Abstract
Two-component signal transduction systems, composed of sensor kinase (SK) and response regulator (RR) proteins, allow bacterial cells to adapt to changes such as environmental flux or the presence of a host. RscS is an SK required for Vibrio fischeri to initiate a symbiotic partnership with the Hawaiian squid Euprymna scolopes, likely due to its role in controlling the symbiosis polysaccharide (syp) genes and thus biofilm formation. To determine which RR(s) functions downstream of RscS, we performed epistasis experiments with a library of 35 RR mutants. We found that several RRs contributed to RscS-mediated biofilm formation in V. fischeri. However, only the syp-encoded symbiosis regulator SypG was required for both biofilm phenotypes and syp transcription induced by RscS. These data support the hypothesis that RscS functions upstream of SypG to induce biofilm formation. In addition, this work also revealed a role for the syp-encoded RR SypE in biofilm formation. To our knowledge, no other study has used a large-scale epistasis approach to elucidate two-component signaling pathways. Therefore, this work both contributes to our understanding of regulatory pathways important for symbiotic colonization by V. fischeri and establishes a paradigm for evaluating two-component pathways in the genomics era.
Bacteria utilize two-component signal transduction pathways as “reflex” systems to sense and adapt to given environmental stimuli (9, 33, 38). Signaling via these systems is initiated by sensor kinase (SK) proteins, which autophosphorylate in response to a specific environmental cue. The phosphoryl group is subsequently transferred to a given response regulator (RR) or, in some cases, multiple RRs. The RR then promotes a given cellular output, often via the transcriptional activation of a subset of genes.
Two-component signaling systems are well suited to allow communication between symbiotic partners, mediating smooth transitions into, as well as maintenance within, such associations. Previous studies established that multiple two-component pathways are required for the initiation and maintenance of the symbiosis between the marine bioluminescent bacterium Vibrio fischeri and the Hawaiian squid Euprymna scolopes (19, 26, 36). At least 14 of the 40 putative RRs within the V. fischeri genome are required for efficient colonization. These include the luminescence regulator LuxO (15-17), the metabolic regulator ArcA (3), the global regulator GacA (39), the extracellular polysaccharide regulator SypG (10), and several less-characterized RRs (10).
Previous work also identified an SK protein, RscS (regulator of symbiotic colonization-sensor), that was required for symbiotic initiation (37). As an SK, RscS would be predicted to exert its influence through an RR protein. However, efforts to understand the RscS pathway were initially stymied by the lack of bioinformatic evidence. Frequently, sensor-regulator partners are encoded adjacently within the genome, often within or near the locus that they regulate. RscS is an orphan sensor, however, as it is not encoded adjacent to a predicted RR gene (37). In addition, while RscS is encoded within a locus of genes that function in glycerol metabolism, mutations in RscS do not alter the ability of V. fischeri wild-type strain ES114 to grow in media containing glycerol as the sole carbon source (37). Finally, general functional cues were also lacking; a disruption of rscS did not lead to defects in a number of phenotypes tested in culture.
Recently, however, we established that RscS activates the expression of the symbiosis polysaccharide (syp) cluster of genes (42). This cluster is composed of 18 genes organized into at least five putative operons (A to E, F to H, I to L, M to O, and P to R) (43; E. A. Hussa, K. Geszvain, and K. L. Visick, unpublished data). The syp gene products are predicted to function in polysaccharide synthesis and transport, and most are required for the initiation of symbiotic colonization (43). The overexpression of rscS results in the induction of syp transcription and syp-dependent biofilm formation (42).
With this discovery of culture phenotypes associated with RscS, it is now possible to use epistasis experiments to uncover the RR or RRs that function downstream of RscS. Recently, we identified and disrupted 35 of 40 putative RRs in the V. fischeri genome (10). Twelve of these genes were required for competitive colonization, a phenotype expected for the RR that functions downstream of RscS. However, many of these 12 RRs have established homologs and functions unrelated to those of RscS. For example, FlrC regulates flagellar synthesis (10, 12), whereas RscS does not appear to control motility (37). Furthermore, 9 of the 12 RRs are linked to a putative SK in the genome. These observations decreased, but did not eliminate, the potential of these RRs to relay the signal from RscS.
One candidate partner already known to affect syp transcription is SypG, a syp-encoded regulator that is also required for colonization (10, 43). SypG is predicted to be an RR in the NtrC-like family of σ54-dependent transcriptional activators. Like RscS, multicopy expression of sypG results in enhanced syp transcription and biofilm formation, as measured by glass attachment and pellicle formation (43); however, the pellicles are not as robust as those formed by RscS-overexpressing cells. In addition, the overexpression of SypG does not induce wrinkled-colony formation, another hallmark of RscS overexpression (42). Furthermore, the sypG gene is adjacent to two genes, sypE and sypF, that encode putative SK and RR proteins, respectively; thus, it is not clear whether SypG functions directly downstream of SypF or whether a more complicated regulatory scheme exists. Despite these complexities, the hypothesis that SypG functions downstream of RscS to control syp expression remained viable.
In this work, we surveyed putative V. fischeri RRs to identify those that function downstream of RscS. We report that several RRs contributed to RscS-mediated biofilm formation. However, the loss of sypG alone abrogated both RscS-mediated biofilm formation and syp transcription. These results thus identify SypG as being a critical link between the symbiosis regulator RscS and the processes that it controls and suggest that RscS and SypG function within the same signal transduction pathway.
MATERIALS AND METHODS
Strains and media.
Plasmids and V. fischeri strains utilized in this study are listed in Tables 1 and 2, respectively. The parental V. fischeri strain used in this work was ES114, a strain isolated from E. scolopes (1). All derivatives were generated by conjugation, as previously described (4, 18, 37). Escherichia coli strains Tam1 λpir (Active Motif, Carlsbad, CA), DH5α, and Top10 (Invitrogen, Carlsbad, CA) were used for cloning and conjugative purposes. V. fischeri strains were grown in either the complex medium LBS (with 0.3% glycerol, where indicated) (8, 32) or HMM (27) containing either 0.3% tryptone (HMM-T) or 0.3% Casamino Acids and 0.2% glucose (43), where indicated. The following antibiotics were added to V. fischeri growth media, as necessary, at the indicated concentrations: chloramphenicol (Cm) at 2.5 μg/ml, erythromycin (Em) at 5 μg/ml, and tetracycline (Tc) at 5 μg/ml in LBS or 30 μg/ml in HMM (or HMM-T). The following antibiotics were added to E. coli growth media where necessary, at the indicated concentrations: Cm at 25 μg/ml, kanamycin (Kan) at 50 μg/ml, Tc at 15 μg/ml, or ampicillin (Ap) at 100 μg/ml.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pCLD1 | Cmr derivative of pEVS107; Kanr Cmr | This study |
| pCLD6 | pCLD1 + 3.2-kb ApaI/SpeI fragment from pEAH41 containing the psypA-lacZ fusion; Kanr Cmr | This study |
| pCLD19 | Delivery vector carrying sequences flanking sypE | This study |
| pCLD46 | pVSV105 carrying the rscS1 allele; Cmr | This study |
| pCLD48 | pVSV105 carrying wild-type sypE; Cmr | This study |
| pCR2.1-TOPO | Commercial cloning vector; Apr Kanr | Invitrogen, Carlsbad, CA |
| pEAH41 | pTMO82 carrying psypA upstream of lacZ; Apr Kanr | This study |
| pEAH73 | pKV69 carrying wild-type sypG; Cmr Tcr | This study |
| pES420 | Mobilizable suicide vector, Emr | 21a |
| pEVS107 | Mini-Tn7 delivery plasmid; mob; Kanr Emr | 18 |
| pKG11 | pKV69 carrying rscS1 allele; Cmr Tcr | 42 |
| pKV69 | Mobilizable vector; Cmr Tcr | 37 |
| pKV276 | pEAH73 with D53E mutation in sypG; Cmr Tcr | This study |
| pTMO82 | pCR2.1-TOPO carrying promoterless lacZ; Apr Kanr | This study |
| pVSV105 | Mobilizable vector; Cmr | 6 |
TABLE 2.
V. fischeri strains used in this study
| Straina | Characteristic(s) | Reference or source |
|---|---|---|
| Without reporter | ||
| ES114 | Wild type | 1 |
| KV1548 | VF2120 (arcA)::pEAH1 | 10 |
| KV1585 | VF1570::pKV174 | 10 |
| KV1592 | VFA1024 (sypE)::pEAH7 | 10 |
| KV1593 | VFA0179::pKV178 | 10 |
| KV1594 | VF1401::pKV177 | 10 |
| KV1595 | VF1396::pKV176 | 10 |
| KV1596 | VFA0561::pKV175 | 10 |
| KV1612 | VFA1017::pKV179 | 10 |
| KV1640 | VFA0041::pTMB26 | 10 |
| KV1641 | VF1054::pAIA1 | 10 |
| KV1650 | VFA0266::pTMB27 | 10 |
| KV1651 | VF1988::pTMB28 | 10 |
| KV1654 | VFA1012::pTMB31 | 10 |
| KV1655 | VF2343::pTMB32 | 10 |
| KV1665 | VF1909::pTMB33 | 10 |
| KV1666 | VFA1026 (sypG)::pAIA4 | 10 |
| KV1668 | VFA0211::pAIA6 | 10 |
| KV1672 | VFA0181::pTMB34 | 10 |
| KV1714 | VFA0795::pEAH10 | 10 |
| KV1715 | VF0454::pEAH11 | 10 |
| KV1727 | VF0526::pEAH4 | 10 |
| KV1730 | VF0095::pKV180 | 10 |
| KV1787 | ΔsypG | 10 |
| KV1809 | VF1854::pEAH26 | 10 |
| KV2164 | VF2374::pEAH24 | 10 |
| KV2165 | VFA0216::pEAH25 | 10 |
| KV2191 | VF0937 (luxO)::pAIA3 | 10 |
| KV2501 | VF1689::pAIA2 | 10 |
| KV2503 | VFA0103::pAIA5 | 10 |
| KV2505 | VFA0802::pKV207 | 10 |
| KV2507 | VF0114::pKV209 | 10 |
| KV2509 | VFA0698::pKV214 | 10 |
| KV2510 | VF1833::pKV215 | 10 |
| KV2636 | VF1148::pTMB30 | 10 |
| KV2637 | VF1879::pKV216 | 10 |
| KV2874 | VFA0732::pKV208 | 10 |
| KV3299 | ΔsypE | This study |
| With reportera | ||
| KV3001 | ES114 att Tn7::psypA-lacZ | This study |
| KV3395 | KV1548 att Tn7::psypA-lacZ | This study |
| KV3396 | KV1585 att Tn7::psypA-lacZ | This study |
| KV3398 | KV1592 att Tn7::psypA-lacZ | This study |
| KV3399 | KV1593 att Tn7::psypA-lacZ | This study |
| KV3397 | KV1594 att Tn7::psypA-lacZ | This study |
| KV3400 | KV1595 att Tn7::psypA-lacZ | This study |
| KV3419 | KV1596 att Tn7::psypA-lacZ | This study |
| KV3420 | KV1612 att Tn7::psypA-lacZ | This study |
| KV3421 | KV1640 att Tn7::psypA-lacZ | This study |
| KV3422 | KV1641 att Tn7::psypA-lacZ | This study |
| KV3429 | KV1650 att Tn7::psypA-lacZ | This study |
| KV3430 | KV1651 att Tn7::psypA-lacZ | This study |
| KV3423 | KV1654 att Tn7::psypA-lacZ | This study |
| KV3424 | KV1655 att Tn7::psypA-lacZ | This study |
| KV3425 | KV1665 att Tn7::psypA-lacZ | This study |
| KV3426 | KV1666 att Tn7::psypA-lacZ | This study |
| KV3427 | KV1668 att Tn7::psypA-lacZ | This study |
| KV3428 | KV1672 att Tn7::psypA-lacZ | This study |
| KV3431 | KV1714 att Tn7::psypA-lacZ | This study |
| KV3432 | KV1715 att Tn7::psypA-lacZ | This study |
| KV3532 | KV1727 att Tn7::psypA-lacZ | This study |
| KV3433 | KV1730 att Tn7::psypA-lacZ | This study |
| KV3232 | KV1787 att Tn7::psypA-lacZ | This study |
| KV3508 | KV1809 att Tn7::psypA-lacZ | This study |
| KV3521 | KV2164 att Tn7::psypA-lacZ | This study |
| KV3509 | KV2165 att Tn7::psypA-lacZ | This study |
| KV3510 | KV2191 att Tn7::psypA-lacZ | This study |
| KV3518 | KV2501 att Tn7::psypA-lacZ | This study |
| KV3519 | KV2503 att Tn7::psypA-lacZ | This study |
| KV3520 | KV2505 att Tn7::psypA-lacZ | This study |
| KV3533 | KV2507 att Tn7::psypA-lacZ | This study |
| KV3534 | KV2509 att Tn7::psypA-lacZ | This study |
| KV3535 | KV2510 att Tn7::psypA-lacZ | This study |
| KV3547 | KV2636 att Tn7::psypA-lacZ | This study |
| KV3548 | KV2637 att Tn7::psypA-lacZ | This study |
| KV3522 | KV2874 att Tn7::psypA-lacZ | This study |
| KV3620 | KV3299 att Tn7::psypA-lacZ | This study |
Derivatives of these strains containing either pKG11 or pKV69 (or pEAH73, pKV276, pVSV105, pCLD48, or pCLD46, where indicated) were constructed and utilized as a part of this study.
Molecular techniques.
The mini-Tn7-based reporters utilized within this study were generated by PCR amplification of the sypA promoter region using oligonucleotides VFA1019intR (TTTTTCGTACGTGATGGGAAATGACGTTGTG) and VFA1020per (CCGATGGCGTCCATATCAC) (MWG, High Point, NC). The product was then cloned into pTMO82, a derivative of pCR 2.1 TOPO (Invitrogen, Carlsbad, CA) carrying a promoterless copy of lacZ, via standard techniques. The resulting plasmid (pEAH41) expresses lacZ via the sypA promoter. The sypA-lacZ transcriptional fusion was then digested out of pEAH41 with ApaI and SpeI (New England Biolabs, Beverly, MA) and cloned into the mini-Tn7 transposon within similarly digested plasmid pCLD1, a Cmr derivative of pEVS107 (18). SypG overexpression vector pEAH73 is a derivative of pKV69 (Table 1) carrying sypG amplified from the V. fischeri genome using primers VFA1025RTF (GCTACACTTTCACTAGACGC) and SypG His R (GGTACCTCATTCCGATTCTTCATAG), obtained from MWG (High Point, NC).
Generation of a sypE deletion.
We constructed the sypE deletion strain (ΔsypE) as follows: we amplified and cloned sequences 2 kb upstream and downstream of sypE into pCR2.1-TOPO and pESY20, respectively. The resulting plasmids were ligated together to produce a composite plasmid, pCLD19, that contained sequences flanking sypE but that lacked sypE itself. pCLD19 was introduced into ES114 by conjugation and selection for Em resistance (Emr). Subsequent passage of the resulting colonies on Em allowed the identification of Emr stable cells; in these cells, a single recombination event had occurred, promoting the integration of the entire plasmid into the chromosome. These cells were subsequently passaged nonselectively to identify Em-sensitive cells in which a second recombination event had occurred, leaving behind either ΔsypE or wild-type sequences. One ΔsypE strain, KV3299, was identified using a PCR approach and subsequently confirmed by Southern analysis using sypE and flanking DNA as a probe.
Generation of the D53E mutation in sypG.
To obtain the D53E mutation in SypG, we performed site-directed mutagenesis using the Change-IT kit (USB, Cleveland, OH). Plasmid pEAH73 (Table 1) served as the template for primers SypG D53E-phosph (CCACATTTGGTGATTCTCGAGTTGAAACTGCCAGATATGTCAG) and Phos-lacZ-up-rev (CCTGTGTGAAATTGTTATCCG). Because the base change (in boldface type) introduces an XhoI site, we screened clones resulting from the mutagenesis with XhoI. A plasmid containing the XhoI site, pKV276, was subsequently identified, and the mutation was confirmed through sequence analysis of the sypG coding region using the Genomics Core Facility at the Center for Genetic Medicine at Northwestern University.
Glass attachment and pellicle assays.
Strains were grown overnight in HMM containing 0.3% Casamino Acids and 0.2% glucose as well as 30 μg/ml Tc to select for plasmid pKV69 or pKG11. Cultures were then diluted to an optical density at 600 nm (OD600) of 0.1 in the same medium and allowed to grow in static culture in either borosilicate glass culture tubes (for glass attachment assays) or 12-well culture plates (for pellicle assays) for 48 h at 22°C (or 28°C, in the case of experiments conducted with pEAH73). To assay attachment to glass, cultures were stained for 15 min with a 1% crystal violet (CV) solution and subsequently rinsed with deionized water, dried by aspiration, and photographed. Staining was then quantitated by adding 2 ml of 100% ethanol to stained tubes containing 1g of 1-mm glass beads. The tubes were then vortexed until stained material was completely removed from the tube surface. Stained material was quantitated by measuring the OD600 (22). The strength of pellicle formation was assessed by drawing a sterile toothpick through the culture surface; pellicles were scored using a scale of “−” to “+++,” representing the amount of resistance encountered by the toothpick. A score of “−” was assigned where no pellicle was detected; “+” represents a very thin, easily disrupted pellicle; “++” represents a more cohesive (less easily disrupted) pellicle; and “+++” represents a thick pellicle that was difficult to disrupt.
β-Galactosidase assays.
Samples were grown for 21 h in HMM-T. β-Galactosidase assays were conducted as described previously (21). The total amount of protein in each sample was assayed using Lowry assays (14), and β-galactosidase activity per mg protein in each sample was calculated.
RESULTS
Dependence of RscS on V. fischeri RRs for surface attachment.
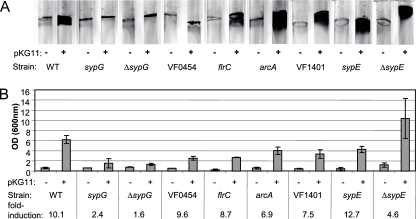
To identify RRs that function downstream of RscS, we introduced the RscS overexpression plasmid pKG11 or a vector control (pKV69) into each of 35 different RR mutants (10). Because RscS mediates biofilm formation in V. fischeri, we first assayed these strains for their abilities to attach to a glass surface using a CV stain to visualize biofilm-associated cells and cellular materials (see Materials and Methods). In control experiments, we found that the overexpression of RscS in wild-type cells enhanced the appearance of CV-stainable biofilm material (Fig. 1A) by 10-fold relative to the vector control (Fig. 1B). Upon a similar examination of the 35 RR mutant strains, we found that 29 of these mutants exhibited no significant differences in RscS-mediated glass attachment compared to vector controls (data not shown).
FIG. 1.
RscS-mediated attachment to a glass surface in RR mutants. (A) Wild-type (WT) and RR mutant strains of V. fischeri carrying either RscS overexpression vector pKG11 (+) or the vector control, pKV69 (−), were grown statically in HMM containing glucose and Casamino Acids and stained with CV to visualize surface-attached material (representative photographs from an experiment conducted in triplicate). (B) Stain was removed by agitation with 1-mm glass beads and ethanol and quantitated by spectrophotometry. Induction represents the OD600 of a given strain carrying pKG11 divided by that of the same strain carrying the vector control.
Of the remainder, only the RscS-overexpressing sypG mutant exhibited a biofilm phenotype indistinguishable from that of the vector control (Fig. 1). Because this phenotype is similar to the loss of RscS-induced biofilm formation that occurs upon the disruption of structural genes such as sypN, which encodes a putative glycosyltransferase (42, 43), we repeated our experiments using an in-frame deletion strain, ΔsypG. We found that the ΔsypG mutation also eliminated RscS-mediated attachment (Fig. 1). Additionally, the co-overexpression of both SypG and RscS in the ΔsypG mutant strain restored the glass attachment phenotype (data not shown). Thus, SypG functions downstream of RscS to facilitate attachment to a glass surface.
RscS-mediated attachment to glass was also altered by the loss of any of five other RRs (Fig. 1A). The loss of either VF0454, a putative homolog of the polysaccharide regulator VpsR (41), or the flagellar regulator FlrC resulted in decreased CV staining (2.5- and 2.3-fold, respectively) (Fig. 1). Mutations in arcA, VF1401, or sypE altered the pattern, but not the overall level, of staining (Fig. 1).
Because sypE is embedded in the syp cluster two genes upstream of sypG, we further assessed its specific role by constructing an in-frame deletion. The overexpression of RscS in the ΔsypE mutant resulted in a slightly diffuse pattern of CV-stained material, which did not differ quantitatively from sypE+ cells (Fig. 1B). These results confirmed a minor role for SypE in RscS-mediated attachment to glass. Throughout the remainder of this work, we limited our studies to the ΔsypE strain.
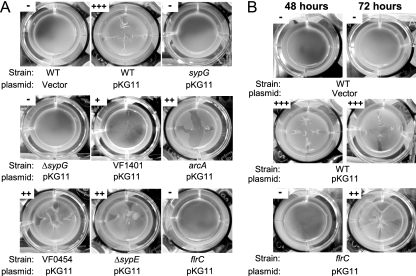
Dependence of RscS on V. fischeri RRs for pellicle formation.
Wild-type V. fischeri strains carrying pKG11 produced strong pellicles at the air-liquid interface of statically grown minimal medium cultures, while vector controls formed no detectable pellicle (Fig. 2A) (42). To identify the V. fischeri RRs that promote RscS-mediated pellicle formation, we grew the RR mutant strains carrying pKG11 or the vector control in minimal medium (HMM) for 48 h in static culture and assessed surface aggregation by dragging a sterile toothpick through the culture surface (see Materials and Methods for a description of scoring). While 29 of the 35 RR mutant strains exhibited strong pellicle formation similar to that of the wild-type strain when multicopy RscS was present (data not shown), 6 displayed decreased pellicle formation or lacked pellicles entirely (Fig. 2). In particular, as observed with glass attachment assays, both sypG mutations (vector integration and in-frame deletion) abrogated RscS-mediated pellicle formation (Fig. 2A).
FIG. 2.
RscS-mediated pellicle formation in RR mutants. Wild-type (WT) and RR mutant strains of V. fischeri carrying either pKG11 or a vector control (pKV69) were grown statically in HMM containing glucose and Casamino Acids for 48 h (A and B, as indicated) or 72 h (B). Pellicle formation was assessed by dragging a sterile toothpick through the culture surface and scored as described in Materials and Methods. Photographs are representative of samples from experiments conducted in triplicate.
Disruption of five additional putative RRs resulted in decreased pellicle formation in RscS-overexpressing strains. Mutations in VF1401, arcA, VF0454, and sypE resulted in pellicles that were less dense and/or cohesive than those of RscS-overexpressing wild-type cells (Fig. 2A). The flrC mutant strain exhibited no detectable pellicle formation after 48 h (Fig. 2A). Previous reports suggested that biofilm formation is often delayed in nonmotile strains of bacteria (13). We therefore assayed pellicle formation after 72 h of incubation and found that pellicle formation by the flrC mutant increased; however, these pellicles remained less dense and less cohesive than those formed by the wild-type strain after either 48 or 72 h of incubation (Fig. 2B). These data indicate that flrC and/or motility influences RscS-mediated pellicle formation. Increased incubation time did not enhance pellicle formation by other RR mutant strains. Importantly, the sypG mutants did not form pellicles regardless of incubation time, suggesting that sypG is absolutely required for this RscS-induced phenotype.
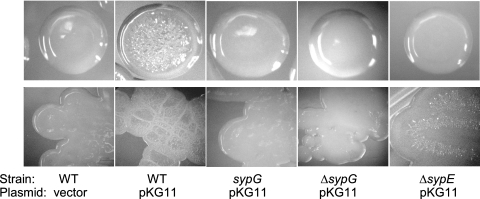
Dependence of RscS on V. fischeri RRs for wrinkled-colony morphology.
Previous studies indicated that RscS-overexpressing V. fischeri cells formed colonies with a dry, wrinkled morphology (42). To identify the RR or RRs that control RscS-mediated wrinkled-colony formation, we examined the colonies formed on solid medium by each RR mutant carrying the vector control or pKG11. All mutants carrying the vector control formed smooth colonies that resembled those formed by the wild-type strain (Fig. 3 and data not shown). Wrinkled-colony formation induced by pKG11 occurred normally in 33 of the 35 RR mutants (data not shown). Only two mutations, in sypG and sypE, abrogated or reduced RscS-mediated wrinkled-colony formation.
FIG. 3.
RscS-mediated wrinkled-colony morphology in RR mutants. Wild-type (WT) and RR mutant strains of V. fischeri carrying either pKG11 or a vector control were streaked onto solid, complex medium (LBS with 0.3% glycerol and Tc) and allowed to grow for 3 days at room temperature. Photographs of individual colonies (top row) and also the heavy part of the streak (bottom row) were taken. Photographs are representative of at least three independent platings.
Both sypG mutants exhibited completely smooth-colony morphology (Fig. 3). The wrinkling phenotype was restored when RscS and SypG were coexpressed in the ΔsypG mutant strain (data not shown), indicating that this RscS-mediated phenotype requires SypG. In contrast, the ΔsypE mutation resulted in a partial loss of RscS-mediated wrinkling. Individual sypE mutant colonies appeared smooth; however, the heavy part of the streak exhibited some dryness and wrinkling (Fig. 3). Thus, sypE plays an important role in, but is not completely required for, RscS-dependent wrinkling.
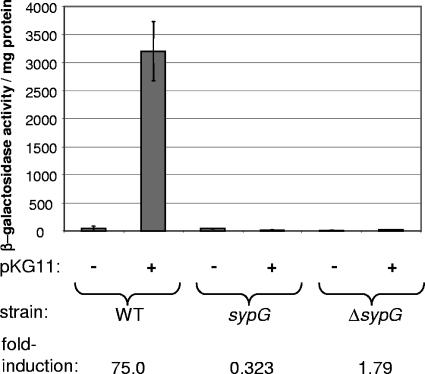
Dependence of RscS on V. fischeri RRs for syp transcription.
All of the RscS-mediated biofilm phenotypes described above require the syp locus (42). Therefore, we assessed the effects of individual RR mutations on RscS-mediated syp transcription, measured via single-copy sypA promoter-lacZ fusions. Consistent with our previous studies (42), the multicopy expression of RscS from pKG11 caused a significant (75-fold) increase in sypA reporter activity in an otherwise wild-type background (Fig. 4).
FIG. 4.
RscS-mediated induction of syp transcription in RR mutants. Wild-type (WT) and RR mutant sypA reporter strains carrying either pKG11 or a vector control were grown with shaking in HMM-T at 22°C overnight. The level of transcription of the sypA reporter is reported as units of β-galactosidase activity per mg protein.
Of the 35 RR mutants tested, 34 exhibited RscS-induced reporter activity at or above the level of the wild-type control (data not shown). The only exception was sypG: when RscS was overexpressed, the sypG vector integration and deletion mutants both exhibited significantly less reporter activity than did the wild-type strain (Fig. 4). Thus, not only is SypG the only RR required for all RscS-mediated biofilm phenotypes, it is also the only RR required for RscS to induce the expression of the syp cluster.
sypG overexpression in a sypE mutant mimics rscS overexpression.
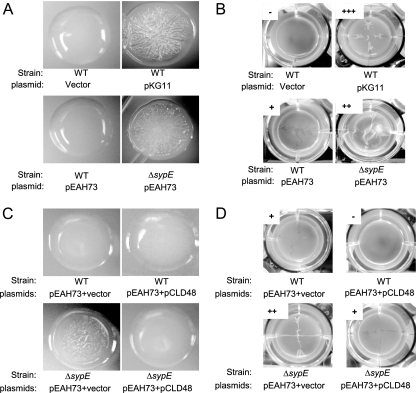
Our results are consistent with the hypothesis that RscS functions upstream of SypG to induce syp transcription and biofilm formation. If the two regulators function together, then it might be expected that the two genes would induce similar phenotypes. Indeed, when overexpressed, SypG and RscS each induce syp transcription (43, 44). However, SypG overexpression, while promoting attachment to test tubes following growth under either static or shaking conditions (43), does not appear to cause wrinkled-colony formation or the production of strong pellicles (Fig. 5A and B, respectively). Based on these results, we formulated two hypotheses. First, SypG overexpressed in the absence of its SK might not be sufficiently activated (via phosphorylation) to induce the transcription of the genes necessary for these phenotypes. Second, RscS could signal through more than one RR, either by activating an additional positive regulator or by inactivating a negative regulator, to induce the observed biofilm phenotypes. We favor the latter possibility, as our data thus far do not reveal another strong positive regulator of biofilm formation.
FIG. 5.
SypG-mediated phenotypes in the absence of SypE. (A and C) Wild-type (WT) and ΔsypE mutant strains carrying the indicated plasmids were streaked onto solid, complex medium (LBS with 0.3% glycerol and Tc) and allowed to grow for 3 days at room temperature. Photographs are representative of samples from at least two independent platings. (B and D) Wild-type and ΔsypE mutant strains carrying the indicated plasmids were grown statically in HMM containing glucose and Casamino Acids at either 22°C (strains carrying the vector control and pKG11) or 28°C (strains carrying pEAH73). Pellicle formation was assessed by dragging a sterile toothpick through the culture surface and scored as described in Materials and Methods. Experiments were conducted in triplicate.
To distinguish these hypotheses, we sought a constitutively active allele of sypG through mutagenesis of the putative site of phosphorylation, aspartate 53. In other RRs such as CheY, NtrC, and LuxO, the substitution of Glu for Asp at that position has resulted in enhanced activity (7, 29, 30). The D53E substitution in SypG in fact resulted in a 3.5-fold increase in the SypG-mediated induction of the sypA promoter-lacZ reporter relative to that of wild-type SypG when expressed from a multicopy plasmid (data not shown). This allele did not, however, result in the appearance of wrinkled colonies or enhanced pellicles in wild-type V. fischeri.
Further attempts to clarify the roles of the various syp regulators, however, yielded an unexpected result: the overexpression of wild-type SypG (from pEAH73) in a ΔsypE mutant strain resulted in wrinkled colonies (Fig. 5A). These wrinkled colonies resembled those formed by RscS-overexpressing wild-type cells. Furthermore, we found that the ΔsypE strain carrying pEAH73 was capable of forming thick pellicles (Fig. 5B). To confirm that SypE inhibits SypG-mediated phenotypes, we complemented the ΔsypE mutation with sypE expressed from pCLD48, which is compatible with the SypG expression vector pEAH73. The co-overproduction of SypG and SypE in the ΔsypE strain restored smooth-colony morphology and weak pellicle formation (Fig. 5C and D, respectively), phenotypes similar to those of wild-type V. fischeri carrying pEAH73 and a vector control (pVSV105). Furthermore, the co-overproduction of SypE and SypG in the wild-type strain eliminated the formation of weak pellicles induced by the overexpression of SypG alone (Fig. 5D). These data reveal that phenotypes induced by SypG overexpression can mimic those induced by RscS, a result fully consistent with the hypothesis that the two regulators function together. They also support a model in which SypE is antagonistic to SypG, demonstrating the complexity of control over syp-dependent biofilm formation in V. fischeri.
DISCUSSION
The V. fischeri SK RscS was discovered as a determinant of colonization of the host squid, Euprymna scolopes (37). RscS regulates the expression of the syp cluster of genes and promotes cell-cell aggregation outside the squid light organ, an early event in colonization (42). Importantly, this biofilm-like behavior correlates with cell-cell aggregation phenotypes observed in cells overexpressing RscS in culture (42). Until now, the identity of the downstream RR(s) in the two-component pathway represented by RscS has remained unclear.
To address this problem, we conducted epistasis experiments to identify the regulator(s) that functions downstream of RscS. Our data indicate that a mutation of six different RRs diminished RscS-mediated liquid biofilm phenotypes (i.e., pellicle formation and attachment to a glass surface): arcA, sypE, sypG, flrC, VF1401, and VF0454. Of these, only the loss of sypG completely eliminated the enhancement of liquid biofilms by RscS. The disruption of sypG also eliminated RscS-mediated wrinkled-colony formation. Only one other mutation, in the other syp cluster RR gene, sypE, diminished (but did not eliminate) the ability of RscS to promote wrinkling. Finally, the mutation of sypG alone eliminated the ability of RscS to induce the expression of a syp reporter.
Our data thus provide compelling genetic evidence that RscS and SypG function within the same two-component pathway. First, both RscS and SypG are required for the initiation of squid colonization (10, 37). Second, the overexpression of either RscS or SypG induces syp transcription and biofilm phenotypes (42, 43). Third, SypG is the only one of the 35 putative RRs tested that is required for all known functions of RscS.
Very few studies have established partnerships between orphan SKs and RRs. Some examples include UvrY and BarA in E. coli (23), ArcB and RssB in E. coli (20), DosT and DosR in Mycobacterium tuberculosis (25, 28), and CenK and CenR in Caulobacter crescentus (31). Often, in vitro phosphotransfer studies are utilized as evidence for such partnerships (31). Despite repeated attempts, both RscS and SypG proved difficult to purify, resulting in either low yields of protein (in the case of RscS) or aggregated, possibly unfolded protein (SypG) (E. A. Hussa and K. L. Visick, unpublished data). Thus, phosphotransfer experiments could not be performed. Due to the lack of biochemical data, it remains formally possible that RscS does not directly donate a phosphoryl group to SypG. There are at least four additional putative V. fischeri RRs that we were unable to disrupt in our previous mutagenesis study (10), one or more of which may function in the RscS pathway. Also, there is an additional V. fischeri RR, GacA, which was not considered in this study; mutation of GacA results in a growth yield defect that makes biofilm-related phenotypes difficult to study (39). However, the genetic evidence presented here demonstrates that RscS activity requires SypG; therefore, we assert that these additional RRs are likely not major components of the RscS pathway. There are also examples in which alternative paths of phosphotransfer are followed, such as the E. coli Rcs system, in which the RcsC kinase donates a phosphoryl group to an Hpt domain within an intermediate protein, RcsD (34). Intriguingly, encoded just upstream of sypG is a hybrid SK, SypF. However, unlike the loss of sypG, the disruption of sypF does not eliminate RscS-mediated wrinkled-colony formation (C. L. Darnell and K. L. Visick, unpublished data). Therefore, we think that it is unlikely that a substantial amount of RscS-initiated activation occurs via SypF.
It is abundantly clear, however, that the regulation of syp-dependent biofilms is complex. In support of this idea, in this study, we determined that the overexpression of SypG in a sypE deletion strain results in the formation of wrinkled colonies and pellicles mimicking those produced by wild-type strains overexpressing RscS. We interpret these data as further support of our model that RscS and SypG function in the same pathway. However, these results generate additional questions. For example, if SypE is required for the full expression of RscS-mediated biofilm phenotypes, then why does its loss allow biofilm formation to be induced by SypG overexpression?
SypE is not a typical RR: its phosphate-accepting receiver (REC) domain is centrally located and is not adjacent to a known DNA binding domain. Instead, C terminal to the REC domain is a putative protein phosphatase 2C domain (2) predicted to function as a serine phosphatase. N terminal to the REC domain is a region with weak similarity to the Bacillus subtilis RsbW protein, which acts as a serine kinase (5). If these domains function as predicted, it is possible that the phosphorylation of SypE could modulate its ability to serve as either a phosphatase or a kinase. Thus, we hypothesize that RscS may also function upstream of SypE in a manner that negates its antagonism of SypG-mediated phenotypes. In this model, the overexpression of RscS would both activate SypG and inactivate SypE, ultimately inducing wrinkled-colony formation. In contrast, the overexpression of SypG alone would not be sufficient to prevent SypE-mediated antagonism; thus, wrinkled-colony formation occurs only in the absence of SypE. A further understanding of how SypE functions awaits specific, mechanistic characterization of this protein and its unusual domains through mutagenesis studies.
This work also revealed roles for four additional RRs in V. fischeri biofilm formation (i.e., glass attachment and pellicle formation): FlrC, ArcA, VF0454, and VF1401. The first three of these RRs were previously shown to be involved in biofilm formation in other bacteria (10, 11, 13, 24, 35). In particular, VF0454 encodes a protein with high sequence identity to the Vibrio cholerae exopolysaccharide regulator VpsR; a distinct study from our laboratory has also uncovered an important role for VF0454 in biofilm formation (3a). Little is known about the final V. fischeri biofilm regulator, VF1401, except that it is required for competitive colonization and belongs to the family of σ54-dependent transcriptional activators (10). At this time, we cannot determine whether these four RRs function specifically downstream of RscS or whether they have independent effects on biofilm formation, as wild-type strains of V. fischeri (i.e., strains not overexpressing syp) do not form robust biofilms in culture (40, 42, 43).
In summary, we have shown that the SK RscS functions primarily upstream of the RR SypG as a major two-component pathway involved in V. fischeri biofilm formation. This RscS/SypG-mediated signaling, however, is likely to be complex, involving at least one additional syp regulator, SypE, which appears to severely inhibit SypG-mediated biofilm formation. In addition, we have uncovered at least four other V. fischeri two-component regulators that feed into biofilm formation: FlrC, VF0454, ArcA, and VF1401. This work has thus provided a basis for understanding the complex control of biofilm formation in V. fischeri.
Acknowledgments
We thank Therese O'Shea for construction of plasmid pTMO82 and Kati Geszvain and Jenny Wu for aid in phenotype screening in the early stages of this work. We also thank Steve Johnston for collaboration in the yeast two-hybrid analysis. Additionally, we are grateful to Alan Wolfe and members of the Visick laboratory for providing useful insight into the manuscript.
This work was funded by the NIH grant GM59690, awarded to K.L.V.
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 1723701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bork, P., N. P. Brown, H. Hegyi, and J. Schultz. 1996. The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 51421-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bose, J. L., U. Kim, W. Bartkowski, R. P. Gunsalus, A. M. Overley, N. L. Lyell, K. L. Visick, and E. V. Stabb. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65538-553. [DOI] [PubMed] [Google Scholar]
- 3a.Darnell, C. L., E. A. Hussa, and K. L. Visick. 9 May 2008. The putative hybrid sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J. Bacteriol. doi: 10.1128/JB.00197-08. [DOI] [PMC free article] [PubMed]
- 4.DeLoney, C. R., T. M. Bartley, and K. L. Visick. 2002. Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J. Bacteriol. 1845121-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 1761813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn, A. K., D. S. Millikan, D. M. Adin, J. L. Bose, and E. V. Stabb. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31665-677. [DOI] [PubMed] [Google Scholar]
- 8.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 1766986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoch, J. A., and T. J. Silhavy (ed.). 1995. Two-component signal transduction. ASM Press, Washington, DC.
- 10.Hussa, E. A., T. M. O'Shea, C. L. Darnell, E. G. Ruby, and K. L. Visick. 2007. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J. Bacteriol. 1895825-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junker, L. M., J. E. Peters, and A. G. Hay. 2006. Global analysis of candidate genes important for fitness in a competitive biofilm using DNA-array-based transposon mapping. Microbiology 1522233-2245. [DOI] [PubMed] [Google Scholar]
- 12.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28501-520. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi, K. 2007. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J. Bacteriol. 1894920-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 15.Lupp, C., and E. G. Ruby. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 1863873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupp, C., and E. G. Ruby. 2005. Vibrio fischeri utilizes two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 1873620-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50319-331. [DOI] [PubMed] [Google Scholar]
- 18.McCann, J., E. V. Stabb, D. S. Millikan, and E. G. Ruby. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 695928-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McFall-Ngai, M. J., and E. G. Ruby. 2000. Developmental biology in marine invertebrate symbioses. Curr. Opin. Microbiol. 3603-607. [DOI] [PubMed] [Google Scholar]
- 20.Mika, F., and R. Hengge. 2005. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev. 192770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, New York, NY.
- 21a.O'Shea, T. M., A. H. Klein, K. Geszvain, A. J. Wolfe, and K. L. Visick. 2006. Diguanylate cyclases control magnesium-dependent motility of Vibrio fischeri. J. Bacteriol. 1888196-8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 23.Pernestig, A. K., O. Melefors, and D. Georgellis. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276225-231. [DOI] [PubMed] [Google Scholar]
- 24.Reid, D. W., and S. M. Kirov. 2004. Iron, Pseudomonas aeruginosa and cystic fibrosis. Microbiology 150516; discussion, 516-518. [DOI] [PubMed] [Google Scholar]
- 25.Roberts, D. M., R. P. Liao, G. Wisedchaisri, W. G. Hol, and D. R. Sherman. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 27923082-23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruby, E. G. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50591-624. [DOI] [PubMed] [Google Scholar]
- 27.Ruby, E. G., and K. H. Nealson. 1977. Pyruvate production and excretion by the luminous marine bacteria. Appl. Environ. Microbiol. 34164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saini, D. K., V. Malhotra, and J. S. Tyagi. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 56575-80. [DOI] [PubMed] [Google Scholar]
- 29.Sanders, D. A., B. L. Gillece-Castro, A. L. Burlingame, and D. E. Koshland, Jr. 1992. Phosphorylation site of NtrC, a protein phosphatase whose covalent intermediate activates transcription. J. Bacteriol. 1745117-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders, D. A., B. L. Gillece-Castro, A. M. Stock, A. L. Burlingame, and D. E. Koshland, Jr. 1989. Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J. Biol. Chem. 26421770-21778. [PubMed] [Google Scholar]
- 31.Skerker, J. M., M. S. Prasol, B. S. Perchuk, E. G. Biondi, and M. T. Laub. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 34.Takeda, S., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC→YojN→RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40440-450. [DOI] [PubMed] [Google Scholar]
- 35.Vilain, S., P. Cosette, G. A. Junter, and T. Jouenne. 2002. Phosphate deprivation is associated with high resistance to latamoxef of gel-entrapped, sessile-like Escherichia coli cells. J. Antimicrob. Chemother. 49315-320. [DOI] [PubMed] [Google Scholar]
- 36.Visick, K. L., and E. G. Ruby. 2006. Vibrio fischeri and its host: it takes two to tango. Curr. Opin. Microbiol. 9632-638. [DOI] [PubMed] [Google Scholar]
- 37.Visick, K. L., and L. M. Skoufos. 2001. A two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26369-376. [DOI] [PubMed] [Google Scholar]
- 39.Whistler, C. A., and E. G. Ruby. 2003. GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J. Bacteriol. 1857202-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolfe, A. J., D. S. Millikan, J. M. Campbell, and K. L. Visick. 2004. Vibrio fischeri σ54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 702520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 1831716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yip, E. S., K. Geszvain, C. R. DeLoney-Marino, and K. L. Visick. 2006. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 621586-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yip, E. S., B. T. Grublesky, E. A. Hussa, and K. L. Visick. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol. Microbiol. 571485-1498. [DOI] [PubMed] [Google Scholar]