Abstract
Erwinia carotovora subsp. carotovora produces an array of extracellular proteins (i.e., exoproteins), including plant cell wall-degrading enzymes and Harpin, an effector responsible for eliciting hypersensitive reaction. Exoprotein genes are coregulated by the quorum-sensing signal, N-acyl homoserine lactone, plant signals, an assortment of transcriptional factors/regulators (GacS/A, ExpR1, ExpR2, KdgR, RpoS, HexA, and RsmC) and posttranscriptional regulators (RsmA, rsmB RNA). rsmB RNA production is positively regulated by GacS/A, a two-component system, and negatively regulated by HexA (PecT in Erwinia chrysanthemi; LrhA [LysR homolog A] in Escherichia coli) and RsmC, a putative transcriptional adaptor. While free RsmA, an RNA-binding protein, promotes decay of mRNAs of exoprotein genes, binding of RsmA with rsmB RNA neutralizes the RsmA effect. In the course of studies of GacA regulation, we discovered that a locus bearing strong homology to the flhDC operon of E. coli also controls extracellular enzyme production. A transposon insertion FlhDC− mutant produces very low levels of pectate lyase, polygalacturonase, cellulase, protease, and E. carotovora subsp. carotovora Harpin (HarpinEcc) and is severely attenuated in its plant virulence. The production of these exoproteins is restored in the mutant carrying an FlhDC+ plasmid. Sequence analysis and transcript assays disclosed that the flhD operon of E. carotovora subsp. carotovora, like those of other enterobacteria, consists of flhD and flhC. Complementation analysis revealed that the regulatory effect requires functions of both flhD and flhC products. The data presented here show that FlhDC positively regulates gacA, rsmC, and fliA and negatively regulates hexA (lrhA). Evidence shows that FlhDC controls extracellular protein production through cumulative effects on hexA and gacA. Reduced levels of GacA and elevated levels of HexA in the FlhDC− mutant are responsible for the inhibition of rsmB RNA production, a condition conducive to the accumulation of free RsmA. Indeed, studies with an RsmA− FlhDC− double mutant and multiple copies of rsmB+ DNA establish that the negative effect of FlhDC deficiency is exerted via RsmA. The FlhDC-mediated regulation of fliA has no bearing on exoprotein production in E. carotovora subsp. carotovora. Our observations for the first time establish a regulatory connection between FlhDC, HexA, GacA, and rsmB RNA in the context of the exoprotein production and virulence of E. carotovora subsp. carotovora.
Erwinia carotovora subsp. carotovora (Ecc), a member of the Enterobacteriaceae family, causes soft-rotting disease on a wide variety of plants worldwide. A consortium of plant cell wall-degrading extracellular enzymes comprising pectate lyase (Pel), polygalacturonase (Peh), protease (Prt), and cellulase (Cel) contribute to its plant virulence (4, 15, 19, 61, 71). Among them, extracellular pectinases, including Pel and Peh, play a crucial role in tissue maceration and cell death. In addition, motility and some effectors secreted through the type III secretion system also augment virulence of E. carotovora subsp. carotovora (12, 37, 65).
The regulation of the extracellular enzymes and proteins including Harpin, the elicitor of the hypersensitive response, in E. carotovora subsp. carotovora has been extensively studied, and many regulatory genes and factors have been identified. These extracellular proteins are coregulated by plant signals, quorum-sensing signals (39, 62), as well as by an assortment of transcriptional factors and posttranscriptional factors (Fig. 1), including the RsmA-rsmB system (13, 23, 48), RsmC (HexY [25, 67]), the GacS/GacA two-component system (20, 28, 30), KdgR (49), RpoS (9, 57), ExpR1 (22), ExpR2 (VirR [8, 21]), Hor (70), and HexA (34, 56). Of these regulators, the posttranscriptional system comprising RsmA and rsmB RNA is absolutely critical in the expression of exoprotein genes. Indeed, many of the transcriptional regulators and the quorum-sensing signal, N-acyl homoserine lactone (AHL), controlling exoprotein production actually act via RsmA-rsmB RNA (Fig. 1). In this system, RsmA, a small RNA-binding protein, promotes mRNA decay (13, 23). rsmB specifies an untranslated regulatory RNA that binds RsmA and neutralizes its negative regulatory effect (48). GacS, the putative sensor kinase, and GacA, the cognate response regulator, members of a widely occurring two-component system, control exoprotein production in E. carotovora subsp. carotovora, mainly via regulating rsmB (20, 28). HexA negatively controls exoprotein and AHL production, as well as motility (34, 56). Our studies demonstrated that the HexA effect on exoprotein production occurs via rsmB (56). In addition, RsmC, a putative transcriptional adaptor, affects exoproteins by modulating the levels of rsmB RNA (25), although the underlying regulatory mechanism remains unknown. KdgR, an IcII-like protein, negatively controls exoproteins by inhibiting the transcription of rsmB by a novel “road-block” mechanism (49). RpoS, an alternate sigma factor, negatively affects the production of exoprotein by stimulating rsmA transcription (57). The two LuxR homologs ExpR1 and ExpR2 activate rsmA transcription in the absence of AHL (21, 22). However, ExpR2, but not ExpR1, severely reduces exoprotein production and attenuates virulence, and its effects are neutralized by AHL.
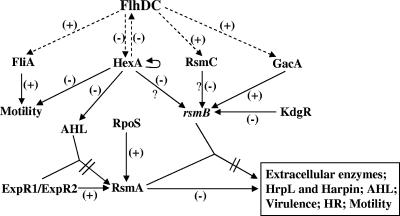
FIG. 1.
Model depicting the regulatory network controlling the production of extracellular enzymes, HrpL, Harpin, and AHL, as well as motility, pathogenicity, and the hypersensitive response in E. carotovora subsp. carotovora (see the text for the details). The regulatory steps indicated by broken lines with arrows are based upon the results presented in this report.
Our findings disclosed a crucial role of GacA in rsmB expression (20). However, gacA expression itself is regulated, as is apparent from the effects of cultural conditions such as medium composition and growth phase. Surprisingly, despite extensive studies of the GacS/A-mediated regulation in various bacteria (14, 20, 28, 35), little is known of genes and environmental factors controlling gacS/A expression. One objective of the present study was to identify regulators controlling the production of GacA. For this, we used a mutagenesis/selection strategy that led to the isolation of mutants that no longer responded to the presence of gacA carrying its native promoter. Subsequent studies revealed that one such class of mutants resulted from inactivation of the flhDC operon.
Flagellar motility is an accessory virulence determinant in animal pathogens, as well as in several plant pathogens, including E. carotovora (31, 37, 40, 44, 52, 53, 59). The FlhDC complex, comprising the products of flhD and flhC, is the master regulatory operon controlling the expression of flagellar genes in Escherichia coli and Salmonella enterica serovar Typhimurium (5, 7, 18, 53). The flagellum-chemotaxis regulon in E. coli and serovar Typhimurium comprises more than 50 genes organized into at least 14 operons. The transcription of these flagellar genes is organized in a three-tier hierarchy: class I, class II, and class III. flhD and flhC comprise the class I genes, and the products are known to form an FlhD4C2 hexamer complex (72). Both FlhD and FlhC subunits are essential for effective transcription. FlhC protein is the DNA-binding component, and its function is strengthened by FlhD. Claret and Hughes (18) showed that reconstituted FlhD2C2 (= FlhD4C2) complex from purified FlhD and FlhC subunits increases the specificity of DNA binding and also increases the stability of the resultant DNA interaction in vitro. The action of FlhD would ensure that FlhC efficiently locates its multiple target genes and stabilizes the FlhC-DNA complex. FlhDC complex binds promoter regions of the class II genes and activates their transcription. The class II flagellar regulon includes genes that encode proteins for the basal body and the hook of the flagellum, as well as two regulators, FliA and FliM. FliA is an alternative sigma factor (σ28) specific for the flagellar regulon, and FlgM acts as an anti-sigma factor that inhibits FliA-dependent transcription by stripping FliA from the core of RNA polymerase, as well as by preventing the sigma-core interaction (2, 3, 10, 43, 63, 68). FliA is required for the expression of some class II genes and all class III genes. The class III genes encode components for assembly of the flagellar filament, chemotaxis proteins, and motor activity (1, 17, 18, 53).
In addition to the flagellum-chemotaxis regulon for swimming motility, the flagellar master operon FlhDC controls genes for virulence, the type III secretion system, and extracellular enzyme production. For example, it is required for the expression of the extracellular phospholipase gene, as well as swarming motility in Serratia liquefaciens (33). Bleves et al. (7) reported that the yop regulon is upregulated in an FlhDC− mutant in Yersinia enterocolitica. The genes of the yop regulon encode proteins for the Yop virulon, which are secreted via the Yop secretion apparatus designated as the type III secretion system. Yop proteins enable bacteria in close contact with target cells to inject bacterial toxic proteins directly into the cytosol of the target cells. Kapatral et al. (41) determined that FlhDC regulates the expression of genes for enzymes involved in the synthesis and degradation of carbamoylphosphate in Y. enterocolitica. In addition to motility, FlhDC is also required for lipolysis, extracellular hemolysis, and full virulence in the insect pathogenic bacterium Xenorhabdus nematophilus (32). Remarkably, a recent study by of Park and Forst (60) showed that the FlhDC effect on extracellular enzymes in X. nematophilus occurs via FliA. FlhD alone is found to regulate the cell division rate in E. coli (64).
As stated above, in E. carotovora subsp. carotovora the FlhDC operon plays an important role in pathogenicity and controls motility. Hossain et al. (37) reported that the nonmotile state in ΔfliC and ΔmotA mutants of the E. carotovora subsp. carotovora strain EC1 reduced their ability to cause soft-rot disease on Chinese cabbage, but the mutations had no deleterious effect on the levels of major extracellular enzymes. In addition, Matsumoto et al. (52) showed that the FlhC− and FlhD− mutants of E. carotovora subsp. carotovora strain EC1-N caused a severe reduction in the transcript levels of fliC and fliA, flagellum synthesis, and virulence in Chinese cabbage and potato. However, how the FlhDC master regulon affected virulence in E. carotovora subsp. carotovora remained unknown. We report here the characteristics of the flhDC operon of E. carotovora subsp. carotovora strain Ecc71 and the effects of this operon on various global regulators known to control extracellular protein production and motility. We show that (i) FlhDC is required for the production of major extracellular enzymes and E. carotovora subsp. carotovora Harpin (HarpinEcc), as well as virulence in E. carotovora subsp. carotovora; (ii) as a master regulator, FlhDC controls the expression of several key regulatory genes and sigma factor genes such as gacA, rsmC, hexA, fliA, and hrpL; (iii) FlhDC activates regulatory rsmB RNA production via its effects on GacA and HexA; and (iv) the sigma factor FliA does not affect extracellular protein production.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids in this study are described in Table 1. E. carotovora subsp. carotovora strains Ecc71 and AC5006 were maintained on LB agar. The strains carrying antibiotic markers were maintained on LB agar containing appropriate antibiotics.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. carotovora subsp. carotovora | ||
| Ecc71 | Wild type | 73 |
| AC5006 | Lac- derivative of Ecc71 | 58 |
| AC5047 | Nalr derivative of AC5006 | 13 |
| AC5070 | Kmr, RsmA- derivative of AC5047 | 13 |
| AC5140 | Kmr, FlhDC- derivative of Ecc71 | This study |
| AC5141 | Kmr, FlhDC- derivative of AC5006 | This study |
| AC5142 | Kmr Spr, FlhDC- RsmC- derivative of AC5006 | This study |
| AC5143 | Kmr Gmr, FlhDC- GacA- derivative of Ecc71 | This study |
| AC5144 | Kmr Spr, FlhDC- RsmA- derivative of AC5006 | This study |
| AC5145 | Kmr Spr, FlhDC- HexA- derivative of Ecc71 | This study |
| E. coli DH5α | φ80lacZΔM15 Δ(lacZYA-argF)U169 hsdR17 recA1 endA1 thi-1 | Gibco BRL |
| Plasmids | ||
| p34S-Gm | Gmr, source of Gmr cassette | 27 |
| pBluescript SK(+) | Apr | Stratagene |
| pCL1920 | Spr Smr | 46 |
| pCL1920Gmr | Spr Gmr, Gmr cassette in pCL1920 | This study |
| pMP220 | Tcr, promoter-probe vector | 69 |
| pRK415 | Tcr | 42 |
| pRK2013 | Kmr, Mob+, Tra+ | 29 |
| pUT mini-Tn5-Sp | A lambda-pir vector containing mini-Tn5-Sp transposon | 26 |
| pUT mini-Tn5-Km | A lambda-pir vector containing mini-Tn5-Km transposon | 26 |
| pAKC781 | Apr, peh-1 DNA in pBluescript SK(+) | 47 |
| pAKC783 | Apr, pel-1 DNA in pBluescript SK(+) | 47 |
| pAKC882 | Apr, rsmA coding region in pT7-7 | 55 |
| pAKC924 | Apr, hrpNEcc DNA in pBluescript SK(+) | 24 |
| pAKC975 | Spr, rsmC DNA in pCL1920 | 25 |
| pAKC980 | Spr Tcr, inactivated rsmC DNA in pRK415 | 25 |
| pAKC983 | Spr Tcr, inactivated hexA DNA in pRK415 | 56 |
| pAKC985 | Kmr, hexA DNA in pDK6 | 56 |
| pAKC1004 | Spr, rsmB DNA in pCL1920 | 48 |
| pAKC1111 | Tcr, gacADC3000 DNA in pLARF5 | 14 |
| pAKC1034 | Apr, 200-bp celV fragment in pGEM-T Easy | 49 |
| pAKC1049 | Spr, plac-rsmB+ in pCL1920 | 51 |
| pAKC1049Gmr | Spr, Gmr,Gmr cassette in pAKC1049 | This study |
| pAKC1057 | Gmr Tcr, inactivated gacA DNA in pRK415 | 20 |
| pAKC1071 | Apr, hrpL DNA in pBluescript SK(+) | 11 |
| pAKC1240 | Apr Kmr, 6.5-kb ClaI fragment containing mini-Tn5-Kmr inactivated flhDC in pBluescript SK(+) | This study |
| pAKC1241 | Spr, plac-flhD in pCL1920 | This study |
| pAKC1242 | Spr, plac-flhDC in pCL1920 | This study |
| pAKC1243 | Tcr, gacA-lacZ in pMP220 | This study |
| pAKC1244 | Tcr, rsmC-lacZ (containing DNA segment of −684 nt to +57 nt from the putative translational start site) in pMP220 | This study |
| pAKC1245 | Spr, plac-gacA in pCL1920 | This study |
| pAKC1246 | Spr, plac-fliA in pCL1920 | This study |
| pAKC1249 | Tcr Kmr, inactivated flhDC in pRK415 from pAKC1240 | This study |
| pAKC1250 | Tcr, rsmC-lacZ (containing DNA segment of −323 nt to +57 nt from the putative translational start site) in pMP220 | This study |
nt, nucleotide.
The compositions of LB, nutrient gelatin, and minimal salts media have been described in previous publications (13, 58). When required, antibiotics were supplemented as follows: ampicillin (Ap), 100 μg/ml; gentamicin (Gm), 10 μg/ml; kanamycin (Km), 50 μg/ml; spectinomycin (Sp), 50 μg/ml; and tetracycline (Tc), 10 μg/ml. Media were solidified by using 1.5% (wt/vol) agar.
The composition of media for agarose plate assays for enzymatic activities was described by Chatterjee et al. (13).
Extracellular enzyme assays.
Bacterial cells were grown in minimal salts medium supplemented with sucrose (0.5% [wt/vol]) without or with appropriate drugs to a Klett value of ca. 250, and the cultural supernatants were used for assays. The quantitative extracellular Pel enzymatic assay, as well as the semiquantitative Pel, Peh, Prt, and Cel agarose plate assays, was performed according to previously published procedures (13, 16). Briefly, the reaction mixture for quantitative Pel assays contained 0.24 ml of substrate (0.575% polygalacturonic acid [pH 5.5]), 0.26 ml of reaction buffer (230 mM Tris-HCl [pH 8.5], 0.78 mM CaCl2), and enzyme or water in a total volume of 0.6 ml. The Pel specific activities were measured at an A235 in a spectrophotometer. The Pel activities are expressed as units/per ml/A600. One unit of Pel activity is defined as the amount of enzyme that produces 1 μmol of unsaturated digalacturonic acid equivalent per min at 30°C. For semiquantitative assays of Pel, Peh, Cel, and Prt, wells were made in agarose media with a number 2 cork borer, and the bottoms were sealed with molten agarose (0.8% [wt/vol]). Samples were applied to the wells, and the plates were incubated at 28°C. After 16 to 18 h, Pel and Peh assay plates were developed with 4 N HCl, and the Cel assay plates were developed with Congo red and NaCl solutions. Halos around the wells due to protease activity became visible in Prt assay plates within 24 to 36 h without any further treatment.
Plant tissue maceration.
The celery petiole assay was performed as described by Murata et al. (58). The extent of tissue maceration was estimated visually.
Determination of nucleotide sequences of Ecc71 flhDC and sequence alignment.
The chromosomal DNA of the FlhDC− mutant AC5140 containing mini-Tn5-Kmr fragment was digested with ClaI and ligated with ClaI-digested pBluescript SK(+). The ligated DNAs were electroporated into DH5α and selected on LB plus Ap and Km agar to yield pAKC1240. The nucleotide sequence of flhDC was determined from pAKC1240 using primers of transposon sequences. Nucleotide sequencing was performed at the DNA Core Facility of University of Missouri-Columbia. The amino acid sequences of FlhDC of different bacteria were obtained from GenBank. Sequence alignment was performed by using CLUSTAL W (www.expasy.ch), and default parameters were used.
DNA techniques.
Standard procedures were used in the isolation of plasmids and chromosomal DNA, gel electrophoresis, DNA ligation, transformation, and electroporation (66). Restriction and modification enzymes were obtained from Promega Biotec (Madison, WI). Prime-a-Gene DNA labeling system (Promega Biotec) was used for labeling DNA probes.
Construction of FlhDC- mutants.
The insertion DNA containing inactivated flhDC DNA in pAKC1240 was transcloned into pRK415 to yield pAKC1249. The FlhDC- mutants AC5140 and AC5141 were constructed by marker exchange of Ecc71 and AC5006 with pAKC1249, respectively. The FlhDC- RsmC- mutant AC5142 was obtained by marker exchange of AC5141 with pKC980. The FlhDC- GacA- mutant AC5143, and the FlhDC- HexA- mutant AC5145 was obtained by marker exchange of AC5140 with pAKC1057 and pAKC983, respectively. The procedures for marker exchange have been described in Chatterjee et al. (13). The FlhDC- RsmA- mutant AC5144 was constructed by inactivating rsmA in AC5141 using mini-Tn5-Spr. Inactivation of target genes in mutants were confirmed by Northern blot analysis.
Construction of plac-flhD, plac-flhDC, plac-gacA, and plac-fliA plasmids, as well as rsmC-lacZ and gacA-lacZ fusions.
DNA fragments containing coding regions of flhD, gacA, and fliA of Ecc71 were PCR amplified by using the primer pairs 71flhD1-71flhD2, 71gacA1-71gacA2, and 71fliA1-71fliA2, respectively (see Table 2 for the primer sequences). The amplified DNA fragments were cloned into pCL1920 to yield pAKC1241, pAKC1245, and pAKC1246. The plac-flhDC plasmid pAKC1242 was constructed by cloning a SalI fragment containing flhDC+ DNA behind the lac promoter in pCL1920. To construct rsmC-lacZ and gacA-lacZ fusions, PCR-amplified DNA fragments containing gacA and rsmC upstream DNAs by using the primer pairs 71gacAZ1-71gacAZ2 and 71rsmCZ1-71rsmCZ2 (Table 2) were cloned into pMP220 to yield pAKC1243 and pAKC1244, respectively.
TABLE 2.
Primers used for PCR amplification
| Primer | Sequence (5′-3′) |
|---|---|
| 71flhD1 | GATGGATTCATAGCCTGTCGGGATGGGAAATATG |
| 71flhD2 | GATCTGCAGTCTCCGCCATTACTTATGCCC |
| 71flhC1 | GATGGATTCGGGCATAAGTAATGGCGGAGA |
| 71flhC2 | GATAAGCTTACAGGCTCAGACTGCGTGTT |
| 71gacA1 | GATGGATCCGGAGAATTATTCTTTGATTAGCG |
| 71gacA2 | GATAAGCTTGCCGACGCATCGAAATCTTCAC |
| 71fliA1 | TGCGGATCCACGCTATTCAGGCGATTGGCTACC |
| 71fliA2 | TGCAAGCTTCGCAGCGCAATTAAACATCGTTC |
| 71gacAZ1 | TGCAGATCTGATGCGGTGAGCAATAGTGCT |
| 71gacAZ2 | TGCGAATTCGGTCATCAACAAGAAAAACGCTAAT |
| 71rsmCZ1 | TGCAGATCTGAATTATCAGTGCTGTTATAATGTC |
| 71rsmCZ2 | TGCTCTAGACTGAACTGGTTGAGAAAGCATGCC |
Northern blot and Western blot analyses.
Bacterial cultures were grown at 28°C in minimal salts medium supplemented with sucrose (0.5% [wt/vol]). Cells were collected while cultures reached a Klett value of ca. 220. RNA isolation and Northern blot analysis were performed as described by Liu et al. (47). The probes used were the 183-bp NdeI-SalI fragment of rsmA from pAKC882, the 314-bp EcoRV-KpnI fragment of pel-1 from pAKC 783, the 743-bp HindIII fragment of peh-1 from pAKC781, the 200-bp EcoRI fragment of celV from pAKC1034, the 779-bp EcoRV-SmaI fragment of hrpN from pAKC924, the 321-bp BamHI-HindIII fragment of rsmB from pAKC1004, the 304-bp EcoRV-KpnI fragment of rsmC from pAKC975, the 950-bp EcoRI-SalI fragment of hexA from pAKC985, the 287-bp BglII-EcoRV fragment of hrpL from pAKC1071. and the 811-bp BamHI-HindIII fragment of fliA from pAKC1246. The flhD, flhC, and gacA probes were PCR amplified using the primer pairs 71flhD1-71flhD2, 71flhC1-71flhC2, and 71gacA1-71gacA2, respectively. For Western blot analysis, bacterial cells were collected, suspended in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer (66), and boiled. The protein concentrations were determined by using a CB-X protein assay kit (Geno Technology, Inc., St. Louis, MO) according to the manufacturer's specifications. Western blot analysis of the total bacterial protein was performed as described by Mukherjee et al. (55). The antisera raised against HarpinEch (6) were used as probes.
β-Galactosidase assays.
Bacterial constructs were grown at 28°C in minimal salts medium plus sucrose and Tc to a Klett value of ca. 200. The β-galactosidase assays were performed according to the method of Miller (54).
Extracellular enzyme assays, Northern blot and Western blot analyses, β-galactosidase assays, and pathogenicity tests were performed at least twice, and the results were reproducible.
RESULTS AND DISCUSSION
E. carotovora subsp. carotovora flhDC is closely related to enterobacterial genes.
To identify gene(s) regulating GacA, we screened a library of Tn5-insertion mutants of Ecc71. Since GacA is a positive regulator of exoprotein production, a mutant downregulated in GacA expression was predicted to produce reduced levels of exoproteins. We mutagenized with mini-Tn5-Kmr, a GacA- mutant of Ecc71 carrying a plasmid containing gacA+ DNA of Pseudomonas syringae pv. tomato strain DC3000. Five colonies that did not produce protease on nutrient gelatin agar were selected, and chromosomal DNA fragments carrying mini-Tn5-Kmr were cloned from these mutants and sequenced using the mini-Tn5 primers. The sequence data revealed that in one of those mutants the Tn5 insertion is located 80 bases upstream of an open reading frame that encodes a homolog of the flagellar transcriptional activator, FlhD. The flagellar transcriptional activator gene, flhC, possessing a putative start site three bases after the stop codon of flhD, is also present in the mini-Tn5 containing fragment cloned from this mutant. The plasmid containing this fragment was used for marker exchange with the E. carotovora subsp. carotovora strains Ecc71 and AC5006 to yield AC5140 and AC5141, respectively. Northern blot analysis with flhD or flhC DNA as probes revealed that Ecc71 produced ∼1.1-kb transcripts (Fig. 2A, lane 1), whereas with those two probes no hybridization signal was detected with RNA of AC5140 (Fig. 2A, lane 2) or the RNA from AC5141 (data not shown). These results indicated that flhD and flhC are components of one transcriptional unit and that the mutants AC5140 and AC5141 are FlhDC-. The Tn5 insertions in the other four mutants are located in ahlI, a gene that encodes AHL synthase.
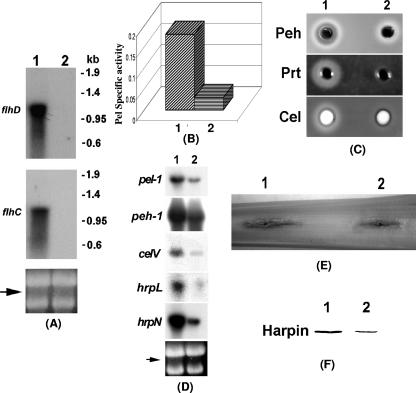
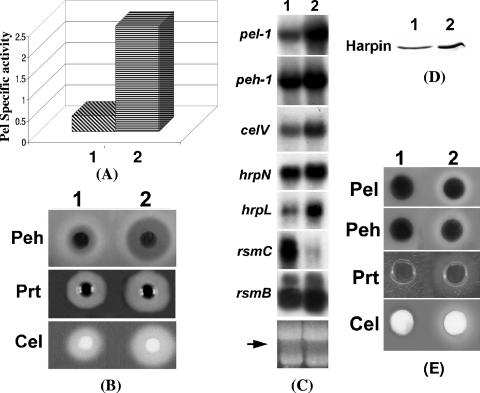
FIG. 2.
Characteristics of an FlhDC- mutant (1). Ecc71 and (2) its FlhDC- mutant AC5140. (A) Northern blot analysis of flhD and flhC; (B) Pel activities; (C) agarose plate assays of Peh, Prt, and Cel activities; (D) Northern blot analysis of pel-1, peh-1, celV, hrpL, and hrpN. For panels A and D, each lane contained 15 μg of total RNA. The arrows show the levels of total RNA as revealed by ethidium bromide staining of denatured agarose gel. (E) Soft-rot disease symptoms in celery petiole; (F) Western blot analysis of HarpinEcc protein. Each lane contained 20 μg of total protein.
flhD and flhC genes are linked together and share high homology in different bacteria. As an example, the translational start site of flhC, as in E. carotovora subsp. carotovora, is three bases behind the stop codon of flhD in both E. coli and S. enterica serovar Typhimurium. The sequence alignment results revealed that the deduced amino acid sequences of Ecc71 FlhD and FlhC have strong homology with previously reported FlhD and FlhC proteins of various enterobacterial species. The percent identities of E. carotovora subsp. carotovora FlhD and FlhC with the cognate proteins, shown parenthetically in that order, are as follows: E. carotovora subsp. atroseptica strain SCRI1043 (accession number YP_049786 [97% identical] and accession number YP_049787 [99% identical]), Serratia marcescens (accession number O85806 [83% identical] and accession number O85807 [88% identical]), Serratia proteamaculans strain 568 (accession number YP_001479218 [82% identical] and accession number YP_001479217 [89% identical]), Y. enterocolitica subsp. enterocolitica strain 8081 (accession number YP_001006784 [81% identical] and accession number YP_001006783 [87% identical]), E. coli strain EDL933 (accession number NP_288329 [76% identical] and accession number NP_288328 [83% identical]), and S. enterica serovar Typhimurium strain LT2 (accession number NP_460882 [74% identical] and accession number NP_460881 [82% identical]).
FlhDC positively controls extracellular protein production and pathogenicity. (i) Effects on extracellular enzymes.
To examine the effects of FlhDC on extracellular enzyme production, the culture supernatants of the FlhDC- mutant AC5140 and its parent strain, Ecc71, were assayed to determine their enzymatic activities, and the cells were used for the extraction of total RNAs for transcript assays. The levels of extracellular Pel (Fig. 2B) and Peh, Prt, and Cel (Fig. 2C) produced by AC5140 were much lower than those produced by Ecc71. Similarly, the transcript levels of pel-1, peh-1, and celV (Fig. 2D) of the FlhDC- mutant also were lower than those of the parent.
(ii) Effects on pathogenicity.
A positive correlation between the levels of extracellular enzymes and virulence of E. carotovora subsp. carotovora has been established (4, 59, 71). Since extracellular enzyme production is suppressed in the FlhDC- mutant AC5140, a reduced level of virulence of the mutant was expected. The pathogenicity test in celery petioles (Fig. 2E) showed that indeed the degree of maceration caused by AC5140 was reduced compared to that caused by Ecc71.
(iii) Effects on the sigma factor HrpL that controls effector (Harpin) production.
We have shown that extracellular enzymes secreted by the type I and type II secretion systems, as well as proteins (effectors) secreted by the type III system, are coregulated in E. carotovora subsp. carotovora (11, 12, 20, 22, 24, 25, 4, 49, 55, 57). These findings raised the possibility that FlhDC in E. carotovora subsp. carotovora could control the expression of effector genes as well as the type III secretion system. To test this hypothesis, we first compared the transcript levels of hrpL, the gene for an alternate sigma factor known to control the expression of genes that encode effectors secreted via the type III pathway, as well as genes specifying the type III secretion machinery. The results (Fig. 2D) revealed that the level of hrpL transcript was much reduced in the FlhDC- mutant (lane 2) compared to the parent (lane 1). The expression of E. carotovora subsp. carotovora hrpN, the gene that encodes HarpinEcc, as well as the levels of HarpinEcc protein, was much lower in AC5140 (Fig. 2D and F, lane 2) compared to Ecc71 (Fig. 2D and F, lane 1). These findings strengthen the notion that FlhDC belongs to the global regulatory network that controls exoprotein production in E. carotovora subsp. carotovora through the type I, type II, and type III secretion systems.
(iv) Multiple copies of flhDC restore exoprotein production in the FlhDC- mutant.
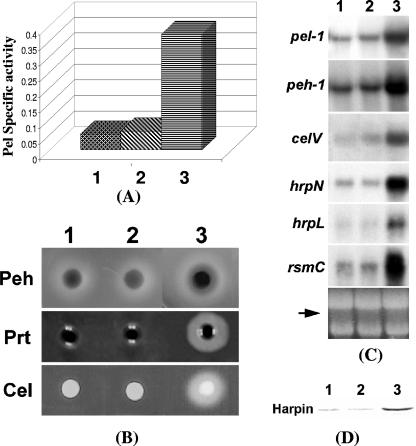
To test whether multiple copies of flhD or flhDC could restore the extracellular enzyme production and expression of the cognate genes, we cloned flhD+ DNA and flhDC+ DNA behind the lac promoter in vector pCL1920 to yield pAKC1241 and pAKC1242, respectively. Extracellular enzyme levels (Fig. 3A, sample 2, and Fig. 3B, lane 2), as well as the transcript levels of pel-1, peh-1, and celV (Fig. 3C, lane 2), of the FlhDC- mutant carrying flhD+ plasmid were similar to that of the mutant carrying the cloning vector (Fig. 3A, sample 1; Fig. 3B, lane 1; Fig. 3C, lane 1). Similarly, a plasmid carrying flhC driven by the lac promoter of pCL1920 failed to restore extracellular protein production in the FlhDC- mutant (data not shown). In contrast, the levels of transcripts of pel-1, peh-1, and celV (Fig. 3C, lane 3) and extracellular enzyme levels (Fig. 3A, sample 3, and Fig. 3B, lane 3) in the FlhDC- mutant were restored by the flhDC+ DNA. These results demonstrate that the regulatory function requires both flhD and flhC products. In addition to these exracllular enzymes, multiple copies of flhDC also restored the levels of hrpL and hrpN transcripts (Fig. 3C, lane 3), as well as the HarpinEcc protein (Fig. 3D, lane 3) in the mutant.
FIG. 3.
Reversal of the pleiotropic phenotype of the FlhDC- mutant by flhDC+ DNA. (A) Pel activities; (B) agarose plate assays of Peh, Prt, and Cel activities; (C) Northern blot analysis of pel-1, peh-1, celV, hrpN, hrpL, and rsmC. Each lane contained 15 μg of total RNA. The arrow shows the levels of total RNA as revealed by ethidium bromide staining of denatured agarose gel. (D) Western blot analysis of HarpinEcc of the FlhDC- mutant AC5140 carrying pCL1920 (cloning vector) (lane 1), pAKC1241 (flhD+) (lane 2), or pAKC1242 (flhDC+) (lane 3). Each lane contained 20 μg of total protein.
FlhDC controls the expression of several regulatory genes.
To understand whether FlhDC controls the expression of exoprotein genes via other regulatory genes, we examined the effects of FlhDC deficiency on expression of several global regulatory genes whose products are known or are predicted to control exoprotein production, motility, and virulence. We present below the evidence for FlhDC effects on the expression of gacA, hexA, rsmC, and fliA.
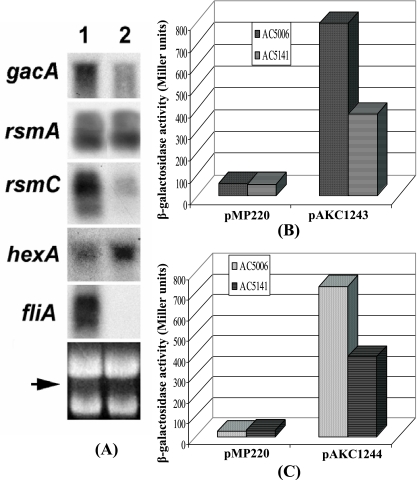
(i) Positive regulation of gacA.
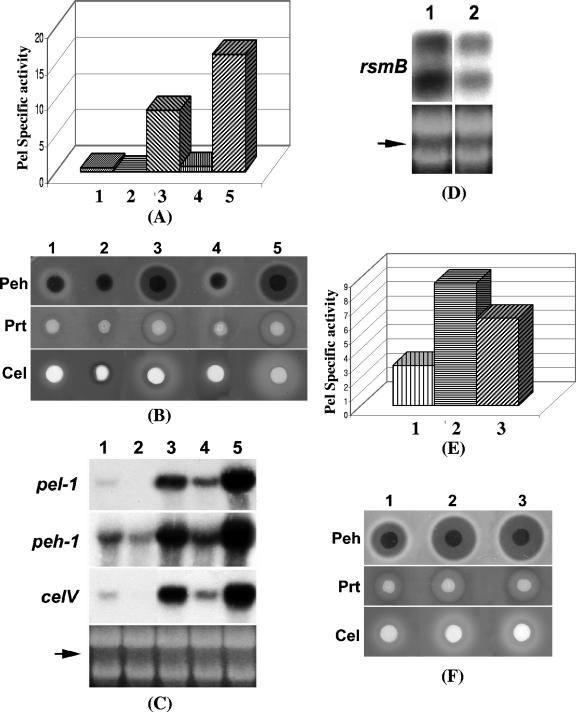
A Northern blot analysis (Fig. 4A) revealed that the transcript levels of gacA were much reduced in the FlhDC- mutant compared to those in Ecc71. In addition, β-galactosidase assay data (Fig. 4B) demonstrated that expression of a gacA-lacZ fusion plasmid pAKC1243 was lower in the FlhDC- mutant than in the parent. It has been well established that FlhDC complex activates transcription of class II flagellar genes by binding within the promoter regions of target genes and interacting with the C-terminal region of the α subunit of RNA polymerase (50). The consensus FlhDC binding sequences have been identified as GCAATAA and TTATTCC with several variations (17). However, sequence analysis revealed that only one element (TTATTCC) of the FlhDC binding sequence occurs upstream of gacA. The significance of this sequence in FlhDC-mediated positive regulation of gacA expression is not known.
FIG. 4.
FlhDC controls regulatory genes for exoprotein production and motility. (A) Northern blot analysis of gacA, rsmA, rsmC, hexA, and fliA in Ecc71 (lane 1) and its FlhDC- mutant AC5140 (lane 2). Each lane contained 15 μg of total RNA. The arrow shows the levels of total RNA as revealed by ethidium bromide staining of denatured agarose gel. (B and C) β-Galactosidase activities of transcriptional gacA-lacZ fusion pAKC1243 and rsmC-lacZ fusion pAKC1244 in AC5006 and AC5141, respectively.
(ii) Negative regulation of hexA.
A Northern blot analysis (Fig. 4A) revealed that the FlhDC- mutant produced higher levels of hexA transcript than the parent. In addition, a potential FlhDC binding site (GAAATAA N16 TTATTGA) was found 202 bases upstream of the putative translational start site of the hexA gene. Thus, one possible explanation for the negative effects of FlhDC on hexA expression is that FlhDC binds sequences upstream of hexA and represses its expression.
The effects of FlhDC on exoprotein production are channeled via GacA and HexA, and the evidence for this is discussed below.
(iii) Positive regulation of rsmC.
Northern blot analysis results (Fig. 4A) revealed that the transcript level of rsmC was severely reduced in the FlhDC- mutant AC5140 compared to the level in Ecc71. Expression of a transcriptional rsmC-lacZ fusion plasmid pAKC1244 was lower in the FlhDC- mutant AC5141 than in the parent AC5006 (Fig. 4C). Furthermore, our data demonstrate that AC5140 carrying the flhDC+ plasmid pAKC1242 (Fig. 3C, lane 3), but not the flhD+ plasmid pAKC1241 (Fig. 3C, lane 2), produces higher levels of rsmC RNA than the mutant carrying the vector (Fig. 3C, lane 1). A putative FlhDC binding site (GCATAAA N8 TTATTCA) very similar to the consensus sequence was found 564 bp upstream of the predicted rsmC translational start site. A transcriptional rsmC-lacZ fusion plasmid pAKC1250 which lacks this putative FlhDC binding sequence produced similar β-galactosidase activities in AC5141 and AC5006 (data not shown). These data suggested that FlhDC binds to this putative FlhDC binding sequence and activates the expression of rsmC.
To examine the possibility that FlhDC controls exoprotein production via regulating rsmC expression, we made an FlhDC- RsmC- double mutant AC5142 by marker exchange of AC5141 with pAKC980. In this plasmid rsmC is inactivated by inserting an omega (Spr) fragment (25). We should recall that RsmC- mutants of Ecc71 produce much higher levels of exoproteins than the parent (25). Extracellular enzyme assay results revealed that inactivation of RsmC in the FlhDC- mutant did not restore extracellular enzyme production (data not shown). Thus, RsmC deficiency in the FlhDC- background has no bearing on exoprotein production. The basis for this response is currently under investigation.
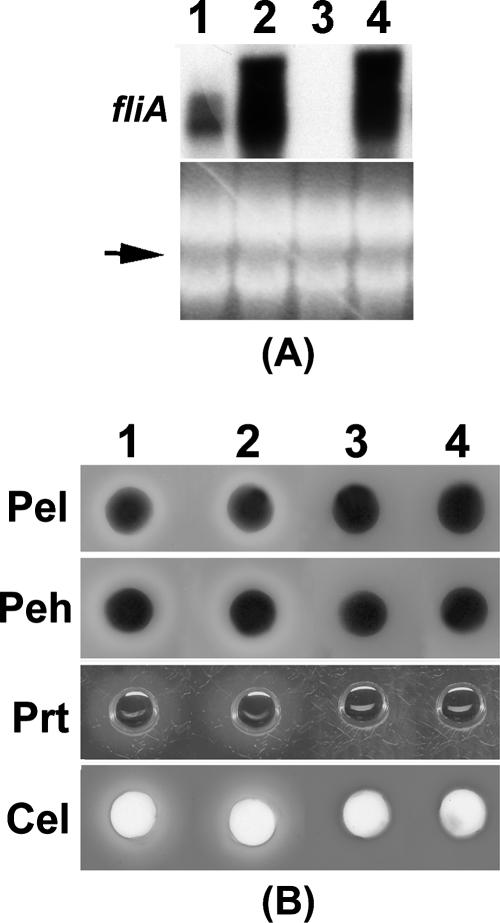
(iv) Positive regulation of fliA.
A Northern blot analysis revealed that fliA RNA was not detectable in the FlhDC- mutant, whereas high levels of fliA transcript were produced by the parent (Fig. 4A). This result was expected since FlhDC is required for the expression of fliA in other enterobacteria (17, 50), as well as in E. carotovora subsp. carotovora strain EC1N (52).
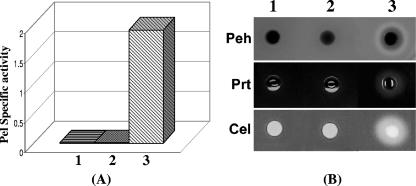
FlhDC effect on extracellular protein production is mediated via gacA and hexA.
The data presented above demonstrated that FlhDC affects the expression of global regulatory genes gacA and hexA. To examine whether the FlhDC effect on extracellular proteins is mediated via GacA, we compared the effects of plac-flhDC plasmid pAKC1242 and plac-gacA plasmid pAKC1245 in the FlhDC- mutant AC5140. The levels of Pel, Peh, Cel, Prt, and HarpinEcc in AC5140 carrying the plac-gacA plasmid pAKC1245 (Fig. 5A, sample 2; Fig. 5B, lane 2; Fig. 5D, lane 2) were higher than in AC5140 carrying plac-flhDC plasmid pAKC1242 (Fig. 5A, sample 1; Fig. 5B, lane 1; Fig. 5D, lane 1). The levels of the cognate transcripts also were higher in AC5140/pAKC1245 (Fig. 5C, lane 2) than in AC5140/pAKC1242 (Fig. 5C, lane 1). This difference most likely results from the differential regulatory effects of FlhDC and GacA. As shown above, FlhDC positively regulates both rsmC and gacA expression, whereas GacA has no apparent effect on rsmC (20; also see below). Since the expression of rsmC is positively regulated by FlhDC and the FlhDC- mutant (AC5140) is rsmC+, rsmC expression in the presence of flhDC+ plasmid would be activated, leading to inhibition of rsmB expression. As expected, Northern blot results revealed that multiple copies of flhDC plasmid stimulate the transcript levels of rsmC in AC5140 (Fig. 5C, lane 1), whereas GacA has no effect on rsmC expression (Fig. 5C, lane 2). Consistent with the differential regulatory effects of GacA and FlhDC on rsmB expression are the findings that AC5140 carrying the plac-gacA plasmid pAKC1245 produces higher levels of rsmB RNA (Flg. 5C, lane 2) than AC5140 carrying the plac-flhDC plasmid pAKC1242 (Fig. 5C, lane 1).
FIG. 5.
Reversal of the pleiotropic phenotype of the FlhDC- mutant by gacA+ DNA, as well as by HexA deficiency. (A) Pel activities; (B) agarose plate assays of Peh, Prt, and Cel activities; (C) Northern blot analysis of pel-1, peh-1, celV, hrpN, hrpL, rsmB, and rsmC. Each lane contained 15 μg of total RNA. The arrow shows the levels of total RNA as revealed by ethidium bromide staining of denatured agarose gel. (D) Western blot analysis of HarpinEcc in the FlhDC- mutant AC5140 carrying pAKC1242 (flhDC+) (lane 1) or pAKC1245 (gacA+) (lane 2). Each lane contained 20 μg of total protein. (E) Agarose plate assays of Pel, Peh, Prt, and Cel activities in FlhDC- HexA+ strain AC5140 (column 1) and FlhDC- HexA- strain AC5145 (column 2).
HexA was previously determined to negatively control exoprotein production via its effect on rsmB RNA (56). To test whether FlhDC controls exoprotein production via its effect on HexA, we examined extracellular enzyme production in an FlhDC- HexA- mutant. The results revealed that the absence of HexA in FlhDC- background partially restored extracellular enzyme production (Fig. 5E). These results suggest that the FlhDC effect on exoprotein production is partly mediated through hexA.
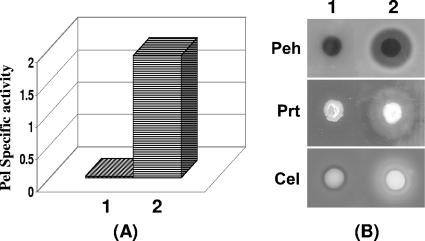
FlhDC restores phenotypes in FlhDC- mutant but not in GacA- mutant.
Extrapolating from the observations described above, we postulated that FlhDC controls gacA which, in turn, activates rsmB transcription to modulate exoprotein gene expression. This hypothesis places GacA below FlhDC in the regulatory network. If true, FlhDC should have no effect in GacA-deficient bacteria. To verify this possibility, we made FlhDC- GacA- double mutant AC5143 and transferred plac-flhDC plasmid pAKC1242 and the cloning vector pCL1920 into this double mutant. AC5143 carrying the plac-gacA plasmid pAKC1245 was used as a positive control. The plac-flhDC plasmid pAKC1242 did not restore extracellular enzyme production in the FlhDC- GacA- mutant (Fig. 6A, sample 2, and Fig. 6B, lane 2), whereas AC5143 carrying the plac-gacA plasmid pAKC1245 produced extracellular Pel, Peh, Prt, and Cel (Fig. 6A, sample 3, and Fig. 6B, lane 3). These results further prove that FlhDC is located at the top of the FlhDC-GacA-exoprotein regulatory pathway.
FIG. 6.
Restoration of extracellular enzyme production by gacA in an FlhDC- GacA- double mutant, AC5143. (A and B) Pel activities (A) and agarose plate assays of Peh, Prt, and Cel activities (B) in AC5143 carrying the cloning vector pCL1920 (column 1), the flhDC+ plasmid pAKC1242 (column 2), or the gacA+ plasmid pAKC1245 (column 3).
GacA and HexA control rsmB RNA production.
The data presented above proved that the FlhDC controls extracellular protein production through cumulative effects on hexA and gacA. Previous studies have established that GacA positively regulates rsmB expression (20) and, on the other hand, HexA suppresses rsmB RNA production (56). Thus, we propose that in the FlhDC- mutant, low levels of GacA and high levels of HexA result in low levels rsmB RNA. rsmB RNA binds RsmA protein and the RsmA-rsmB RNA complex loses its ability to promote decay of target gene mRNAs. Although FlhDC has no direct effect on expression of rsmA (Fig. 4A), low levels of rsmB RNA presumably result in an increase in the pool of free RsmA which, in turn, causes decay of the mRNAs of exoprotein genes and consequently suppresses the exoprotein production. To test the hypothesis, we first established that rsmB expression driven by the lac promoter could restore exoprotein production in the FlhDC- mutant. We transferred the plac-rsmB pasmid pAKC1049 and the cloning vector into the FlhDC- mutant AC5141. These constructs were grown in minimal salts plus sucrose medium for extracellular enzyme assay. The assay results revealed that pAKC1049 restored the extracellular Pel, Peh, Prt, and Cel production in the FlhDC- mutant (Fig. 7A, sample 2, and Fig. 7B, lane 2).
FIG. 7.
Reversal of extracellular enzyme production of the FlhDC- mutant by rsmB+ DNA. (A and B) Pel activities (A) and agarose plate assays of Peh, Prt, and Cel activities (B) in FlhDC- mutant AC5141 carrying pCL1920 (column 1) or pAKC1049 (rsmB+) (column 2).
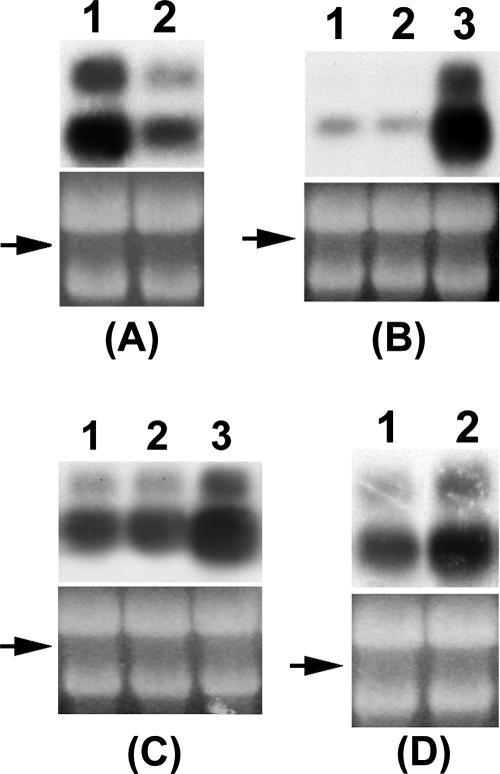
The findings of mutant studies (Fig. 8) further demonstrate that FlhDC controls rsmB RNA levels through GacA and HexA. For example, the inactivation of flhDC or gacA severely reduces rsmB RNA levels (Fig. 8A, lane 2, and Fig. 8B, lane 1), indicating their positive regulation. In addition, multiple copies of flhDC plasmid restored rsmB transcript levels in an FlhDC- GacA+ strain AC5140 (Fig. 8C, lane 3). In contrast, in the FlhDC- GacA- mutant, FlhDC does not stimulate rsmB RNA production (Fig. 8B, lane 2), whereas GacA reverses the negative effect in this mutant (Fig. 8B, lane 3).
FIG. 8.
FlhDC controls rsmB RNA via their effects on gacA and hexA. Northern blot analysis of rsmB RNA in wild-type strain Ecc71 (A1) and its FlhDC- mutant AC5140 (A2); in FlhDC- GacA- strain AC5143 carrying pCL1920 (B1), pAKC1242 (flhDC+) (B2), or pAKC1245 (gacA+) (B3); in FlhDC- GacA+ strain AC5140 carrying pCL1920 (C1), pAKC1241 (flhD+) (C2), or pAKC1242 (C3); and in FlhDC- HexA+ strain AC5140 (D1) and FlhDC- HexA- strain AC5145 (D2). Each lane contained 10 μg of total RNA. The arrows show the levels of total RNA as revealed by ethidium bromide staining of denatured agarose gel.
The inhibition of rsmB RNA production by HexA in the FlhDC+ wild-type strain Ecc71 (56) could result in two ways. It could be mediated through the negative effect of HexA on flhDC expression; the latter is required for rsmB expression via GacA as documented above. LrhA, a HexA homolog negatively regulates expression of flhDC in E. coli (45). We also have observed a similar response in E. carotovora subsp. carotovora (data not shown).
However, we also considered the possibility that HexA may affect rsmB expression by a pathway different from FlhDC-GacA-RsmB. If so, rsmB RNA production would be elevated in a HexA- FlhDC- strain compared to a HexA+ FlhDC- strain. The data in Fig. 8D reveal that indeed is the case. Thus, rsmB RNA production in E. carotovora subsp. carotovora is controlled by HexA via the FlhDC pathway, as well as by an FlhDC-independent pathway. We have initiated studies to clarify the events associated with the latter pathway.
The primary action of rsmB RNA is to bind RsmA and thereby neutralize its negative effects on gene expression. We therefore predicted that reduced levels of rsmB RNA in the FlhDC- mutant resulted in RsmA-promoted decay of transcripts of exoprotein genes. To test this possibility, we compared mRNA stabilities of pel-1, peh-1, and hrpL genes in Ecc71 and its FlhDC- mutant AC5140. The results (Fig. 9) clearly demonstrate that the mRNAs of these genes were more stable in the parent (lanes 1 to 7) than in the FlhDC- mutant (lanes 8 to 14). In addition, we constructed an FlhDC- RsmA- mutant (AC5144) by inactivating rsmA in AC5141 (FlhDC- RsmA+) using mini-Tn5-Spr. This mutant was grown in minimal salts plus sucrose medium along with its parents and an FlhDC+ RsmA- strain AC5070 for extracellular enzyme assay. The results revealed that RsmA deficiency in the FlhDC- mutant restored the enzyme production (Fig. 10A, sample 3, and Fig. 10B, lane 3). The levels of extracellular Pel, Peh, Prt, and Cel were higher than those of AC5006 (FlhDC+ RsmA+) (Fig. 10A, sample 1, and Fig. 10B, lane 1) and AC5141 (FlhDC- RsmA+) (Fig. 10A, sample 2, and Fig. 10B, lane 2) but slightly lower than those of AC5070 (FlhDC+ RsmA-) (Fig. 10A, sample 5, and Fig. 10B, lane 5). Similar results were observed with transcript levels of pel-1, peh-1, and celV of these strains (Fig. 10C). The difference between the FlhDC- RsmA- mutant (AC5144) and the FlhDC+ RsmA- mutant (AC5070) may be due to the levels of rsmB RNA. Northern blot analysis revealed that rsmB transcript levels of AC5144 (Fig. 10D, lane 2) were lower than that of AC5070 (Fig. 10D, lane 1). We have previously reported that overexpression of rsmB has some stimulatory effect in the RsmA- background (48). To further test this, we transferred a plac-rsmB plasmid pAKC1049Gmr or the cloning vector into AC5144. The assay results showed that AC5144 (FlhDC- RsmA-) carrying the plac-rsmB plasmid (Fig. 10E, sample 2, and Fig. 10F, lane 2) produced levels of extracellular enzymes comparable to those produced by the FlhDC+ RsmA- strain AC5070 (Fig. 10E, sample 3, and Fig. 10F, lane 3); these levels were higher than those of AC5144 carrying the vector (Fig. 10E, sample 1, and Fig. 10F, lane 1).
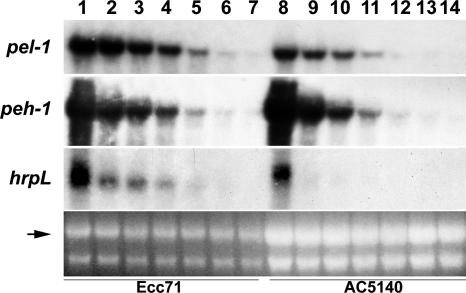
FIG. 9.
Stabilities of pel-1, peh-1, and hrpL transcripts in Ecc71 (lanes 1 to 7) and its FlhDC- mutant AC5140 (lanes 8 to 14). Samples were collected at 0, 2.5, 5, 7.5, 10, 12.5, and 15 min after the addition of rifampin. For lanes 1 to 7, each lane contained 15 μg of total RNA, and for lanes 8 to 14, each lane contained 30 μg of total RNA. The arrow shows the levels of total RNA as revealed by ethidium bromide staining of denatured agarose gel.
FIG. 10.
RsmA is responsible for the pleiotropic phenotype resulting from FlhDC deficiency. (A to C) Pel activities (A), agarose plate assays of Peh, Prt, and Cel activities (B), and pel-1, peh-1, and celV transcripts (C) in FlhDC+ RsmA+ strain AC5006 (column 1), FlhDC- RsmA+ strain AC5141 (column 2), FlhDC- RsmA- strain AC5144 (column 3), FlhDC+ RsmA+ strain AC5047 (column 4), and FlhDC+ RsmA- strain AC5070 (column 5). (D) rsmB RNA levels in AC5070 (column 1) and AC5144 (column 2). (E and F) Pel, Peh, Prt, and Cel activities in AC5144 carrying the cloning vector pCL1920Gmr (column 1) or the rsmB+ plasmid pAKC1049Gmr (column 2) and AC5070 carrying pCL1920Gmr (column 3).
FliA does not affect extracellular enzyme production in E. carotovora subsp. carotovora.
FliA is a flagellar sigma factor that is required for the expression of class III flagellar genes. However, it has also been shown that FliA controls several extracellular enzymes such as lipase (XlpA) and protease (XrtA) in Xenorhabdus nematophila (60). To test the effect of FliA on extracellular enzymes of E. carotovora subsp. carotovora, we transferred a plac-fliA plasmid pAKC1246 into AC5006 and its FlhDC- mutant. Northern analysis showed that fliA in this plasmid was expressed in both AC5006 and the FlhDC- mutant (Fig. 11A, lanes 2 and 4). However, the enzyme assay results revealed that pAKC1246 had no effect on extracellular enzyme production in AC5006 (Fig. 11B, lanes 1 and 2), and it did not restore the extracellular enzyme levels in the FlhDC- mutant (Fig. 11B, lanes 3 and 4). These results collectively demonstrate that FliA is not required for extracellular enzyme/protein production in E. carotovora subsp. carotovora, and the effects of FlhDC on extracellular enzymes/proteins are not mediated through FliA. The consensus sequence for FliA binding has been identified as TAAAGTTT N11 GCCGATAA (17, 36, 38). The absence of these sequences in the promoter regions of rsmA, rsmB, or hrpL or exoprotein genes further supports that hypothesis.
FIG. 11.
FliA does not control exoprotein production in E. carotovora subsp. carotovora. (A) Northern blot analysis of fliA and (B) agarose plate assays of Pel, Peh, Prt, and Cel activities in AC5006 (FlhDC+) carrying the cloning vector pCL1920 (column 1) or the fliA+ plasmid pAKC1246 (column 2), as well as AC5141 (FlhDC-) carrying pCL1920 (column 3) or pAKC1246 (column 4).
Conclusions.
The findings presented here now provide us with a clearer picture of the regulatory network controlling exoprotein production in E. carotovora subsp. carotovora. As depicted in Fig. 1, we propose that FlhDC, one of the major regulatory components, is located at the top of the network. This master regulator controls the production of FliA, HexA, RsmC, and GacA. These four regulators, in turn, control genes required for bacterial locomotion, exoprotein production, or both. Based upon the available evidence, it is likely that FliA controls the genes for flagellum formation and bacterial movement. In contrast, HexA controls bacterial locomotion by regulating FliA production (34), most likely through its negative effect on FlhDC and exoprotein production by regulating rsmB RNA levels via FlhDC, as well as an FlhDC-independent pathway. GacA and RsmC actions are directed primarily toward rsmB RNA production (20, 25). The mechanisms underlying these two regulatory events are not yet known. Our working hypotheses postulates that phosphorylated GacA (GacA∼P) directly activates transcription of the rsmB promoter and that RsmC interacts with another regulator, possibly FlhDC, to reduce the levels of transcription of gacA and other targets of FlhDC. A plausible teleological significance of FlhDC-mediated regulation of negative and positive regulators is apparent from the following: as a negative regulator, HexA controls many phenotypes, including bacterial movement, exoprotein production, and the quorum-sensing system (34, 56). On the other hand, FlhDC is a positive regulator of most of these traits. Under conditions that are not conducive to the expression of these traits, bacteria produce HexA, which represses not only these traits but also prevents flhDC expression that could potentially counteract the responses triggered by HexA. It is also significant that hexA expression is positively autoregulated (34), ensuring the maintenance of an adequate HexA pool size. Activation of flhDC expression by environmental conditions, as yet undefined in E. carotovora subsp. carotovora, would reduce HexA pool and increase GacA and FliA levels, resulting in the activation of genes for motility and rsmB RNA controlling exoprotein production. How RsmC fits into this model would become clear as we gain a better understanding of its target and its regulatory mechanism. A well-orchestrated regulation of flhDC, hexA, gacA, fliA, rsmC, and rsmB is apparently geared toward efficient and timely expression of environmentally significant traits under appropriate conditions.
Acknowledgments
This study was supported by the Research Board grant and the Food for the 21st Century program of the University of Missouri.
We thank J. E. Schoelz for reviewing the manuscript.
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5160-165. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, P. D., J. E. Karlinsey, C. Aldridge, C. Birchall, D. Thompson, J. Yagasaki, and K. T. Hughes. 2006. The flagellar-specific transcription factor, sigma28, is the type III secretion chaperone for the flagellar-specific anti-sigma28 factor FlgM. Genes Dev. 202315-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barembruch, C., and R. Hengge. 2007. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 6576-89. [DOI] [PubMed] [Google Scholar]
- 4.Barras, F., F. Van Gijsegem, and A. K. Chatterjee. 1994. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu. Rev. Phytopathol. 32201-234. [Google Scholar]
- 5.Bartlett, D. H., B. B. Frantz, and P. Matsumura. 1988. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5′ consensus sequences of operons under FlbB and FlaI control. J. Bacteriol. 1701575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer, D. W., Z. M. Wei, S. V. Beer, and A. Collmer. 1995. Erwinia chrysanthemi HarpinEch: an elicitor of the hypersensitive response that contributes to soft-rot pathogenesis. Mol. Plant-Microbe Interact. 8484-491. [DOI] [PubMed] [Google Scholar]
- 7.Bleves, S., M. N. Marenne, G. Detry, and G. R. Cornelis. 2002. Up-regulation of the Yersinia enterocolitica yop regulon by deletion of the flagellum master operon flhDC. J. Bacteriol. 1843214-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burr, T., A. M. Barnard, M. J. Corbett, C. L. Pemberton, N. J. Simpson, and G. P. Salmond. 2006. Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol. Microbiol. 59113-125. [DOI] [PubMed] [Google Scholar]
- 9.Calcutt, M. J., M. Becker-Hapak, M. Gaut, J. Hoerter, and A. Eisenstark. 1998. The rpoS gene of Erwinia carotovora: gene organization and functional expression in Escherichia coli. FEMS Microbiol. Lett. 159275-281. [DOI] [PubMed] [Google Scholar]
- 10.Chadsey, M. S., and K. T. Hughes. 2001. multipartite interaction between Salmonella transcription factor sigma28 and its anti-sigma factor FlgM: implications for sigma28 holoenzyme destabilization through stepwise binding. J. Mol. Biol. 306915-929. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee, A., Y. Cui, and A. K. Chatterjee. 2002. Regulation of Erwinia carotovora hrpL(Ecc) (sigma-L(Ecc)), which encodes an extracytoplasmic function subfamily of sigma factor required for expression of the HRP regulon. Mol. Plant-Microbe Interact. 15971-980. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee, A., Y. Cui, S. Chaudhuri, and A. K. Chatterjee. 2002. Identification of regulators of hrp/hop genes of Erwinia carotovora ssp. carotovora and characterization of HrpLEcc (SigmaLEcc), an alternative sigma factor. Mol. Plant Pathol. 3359-370. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee, A., Y. Cui, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl. Environ. Microbiol. 611959-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee, A., Y. Cui, H. Yang, A. Collmer, J. R. Alfano, and A. K. Chatterjee. 2003. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant-Microbe Interact. 161106-1117. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee, A. K., C. K. Dumenyo, Y. Liu, and A. Chatterjee. 2000. Erwinia: genetics of pathogenicity factors, p. 236-260. In J. Lederberg (ed.), Encyclopedia of microbiology, 2nd ed., vol. 2. Academic Press, Inc., New York, NY. [Google Scholar]
- 16.Chatterjee, A. K., K. K. Thurn, and D. J. Tyrell. 1985. Isolation and characterization of Tn5 insertion mutants of Erwinia chrysanthemi that are deficient in polygalacturonate catabolic enzymes oligogalacturonate lyase and 3-deoxy-d-glycero-2,5-hexodiulosonate dehydrogenase. J. Bacteriol. 162708-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claret, L., and C. Hughes. 2000. Functions of the subunits in the FlhD(2)C(2) transcriptional master regulator of bacterial flagellum biogenesis and swarming. J. Mol. Biol. 303467-478. [DOI] [PubMed] [Google Scholar]
- 19.Collmer, A., and N. T. Keen. 1986. The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 24383-409. [Google Scholar]
- 20.Cui, Y., A. Chatterjee, and A. K. Chatterjee. 2001. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and HarpinEcc. Mol. Plant-Microbe Interact. 14516-526. [DOI] [PubMed] [Google Scholar]
- 21.Cui, Y., A. Chatterjee, H. Hasegawa, and A. K. Chatterjee. 2006. Erwinia carotovora subspecies produce duplicate species of ExpR, LuxR homologs that activate rsmA transcription but differ in their interactions with N-acylhomoserine lactone signals. J. Bacteriol. 1884715-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui, Y., A. Chatterjee, H. Hasegawa, V. Dixit, N. Leigh, and A. K. Chatterjee. 2005. ExpR, a LuxR homolog of Erwinia carotovora subspecies carotovora, activates transcription of rsmA which specifies a global regulatory RNA-binding protein. J. Bacteriol. 1874792-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui, Y., A. Chatterjee, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J. Bacteriol. 1775108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui, Y., L. Madi, A. Mukherjee, C. K. Dumenyo, and A. K. Chatterjee. 1996. The RsmA- mutants of Erwinia carotovora subsp. carotovora strain Ecc71 overexpress hrpNEcc and elicit a hypersensitive reaction-like response in tobacco leaves. Mol. Plant-Microbe Interact. 9565-573. [DOI] [PubMed] [Google Scholar]
- 25.Cui, Y., A. Mukherjee, C. K. Dumenyo, Y. Liu, and A. K. Chatterjee. 1999. rsmC of the soft-rotting bacterium Erwinia carotovora subsp. carotovora negatively controls extracellular enzyme and HarpinEcc production and virulence by modulating the levels of regulatory RNA (rsmB) and RNA binding protein (RsmA). J. Bacteriol. 1816042-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 1726568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 642710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eriksson, A. R., R. A. Andersson, M. Pirhonen, and E. T. Palva. 1998. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 11743-752. [DOI] [PubMed] [Google Scholar]
- 29.Figurski, D. H., and D. R. Helinski. 1979. Rsmlication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 761648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frederick, R. D., J. Chiu, J. L. Bennetzen, and A. K. Handa. 1997. Identification of a pathogenicity locus, rpfA, in Erwinia carotovora subsp. carotovora that encodes a two-component sensor-regulator protein. Mol. Plant-Microbe Interact. 10407-415. [DOI] [PubMed] [Google Scholar]
- 31.Gauger, E. J., M. P. Leatham, R. Mercado-Lubo, D. C. Laux, T. Conway, and P. S. Cohen. Role of motility and the flhDC operon in Escherichia coli MG1655 colonization of the mouse intestine. Infect. Immun. 753315-3324. [DOI] [PMC free article] [PubMed]
- 32.Givaudan, A., and A. Lanois. 2000. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J. Bacteriol. 182107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Givskov, M., L. Eberl, G. Christiansen, M. J. Benedik, and S. Molin. 1995. Induction of phospholipase- and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol. Microbiol. 15445-454. [DOI] [PubMed] [Google Scholar]
- 34.Harris, S. J., Y. L. Shih, S. D. Bentley, and G. P. C. Salmond. 1998. The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence determinants. Mol. Microbiol. 28705-717. [DOI] [PubMed] [Google Scholar]
- 35.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 141351-1363. [DOI] [PubMed] [Google Scholar]
- 36.Helmann, J. D., and M. J. Chamberlin. 1987. DNA sequence analysis suggests that expression of flagellar and chemotaxis genes in Escherichia coli and Salmonella typhimurium is controlled by an alternative sigma factor. Proc. Natl. Acad. Sci. USA 846422-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hossain, M. M., S. Shibata, S.-I. Aizawa, and S. Tsuyumu. 2005. Motility is an important determinant for pathogenesis of Erwinia carotovora subsp. carotovora. Physiol. Mol. Plant Pathol. 66134-143. [Google Scholar]
- 38.Ide, N., T. Ikebe, and K. Kutsukake. 1999. Reevaluation of the promoter structure of the class 3 flagellar operons of Escherichia coli and Salmonella genes. Genet. Syst. 74113-116. [DOI] [PubMed] [Google Scholar]
- 39.Jones, S., B. Yu, N. J. Bainton, M. Birdsall, B. W. Bycroft, S. R. Chhabra, A. J. R. Cox, P. Golby, P. J. Reeves, S. Stephens, M. K. Winson, G. P. C. Salmond, G. S. A. B. Stewart, and P. Williams. 1993. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 122477-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291605-614. [DOI] [PubMed] [Google Scholar]
- 41.Kapatral, V., J. W. Campbell, S. A. Minnich, N. R. Thomson, P. Matsumura, and B. M. Prüβ. 2004. Gene array analysis of Yersinia enterocolitica FlhD and FlhC: regulation of enzymes affecting synthesis and degradation of carbamoylphosphate. Microbiology 1502289-2300. [DOI] [PubMed] [Google Scholar]
- 42.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70191-197. [DOI] [PubMed] [Google Scholar]
- 43.Kutsukake, K., and T. Iino. 1994. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J. Bacteriol. 1763598-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lane, M. C., V. Lockatell, G. Monterosso, D. Lamphier, J. Weinert, J. R. Hebel, D. E. Johnson, and H. L. Mobley. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immun. 737644-7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility, and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45521-532. [DOI] [PubMed] [Google Scholar]
- 46.Lerner, C. G., and M. Inouye. 1990. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 184631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, Y., A. Chatterjee, and A. K. Chatterjee. 1994. Nucleotide sequence and expression of a novel pectate lyase gene (pel-3) and a closely endopolygalacturonase gene (peh-1) of Erwinia carotovora subsp. carotovora 71. Appl. Environ. Microbiol. 602545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, Y., Y. Cui, A. Mukherjee, and A. K. Chatterjee. 1998. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol. Microbiol. 29219-234. [DOI] [PubMed] [Google Scholar]
- 49.Liu, Y., G.-Q. Jiang, Y. Cui, A. Mukherjee, W.-L. Ma, and A. K. Chatterjee. 1999. kdgREcc negatively regulates genes for pectinases, cellulase, protease, harpinEcc, and a global RNA regulator in Erwinia carotovora subsp. carotovora. J. Bacteriol. 1812411-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu, X., N. Fujita, A. Ishihama, and P. Matsumura. 1995. The C-terminal region of the alpha subunit of Escherichia coli RNA polymerase is required for transcriptional activation of the flagellar level II operons by the FlhD/FlhC complex. J. Bacteriol. 1775186-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma, W., Y. Cui, Y. Liu, C. K. Dumenyo, A. Mukherjee, and A. K. Chatterjee. 2001. Molecular characterization of global regulatory RNA species that control pathogenicity factors in Erwinia amylovora and Erwinia herbicola pv. gypsophilae. J. Bacteriol. 1831870-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsumoto, H., M. Umehara, H. Muroi, Y. Yoshitake, and S. Tsuyumu. 2003. Homolog of FlhDC, a master regulator for flagellum synthesis: required for pathogenicity in Erwinia carotovora subsp. carotovora. J. Gen. Plant Pathol. 69189-193. [Google Scholar]
- 53.McCarter, L. L. 2006. Regulation of flagella. Curr. Opin. Microbiol. 9180-186. [DOI] [PubMed] [Google Scholar]
- 54.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 55.Mukherjee, A., Y. Cui, Y. Liu, and A. K. Chatterjee. 1997. Molecular characterization and expression of the Erwinia carotovora hrpNEcc gene, which encodes an elicitor of the hypersensitive reaction. Mol. Plant-Microbe Interact. 10462-471. [DOI] [PubMed] [Google Scholar]
- 56.Mukherjee, A., Y. Cui, W. Ma, Y. Liu, and A. K. Chatterjee. 2000. hexA of Erwinia carotovora ssp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator of extracellular proteins, plant virulence and the quorum-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Environ. Microbiol. 2203-215. [DOI] [PubMed] [Google Scholar]
- 57.Mukherjee, A., Y. Cui, W.-L. Ma, Y. Liu, A. Ishihama, A. Eisenstark, and A. K. Chatterjee. 1998. RpoS (Sigma-S) controls expression of rsmA, a global regulator of secondary metabolites, harpin, and extracellular proteins in Erwinia carotovora. J. Bacteriol. 1803629-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murata, H., J. L. McEvoy, A. Chatterjee, A. Collmer, and A. K. Chatterjee. 1991. Molecular cloning of an aepA gene that activates production of extracellular pectolytic, cellulolytic, and proteolytic enzymes in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 4239-246. [Google Scholar]
- 59.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 241109-1117. [DOI] [PubMed] [Google Scholar]
- 60.Park, D., and S. Forst. 2006. Co-regulation of motility, exoenzyme and antibiotic production by the EnvZ-OmpR-FlhDC-FliA pathway in Xenorhabdus nematophila. Mol. Microbiol. 611397-1412. [DOI] [PubMed] [Google Scholar]
- 61.Pérombelon, M. C. M. 2002. Potato diseases caused by soft-rot erwinias: an overview of pathogenesis. Plant Pathol. 511-12. [Google Scholar]
- 62.Pirhonen, M., D. Flego, R. Heikinheimo, and E. T. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 122467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pons, T., B. González, F. Ceciliani, and A. Galizzi. 2006. FlgM anti-sigma factors: identification of novel members of the family, evolutionary analysis, homology modeling, and analysis of sequence-structure-function relationships. J. Mol. Model. 12973-983. [DOI] [PubMed] [Google Scholar]
- 64.Prüss, B. M., and P. Matsumura. 1996. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J. Bacteriol. 178668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rantakari, A., O. Virtaharju, S. Vähämiko, S. Taira, E. T. Palva, H. T. Saarilahti, and M. Romantschuk. 2001. Type III secretion contributes to the pathogenesis of the soft-rot pathogen Erwinia carotovora: partial characterization of the hrp gene cluster. Mol. Plant-Microbe Interact. 14962-968. [DOI] [PubMed] [Google Scholar]
- 66.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 67.Shih, Y. L., S. J. Harris, G. Borner, M. M. Rivet, and G. P. Salmond. 1999. The hexY genes of Erwinia carotovora ssp. carotovora and ssp. atroseptica encode novel proteins that regulate virulence and motility co-ordinately. Environ. Microbiol. 1535-547. [DOI] [PubMed] [Google Scholar]
- 68.Sorenson, M. K., S. S. Ray, and S. A. Darst. 2004. Crystal structure of the flagellar sigma/anti-sigma complex sigma(28)/FlgM reveals an intact sigma factor in an inactive conformation. Mol. Cell 14127-138. [DOI] [PubMed] [Google Scholar]
- 69.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 927-39. [DOI] [PubMed] [Google Scholar]
- 70.Thomson, N. R., A. Cox, B. W. Bycroft, G. S. Stewart, P. Williams, and G. P. C. Salmond. 1997. The rap and hor proteins of Erwinia, Serratia, and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol. Microbiol. 26531-544. [DOI] [PubMed] [Google Scholar]
- 71.Thomson, N. R., J. D. Thomas, and G. P. C. Salmond. 1999. Virulence determinants in the bacterial phytopathogen Erwinia. Methods Microbiol. 29347-426. [Google Scholar]
- 72.Wang, S., R. T. Fleming, E. M. Westbrook, P. Matsumura, and D. B. McKay. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355798-808. [DOI] [PubMed] [Google Scholar]
- 73.Zink, R. T., R. J. Kemble, and A. K. Chatterjee. 1984. Transposon Tn5 mutagenesis in Erwinia carotovora subsp. carotovora and Erwinia carotovora subsp. atroseptica. J. Bacteriol. 157809-814. [DOI] [PMC free article] [PubMed] [Google Scholar]