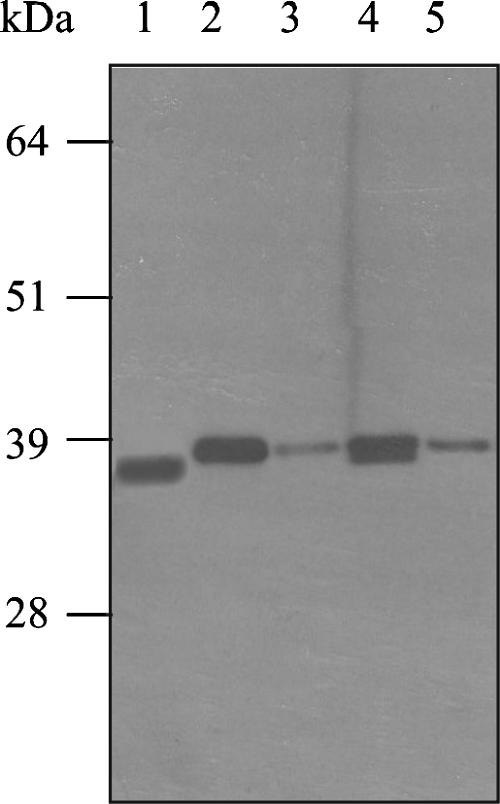
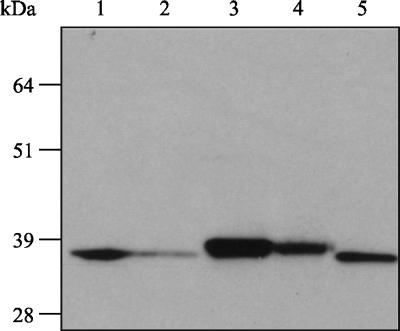
In an experiment where the lsp and lsp/eep mutants were compared directly with the wild-type strain, whole-cell lysates of the lsp mutant and lsp/eep double mutant, grown to the onset of stationary phase (approximately 8 h), revealed the presence of a single MtuA protein that corresponded to full-length MtuA (Fig. 3). In similar extracts prepared from cultures grown to late stationary phase (24 h), the lsp mutant displayed two MtuA protein bands (as previously reported for the membrane fraction [Fig. 2B]) However, in equivalent extracts prepared from the lsp/eep double mutant, only the higher-molecular-weight protein band was observed, indicating that the presence of the lower-molecular-weight form of MtuA required intact (functional) eep. In whole-cell extracts from the single eep mutant, MtuA was detected only at a size consistent with that present in the wild-type strain (Fig. 4), indicating that any cleavage of MtuA due to Eep was detected only in the absence of Lsp.
FIG. 3.
Presence of the different forms of MtuA in the lsp mutant and the lsp/eep mutant. Lane 1, wild-type whole-cell lysate at 24 h; lane 2, lsp mutant at start of stationary phase; lane 3, lsp/eep mutant at start of stationary phase; lane 4, lsp mutant at 24 h; lane 5, lsp/eep mutant at 24 h. Molecular masses are indicated in kDa.
FIG. 4.
MtuA is processed normally in an individual eep insertion mutant. Lane 1, wild-type 24-h whole-cell lysate; lane 2, eep mutant 24-h whole-cell lysate; lane 3, lsp mutant at 24 h; lane 4, lsp/eep mutant at 24 h; lane 5, wild-type 24-h whole-cell lysates. Molecular masses are indicated in kDa.
DISCUSSION
Using the experimentally verified G+Lpp search pattern (37, 38), the genome of S. uberis was shown to contain 31 ORFs encoding putative lipoproteins. Additional ORFs identified using the Prosite search pattern PS00013 were all considered to be false positives due to incorrect/unsuitable positioning of the lipobox motif and/or the absence of an appropriate lipoprotein signal peptide sequence as predicted by SignalP.
Processing of lipoproteins in gram-positive bacteria has been shown to require the activity of two enzymes, lipoprotein diacylglyceryl transferase (Lgt) and lipoprotein signal peptidase (Lsp). In gram-negative bacteria a third enzyme has been described in the lipoprotein processing pathway: the apolipoprotein transferase, Lnt (17). This enzyme is responsible for the correct localization of lipoproteins to the outer membrane. This protein has not been described in gram-positive bacteria, and a homologue encoded by the genome of S. uberis could not be found. Modification of lipoproteins at the cysteine of the LXXC lipobox by Lgt has been shown to be a prerequisite for the removal of the signal peptide by Lsp in some species (44). The role of Lsp in the cleavage of lipoprotein signal peptides in S. uberis was indicated by the detection of full-length MtuA following growth of the organism in the presence of globomycin (a specific inhibitor of Lsp-like activities) (11, 21). This was confirmed by similar observations in a mutant strain in which the lsp homologue (SUB0729) had been insertionally inactivated. However, in the absence of Lsp, late-stationary-phase cells revealed an additional MtuA protein, smaller than the full-length protein but larger than that cleaved by Lsp in the wild-type strain, implying that a further activity was able to cleave this lipoprotein at an alternative position. In all cases the MtuA protein remained cell associated and localized predominantly within the membrane-enriched fraction. Consequently, it was reasonable to deduce that anchoring via lipidation of the cysteine residue within the LXXC lipobox remained unaltered. Cleavage of lipoprotein signal peptides in the absence of Lsp has been reported for several other gram-positive bacteria, including B. subtilis, Lactococcus lactis, and S. suis (10, 41, 46); however, no explanation has been offered to account for this phenomenon.
The enzyme Eep (enhanced expression of pheromone) has been implicated in cleavage of the signal peptide region of bacterial lipoproteins. Eep was characterized in E. faecalis (3) and shown to cleave the signal peptides of lipoproteins to yield octapeptides that act as bacterial pheromones, inducing conjugation between different strains of E. faecalis.
The transfer-associated region of the staphylococcal conjugative plasmid pSK41 encodes a lipoprotein (TraH) the signal peptide of which includes a region of high homology (seven out of eight residues identical) to the enterococcal pheromone cAD1. Cells responsive to cAD1 signaling also respond to S. aureus (and recombinant E. coli) expressing TraH (13). In E. coli, in the absence of Lsp, the TraH-derived peptide pheromone was not produced, indicating a role for Lsp in its production (5). The data provided from the study of Eep in E. faecalis and that from the E. coli lsp mutant together suggested that these two peptidases were responsible for correctly processing the signal peptides of certain lipoproteins to produce bacterial peptide pheromones.
Consequently, we hypothesized that cleavage of MtuA, observed in the absence of Lsp, may be due to the activity from an Eep-like protein. Eep appears to be highly conserved in streptococci and lactococci, as determined by BLAST alignment (1). The in silico translation product of Eep in the single eep mutant was predicted to be interrupted at amino acid 120. This point is in the middle of the predicted intracellular region between the first and second transmembrane domains, and the resultant protein was not considered likely to be functional due to the absence of the remaining three transmembrane domains. The function of these domains is unknown, but they are predicted to be regions essential for the activity of the peptidase (7). The predicted translation product of the insertionally inactivated eep in the lsp mutant background was restricted to only the first 12 amino acids of the sequence.
In the absence of both Lsp and Eep, MtuA was present only as a full-length gene product, strongly implicating Eep in the cleavage of the protein. In the absence of Eep alone, MtuA was detected only as the mature protein, similar to that detected in the wild-type strain, indicating that Eep was likely to cleave the protein in the region removed by Lsp (the signal peptide). Such an observation would be consistent with the previously reported activities for the metallopeptidase Eep.
A reduced amount of MtuA was detected within the lsp/eep mutant compared to that in the wild type and/or each individual mutant strain; this could not be attributed to reduced growth rate, as cell densities similar to those of the wild type were achieved. The signal peptides of MtuA from S. uberis and cCF10, cOB1, cAM1, and cAD1 of E. faecalis were compared using Clustal alignments; all sequences aligned and contained lipoprotein processing determinants, i.e., a lipobox, but no other common determinant, indicative of an Eep cleavage motif, could be found.
Alternative processing of lipoproteins in the presence of an lsp mutation has been described for B. subtilis, L. monocytogenes, and L. lactis (32, 41, 46), but the mechanism by which these lipoproteins are alternatively processed was not identified in any of the studies. L. monocytogenes strain EGDe contains a protein that is a member of the M50 metallopeptidases (encoded by lmo1318 [29]) and shares 49% and 41% amino acid sequence identity with Eep of E. faecalis and that of S. uberis, respectively. The metal ion binding site is conserved. YluC of B. subtilis has been characterized as an RIP peptidase and degrades the anti-sigma factor RsiW through intramembrane proteolysis (35). This protein has the highest homology in the B. subtilis genome to Eep: amino acid identities of 44% and 39% for E. faecalis and S. uberis Eep, respectively. The protein with the highest homology to Eep in L. lactis is encoded by the ORF L181494 (6); this shows homologies of 54% and 52% with E. faecalis and S. uberis, respectively. These levels of homology to Eep suggest that the protein YluC, and gene products of L181494 and lmo1318, may constitute a conserved activity for alternatively processing lipoproteins.
Interestingly, the aberrant processing of MtuA in the absence of Lsp and/or Eep did not alter the ability of S. uberis to grow in skimmed bovine milk, suggesting that this metal transporter was functional despite retaining all or part of its signal peptide. The consequences of aberrant lipoprotein processing in other bacteria appear varied. A mutant of L. lactis lacking Lsp remained viable and able to grow in skimmed milk (46). The absence of Lsp in S. suis did not alter growth rates in nutrient-rich medium, and the mutant retained its virulence in pigs (10). In contrast, an lsp mutant of L. monocytogenes exhibited reduced efficiency for phagosomal escape during infection of eukaryotic cells and showed an attenuation in virulence (32). In M. tuberculosis growth of an lsp mutant was not affected in vitro, but virulence was attenuated in a mouse model of infection (34).
In conclusion, we have shown that alternative processing of lipoproteins, similar to that demonstrated for other gram-positive bacteria, occurs in S. uberis in the absence of Lsp activity. The alternative cleavage of a representative lipoprotein MtuA was ablated in a strain in which both Lsp and the metallopeptidase Eep were absent, indicating that both Lsp and Eep in conjunction are likely to be responsible for processing lipoproteins in this bacterium. The consequences of these findings for the pathogenesis of S. uberis within the bovine mammary gland remain a subject worthy of further investigation.
Supplementary Material
Acknowledgments
We acknowledge the financial support of BBSRC for this research.
We are grateful to the Sanger Institute for supplying genomic sequences (http://www.sanger.ac.uk/Projects/S_uberis/) and annotation data (M. Holden, personal communication) prior to formal publication. We also thank Shunichi Miyakoshi, Sankyo Co., Ltd., Japan, for the kind donation of globomycin.
Footnotes
Published ahead of print on 9 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 1841880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, F. Y., M. C. Sulavik, and D. B. Clewell. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 1815915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 5.Berg, T., N. Firth, and R. A. Skurray. 1997. Enterococcal pheromone-like activity derived from a lipoprotein signal peptide encoded by a Staphylococcus aureus plasmid. Adv. Exp. Med. Biol. 4181041-1044. [DOI] [PubMed] [Google Scholar]
- 6.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100391-398. [DOI] [PubMed] [Google Scholar]
- 8.Chandler, J. R., and G. M. Dunny. 2004. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides 251377-1388. [DOI] [PubMed] [Google Scholar]
- 9.Clewell, D. B., F. Y. An, S. E. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35246-247. [DOI] [PubMed] [Google Scholar]
- 10.De Greeff, A., A. Hamilton, I. C. Sutcliffe, H. Buys, L. Van Alphen, and H. E. Smith. 2003. Lipoprotein signal peptidase of Streptococcus suis serotype 2. Microbiology 1491399-1407. [DOI] [PubMed] [Google Scholar]
- 11.Dev, I. K., R. J. Harvey, and P. H. Ray. 1985. Inhibition of prolipoprotein signal peptidase by globomycin. J. Biol. Chem. 2605891-5894. [PubMed] [Google Scholar]
- 12.Dev, I. K., and P. H. Ray. 1984. Rapid assay and purification of a unique signal peptidase that processes the prolipoprotein from Escherichia coli B. J. Biol. Chem. 25911114-11120. [PubMed] [Google Scholar]
- 13.Firth, N., P. D. Fink, L. Johnson, and R. A. Skurray. 1994. A lipoprotein signal peptide encoded by the staphylococcal conjugative plasmid pSK41 exhibits an activity resembling that of Enterococcus faecalis pheromone cAD1. J. Bacteriol. 1765871-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flannagan, S. E., and D. B. Clewell. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44803-817. [DOI] [PubMed] [Google Scholar]
- 15.Frishman, D., K. Albermann, J. Hani, K. Heumann, A. Metanomski, A. Zollner, and H. W. Mewes. 2001. Functional and structural genomics using PEDANT. Bioinformatics 1744-57. [DOI] [PubMed] [Google Scholar]
- 16.Frishman, D., M. Mokrejs, D. Kosykh, G. Kastenmuller, G. Kolesov, I. Zubrzycki, C. Gruber, B. Geier, A. Kaps, K. Albermann, A. Volz, C. Wagner, M. Fellenberg, K. Heumann, and H. W. Mewes. 2003. The PEDANT genome database. Nucleic Acids Res. 31207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuda, A., S. Matsuyama, T. Hara, J. Nakayama, H. Nagasawa, and H. Tokuda. 2002. Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J. Biol. Chem. 27743512-43518. [DOI] [PubMed] [Google Scholar]
- 18.Hill, A. W., and J. A. Leigh. 1989. DNA fingerprinting of Streptococcus uberis: a useful tool for epidemiology of bovine mastitis. Epidemiol. Infect. 103165-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillerton, J. E., and E. A. Berry. 2005. Treating mastitis in the cow—a tradition or an archaism. J. Appl. Microbiol. 981250-1255. [DOI] [PubMed] [Google Scholar]
- 20.Innis, M. A., M. Tokunaga, M. E. Williams, J. M. Loranger, S. Y. Chang, S. Chang, and H. C. Wu. 1984. Nucleotide sequence of the Escherichia coli prolipoprotein signal peptidase (lsp) gene. Proc. Natl. Acad. Sci. USA 813708-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inukai, M., M. Takeuchi, K. Shimizu, and M. Arai. 1978. Mechanism of action of globomycin. J. Antibiot. (Tokyo) 311203-1205. [DOI] [PubMed] [Google Scholar]
- 22.Isaki, L., M. Kawakami, R. Beers, R. Hom, and H. C. Wu. 1990. Cloning and nucleotide sequence of the Enterobacter aerogenes signal peptidase II (lsp) gene. J. Bacteriol. 172469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, C. L. 2006. Ph.D. thesis. University of Sussex, Brighton, United Kingdom.
- 24.Jones, C. L., P. Monaghan, T. R. Field, A. J. Smith, P. N. Ward, and J. A. Leigh. 2004. Localization of MtuA, an LraI homologue in Streptococcus uberis. J. Appl. Microbiol. 97149-157. [DOI] [PubMed] [Google Scholar]
- 25.Jones, D. T. 2007. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23538-544. [DOI] [PubMed] [Google Scholar]
- 26.Jongeneel, C. V., J. Bouvier, and A. Bairoch. 1989. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 242211-214. [DOI] [PubMed] [Google Scholar]
- 27.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 3381027-1036. [DOI] [PubMed] [Google Scholar]
- 28.Khandavilli, S., K. A. Homer, J. Yuste, S. Basavanna, T. Mitchell, and J. S. Brown. 2008. Maturation of Streptococcus pneumoniae lipoproteins by a type II signal peptidase is required for ABC transporter function and full virulence. Mol. Microbiol. 67541-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lalioui, L., E. Pellegrini, S. Dramsi, M. Baptista, N. Bourgeois, F. Doucet-Populaire, C. Rusniok, M. Zouine, P. Glaser, F. Kunst, C. Poyart, and P. Trieu-Cuot. 2005. The SrtA sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect. Immun. 733342-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munoa, F. J., K. W. Miller, R. Beers, M. Graham, and H. C. Wu. 1991. Membrane topology of Escherichia coli prolipoprotein signal peptidase (signal peptidase II). J. Biol. Chem. 26617667-17672. [PubMed] [Google Scholar]
- 32.Reglier-Poupet, H., C. Frehel, I. Dubail, J. L. Beretti, P. Berche, A. Charbit, and C. Raynaud. 2003. Maturation of lipoproteins by type II signal peptidase is required for phagosomal escape of Listeria monocytogenes. J. Biol. Chem. 27849469-49477. [DOI] [PubMed] [Google Scholar]
- 33.Regue, M., J. Remenick, M. Tokunaga, G. A. Mackie, and H. C. Wu. 1984. Mapping of the lipoprotein signal peptidase gene (lsp). J. Bacteriol. 158632-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sander, P., M. Rezwan, B. Walker, S. K. Rampini, R. M. Kroppenstedt, S. Ehlers, C. Keller, J. R. Keeble, M. Hagemeier, M. J. Colston, B. Springer, and E. C. Bottger. 2004. Lipoprotein processing is required for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 521543-1552. [DOI] [PubMed] [Google Scholar]
- 35.Schobel, S., S. Zellmeier, W. Schumann, and T. Wiegert. 2004. The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 521091-1105. [DOI] [PubMed] [Google Scholar]
- 36.Smith, A. J., P. N. Ward, T. R. Field, C. L. Jones, R. A. Lincoln, and J. A. Leigh. 2003. MtuA, a lipoprotein receptor antigen from Streptococcus uberis, is responsible for acquisition of manganese during growth in milk and is essential for infection of the lactating bovine mammary gland. Infect. Immun. 714842-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutcliffe, I. C., and D. J. Harrington. 2002. Pattern searches for the identification of putative lipoprotein genes in Gram-positive bacterial genomes. Microbiology 1482065-2077. [DOI] [PubMed] [Google Scholar]
- 38.Sutcliffe, I. C., and D. J. Harrington. 2004. Putative lipoproteins of Streptococcus agalactiae identified by bioinformatic genome analysis. Antonie van Leeuwenhoek 85305-315. [DOI] [PubMed] [Google Scholar]
- 39.Taylor, D. L., P. N. Ward, C. D. Rapier, J. A. Leigh, and L. D. Bowler. 2003. Identification of a differentially expressed oligopeptide binding protein (OppA2) in Streptococcus uberis by representational difference analysis of cDNA. J. Bacteriol. 1855210-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tjalsma, H., V. P. Kontinen, Z. Pragai, H. Wu, R. Meima, G. Venema, S. Bron, M. Sarvas, and J. M. van Dijl. 1999. The role of lipoprotein processing by signal peptidase II in the Gram-positive eubacterium Bacillus subtilis. Signal peptidase II is required for the efficient secretion of alpha-amylase, a non-lipoprotein. J. Biol. Chem. 2741698-1707. [DOI] [PubMed] [Google Scholar]
- 42.Tjalsma, H., G. Zanen, G. Venema, S. Bron, and J. M. van Dijl. 1999. The potential active site of the lipoprotein-specific (type II) signal peptidase of Bacillus subtilis. J. Biol. Chem. 27428191-28197. [DOI] [PubMed] [Google Scholar]
- 43.Tokunaga, M., J. M. Loranger, S. Y. Chang, M. Regue, S. Chang, and H. C. Wu. 1985. Identification of prolipoprotein signal peptidase and genomic organization of the lsp gene in Escherichia coli. J. Biol. Chem. 2605610-5615. [PubMed] [Google Scholar]
- 44.Tokunaga, M., H. Tokunaga, and H. C. Wu. 1982. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc. Natl. Acad. Sci. USA 792255-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Roosmalen, M. L., N. Geukens, J. D. Jongbloed, H. Tjalsma, J. Y. Dubois, S. Bron, J. M. van Dijl, and J. Anne. 2004. Type I signal peptidases of Gram-positive bacteria. Biochim. Biophys. Acta 1694279-297. [DOI] [PubMed] [Google Scholar]
- 46.Venema, R., H. Tjalsma, J. M. van Dijl, A. de Jong, K. Leenhouts, G. Buist, and G. Venema. 2003. Active lipoprotein precursors in the Gram-positive eubacterium Lactococcus lactis. J. Biol. Chem. 27814739-14746. [DOI] [PubMed] [Google Scholar]
- 47.von Heijne, G. 1989. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2531-534. [DOI] [PubMed] [Google Scholar]
- 48.Ward, P. N., T. R. Field, W. G. Ditcham, E. Maguin, and J. A. Leigh. 2001. Identification and disruption of two discrete loci encoding hyaluronic acid capsule biosynthesis genes hasA, hasB, and hasC in Streptococcus uberis. Infect. Immun. 69392-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamagata, H. 1983. Temperature-sensitive prolipoprotein signal peptidase in an Escherichia coli mutant: use of the mutant for an efficient and convenient assay system. J. Biochem. (Tokyo) 931509-1515. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, X. J., and H. C. Wu. 1992. Nucleotide sequence of the Staphylococcus aureus signal peptidase II (lsp) gene. FEBS Lett. 29980-84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.